 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ciprofloxacin as a tryptophan mimic within an antimicrobial peptide†
John R. F. B.
Connolly
 *a,
Deirdre
Fitzgerald-Hughes
*a,
Deirdre
Fitzgerald-Hughes
 b,
Marc
Maresca
b,
Marc
Maresca
 c,
Jimmy
Muldoon
c,
Jimmy
Muldoon
 d and
Marc
Devocelle
d and
Marc
Devocelle
 a
a
aDepartment of Chemistry, RCSI University of Medicine and Health Sciences, Dublin, Ireland. E-mail: johnrfbconnolly@gmail.com
bDepartment of Microbiology, RCSI University of Medicine and Health Sciences, Dublin, Ireland
cAix Marseille Univ, CNRS, Centrale Med, ISM2, 13013, Marseille, France
dSchool of Chemistry, University College Dublin, Dublin, Ireland
First published on 13th August 2024
Abstract
Ciprofloxacin has been used to replace tryptophan at positions 3 and 6 in the antimicrobial peptide (AMP) Bac8c. Bac8c(Cip3,6) showed comparable antimicrobial activity but increased selectivity toward some Gram-negative bacteria with MIC values of ≤6.25 μM. Bac8c(Cip3,6) was also non-cytotoxic to cultured HepG2 cells at antimicrobial concentrations, with a CC50 >300 μM.
Antimicrobial resistance (AMR) has become a global health threat with an estimated 10 million deaths predicted by 2050.1 Antimicrobial peptides (AMPs) are of particular interest to combat this challenge due to their natural origins, alternative and multimodal mechanisms of action and a tendency for low promotion of AMR compared to conventional antibiotics.2,3 AMPs are a diverse group of peptides with a variety of lengths (usually 12–50) and sequences of amino acids, charge, function and mechanism of action. Despite these diverse structures, most AMPs share similar characteristics, such as the ability to form an amphiphilic structure and an overall cationic charge, which allows stronger and more selective interactions with anionic bacterial membranes compared to human cell membranes.3 Consequently, conjugation of small biomolecules and peptide mimetics of AMPs has been applied to exploit these advantageous properties while increasing effective and selective killing of bacteria.4
One group of antibiotics which are increasingly associated with AMR and has been used as a peptide conjugate, are fluoroquinolones (FQ).5 Peptides used in conjugation with FQs include cell penetrating peptides (CPPs),6,7 AMPs8,9 and homopoly(α-amino acids) for physicochemical property modifications,10 with variable results. Further modification of peptides has also produced peptidomimetics, molecules designed to mimic a peptide, usually containing non-peptide moieties.11 One amino acid which has been modified or replaced for the purposes of a peptidomimetic is tryptophan (Trp), due to its aromatic nature and unique contributions to membrane interactions and subsequently antimicrobial activity.12–15 Mimics of Trp have been used for several applications including fluorescence16–18 and some mechanistic studies.19
These peptide conjugate and mimetic approaches have been developed relatively independently to date. To bridge this gap between peptide mimetic and conjugate, molecular hybridisation, the combining of two or more pharmacophores into a single entity, has been employed to produce a series of hybrid mimetic peptides. Peptides containing an FQ as a replacement for Trp have been proposed and synthesised to address this and may offer combined advantages of both peptide conjugate and mimic. Ciprofloxacin (Cip) and Bac8c, a FQ and AMP respectively, were chosen as candidates for this research. Ciprofloxacin is a broad spectrum FQ which has been used clinically for decades and has a well understood mechanism of action as well as pharmacokinetic/pharmacodynamic profiles.20 Bac8c (RIWVIWRR-NH2) is an eight residue synthetic AMP also with a broad spectrum of activity and well documented mechanism of action.21 Analogues of Bac8c have also been previously reported including amino acid alteration and conjugation.22–24 Crucially, Bac8c contains two Trp residues regularly interspersed within the sequence, lending itself to systematic replacement with a ciprofloxacin unit to assess its impact on the antimicrobial activity and mechanism of action in comparison to the parent peptide.
