Open Access Article
Open Access ArticleBiobased, biodegradable but not bio-neutral: about the effects of polylactic acid nanoparticles on macrophages†
Véronique
Collin-Faure
a,
Marianne
Vitipon
a,
Hélène
Diemer
bc,
Sarah
Cianférani
bc,
Elisabeth
Darrouzet
a and
Thierry
Rabilloud
 *a
*a
aChemistry and Biology of Metals, Univ. Grenoble Alpes, CNRS UMR5249, CEA, IRIG-LCBM, F-38054 Grenoble, France. E-mail: thierry.rabilloud@cnrs.fr
bLaboratoire de Spectrométrie de Masse BioOrganique (LSMBO), IPHC UMR 7178, Université de Strasbourg, CNRS, 67087 Strasbourg, France
cInfrastructure Nationale de Protéomique ProFI – FR2048, 67087 Strasbourg, France
First published on 29th July 2024
Abstract
Plastics are persistent pollutants because of their slow degradation, which suggests that they may lead to cumulative and/or delayed adverse effects due to their progressive accumulation over time. Macroplastics produced by human activity are released in the environment, where they degrade into micro- and nanoplastics that are very easily uptaken by a wide variety of organisms, including humans. Microplastics and nanoplastics being particulates, they are handled in the body by specialized cells such as macrophages (or their evolutionary counterparts), where they can elicit a variety of responses. One solution to alleviate the problems due to biopersistence, such as accumulation over life, would be to use biodegradable plastics. One of the emerging biodegradable plastics being polylactide, we decided to test the responses of macrophages to polylactide nanoparticles using a combination of untargeted proteomics and targeted validation experiments. Proteomics showed important adaptive changes in the proteome in response to exposure to polylactide nanoparticles. These changes affected, for example, mitochondrial, cytoskeletal and lysosomal proteins, but also proteins implicated in immune functions or redox homeostasis. Validation experiments showed that many of these changes were homeostatic, with no induced oxidative stress and no gross perturbation of the mitochondrial function. However, polylactide particles altered the immune functions such as phagocytosis (−20%) or cytokine production (2-fold increase for TNF production), which may translate into a decreased ability of macrophages to respond to bacterial infections. Furthermore, polylactide particles also induced moderate cross-toxicity with some quinones such as phenanthrene quinone, a combustion by-product that is a suspected carcinogen.
Environmental significancePlastic pollution is a really concerning topic, inter alia, because of the degradation of plastics in nanoparticles that are easily uptaken by living organisms and poorly degradable. Thus, there is a trend toward the development of biobased and biodegradable plastics, and polyhydroxyalkanoates are among the most promising ones. However, they too can fragment into nanoparticles and show effects on living cells. Consequently, we have studied the effect of polylactide nanoparticles on macrophages, i.e. professional phagocytes conserved in evolution, by a combination of proteomic and targeted approaches. No inflammatory response was observed after exposure to polylactide. However, plastic-exposed cells showed a decreased ability to respond to a bacterial stimulus, suggesting slightly impaired immune functions of macrophages after exposure to plastics. |
1. Introduction
The wide use of plastics in very diverse areas (e.g. packaging, automotive, textile, electronics, to name just a few) translates into huge annual production figures, i.e. close to a gigaton per year.1 Unfortunately a huge proportion of these plastics, estimated to be 500 Mt per year, is released in the environment,2 where it has deleterious effects that are more and more documented in detail, on a wide range of marine taxa,3e.g. sea birds.4 This pollution was first documented in aquatic marine environments,5–8 but is now found in marine sediments,9,10 freshwater environments,11–13 and also in terrestrial ones.14,15Although plastics are chemically resistant and degrade very slowly, with half-lives in the environment amounting to decades,16 they progressively fragment, first into microplastics (lesser than 5 mm in size) and then into nanoplastics (less than 1 μm in size). These nanoplastics are far more difficult to detect in the environment, and their effects are much less known. It is however anticipated that the smaller they are, the more easily they will cross biological barriers and be able to disturb the homeostasis of living organisms. This hypothesis has prompted extensive research on the effects of micro- and nanoplastics on a wide variety of living organisms from various phylla, ranging from worms17,18 to molluscs,19–21 crustaceans,22,23 insects,24–26 and then vertebrates such as fish27–29 and of course mammals30 including human models (e.g. in ref. 31–35).
Multicellular organisms defend themselves against particles, including plastic particles, by a series of mechanisms. The first line of defense is represented by biological barriers (e.g. intestinal or epidermal barriers) and research has been devoted to understand how these barriers interact with plastic particles. When translocation across these barriers occurs,31 then a second line of barriers comes into play, and is represented by professional phagocytes. This cell type is encountered in invertebrates (annelid coelomocytes, insect hemocytes) as well as in vertebrates (macrophages, neutrophils). Indeed, it has been shown that this cell type does respond to plastic particles.18,26,36–42
In view of these deleterious effects and of the fact that fabrication of plastics consumes fossil resources and increases the concentration of greenhouse effect gases, the development of biobased and biodegradable plastics has been investigated and is a very active area of research. Plastics that are both biobased and biodegradable belong to two main families. The first family is represented by modified polysaccharides (e.g. modified starch or cellulose derivatives), and the second family is represented by polyhydroxyalkanoates, i.e. polymers of natural hydroxyacids. Indeed polyhydroxyalkanoates are produced by a variety of bacteria both as a protective medium and as a chemical energy storage.43–47 One of the simplest polyhydroxyalkanoate, i.e. polylactic acid (PLA), was first used as a material for surgical sutures,48,49 and it was demonstrated that it was biodegradable.50 It also gained popularity by the fact that it could be polymerized as nanospheres for drug encapsulation and controlled release, most often as a copolymer of lactic and glycolic acid.51–53 However, it was also shown that pure PLA is less toxic than the copolymer,54,55 so that PLA is a preferred choice as a model for a safe, biobased and biodegradable plastic available in the nanoplastic format.
Recent work showed that degradation of PLA produced fragmentation and nanoparticles,56 even in domestic use,57 which further increases the probability that living organisms may encounter PLA nanoparticles, and especially if the use of this plastic increases. We thus decided to investigate the responses of macrophages to PLA nanoparticles, using a combination of proteomic and targeted approaches, as previously done with polystyrene nanoparticles.41 In this approach, proteomics is used to gain a wide appraisal of the cell responses, highlighting hypotheses that are then verified by targeted experiments.
2. Materials and methods
2.1. Plastic particles
Polylactic acid (PLA) particles (red polylactic acid, 150 nm, fluorescent red labelled, catalog number #RFiP-600-150, batch number #230612-ip) were purchased from Adjuvatis (Lyon, France) and provided as sterile 3% (w/v) suspensions. Deep red fluorescent labelled polystyrene (PS) particles (SkyBlue particles, 100–300 nm, catalog number #FP-0270-2, batch number #AL01) were purchased from Spherotech and provided as non-sterile 0.25% (w/v) suspensions. The particles were used for both the proteomic experiments and the validation experiments. The polystyrene suspensions were pasteurized overnight at 80 °C before use in cell culture. The PLA suspension was provided sterile by the supplier. The particles were excited at 560 nm and their emission read at 613/18 nm for the PLA particles and n at 695/40 nm for the PS particles. It should be noted that the fluorophore is trapped within the polymer matrix for both PLA58 and PS59 nanoparticles.The particles were diluted at 10 μg mL−1 in 0.001× PBS and characterized by DLS using a Litesizer 500 instrument (Anton Paar, USA) and an Omega cuvette (225288, Anton Paar, USA). An average value was obtained from repeated measurements for each sample (n = 3) and analyzed with the instrument-associated Kaliope software.
2.2. Cell culture
The J774A.1 cell line (mouse macrophages) was purchased from European cell culture collection (Salisbury, UK). Cells were routinely propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in non-adherent flasks (Cellstar flasks for suspension culture, Greiner Bio One, Les Ulis, France). For routine culture, the cells were seeded at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml and passaged two days later, with a cell density ranging from 800
000 cells per ml and passaged two days later, with a cell density ranging from 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 1
000 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml. For exposure to plastic particles and to limit the effects of cell growth, cells were seeded at 500
000 cells per ml. For exposure to plastic particles and to limit the effects of cell growth, cells were seeded at 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml in 6 or 12 well plates, allowed to settle and recover for 24 hours, and then exposed to the particles at 50 μg ml−1 for 24 hours before harvesting for the experiments. Proteomic experiments were carried out in 6 well plates, and all the other experiments in 12 well plates. The medium volume was adjusted to keep the same height across all cell culture formats. Cells were used at passage numbers from 5 to 15 post-reception from the repository. Cell viability was measured by the propidium iodide method60 or with a SytoxGreen probe (ThermoFisher S7020) using the protocol provided by the supplier.
000 cells per ml in 6 or 12 well plates, allowed to settle and recover for 24 hours, and then exposed to the particles at 50 μg ml−1 for 24 hours before harvesting for the experiments. Proteomic experiments were carried out in 6 well plates, and all the other experiments in 12 well plates. The medium volume was adjusted to keep the same height across all cell culture formats. Cells were used at passage numbers from 5 to 15 post-reception from the repository. Cell viability was measured by the propidium iodide method60 or with a SytoxGreen probe (ThermoFisher S7020) using the protocol provided by the supplier.