The minimum inhibitory concentration (MIC) of the Bac8c-mimetic hybrids, the structures of which are shown in Fig. 1, were assessed against representative bacteria, Escherichia coli (E. coli, ATCC 25922) and Staphylococcus aureus (S. aureus, ATCC 25923), results of which are summarised in Table 1. MIC values for the analogues were dependent on the position of replacement. Comparison of the Bac8c(Cip3) and Bac8c(Cip6) analogues showed a noticeable difference in activity. An 8-fold decrease in MIC was observed against Gram-negative E. coli compared to Gram-positive S. aureus for Bac8c(Cip3), however, the activity of Bac8c(Cip6) was less discriminatory between the two bacteria. Compared to single substitutions, the doubly substituted Bac8c(Cip3,6) analogue showed almost a 4-fold and 10-fold lower MIC against E. coli than Bac8(Cip3) and Bac8c(Cip6) respectively, maintaining the activity of the parent peptide Bac8c against this organism. Interestingly, Bac8(Cip3,6) was less active than Bac8c and ciprofloxacin against Gram-positive S. aureus thereby modifying the broad-spectrum profile of the parent molecules.25 The specificity and low MIC of Bac8c(Cip3,6) towards E. coli warranted further investigation against ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanni, Pseudomonas aeruginosa and Enterobacter cloacae.) pathogens.25 These pathogens represent those associated with the greatest AMR threat and are therefore those for which the need for novel and effective treatment is most pressing. Susceptibility evaluation of these bacteria is important in progressing potential peptide-based therapeutics for priority AMR pathogens of clinical relevance.25 In addition, other Gram-positive (Bacillus cereus, Bacillus subtilis, Listeria monocytogenes and Staphylococcus epidermidis) as well as Gram-negative bacteria (Salmonella enterica and Shigella sonnei) were tested to further evaluate the selectivity of Bac8c(Cip3,6).
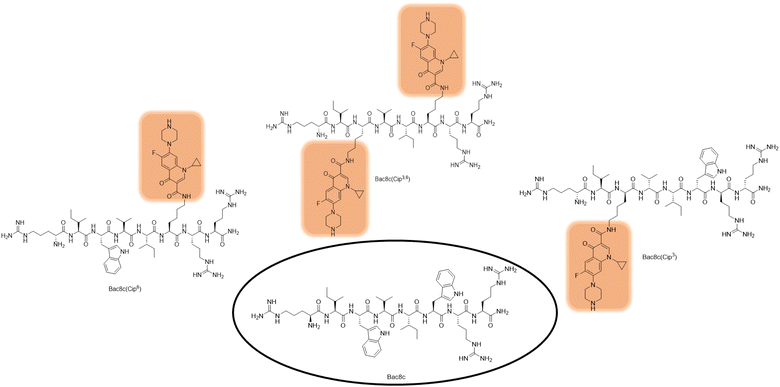 | ||
| Fig. 1 Structures of Bac8c and the Lys(Cip) analogues synthesised Bac8c(Cip3), Bac8c(Cip6) and Bac8c(Cip3,6). | ||
| Candidate | E.coli (μM) | S. aureus (μM) |
|---|---|---|
| Bac8c | 1.7 | 3.4 |
| Bac8c(Cip3) | 22.2 | 177.9 |
| Bac8c(Cip6) | 44.5 | 44.5–89 |
| Bac8c(Cip3,6) | 4.7 | 75.6 |
Tables 2 and 3 show the MICs and minimum bactericidal concentrations (MBCs) found for Bac8c, Bac8c(Cip3,6), ciprofloxacin and a simple mixture of ciprofloxacin and Bac8c in the same proportion as Bac8c(Cip3,6) (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), with the MICs/MBCs presented in the tables for this mixture, as ratios of concentrations of ciprofloxacin and Bac8c respectively. While the parent peptide and fluoroquinolone remained more active against both Gram-positive and Gram-negative bacteria compared to Bac8c(Cip3,6), the spectrum of activity for the latter appeared more targeted towards some Gram-negative bacteria, specifically P. aeruginosa, S. enterica and S. sonnei with MIC values of 6.25 μM in all cases, the same as Bac8c. Ciprofloxacin itself had MICs and MBCs of up to 1000 times lower than Bac8c or Bac8c(Cip3,6) against E. cloacae. The physical mixture of 2
1), with the MICs/MBCs presented in the tables for this mixture, as ratios of concentrations of ciprofloxacin and Bac8c respectively. While the parent peptide and fluoroquinolone remained more active against both Gram-positive and Gram-negative bacteria compared to Bac8c(Cip3,6), the spectrum of activity for the latter appeared more targeted towards some Gram-negative bacteria, specifically P. aeruginosa, S. enterica and S. sonnei with MIC values of 6.25 μM in all cases, the same as Bac8c. Ciprofloxacin itself had MICs and MBCs of up to 1000 times lower than Bac8c or Bac8c(Cip3,6) against E. cloacae. The physical mixture of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ciprofloxacin and Bac8c has MICs and MBCs generally equal to that of ciprofloxacin alone, suggesting that ciprofloxacin is the main active compound at such low concentrations and there is no additive or synergistic effect as seen in previous literature.26,27 Bac8c(Cip3,6) had interesting pathogen selectivity but the activity did not surpass the parent peptide Bac8c. The use of a ciprofloxacin rather than Trp residue may reduce the cost effectiveness of any clinical application. However, there may be additional inherent benefits in using a modified peptide, such as increased metabolic stability or better bioavailability which could contribute to reduced dosage ultimately.28