For assessing the persistence of the particles in the cells, we followed the strategy published previously.42 The cells were seeded in adherent 12-well plates at 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml in DMEM supplemented with 1% horse serum in order to limit their proliferation.61 After adaptation to the medium for 72 hours (D0), the medium was changed and the cells were exposed to the plastic particles for 24 hours at a concentration of 50 or 100 μg ml−1. The particle-containing medium was removed (D1). The remaining cells were cultivated in DMEM supplemented with 1% horse serum for an additional 3 days without splitting and with a culture medium change every 2 days. Wells were harvested every day starting at D1 during this process and analyzed by flow cytometry to measure the cell-associated fluorescence. In order to normalize for the cell number, a cell numeration was performed on the harvested cells in each well. This process allowed us to determine the remaining fraction of particles over time after the initial loading (D1).
000 cells per ml in DMEM supplemented with 1% horse serum in order to limit their proliferation.61 After adaptation to the medium for 72 hours (D0), the medium was changed and the cells were exposed to the plastic particles for 24 hours at a concentration of 50 or 100 μg ml−1. The particle-containing medium was removed (D1). The remaining cells were cultivated in DMEM supplemented with 1% horse serum for an additional 3 days without splitting and with a culture medium change every 2 days. Wells were harvested every day starting at D1 during this process and analyzed by flow cytometry to measure the cell-associated fluorescence. In order to normalize for the cell number, a cell numeration was performed on the harvested cells in each well. This process allowed us to determine the remaining fraction of particles over time after the initial loading (D1).
2.3. Confocal microscopy
J774.A1 cells were seeded onto glass coverslips at a density of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL in DMEM supplemented with 1% horse serum and 1% streptomycin–penicillin. The cells were then incubated overnight at 37 °C with 5% CO2.
000 cells per mL in DMEM supplemented with 1% horse serum and 1% streptomycin–penicillin. The cells were then incubated overnight at 37 °C with 5% CO2.
Subsequently, PLA beads were added to the cells at a concentration of 50 μg ml−1, and further incubation was carried out for 24 hours. The medium was then changed with particle-free medium and the cells were left to recover for 24 hours and 48 hours at 37 °C.
After each incubation period (without exposure, immediately after exposure and after the 24- and 48-hour recovery periods) the cells were fixed with 4% paraformaldehyde for 30 minutes at room temperature and permeabilized with 0.1% Triton X-100 for 20 minutes at room temperature. Following permeabilization, the cells were incubated with Phalloidin Atto 488 (1/500 dilution) and DAPI (1/1000 dilution) for 20 minutes and 5 minutes, respectively, at room temperature. Between each step, the cells were washed three times with PBS (1×).
Confocal microscopy experiments were performed using a Zeiss LMS880 confocal microscope (Carl Zeiss, Jena, Germany) equipped with a 20× objective. Laser tracks were set as follows:
• DAPI: excitation at 405 nm, emission at 410–467 nm.
• Phalloidin Atto 488: excitation at 488 nm, emission at 494–550 nm.
• PLA beads: excitation at 561 nm, emission at 596–632 nm.
Laser settings were optimized using the 24-hour exposed cells labeled with phalloidin and DAPI to maximize fluorescence intensity before imaging each condition. Confocal laser scanning microscopy was employed to visualize the actin structure, nucleus, and PLA beads.
The acquired images were processed using ImageJ software. Specifically, adjustments were made to enhance the quality and uniformity of the images. Green fluorescence intensity in the B1 merged image was increased to ensure consistency across all panels. Similarly, brightness levels in the red channel were uniformly adjusted for all samples, with or without PLA beads.
2.4. Proteomics
Proteomics was carried out essentially as described previously.41 However, the experimental details are given here for the sake of consistency.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 30 minutes) to pellet the nucleic acids. The supernatants were then stored at −20 °C until use.
000g, 30 minutes) to pellet the nucleic acids. The supernatants were then stored at −20 °C until use.
As mentioned above, methylene blue was included in the cell lysis buffer. The other initiator solutions consisted of a 1 M solution of sodium toluene sulfinate in water and in a saturated water solution of diphenyliodonium chloride. The ready-to-use polyacrylamide solution consisted of 1.2 ml of a commercial 40% acrylamide/bis solution (37.5/1) to which 100 μl of diphenyliodonium chloride solution, 100 μl of sodium toluene sulfinate solution and 100 μl of water were added.
To the protein samples (15 μl), 5 μl of acrylamide solution were added and mixed by pipetting in a 500 μl conical polypropylene microtube. 100 μl of water-saturated butanol were then layered on top of the samples, and polymerization was carried out under a 1500 lumen 2700 K LED lamp for 2 hours, during which the initially blue gel solution discolored. At the end of the polymerization period, the butanol was removed, and the gel plugs were fixed for 2 × 1 h with 200 μl of 30% ethanol and 2% phosphoric acid, followed by a 30-minute wash in 30% ethanol. The fixed gel plugs were then stored at −20 °C until use.
Gel plug processing, digestion, peptide extraction and nanoLC-MS/MS were performed as previously described, without the robotic protein handling system and using a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Further details are available in Methods S1.†
For protein identification, the MS/MS data were interpreted using a local Mascot server with MASCOT 2.6.2 algorithm (Matrix Science, London, UK) against an in-house database containing all Mus musculus and Rattus norvegicus entries from UniProtKB/SwissProt (version 2019_10, 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 156 sequences) and the corresponding 25
156 sequences) and the corresponding 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 156 reverse entries. Spectra were searched with a mass tolerance of 10 ppm for MS and 0.05 Da for MS/MS data, allowing a maximum of one trypsin missed cleavage. Trypsin was specified as the enzyme. Acetylation of protein N-termini, carbamidomethylation of cysteine residues and oxidation of methionine residues were specified as variable modifications. Identification results were imported into Proline software version 2.2 (https://www.profiproteomics.fr/proline/) for validation. Peptide spectrum matches (PSMs) with pretty rank equal to one and a length greater than 7 amino acids were retained. False discovery rate was then optimized to be below 1% at PSM level using Mascot adjusted E-value and below 1% at protein level using Mascot standard score.
156 reverse entries. Spectra were searched with a mass tolerance of 10 ppm for MS and 0.05 Da for MS/MS data, allowing a maximum of one trypsin missed cleavage. Trypsin was specified as the enzyme. Acetylation of protein N-termini, carbamidomethylation of cysteine residues and oxidation of methionine residues were specified as variable modifications. Identification results were imported into Proline software version 2.2 (https://www.profiproteomics.fr/proline/) for validation. Peptide spectrum matches (PSMs) with pretty rank equal to one and a length greater than 7 amino acids were retained. False discovery rate was then optimized to be below 1% at PSM level using Mascot adjusted E-value and below 1% at protein level using Mascot standard score.
Mass spectrometry data are available via ProteomeXchange with the identifier PXD048664.
Proteins were considered as significantly different if their p value in the Mann–Whitney U-test against control values was less than 0.05. No quantitative change threshold value was applied. The selected proteins were then submitted to pathway analysis using the DAVID tool,66 with a cutoff value set at a FDR of 0.1.
2.5. Mitochondrial transmembrane potential assay
The mitochondrial transmembrane potential assay was performed essentially as described previously.42 Rhodamine 123 (Rh123) was added to the cultures at an 80 nM final concentration (to avoid quenching67), and the cultures were further incubated at 37 °C for 30 minutes. At the end of this period, the cells were collected, washed in cold PBS containing 0.1% glucose, resuspended in PBS glucose and analyzed for green fluorescence (excitation 488 nm, emission 525 nm) on a Melody flow cytometer. As a positive control, butanedione monoxime (BDM) was added at a 30 mM final concentration together with the Rh123.68 As a negative control, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) was added at 5 μM final concentration together with the Rh123.682.6. Phagocytosis assay
For this assay, the cells were first exposed to the red fluorescent particles. After 24 hours of exposure, the cells were then exposed to 0.5 μm latex beads (carboxylated surface, yellow green-labelled, from Polysciences; excitation 488, nm emission 527/32 nm) for 3 hours. After this second exposure, the cells were collected, rinsed twice with PBS, and analyzed for the two fluorescences (green and red) on a Melody flow cytometer.2.7. Lysosomal assay
For the lysosomal function assay, the LysoSensor method was used, as described previously.42 After exposure to plastic beads, the medium was removed, the cell layer was rinsed with complete culture medium and incubated with 1 μM LysoSensor Green (Molecular Probes) diluted in warm (37 °C) complete culture medium for 1 hour at 37 °C. At the end of this period, the cells were collected, washed in cold PBS containing 0.1% glucose, resuspended in PBS glucose and analyzed for green fluorescence (excitation 488 nm, emission 540 nm) on a Melody flow cytometer.2.8. Cytokine release assays
Cells were first exposed to nanoplastics (50 μg ml−1) for 24 hours. At the end of this exposure period, the culture medium was removed, the cell layer was rinsed with culture medium and fresh medium was added to the wells. In half of the wells LPS (1 ng ml−1) was added. After another 24 hours, the medium was collected and analyzed for proinflammatory cytokines. Tumor necrosis factor (catalog number 558299, BD Biosciences, Le Pont de Claix, France) and interleukin 6 (IL-6) (catalog number 558301, BD Biosciences, Le Pont de Claix) levels were measured using a Cytometric Bead Array Mouse Inflammation Kit (catalog number 558266, BD Biosciences, Le Pont de Claix) and analyzed with FCAP Array software (3.0, BD Biosciences) according to the manufacturer's instructions.2.9. Assay for oxidative stress
For the oxidative stress assay, a protocol based on the oxidation of dihydrorhodamine 123 (DHR123) was used, essentially as described previously.42 After exposure to plastic beads, the cells were treated in PBS containing 500 ng ml−1 DHR123 for 20 minutes at 37 °C. The cells were then harvested, washed in cold PBS containing 0.1% glucose, resuspended in PBS glucose and analyzed for green fluorescence (same parameters as for rhodamine 123) on a Melody flow cytometer. Menadione (applied on the cells for 2 hours prior to treatment with DHR123) was used as a positive control in a concentration range of 25–50 μM.2.10. Cross-toxicity assays
For these assays, the cells were plated at 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml in a 12-well non adherent plate. After 24 hours, 50 μg ml−1 beads were added, followed 6 hours later by a variable concentration of the toxicant to be tested, predissolved in ethanol so that the final ethanol concentration in the culture well did not exceed 1% in volume. Toxicants tested included menadione and 9,10-phenanthrenequinone.
000 cells per ml in a 12-well non adherent plate. After 24 hours, 50 μg ml−1 beads were added, followed 6 hours later by a variable concentration of the toxicant to be tested, predissolved in ethanol so that the final ethanol concentration in the culture well did not exceed 1% in volume. Toxicants tested included menadione and 9,10-phenanthrenequinone.
3. Results
3.1. Plastic bead characterization
Plastic beads were characterized by DLS and electrophoretic mobility in order to determine their average hydrodynamic size and zeta potential. The results are presented in Table 1.| Beads | Average hydrodynamic diameter | Polydispersity index | Zeta potential |
|---|---|---|---|
| The results are expressed as mean ± standard deviation (N = 3). | |||
| PLA | 169 ± 4 nm | 18.5 ± 3.7% | −39.6 ± 0.7 mV |
| PS | 477 ± 16 nm | 22.2 ± 3.1% | −43.9 ± 5.7 mV |
The PS beads proves larger in size through these measurements than the nominal diameter given by the supplier (0.1–0.3 μm, mean 0.26 μm). This larger size however did not prevent a bead per cell number ratio close to 1000 beads per cell (at 50 μg ml−1) for the largest beads and higher than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 beads per cell for the smaller ones.
000 beads per cell for the smaller ones.
3.2. Viability of the plastic-treated cells
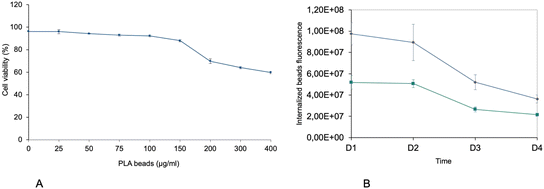
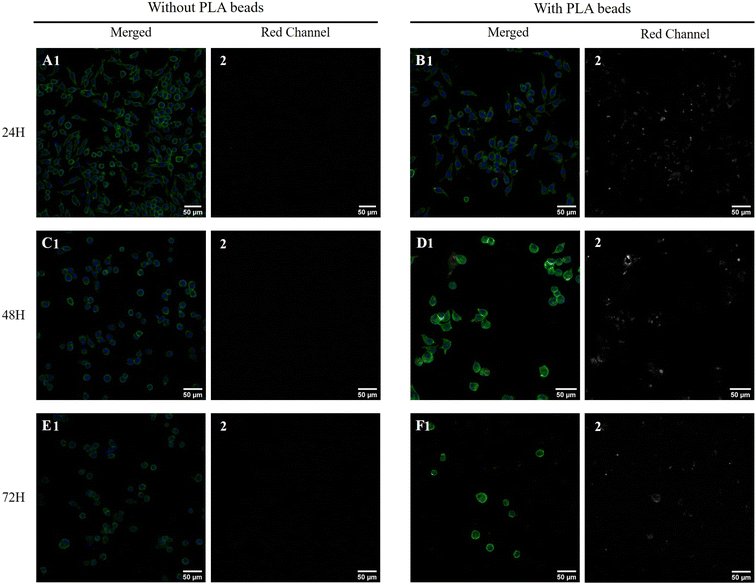
First, the toxic effects of the PLA beads on J774A.1 cells was determined after 24 hours of exposure. The results, shown in Fig. 1A, demonstrated a very low toxicity of the PLA beads, with an LD50 that reached 400 μg ml−1. As we planned to use polystyrene beads as positive controls for non-degradable plastics in subsequent experiments, we decided to use a dose that was half of that of the LD20 (100 μg ml−1) for the PS beads, i.e. 50 μg ml−1. This dose showed very low toxicity for the PLA beads, but was however the first concentration for which the viability was slightly but statistically significantly lower than that of unexposed, control cells. As the particles were labelled, we could follow their internalization and their degradation. As shown on Fig. 1B, the amount of internalized PLA particles increased with increasing concentration in the medium. However, the amount of intracellular fluorescence rapidly decreased over time, suggesting a rather fast degradation of the particles. This was further confirmed by confocal microscopy, as shown in Fig. 2. Disappearance of the particles and appearance of a fuzzy intracytoplasmic fluorescence was observed, indicating degradation of the particles. These results are in sharp contrast with those obtained for PS nanoparticles,42 which showed no degradation in the cells for more than a week.3.3. Analysis of the proteomic results
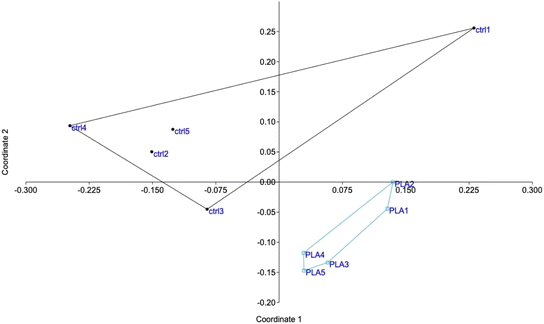
The shotgun proteomic analysis was able to detect and quantify 2869 proteins (Table S1†). A first global analysis of the complete protein list by principal coordinates analysis showed that the two groups (control and particle-treated) appeared separated on the diagram (Fig. 3), indicating significant changes in the proteome, even if the chosen concentration was quite remote from the toxicity threshold. The fact that the two proteomes were significantly different, on a statistical point of view was further substantiated by an analysis of similarities,69 which led to a Bonferroni-corrected post hoc p value of 0.009.Proteins modulated by the internalization of PLA particles were selected on the basis of a Mann–Whitney U test ≤2 in the comparison of plastic-treated cells compared to unexposed controls. This resulted in the selection of 346 modulated proteins (Table S2†). In order to gain further insight into the significance of the observed changes, this list of modulated proteins was used to perform pathway analyses by the DAVID software, and the results are shown in Table S3.† Some of the pathways highlighted by this analysis indicated a global stress response (e.g. translation, nucleotide binding, carbon metabolism, endoplasmic reticulum), which is expected for any cellular stress, while other pathways appeared more specific for cellular internalization of particles (e.g. mitochondria, lysosomes).
As these pathway analysis software proceed by aggregation of proteins sharing the same annotations, they require a minimum number of proteins to build a cluster. Thus, the list of modulated proteins was manually combed in addition to this global analysis in order to retrieve isolated protein changes, which are discarded by pathway analyses but may be of interest in the frame of macrophage physiology. This comprehensive analysis of the proteomic results led us to perform validation experiments on several functions.
3.4. Mitochondria and endoplasmic reticulum
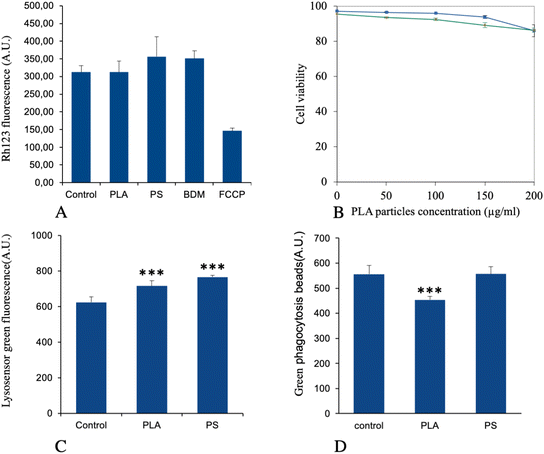
The list of the mitochondrial proteins modulated by cell treatment with PLA nanoparticles is presented in Table S4† and included 41 proteins, of which 32 were increased in abundance in response to PLA nanoparticles. Among them, 11 subunits of the respiratory chain were highlighted (8 increases, 3 decreases). Although the changes observed in protein abundances were usually of low magnitude for each protein, the changes occurring on several subunits of the same complexes may suggest a functional alteration. To test this hypothesis, we tested the mitochondrial transmembrane potential as a proxy of mitochondrial function. The results, shown in Fig. 4A, indicated that PLA particles did not induce any change in the mitochondrial transmembrane potential, while PS particles induced a small but not statistically significant increase in the mitochondrial transmembrane potential, as described previously.41,42Regarding endoplasmic reticulum, 30 proteins were modulated by treatment with PLA nanoparticles, as shown in Table S5.† These included proteins involved in the endoplasmic reticulum stress response, such as VapB protein (UniProt Q9QY76) or tyrosine phosphatase Ptpn1 (UniProt P35821). This prompted us to test whether endoplasmic reticulum stress response may play a role in PLA nanoparticle toxicity. To this purpose we used salubrinal, an inhibitor of endoplasmic reticulum stress response,70 which has been shown to counteract the lethal effects of this stress on cells.71 The results, shown in Fig. 4B, indicated no effect of salubrinal on cell survival on treatment with 200 μg ml−1 PLA particles for up to 24 hours.