1 ciprofloxacin and Bac8c has MICs and MBCs generally equal to that of ciprofloxacin alone, suggesting that ciprofloxacin is the main active compound at such low concentrations and there is no additive or synergistic effect as seen in previous literature.26,27 Bac8c(Cip3,6) had interesting pathogen selectivity but the activity did not surpass the parent peptide Bac8c. The use of a ciprofloxacin rather than Trp residue may reduce the cost effectiveness of any clinical application. However, there may be additional inherent benefits in using a modified peptide, such as increased metabolic stability or better bioavailability which could contribute to reduced dosage ultimately.28
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) against Gram-positive bacteria
1) against Gram-positive bacteria
| Candidate | B. cereus (DSM 31) | B. subtilis (ATCC 6633) | E. faecalis (DSM 2570) | E. faecium (DSM 20477) | L. monocytogenes (DSM 20600) | S. epidermidis (DSM 20044) |
|---|---|---|---|---|---|---|
| Bac8c | 50 (50) | 3.12 (3.12) | 12.5 (25) | 12.5 (12.5) | 3.12 (3.12) | 3.12 (3.12) |
| Bac8c(Cip3,6) | 50 (50) | 12.5 (12.5) | 100 (>100) | 50 (100) | 12.5 (12.5) | 12.5 (12.5) |
| Cip | 0.39 (0.39) | 0.097 (0.097) | 3.12 (3.12) | 12.5 (12.5) | 25 (25) | 0.78 (0.78) |
Cip + Bac8c (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
0.39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 0.19 (0.39 0.19 (0.39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.19) 0.19) |
0.097![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 0.048 (0.097 0.048 (0.097![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.048) 0.048) |
3.12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1.56 (3.12 1.56 (3.12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.56) 1.56) |
12.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 6.25 (12.5 6.25 (12.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6.25) 6.25) |
6.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3.12 (6.25 3.12 (6.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.12) 3.12) |
0.78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 0.39 (0.78 0.39 (0.78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.39) 0.39) |
Bac8c(Cip3,6) appeared to be selective for certain Gram-negative bacteria, therefore potential cytotoxicity to eukaryotic cells was investigated. Replacement of a natural amino acid with a non-canonical one could influence eukaryotic cell interactions significantly. As such, cytotoxicity investigation of all candidates discussed in Tables 2 and 3 were performed against cultured human cells, taking human liver cells HepG2 as a model. Fig. 2 shows the cell viability when exposed to increasing concentrations of antimicrobial up to 300 μM. All antimicrobials had a concentration cytotoxic to 50% of cells (CC50) >300 μM, with cell viability being maintained above 90% for Bac8c, Bac8c(Cip3,6) and ciprofloxacin for all concentrations tested up to 300 μM. The 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of ciprofloxacin:Bac8c maintained over 90% cell viability up to 150 μM, after which there was a marked decrease in cell viability to 55% ± 13.3% of negative controls. High concentrations of fluoroquinolones are toxic to mammalian cells, as discussed in the literature reporting their anti-cancer properties and known toxicity/side effects clinically.29–31 For Bac8c(Cip3,6), although the ciprofloxacin was present in the same concentration/ratio as the Bac8c
1 mixture of ciprofloxacin:Bac8c maintained over 90% cell viability up to 150 μM, after which there was a marked decrease in cell viability to 55% ± 13.3% of negative controls. High concentrations of fluoroquinolones are toxic to mammalian cells, as discussed in the literature reporting their anti-cancer properties and known toxicity/side effects clinically.29–31 For Bac8c(Cip3,6), although the ciprofloxacin was present in the same concentration/ratio as the Bac8c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cip mixture, it did not appear to display toxicity to the mammalian cells when bound to the peptide. Furthermore, the Lys-Cip amide bond is stable in several environments. Data presented in Fig. 2 support the replacement of Trp within Bac8c(Cip3,6), as it did not compromise selectivity towards non-eukaryotic cells.
Cip mixture, it did not appear to display toxicity to the mammalian cells when bound to the peptide. Furthermore, the Lys-Cip amide bond is stable in several environments. Data presented in Fig. 2 support the replacement of Trp within Bac8c(Cip3,6), as it did not compromise selectivity towards non-eukaryotic cells.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) against Gram-negative bacteria
1) against Gram-negative bacteria
| Candidate | A. baumannii (DSM 30007) | E. cloacae (DSM 30054) | K. pneumoniae (DSM 30054) | P. aeruginosa (ATCC 9027) | S. enterica (CIP 80.39) | S. sonnei (ATCC 29930) |
|---|---|---|---|---|---|---|
MIC values for 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Cip + Bac8c mixtures are quoted as the concentration of each component in the same ratio respectively, values in bold represent MICs and values in brackets represent MBCs 1 Cip + Bac8c mixtures are quoted as the concentration of each component in the same ratio respectively, values in bold represent MICs and values in brackets represent MBCs |
||||||
| Bac8c | 3.12 (6.25) | 50 (100) | 12.5 (12.5) | 6.25 (6.25) | 6.25 (6.25) | 6.25 (6.25) |
| Bac8c(Cip3,6) | 25 (25) | 50 (50) | 50 (50) | 6.25 (6.25) | 6.25 (6.25) | 6.25 (6.25) |
| Cip | 1.56 (1.56) | 0.048 (0.048) | 0.78 (0.78) | 0.097 (0.097) | 0.097 (0.097) | 0.097 (0.097) |
Cip + Bac8c (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
1.56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 0.78 (1.56 0.78 (1.56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.78) 0.78) |
0.097![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 0.048 (0.097 0.048 (0.097![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.048) 0.048) |
0.78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 0.048 (0.78 0.048 (0.78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.048) 0.048) |
0.097![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 0.048 (0.097 0.048 (0.097![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.048) 0.048) |
0.097![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 0.048 (0.097 0.048 (0.097![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.048) 0.048) |
0.097![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 0.048 (0.097 0.048 (0.097![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.048) 0.048) |
Both Bac8c and Cip are broad-spectrum antimicrobials but, their incorporation into Bac8c(Cip3,6) resulted in a more selective agent which could have inherent benefits. Interestingly Bac8c(Cip3,6) generally keeps the activity of the parent peptide Bac8c against most Gram-negative bacteria within the Enterobacteriaceae family and also P. aeruginosa. More targeted antimicrobials have a lower risk for AMR development than broad-spectrum antibiotics. In addition, targeted agents better support maintenance of a healthy microbiome, important in lowering the risk of opportunistic infections.32 The hybridisation of the two antimicrobials has produced a peptide with similar properties to specifically targeted antimicrobial peptides (STAMPs), something which warrants further investigation.33 Although ciprofloxacin is much more potent than either peptide against the bacteria tested, FQs are associated with increasing AMR mechanisms such as DNA gyrase mutation, upregulation of efflux pumps and downregulation of porins34–36 among clinically important bacteria.20,37,38 These mechanisms are not activated in response to AMPs, where different and multiple targets have been described. In addition, the covalent binding of ciprofloxacin to an AMP in this way may help reduce toxicity/side effects currently causing reduced use for FQs. Moreover, there exists a critical unmet need for narrow-spectrum Gram-negative selective antimicrobial agents. Therefore, modifications of broad-spectrum antimicrobials that favour more targeted killing of Gram-negative organisms may extend their clinical lifespan.