3.5. Lysosomes, phagocytosis
For this important macrophage function, we gathered the proteins selected under the “lysosome” and “actin cytoskeleton” pathways, to which we added some proteins picked manually on the basis of their annotations in the UniProt database, such as the two “engulfment and cell motility” proteins (accession numbers Q8BPU7 and Q8BHL5, respectively). This led to a set of 36 proteins, which is presented in Table S6.† Two of the V-type proton ATPase subunits were present in this list (among the 11 V-type proton ATPase subunits that were detected in the whole proteomic analysis) so that we wanted to investigate using the LysoSensor probe the lysosome acidification in response to treatment with PLA nanoplastics, PS nanoparticles being used as a control for non-degradable plastics. The results, shown in Fig. 4C, indicated a small but statistically significant (p < 0.001) increase in the LysoSensor signal, indicating an increase in the number of functional lysosomes and/or an increase in proton pumping in the existing lysosomes.When the phagocytic capacity of plastic-treated cells was probed, as described in Fig. 4D, a small (−19%) but significant (p < 0.001) decrease was observed for PLA-treated cells, while PS-treated cells were not altered compared to control cells.
3.6. Immunity-related proteins, inflammation
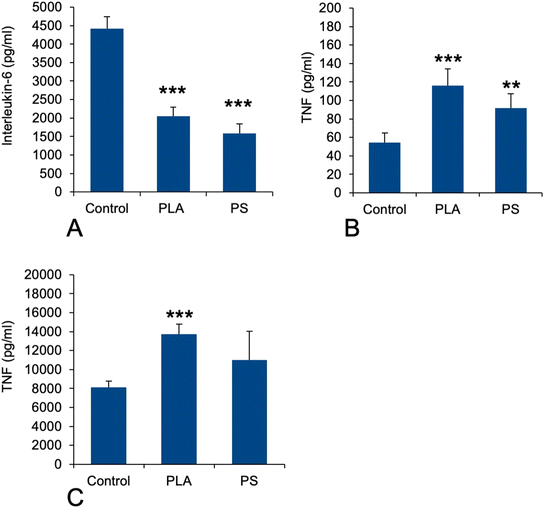
As this function was not detected through the classical pathway analysis, opposite to what was observed for polystyrene particles,41 we hand-picked on the basis of UniProt annotations the proteins that could be associated with this pathway in the shortlist of proteins whose expression was significantly modulated in response to exposure to PLA particles. This led to a set of 22 proteins, which is presented in Table S7.† This relatively low number of proteins may explain why the “innate immunity” annotation was not selected in the classical pathway analysis. Among the selected proteins, several were annotated as regulators of immune response and cytokine secretion (P09581, P10810, P11680, Q60875, Q6P549, Q9D8Y7). This prompted us to investigate the pro-inflammatory response of PLA-exposed macrophages, either in response to PLA alone or to a succession of exposures to PLA and to LPS, by measuring the secretion of TNF-alpha and IL-6. The results, shown in Fig. 5, indicated that the LPS-induced IL-6 secretion was decreased close to twofold upon prior exposure to PLA (and PS) particles (Fig. 5A) No Il-6 secretion could be measured in response to plastic alone. In the case of TNF, the opposite response was observed. A statistically significant increase could be detected for the basal levels after exposure to plastics alone (Fig. 5B) and this increase was more pronounced in response to PLA than to PS. After successive exposure to plastics and LPS, the same trend was observed again, of course at much higher levels (Fig. 5C).3.7. Redox homeostasis-related proteins, oxidative stress
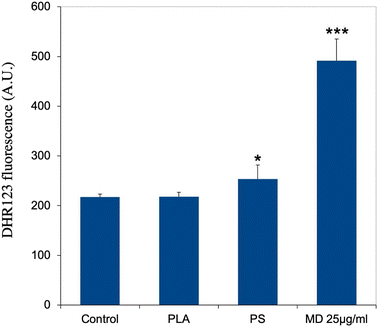
As an increase in oxidative stress was observed in response to polystyrene particles,42 we hand-picked, on the basis of UniProt annotations, proteins that could be associated with redox metabolism at large (excluding central metabolism) in the shortlist of proteins whose expression was significantly modulated in response to exposure to PLA particles. This led to a set of 13 proteins, which is presented in Table S8.† This relatively low number of proteins may explain why the “redox homeostasis” annotation was not selected in the classical pathway analysis.However, we decided to investigate the level of cellular oxidative stress in response to exposure to PLA (and PS nanoparticles). The results, shown in Fig. 6, indicated no increase in oxidative stress in response to PLA nanoparticles, while a small but significant increase was observed in response to PS nanoparticles, as previously described.42
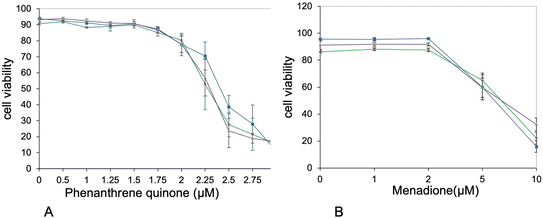
Furthermore, the proteins highlighted in Table S8† included a few proteins included in quinine metabolism, such as aflatoxin B1 aldehyde reductase member 2 and quinone oxidoreductase. This prompted us to seek for cross-toxicity between quinones and plastic beads. To this purpose, we performed comparative cell viability on cells first treated (or not) with nanoplastics, then with variable concentrations of polycyclic quinones. The results, shown on Fig. 7, showed an increased toxicity of phenanthrene quinone on plastic-treated cells, while such increased toxicity was not observed with menadione.
4. Discussion
The wide release of plastics in the environment is more than suspected to have major deleterious consequences on life. These can be due to mechanical effects of macroplastics,3 but microplastics arising from the fragmentation of macroplastics are also known to have deleterious effects, e.g. on oysters19 and on seabirds.4 Microplastics fragment even further into nanoplastics, which have been associated with liver fibrosis in rodents72 and more recently with increased cardiovascular risk in humans.73 These deleterious effects are the indirect consequences of the very low biodegradability of conventional, oil-derived plastics, which makes the particles resulting from their fragmentation long-lasting in the environment and within living organisms. Even though this low biodegradability will make this concern a problem for many years to come, one way not to aggravate it would be to switch from poorly degradable plastics to biodegradable ones, so that the persistent mechanical damage as well as the bioaccumulation issues will be alleviated. Among the few chemical solutions available to produce biodegradable plastics, polymers of organic alkyl hydroxyesters are a promising area of research. Among this class of chemicals, polylactic acid (PLA) represents an interesting choice, as it is both biobased and biodegradable.50 However, it is also known to fragment easily into nanoparticles.56,57 As the global biodegradability in the environment does not necessarily translate into the absence of accumulation in all cell types, it is necessary to obtain data on the internalization, degradation and effects of PLA nanoparticles on cell types of interest. This requires figuring out if more or less transient effects can be expected on certain cell types. Within the cell types of interest in multicellular organisms, macrophages are important to test, as they are present from invertebrates to vertebrates and play a key role in inflammation, which can be deleterious in a long time frame.74First of all, we investigated the acute toxicity of PLA nanoparticles toward macrophages. The LD20 proved to be rather high (150 μg ml−1), i.e. around twice the value observed for polystyrene.75 More importantly, PLA particles were actively degraded in macrophages. Half of the fluorescence had disappeared from cells exposed to PLA nanoparticles 48 hours after the end of exposure. As this disappearance of fluorescence, as measured by flow cytometry, implies degradation of the PLA shell of the nanoparticles, then release of the fluorophore within the cell and excretion of the free fluorophore, the real degradation of the PLA shell is probably even faster than this figure. This shows a very active degradation of the PLA particles within macrophages. This shall not come as a surprise, as PLA has been shown to be very sensitive to proteases (which often also have an esterase activity),76 proteases being quite abundant in lysosomes. This easy degradation of PLA by mammalian enzymes may be attributed to the fact that lactic acid is an alpha hydroxy acid, quite close in its structure to the alpha amino acids that make proteins.
We then investigated the effects of the PLA particles on macrophages immediately after exposure using a proteomic screen. Out of 2869 proteins quantified, the proteomic screen highlighted 346 proteins modulated upon treatment with PLA, covering several pathways.
Interestingly, we did not see any modulation of metabolic proteins, although lactate has been shown to induce major metabolic reprogramming.77 In the same trend, we did not detect a switch to an inflammatory phenotype, as described when macrophages are confronted with a high lactate concentration.78 There can be several explanations to this fact. The first one could just be that immediately after exposure, there is not a sufficient intracellular release of lactate to induce these phenomena. Moreover, as particles are present in the lysosomes and degraded there, a control of the speed of lactate release from the lysosomes to the cytosol may also occur. Finally, regarding the central metabolism, it should be kept in mind that it is heavily regulated by post-translational modifications,79–81 which do not appear easily in a global shotgun proteomic screen as in this work.
Nevertheless we detected changes in the abundances of many proteins related to organelles such as mitochondria, lysosomes and endoplasmic reticulum (ER). Regarding ER proteins, of the 30 that showed significant changes in their abundance, 24 were increased in response to PLA treatment and only 6 decreased. As ER stress may be a factor explaining toxicity, we tested this hypothesis with the drug salubrinal, which is an inhibitor of ER stress70 and has been used to demonstrate the implication of ER stress in toxicity.71 As we did not detect any effect of salubrinal on the cell viability of cells treated with up to 200 μg ml−1 PLA, we can conclude that this pathway is not a major determinant of PLA toxicity in macrophages. In the same trend, we could not detect any sign of mitochondrial depolarization after treatment with PLA nanoparticles, although the abundance of more than 40 mitochondrial proteins is changed, with 32 increases and 9 decreases. It must therefore be concluded that the observed changes are homeostatic and reflect how the cells adapt to the presence of PLA particles.
Regarding lysosomes, 36 proteins were modulated in their abundance, with 23 increases and 13 decreases. Many of the observed decreases are indeed lysosome-associated cytoskeletal proteins, such as tropomyosins, ARP complex subunits, gelsolin and myosin 9. Among the increases were detected two subunits of the lysosomal proton pump. This prompted us to test some lysosomal functions with the LysoSensor probe. As neutral red, Lysosensor probes are pumped in acidic organelles.82 The LysoSensor signal thus reflects both the activity of the proton pump and the integrity of the lysosomes, as damaged lysosomes cannot maintain a proton gradient and thus exhibit a reduced LysoSensor signal.83 We detected indeed a small but significant increase in the intensity of the LysoSensor signal not only for PLA-treated cells but also for PS-treated cells, suggesting that this effect is not specific for a given nanoparticle but more a generic effect linked to particle internalization per se.
We then tested more specific functions of the macrophage, starting with phagocytosis. To this purpose, we first exposed cells to the nanoplastics of interest, and then to a fluorescently labelled test bead for a relatively short time, in order to determine whether plastic-treated cells are still active for phagocytosis. In this case we observed a specific decrease in the phagocytic activity of PLA-treated beads, which did not occur when the cells were pretreated with PS beads, as described earlier.42 A possible explanation for this phenomenon is the induction of the phosphatase SHIP2 (Q6P549) in response to PLA treatment. This phosphatase is known to decrease Fc-gamma-R-mediated phagocytosis in macrophages,84 and the beads that we use to probe phagocytosis are incubated with serum, a process known to lead to opsonization.85
Regarding cytokine production, which is another important specialized function of macrophages, the proteomic analysis provided interesting, and somewhat contradictory, hypotheses. On the one hand, some proteins increasing the inflammatory response were increased in their abundance in response to treatment with PLA. Examples are the M-CSF receptor (P09581) which mediates the pro-inflammatory effects of M-CSF, the ARHGEF2 protein (Q60875), which plays a role in the signal transduction in response to peptidoglycans,86 and Rab-10, which plays a role in the recycling of TLR4 at the membrane and therefore in the response to LPS.87
On the other hand, some proteins acting directly or indirectly as negative regulators of the inflammatory response were also increased in their abundance in response to treatment with PLA. Examples are the SHIP2 protein (Q6P549), which downregulates the signal transduction pathway downstream of the M-CSF receptor, the Tnfaip8l2 protein (Q9D8Y7), which acts as a negative regulator downstream of some TLRs,88 and the TMEM43 protein (Q9DBS1), which acts as a negative regulator of the cGAS/STING interferon response pathway.89 Interestingly, the cGAS/STING pathway has been implicated in the PS particle-induced liver inflammation and fibrosis.72
In view of this complex landscape of responses, we decided to perform simple validation experiments in measuring the secretion of the pro-inflammatory cytokines IL-6 and TNF-alpha in response to exposure to PLA or PS nanoparticles, with and without terminal activation with LPS. The results were not consistent between the two cytokines, indicating that their regulations are not strictly similar. For IL-6, we did not detect any secretion without LPS stimulation, indicating that neither PLA nor PS alone induced its secretion. However, when cells were exposed to plastics and then to LPS, an important reduction in IL-6 secretion was observed. This means in turn that plastic-exposed macrophages are less performing in their antibacterial response compared to plastic-free macrophages. For TNF alpha, the same trend was observed with or without LPS activation, i.e. an increase in TNF secretion upon exposure to plastic, the effect being more pronounced for PLA than for PS. This effect is consistent with the one already described on RAW264.7,40i.e. another mouse macrophage cell line. These diverging results between IL-6 and TNF-alpha showed how complex the inflammatory response to plastics can be. The important point to be underlined here is that although biodegradable, PLA induces at least a transient pro-inflammatory response at the TNF level.
Finally, we investigated the redox metabolism of plastic-treated macrophages. We started with the ROS production, as overproduction of hydrogen peroxide, which is diffusible, may contribute to damage of the adjacent tissues.90 In macrophages, hydrogen peroxide is produced by the combined action of NADPH oxidase, which produces superoxide and superoxide dismutase, which transforms superoxide into hydrogen peroxide. Here again, the proteomic screen revealed inconsistent results. The levels of the NADPH oxidase subunits Ncf1 and Ncf4 were constant, while the level of cytochrome b245 (Q61093), the protein that transfers single electrons to oxygen atoms to produce superoxide increased, and the level of superoxide dismutase decreased. Thus, no clear trend emerged from the proteomic screen. However, targeted validation experiments showed that PLA nanoparticles did not induce any overproduction of ROS in cells, while PS nanoparticles induced a moderate but significant overproduction, as analyzed in more detail previously.42 Thus in this regard, PLA particles may induce less oxidative damage to the surrounding tissue than PS particles.
In the proteins grouped under the “redox homeostasis” header, two proteins drew our attention. First the AKR7A2 oxidoreductase (P45376) and then zeta-crystallin, also known as the QOR quinone oxidoreductase (P47199). The latter protein is different from the more well-known NQO1 quinone reductase (Q64669). Opposite to NQO1, whose catalytic mechanism ensures a two-electron reduction process,91 both QOR (which is known to produce semiquinones92) and AKR7A2 are able to induce redox cycling with their preferential substrate 9,10-phenanthrenequinone.92,93 This redox cycling is known to induce in turn adverse effects such as cell death and malignant transformation.94 We therefore decided to investigate whether treatment of macrophages with PLA could induce a cross-toxicity with 9,10-phenanthrenequinone. As a control, we used menadione, which is not a substrate for at least zeta-crystallin.92 Although the toxicity shown by 9,10-phenanthrenequinone was characterized by high standard deviations, which decreased the statistical significance of the results, we could observe a differential toxicity of 9,10-phenanthrenequinone in cells treated with plastics (either PLA or PS) and control cells. This differential toxicity was not observed with menadione.
5. Conclusions
The first important conclusion of this work on PLA particles is that they are biodegradable within macrophages. However, this does not mean that they do not show immediate consequences on the physiology of macrophages, as shown by the proteome alteration that they induce. Comparison with PS nanoparticles shows that there are both common consequences and specific ones. Among the specific consequences are the inhibition of phagocytosis by the PLA nanoparticles and the absence of detectable oxidative stress. Among the shared ones are the impact on cytokine production, the induction of lysosomal acidification and the cross-toxicity with phenanthrene quinone. Phenanthrene quinone is found in combustion products and is also one of the compounds produced by the metabolic activation of phenanthrene,95 a combustion product itself. This suggests that some nanoplastics may induce cross-effects with chemicals present in, e.g., diesel exhaust particles or cigarette smoke, which may be another mechanism underlying noxious effects of nanoplastics.Data availability
The proteomic data have been deposited in the ProteomeXchange Consortium database and are available through the DOI: https://doi.org/10.6019/PXD048664. The data on targeted experiments are available with the DOI: https://doi.org/10.6019/S-BSST1470 from the EMBL BioStudies database (https://www.ebi.ac.uk/biostudies/studies/S-BSST1470).Author contributions
VCF: methodology, investigation. MV: methodology, investigation, writing (review and editing). HD: methodology, investigation, writing (review and editing). SC: resources, funding acquisition, writing (review and editing). ED: formal analysis, writing (review and editing). TR: conceptualization, formal analysis, funding acquisition, writing (original draft).Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work used the flow cytometry facility and the microscopy facility MuLife of IRIG/DBSCI, funded by CEA Nanobio and by GRAL LabEX (ANR-10-LABX-49-01), a project of the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003) as well as the platforms of the French Proteomic Infrastructure (ProFI) project (grant ANR-10-INBS-08-03). This work was carried out in the frame of the PlasticHeal project, which has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No. 965196. This work was also supported by the ANR Plastox project (grant ANR-21-CE34-0028-04).References
- R. Lehner, C. Weder, A. Petri-Fink and B. Rothen-Rutishauser, Emergence of Nanoplastic in the Environment and Possible Impact on Human Health, Environ. Sci. Technol., 2019, 53, 1748–1765 CrossRef CAS PubMed.
- Y. V. Fan, P. Jiang, R. R. Tan, K. B. Aviso, F. You, X. Zhao, C. T. Lee and J. J. Klemeš, Forecasting plastic waste generation and interventions for environmental hazard mitigation, J. Hazard. Mater., 2022, 424, 127330 CrossRef CAS PubMed.