Conclusions
Ciprofloxacin has been used as a tryptophan mimic within AMP Bac8c to produce a series of novel Bac8c mimetic hybrids which combine peptide conjugation and mimetic approaches. These analogues were investigated for their antimicrobial activity and it was found that activity is site dependent with up to 2-fold difference in MICs for E.coli and S. aureus when comparing substitution at the 3 and 6 positions. One analogue in particular, Bac8c(Cip3,6) showed good activity and selectivity towards the majority of Gram-negative bacteria tested. Bac8c(Cip3,6) also showed no cytotoxicity against epithelial-like cells up to 300 μM. These results could also contribute to a better understanding of the role played by Trp within AMPs. Additional work is needed to further elucidate the mechanism of action of the mimetic hybrid peptide and the contribution of the ciprofloxacin to its antimicrobial activity. The potential evolution of resistance to Bac8c(Cip3,6), or cross-resistance, between Bac8c(Cip3,6) and its parent peptide also warrant further investigation.Author contributions
Conceptualization: JC; methodology: JC, MD.; supervision: MD, DFH.; project administration: MD.; chemical synthesis JC.; biological evaluation: JC, DFH, MM.; analytical analysis: JC, JM.; writing – original draft: JC, MD.; writing – review & editing: JC, MD, MM, DFH, JM.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
The peptide synthesis systems CEM Liberty Blue Microwave Peptide Synthesizer and Shimadzu HPLC were funded through Science Foundation Ireland (SFI) Research Infrastructure Programme, reference 16/RI/3737. The lyophiliser Buchi Lyovapor L-200 was funded through the SFI Frontiers for the Future Programme, reference 19/FFP/6889. Mass Spectrometry systems were funded as part of The Comprehensive Molecular Analysis Platform (CMAP) initiative under The SFI Research Infrastructure Programme in 2019, reference 18/RI/5702, and BiOrbic, the SFI Bioeconomy Research Centre, and with the support of the School of Chemistry and University College Dublin (UCD). The research was funded through Royal College of Surgeons in Ireland STaR PhD reference 20270A06 with support of the Department of Chemistry.Notes and references
- C. J. Murray, et al. , Lancet, 2022, 399, 629–655 CrossRef CAS PubMed.
- P. Das, et al. , Nat. Biomed. Eng., 2021, 1–11 Search PubMed.
- R. W. Scott and G. N. Tew, Curr. Top. Med. Chem., 2017, 17, 576–589 CrossRef CAS PubMed.
- W. Li, F. Separovic, N. M. O’Brien-Simpson and J. D. Wade, Chem. Soc. Rev., 2021, 50, 4932–4973 RSC.
- D. Thompson, J. Xu, J. Ischia and D. Bolton, BJUI Compass, 2024, 5, 5–11 CrossRef PubMed.
- N. Ptaszyńska, et al. , Int. J. Mol. Sci., 2020, 21, 4696 CrossRef PubMed.
- M. Ahmed and S. O. Kelley, ACS Chem. Biol., 2017, 12, 2563–2569 CrossRef CAS PubMed.
- F. Ceccherini, C. Falciani, M. Onori, S. Scali, S. Pollini, G. M. Rossolini, L. Bracci and A. Pini, Med. Chem. Commun., 2016, 7, 258–262 RSC.
- C. A. Rodriguez, E. A. Papanastasiou, M. Juba and B. Bishop, Front. Chem., 2014, 2 DOI:10.3389/fchem.2014.00071.
- S. K. Saraf, M. S. M. Rawat and A. C. Tripathi, IJCTR, 2018, 11, 298–307 CrossRef CAS.
- J. M. Sierra and M. Viñas, Expert Opin. Drug Discovery, 2021, 16, 601–604 CrossRef CAS PubMed.
- X. Bi, C. Wang, W. Dong, W. Zhu and D. Shang, J. Antibiot., 2014, 67, 361–368 CrossRef CAS PubMed.
- X. Bi, C. Wang, L. Ma, Y. Sun and D. Shang, J. Appl. Microbiol., 2013, 115, 663–672 CrossRef CAS PubMed.