- J. G. B. Derraik, The pollution of the marine environment by plastic debris: a review, Mar. Pollut. Bull., 2002, 44, 842–852 CrossRef CAS PubMed.
- H. S. Charlton-Howard, A. L. Bond, J. Rivers-Auty and J. L. Lavers, ‘Plasticosis’: Characterising macro- and microplastic-associated fibrosis in seabird tissues, J. Hazard. Mater., 2023, 450, 131090 CrossRef CAS PubMed.
- K. Enders, R. Lenz, C. A. Stedmon and T. G. Nielsen, Abundance, size and polymer composition of marine microplastics ≥ 10 μm in the Atlantic Ocean and their modelled vertical distribution, Mar. Pollut. Bull., 2015, 100, 70–81 CrossRef CAS PubMed.
- A. Ter Halle, L. Jeanneau, M. Martignac, E. Jardé, B. Pedrono, L. Brach and J. Gigault, Nanoplastic in the North Atlantic Subtropical Gyre, Environ. Sci. Technol., 2017, 51, 13689–13697 CrossRef CAS PubMed.
- J. A. Brandon, A. Freibott and L. M. Sala, Patterns of suspended and salp-ingested microplastic debris in the North Pacific investigated with epifluorescence microscopy, Limnol. Oceanogr. Lett., 2020, 5, 46–53 CrossRef.
- M. Kedzierski, M. Palazot, L. Soccalingame, M. Falcou-Préfol, G. Gorsky, F. Galgani, S. Bruzaud and M. L. Pedrotti, Chemical composition of microplastics floating on the surface of the Mediterranean Sea, Mar. Pollut. Bull., 2022, 174, 113284 CrossRef CAS PubMed.
- E. S. Jones, S. W. Ross, C. M. Robertson and C. M. Young, Distributions of microplastics and larger anthropogenic debris in Norfolk Canyon, Baltimore Canyon, and the adjacent continental slope (Western North Atlantic Margin, U.S.A.), Mar. Pollut. Bull., 2021, 174, 113047 CrossRef PubMed.
- L. Cutroneo, M. Capello, A. Domi, S. Consani, P. Lamare, P. Coyle, V. Bertin, D. Dornic, A. Reboa, I. Geneselli and M. Anghinolfi, Microplastics in the abyss: a first investigation into sediments at 2443-m depth (Toulon, France), Environ. Sci. Pollut. Res., 2022, 29, 9375–9385 CrossRef CAS PubMed.
- T. Mani, A. Hauk, U. Walter and P. Burkhardt-Holm, Microplastics profile along the Rhine River, Sci. Rep., 2015, 5, 17988 CrossRef CAS PubMed.
- C. Scherer, A. Weber, F. Stock, S. Vurusic, H. Egerci, C. Kochleus, N. Arendt, C. Foeldi, G. Dierkes, M. Wagner, N. Brennholt and G. Reifferscheid, Comparative assessment of microplastics in water and sediment of a large European river, Sci. Total Environ., 2020, 738, 139866 CrossRef CAS PubMed.
- L. Weiss, W. Ludwig, S. Heussner, M. Canals, J.-F. Ghiglione, C. Estournel, M. Constant and P. Kerhervé, The missing ocean plastic sink: Gone with the rivers, Science, 2021, 373, 107–111 CrossRef CAS PubMed.
- C. J. Weber, C. Opp, J. A. Prume, M. Koch, T. J. Andersen and P. Chifflard, Deposition and in-situ translocation of microplastics in floodplain soils, Sci. Total Environ., 2021, 152039 Search PubMed.
- H. Golwala, X. Zhang, S. M. Iskander and A. L. Smith, Solid waste: An overlooked source of microplastics to the environment, Sci. Total Environ., 2021, 769, 144581 CrossRef CAS PubMed.
- A. Chamas, H. Moon, J. Zheng, Y. Qiu, T. Tabassum, J. H. Jang, M. Abu-Omar, S. L. Scott and S. Suh, Degradation Rates of Plastics in the Environment, ACS Sustainable Chem. Eng., 2020, 8, 3494–3511 CrossRef CAS.
- L. Lei, M. Liu, Y. Song, S. Lu, J. Hu, C. Cao, B. Xie, H. Shi and D. He, Polystyrene (nano)microplastics cause size-dependent neurotoxicity, oxidative damage and other adverse effects in Caenorhabditis elegans, Environ. Sci.: Nano, 2018, 5, 2009–2020 RSC.
- X. Jiang, Y. Chang, T. Zhang, Y. Qiao, G. Klobučar and M. Li, Toxicological effects of polystyrene microplastics on earthworm (Eisenia fetida), Environ. Pollut., 2020, 259, 113896 CrossRef CAS PubMed.
- R. Sussarellu, M. Suquet, Y. Thomas, C. Lambert, C. Fabioux, M. E. J. Pernet, N. Le Goïc, V. Quillien, C. Mingant, Y. Epelboin, C. Corporeau, J. Guyomarch, J. Robbens, I. Paul-Pont, P. Soudant and A. Huvet, Oyster reproduction is affected by exposure to polystyrene microplastics, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2430–2435 CrossRef CAS PubMed.
- Z. Liu, P. Yu, M. Cai, D. Wu, M. Zhang, Y. Huang and Y. Zhao, Polystyrene nanoplastic exposure induces immobilization, reproduction, and stress defense in the freshwater cladoceran Daphnia pulex, Chemosphere, 2019, 215, 74–81 CrossRef CAS PubMed.
- Y. Song, C. Cao, R. Qiu, J. Hu, M. Liu, S. Lu, H. Shi, K. M. Raley-Susman and D. He, Uptake and adverse effects of polyethylene terephthalate microplastics fibers on terrestrial snails (Achatina fulica) after soil exposure, Environ. Pollut., 2019, 250, 447–455 CrossRef CAS PubMed.
- A. H. D'Costa, Microplastics in decapod crustaceans: Accumulation, toxicity and impacts, a review, Sci. Total Environ., 2022, 832, 154963 CrossRef PubMed.
- O. Pikuda, E. Roubeau Dumont, Q. Chen, J.-R. Macairan, S. A. Robinson, D. Berk and N. Tufenkji, Toxicity of microplastics and nanoplastics to Daphnia magna: Current status, knowledge gaps and future directions, TrAC, Trends Anal. Chem., 2023, 167, 117208 CrossRef CAS.
- A. Muhammad, X. Zhou, J. He, N. Zhang, X. Shen, C. Sun, B. Yan and Y. Shao, Toxic effects of acute exposure to polystyrene microplastics and nanoplastics on the model insect, silkworm Bombyx mori, Environ. Pollut., 2021, 285, 117255 CrossRef CAS PubMed.
- A. T. B. Guimarães, A. S. De Lima Rodrigues, P. S. Pereira, F. G. Silva and G. Malafaia, Toxicity of polystyrene nanoplastics in dragonfly larvae: An insight on how these pollutants can affect bentonic macroinvertebrates, Sci. Total Environ., 2021, 752, 141936 CrossRef PubMed.
- M. Alaraby, D. Abass, J. Domenech, A. Hernández and R. Marcos, Hazard assessment of ingested polystyrene nanoplastics in Drosophila larvae, Environ. Sci.: Nano, 2022, 9, 1845–1857 RSC.
- N. R. Brun, P. van Hage, E. R. Hunting, A.-P. G. Haramis, S. C. Vink, M. G. Vijver, M. J. M. Schaaf and C. Tudorache, Polystyrene nanoplastics disrupt glucose metabolism and cortisol levels with a possible link to behavioural changes in larval zebrafish, Commun. Biol., 2019, 2, 382 CrossRef PubMed.
- I. Brandts, M. Garcia-Ordoñez, L. Tort, M. Teles and N. Roher, Polystyrene nanoplastics accumulate in ZFL cell lysosomes and in zebrafish larvae after acute exposure, inducing a synergistic immune response in vitro without affecting larval survival in vivo, Environ. Sci.: Nano, 2020, 7, 2410–2422 RSC.
- W. Gu, S. Liu, L. Chen, Y. Liu, C. Gu, H. Ren and B. Wu, Single-Cell RNA Sequencing Reveals Size-Dependent Effects of Polystyrene Microplastics on Immune and Secretory Cell Populations from Zebrafish Intestines, Environ. Sci. Technol., 2020, 54, 3417–3427 CrossRef CAS PubMed.
- X. Li, T. Zhang, W. Lv, H. Wang, H. Chen, Q. Xu, H. Cai and J. Dai, Intratracheal administration of polystyrene microplastics induces pulmonary fibrosis by activating oxidative stress and Wnt/β-catenin signaling pathway in mice, Ecotoxicol. Environ. Saf., 2022, 232, 113238 CrossRef CAS PubMed.
- V. Stock, L. Böhmert, E. Lisicki, R. Block, J. Cara-Carmona, L. K. Pack, R. Selb, D. Lichtenstein, L. Voss, C. J. Henderson, E. Zabinsky, H. Sieg, A. Braeuning and A. Lampen, Uptake and effects of orally ingested polystyrene microplastic particles in vitro and in vivo, Arch. Toxicol., 2019, 93, 1817–1833 CrossRef CAS PubMed.
- J. Domenech, A. Hernández, L. Rubio, R. Marcos and C. Cortés, Interactions of polystyrene nanoplastics with in vitro models of the human intestinal barrier, Arch. Toxicol., 2020, 94, 2997–3012 CrossRef CAS PubMed.
- S. Ballesteros, J. Domenech, I. Barguilla, C. Cortés, R. Marcos and A. Hernández, Genotoxic and immunomodulatory effects in human white blood cells after ex vivo exposure to polystyrene nanoplastics, Environ. Sci.: Nano, 2020, 7, 3431–3446 RSC.
- C. Meindl, K. Öhlinger, V. Zrim, T. Steinkogler and E. Fröhlich, Screening for Effects of Inhaled Nanoparticles in Cell Culture Models for Prolonged Exposure, Nanomaterials, 2021, 11, 606 CrossRef CAS PubMed.
- J. Antunes, P. Sobral, M. Martins and V. Branco, Nanoplastics activate a TLR4/p38-mediated pro-inflammatory response in human intestinal and mouse microglia cells, Environ. Toxicol. Pharmacol., 2023, 104, 104298 CrossRef CAS PubMed.
- M. Zhang, J. Li, G. Xing, R. He, W. Li, Y. Song and H. Guo, Variation in the internalization of differently sized nanoparticles induces different DNA-damaging effects on a macrophage cell line, Arch. Toxicol., 2011, 85, 1575–1588 CrossRef CAS PubMed.
- B. Prietl, C. Meindl, E. Roblegg, T. R. Pieber, G. Lanzer and E. Fröhlich, Nano-sized and micro-sized polystyrene particles affect phagocyte function, Cell Biol. Toxicol., 2014, 30, 1–16 CrossRef CAS PubMed.
- V. Paget, S. Dekali, T. Kortulewski, R. Grall, C. Gamez, K. Blazy, O. Aguerre-Chariol, S. Chevillard, A. Braun, P. Rat and G. Lacroix, Specific Uptake and Genotoxicity Induced by Polystyrene Nanobeads with Distinct Surface Chemistry on Human Lung Epithelial Cells and Macrophages, PLoS One, 2015, 10, e0123297 CrossRef PubMed.
- I. Florance, S. Ramasubbu, A. Mukherjee and N. Chandrasekaran, Polystyrene nanoplastics dysregulate lipid metabolism in murine macrophages in vitro, Toxicology, 2021, 458, 152850 CrossRef CAS PubMed.
- Q. Hu, H. Wang, C. He, Y. Jin and Z. Fu, Polystyrene nanoparticles trigger the activation of p38 MAPK and apoptosis via inducing oxidative stress in zebrafish and macrophage cells, Environ. Pollut., 2021, 269, 116075 CrossRef CAS PubMed.
- V. Collin-Faure, B. Dalzon, J. Devcic, H. Diemer, S. Cianférani and T. Rabilloud, Does size matter? A proteomics-informed comparison of the effects of polystyrene beads of different sizes on macrophages, Environ. Sci.: Nano, 2022, 9, 2827–2840 RSC.
- V. Collin-Faure, M. Vitipon, A. Torres, O. Tanyeres, B. Dalzon and T. Rabilloud, The internal dose makes the poison: higher internalization of polystyrene particles induce increased perturbation of macrophages, Front. Immunol., 2023, 14, 1092743 CrossRef CAS PubMed.
- D. G. Lundgren, R. Alper, C. Schnaitman and R. H. Marchessault, Characterization of Poly-Beta-Hydroxybutyrate extracted from different bacteria, J. Bacteriol., 1965, 89, 245–251 CrossRef CAS PubMed.
- R. Bhati, S. Samantaray, L. Sharma and N. Mallick, Poly-β-hydroxybutyrate accumulation in cyanobacteria under photoautotrophy, Biotechnol. J., 2010, 5, 1181–1185 CrossRef CAS PubMed.
- D. Jendrossek and D. Pfeiffer, New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate), Environ. Microbiol., 2014, 16, 2357–2373 CrossRef CAS PubMed.
- L. H. Gracioso, A. Bellan, B. Karolski, L. O. B. Cardoso, E. A. Perpetuo, C. A. O. do Nascimento, R. Giudici, V. Pizzocchero, M. Basaglia and T. Morosinotto, Light excess stimulates Poly-beta-hydroxybutyrate yield in a mangrove-isolated strain of Synechocystis sp, Bioresour. Technol., 2021, 320, 124379 CrossRef CAS PubMed.
- P. Cabecas Segura, R. Onderwater, A. Deutschbauer, L. Dewasme, R. Wattiez and B. Leroy, Study of the Production of Poly(Hydroxybutyrate-co-Hydroxyhexanoate) and Poly(Hydroxybutyrate-co-Hydroxyvalerate-co-Hydroxyhexanoate) in Rhodospirillum rubrum, Appl. Environ. Microbiol., 2022, 88, e0158621 CrossRef PubMed.
- D. E. Cutright and E. E. Hunsuck, Tissue reaction to the biodegradable polylactic acid suture, Oral Surg., Oral Med., Oral Pathol., 1971, 31, 134–139 CrossRef CAS PubMed.
- D. E. Cutright, J. D. Beasley and B. Perez, Histologic comparison of polylactic and polyglycolic acid sutures, Oral Surg., Oral Med., Oral Pathol., 1971, 32, 165–173 CrossRef CAS PubMed.
- R. K. Kulkarni, E. G. Moore, A. F. Hegyeli and F. Leonard, Biodegradable poly(lactic acid) polymers, J. Biomed. Mater. Res., 1971, 5, 169–181 CrossRef CAS PubMed.
- Y. Kawashima, H. Yamamoto, H. Takeuchi, S. Fujioka and T. Hino, Pulmonary delivery of insulin with nebulized DL-lactide/glycolide copolymer (PLGA) nanospheres to prolong hypoglycemic effect, J. Controlled Release, 1999, 62, 279–287 CrossRef CAS PubMed.
- O. Pillai and R. Panchagnula, Polymers in drug delivery, Curr. Opin. Chem. Biol., 2001, 5, 447–451 CrossRef CAS PubMed.
- S. Freiberg and X. X. Zhu, Polymer microspheres for controlled drug release, Int. J. Pharm., 2004, 282, 1–18 CrossRef CAS PubMed.
- M. S. Shive and J. M. Anderson, Biodegradation and biocompatibility of PLA and PLGA microspheres, Adv. Drug Delivery Rev., 1997, 28, 5–24 CrossRef PubMed.
- S. Jesus, M. Schmutz, C. Som, G. Borchard, P. Wick and O. Borges, Hazard Assessment of Polymeric Nanobiomaterials for Drug Delivery: What Can We Learn From Literature So Far, Front. Bioeng. Biotechnol., 2019, 7, 261 CrossRef PubMed.
- M. H. Wolf, O. Gil-Castell, J. Cea, J. C. Carrasco and A. Ribes-Greus, Degradation of Plasticised Poly(lactide) Composites with Nanofibrillated Cellulose in Different Hydrothermal Environments, J. Polym. Environ., 2023, 31, 2055–2072 CrossRef CAS.
- G. Banaei, A. García-Rodríguez, A. Tavakolpournegari, J. Martín-Pérez, A. Villacorta, R. Marcos and A. Hernández, The release of polylactic acid nanoplastics (PLA-NPLs) from commercial teabags. Obtention, characterization, and hazard effects of true-to-life PLA-NPLs, J. Hazard. Mater., 2023, 458, 131899 CrossRef CAS PubMed.
- N. Bhutiani, A. Samykutty, K. M. McMasters, N. K. Egilmez and L. R. McNally, In vivo tracking of orally-administered particles within the gastrointestinal tract of murine models using multispectral optoacoustic tomography, Photoacoustics, 2019, 13, 46–52 CrossRef PubMed.
- T. Behnke, C. Würth, K. Hoffmann, M. Hübner, U. Panne and U. Resch-Genger, Encapsulation of Hydrophobic Dyes in Polystyrene Micro- and Nanoparticles via Swelling Procedures, J. Fluoresc., 2011, 21, 937–944 CrossRef CAS PubMed.
- A. Torres, V. Collin-Faure, D. Fenel, J.-A. Sergent and T. Rabilloud, About the Transient Effects of Synthetic Amorphous Silica: An In Vitro Study on Macrophages, Int. J. Mol. Sci., 2022, 24, 220 CrossRef PubMed.
- B. Dalzon, A. Torres, J. Devcic, D. Fenel, J.-A. Sergent and T. Rabilloud, A Low-Serum Culture System for Prolonged in Vitro Toxicology Experiments on a Macrophage System, Front. Toxicol., 2021, 3, 780778 CrossRef PubMed.
- L. Muller, L. Fornecker, M. Chion, A. Van Dorsselaer, S. Cianférani, T. Rabilloud and C. Carapito, Extended investigation of tube-gel sample preparation: a versatile and simple choice for high throughput quantitative proteomics, Sci. Rep., 2018, 8, 8260 CrossRef PubMed.
- C. Cavazza, V. Collin-Faure, J. Pérard, H. Diemer, S. Cianférani, T. Rabilloud and E. Darrouzet, Proteomic analysis of Rhodospirillum rubrum after carbon monoxide exposure reveals an important effect on metallic cofactor biosynthesis, J. Proteomics, 2022, 250, 104389 CrossRef CAS PubMed.
- T. Lyubimova, S. Caglio, C. Gelfi, P. G. Righetti and T. Rabilloud, Photopolymerization of polyacrylamide gels with methylene blue, Electrophoresis, 1993, 14, 40–50 CrossRef CAS PubMed.