- I. Schiopu, L. Mereuta, A. Apetrei, Y. Park, K.-S. Hahm and T. Luchian, Mol. BioSyst., 2012, 8, 2860 RSC.
- S. Khemaissa, A. Walrant and S. Sagan, Q. Rev. Biophys., 2022, 55, e10 CrossRef CAS PubMed.
- Y. Lv, X. Chen, Z. Chen, Z. Shang, Y. Li, W. Xu, Y. Mo, X. Wang, D. Xu, S. Li, Z. Wang, M. Wu and J. Wang, Toxins, 2022, 14, 428 CrossRef CAS PubMed.
- Z. Ridgway, et al. , Pept. Sci., 2015, 104, 384–394 CrossRef CAS PubMed.
- A. R. D’Souza, M. R. Necelis, A. Kulesha, G. A. Caputo and O. V. Makhlynets, Biomolecules, 2021, 11, 421 CrossRef PubMed.
- A. V. Strizhak, V. Y. Postupalenko, V. V. Shvadchak, N. Morellet, E. Guittet, V. G. Pivovarenko, A. S. Klymchenko and Y. Mély, Bioconjugate Chem., 2012, 23, 2434–2443 CrossRef CAS PubMed.
- C. C. Sanders, Rev. Infect. Dis., 1988, 10, 516–527 CrossRef CAS PubMed.
- E. C. Spindler, J. D. F. Hale, T. H. Giddings, R. E. W. Hancock and R. T. Gill, Antimicrob. Agents Chemother., 2011, 55, 1706–1716 CrossRef CAS PubMed.
- W. Zhou, Y. Du, X. Li and C. Yao, Bioorg. Med. Chem., 2020, 28, 115682 CrossRef CAS PubMed.
- G. Barzan, et al. , ACS Omega, 2022, 7, 16402–16413 CrossRef CAS PubMed.
- M. Zapotoczna, et al. , J. Infect. Dis., 2017, 215, 975–983 CrossRef CAS PubMed.
- L. B. Rice, J. Infect. Dis., 2008, 197, 1079–1081 CrossRef PubMed.
- F. Kampshoff, M. D. P. Willcox and D. Dutta, Antibiotics, 2019, 8, 60 CrossRef CAS PubMed.
- A. G. Khairunnisa, M. H. Waleed, G. K. Zeinab, J. C. Robert, S. Mariusz and T. Istvan, Curr. Drug Delivery, 2015, 12, 108–114 CrossRef PubMed.
- R. Gattu, S. S. Ramesh, S. Nadigar and S. Ramesh, Antibiotics, 2023, 12, 532 CrossRef CAS PubMed.
- T. Kloskowski, et al. , Int. J. Mol. Sci., 2021, 22, 11970 CrossRef CAS PubMed.
- K. Esfandiari Mazandaran, S. A. Mirshokraee, K. Didehban and M. H. Houshdar Tehrani, Iran. J. Pharm. Res., 2019, 18, 1823–1830 Search PubMed.
- J. H. Tanne, BMJ, 2008, 337, 135 CrossRef PubMed.
- C. Zampaloni, et al. , Nature, 2024, 1–6 Search PubMed.
- P. Sarma, S. Mahendiratta, A. Prakash and B. Medhi, Indian J. Pharmacol., 2018, 50, 1–3 CrossRef PubMed.
- J. M. A. Blair, M. A. Webber, A. J. Baylay, D. O. Ogbolu and L. J. V. Piddock, Nat. Rev. Microbiol., 2015, 13, 42–51 CrossRef CAS PubMed.
- L. S. Redgrave, S. B. Sutton, M. A. Webber and L. J. V. Piddock, Trends Microbiol., 2014, 22, 438–445 CrossRef CAS PubMed.
- D. C. Hooper and G. A. Jacoby, Ann. N. Y. Acad. Sci., 2015, 1354, 12–31 CrossRef CAS PubMed.
- A. Rehman, W. M. Patrick and I. L. Lamont, J. Med. Microbiol., 2019, 68, 1–10 CrossRef CAS PubMed.
- A. Shariati, M. Arshadi, M. A. Khosrojerdi, M. Abedinzadeh, M. Ganjalishahi, A. Maleki, M. Heidary and S. Khoshnood, Front. Public Health, 2022, 10, 1025633 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nj01445f |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2024 |