- O. Hammer, D. A. T. Harper and P. D. Ryan, Paleontological statistics software package for education and data analysis, Palaeontol. Electron., 2001, 4, 9 Search PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat. Protoc., 2009, 4, 44–57 CrossRef CAS PubMed.
- S. W. Perry, J. P. Norman, J. Barbieri, E. B. Brown and H. A. Gelbard, Mitochondrial membrane potential probes and the proton gradient: a practical usage guide, BioTechniques, 2011, 50, 98–115 CrossRef CAS PubMed.
- J. F. Keij, C. Bell-Prince and J. A. Steinkamp, Staining of mitochondrial membranes with 10-nonyl acridine orange, MitoFluor Green, and MitoTracker Green is affected by mitochondrial membrane potential altering drugs, Cytometry, 2000, 39, 203–210 CrossRef CAS PubMed.
- K. R. Clarke, Non-parametric multivariate analyses of changes in community structure, Aust. J. Ecol., 1993, 18, 117–143 CrossRef.
- M. Boyce, K. F. Bryant, C. Jousse, K. Long, H. P. Harding, D. Scheuner, R. J. Kaufman, D. Ma, D. M. Coen, D. Ron and J. Yuan, A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress, Science, 2005, 307, 935–939 CrossRef CAS PubMed.
- T. L. Pan, P. W. Wang, Y. C. Hung, C. H. Huang and K. M. Rau, Proteomic analysis reveals tanshinone IIA enhances apoptosis of advanced cervix carcinoma CaSki cells through mitochondria intrinsic and endoplasmic reticulum stress pathways, Proteomics, 2013, 13, 3411–3423 CrossRef CAS PubMed.
- R. Shen, K. Yang, X. Cheng, C. Guo, X. Xing, H. Sun, D. Liu, X. Liu and D. Wang, Accumulation of polystyrene microplastics induces liver fibrosis by activating cGAS/STING pathway, Environ. Pollut., 2022, 300, 118986 CrossRef CAS PubMed.
- R. Marfella, F. Prattichizzo, C. Sardu, G. Fulgenzi, L. Graciotti, T. Spadoni, N. D'Onofrio, L. Scisciola, R. La Grotta, C. Frigé, V. Pellegrini, M. Municinò, M. Siniscalchi, F. Spinetti, G. Vigliotti, C. Vecchione, A. Carrizzo, G. Accarino, A. Squillante, G. Spaziano, D. Mirra, R. Esposito, S. Altieri, G. Falco, A. Fenti, S. Galoppo, S. Canzano, F. C. Sasso, G. Matacchione, F. Olivieri, F. Ferraraccio, I. Panarese, P. Paolisso, E. Barbato, C. Lubritto, M. L. Balestrieri, C. Mauro, A. E. Caballero, S. Rajagopalan, A. Ceriello, B. D'Agostino, P. Iovino and G. Paolisso, Microplastics and Nanoplastics in Atheromas and Cardiovascular Events, N. Engl. J. Med., 2024, 390, 900–910 CrossRef CAS PubMed.
- K. Hamidzadeh, S. M. Christensen, E. Dalby, P. Chandrasekaran and D. M. Mosser, Macrophages and the Recovery from Acute and Chronic Inflammation, Annu. Rev. Physiol., 2017, 79, 567–592 CrossRef CAS PubMed.
- V. Olivier, J. L. Duval, M. Hindié, P. Pouletaut and M. D. Nagel, Comparative particle-induced cytotoxicity toward macrophages and fibroblasts, Cell Biol. Toxicol., 2003, 19, 145–159 CrossRef CAS PubMed.
- D. F. Williams, Enzymic Hydrolysis of Polylactic Acid, Eng. Med., 1981, 10, 5–7 CrossRef.
- G. A. Brooks, Lactate as a fulcrum of metabolism, Redox Biol., 2020, 35, 101454 CrossRef CAS PubMed.
- C. Manosalva, J. Quiroga, A. I. Hidalgo, P. Alarcón, N. Anseoleaga, M. A. Hidalgo and R. A. Burgos, Role of Lactate in Inflammatory Processes: Friend or Foe, Front. Immunol., 2021, 12, 808799 CrossRef CAS PubMed.
- A. Drazic, L. M. Myklebust, R. Ree and T. Arnesen, The world of protein acetylation, Biochim. Biophys. Acta, 2016, 1864, 1372–1401 CrossRef CAS PubMed.
- E. B. Ketema and G. D. Lopaschuk, Post-translational Acetylation Control of Cardiac Energy Metabolism, Front. Cardiovasc. Med., 2021, 8, 723996 CrossRef CAS PubMed.
- Y.-H. Yang, R. Wen, N. Yang, T.-N. Zhang and C.-F. Liu, Roles of protein post-translational modifications in glucose and lipid metabolism: mechanisms and perspectives, Mol. Med., 2023, 29, 93 CAS.
- H.-J. Lin, P. Herman, J. S. Kang and J. R. Lakowicz, Fluorescence Lifetime Characterization of Novel Low-pH Probes, Anal. Biochem., 2001, 294, 118–125 CrossRef CAS PubMed.
- Y. Zhao, Z. Ye, D. Song, D. Wich, S. Gao, J. Khirallah and Q. Xu, Nanomechanical action opens endo-lysosomal compartments, Nat. Commun., 2023, 14, 6645 CrossRef CAS PubMed.
- J. Ai, A. Maturu, W. Johnson, Y. Wang, C. B. Marsh and S. Tridandapani, The inositol phosphatase SHIP-2 down-regulates FcgammaR-mediated phagocytosis in murine macrophages independently of SHIP-1, Blood, 2006, 107, 813–820 CrossRef CAS PubMed.
- J. M. Rubio, A. M. Astudillo, J. Casas, M. A. Balboa and J. Balsinde, Regulation of Phagocytosis in Macrophages by Membrane Ethanolamine Plasmalogens, Front. Immunol., 2018, 9, 1723 CrossRef PubMed.
- Y. Zhao, C. Alonso, I. Ballester, J. H. Song, S. Y. Chang, B. Guleng, S. Arihiro, P. J. Murray, R. Xavier, K. S. Kobayashi and H.-C. Reinecker, Control of NOD2 and Rip2-dependent innate immune activation by GEF-H1, Inflammatory Bowel Dis., 2012, 18, 603–612 CrossRef PubMed.
- D. Wang, J. Lou, C. Ouyang, W. Chen, Y. Liu, X. Liu, X. Cao, J. Wang and L. Lu, Ras-related protein Rab10 facilitates TLR4 signaling by promoting replenishment of TLR4 onto the plasma membrane, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 13806–13811 CrossRef CAS PubMed.
- H. Sun, S. Gong, R. J. Carmody, A. Hilliard, L. Li, J. Sun, L. Kong, L. Xu, B. Hilliard, S. Hu, H. Shen, X. Yang and Y. H. Chen, TIPE2, a negative regulator of innate and adaptive immunity that maintains immune homeostasis, Cell, 2008, 133, 415–426 CrossRef CAS PubMed.
- E. J. Fenech, F. Lari, P. D. Charles, R. Fischer, M. Laétitia-Thézénas, K. Bagola, A. W. Paton, J. C. Paton, M. Gyrd-Hansen, B. M. Kessler and J. C. Christianson, Interaction mapping of endoplasmic reticulum ubiquitin ligases identifies modulators of innate immune signalling, eLife, 2020, 9, e57306 CrossRef CAS PubMed.
- J. E. Klaunig and L. M. Kamendulis, The role of oxidative stress in carcinogenesis, Annu. Rev. Pharmacol. Toxicol., 2004, 44, 239–267 CrossRef CAS PubMed.
- R. Li, M. A. Bianchet, P. Talalay and L. M. Amzel, The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 8846–8850 CrossRef CAS PubMed.
- P. V. Rao, C. M. Krishna and J. S. Zigler, Identification and characterization of the enzymatic activity of zeta-crystallin from guinea pig lens. A novel NADPH:quinone oxidoreductase, J. Biol. Chem., 1992, 267, 96–102 CrossRef CAS PubMed.
- C. A. Shultz, A. M. Quinn, J.-H. Park, R. G. Harvey, J. L. Bolton, E. Maser and T. M. Penning, Specificity of human aldo-keto reductases, NAD(P)H:quinone oxidoreductase, and carbonyl reductases to redox-cycle polycyclic aromatic hydrocarbon diones and 4-hydroxyequilenin-o-quinone, Chem. Res. Toxicol., 2011, 24, 2153–2166 Search PubMed.
- J. L. Bolton, M. A. Trush, T. M. Penning, G. Dryhurst and T. J. Monks, Role of quinones in toxicology, Chem. Res. Toxicol., 2000, 13, 135–160 Search PubMed.
- K. Luo, X. Luo, W. Cao, J. B. Hochalter, V. Paiano, C. J. Sipe, S. G. Carmella, S. E. Murphy, J. Jensen, S. Lam, A. P. Golin, L. Bergstrom, D. Midthun, N. Fujioka, D. Hatsukami and S. S. Hecht, Cigarette smoking enhances the metabolic activation of the polycyclic aromatic hydrocarbon phenanthrene in humans, Carcinogenesis, 2021, 42, 570–577 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4en00335g |
| This journal is © The Royal Society of Chemistry 2024 |