Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Construction of viral protein-based hybrid nanomaterials mediated by a macromolecular glue†
Shuqin
Cao
 ab,
Sandro
Peeters
a,
Sandra
Michel-Souzy
ab,
Sandro
Peeters
a,
Sandra
Michel-Souzy
 a,
Naomi
Hamelmann
a,
Jos M. J.
Paulusse
a,
Naomi
Hamelmann
a,
Jos M. J.
Paulusse
 a,
Liu-lin
Yang
a,
Liu-lin
Yang
 *ac and
Jeroen J. L. M.
Cornelissen
*ac and
Jeroen J. L. M.
Cornelissen
 *a
*a
aLaboratory for Biomolecular Nanotechnology, MESA+ Institute for Nanotechnology, University of Twente, P.O. Box 217, 7500 AE, Enschede, The Netherlands. E-mail: J.J.L.M.Cornelissen@utwente.nl
bState Key Laboratory of Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China
cCollege of Chemistry and Chemical Engineering, Xiamen University, 361005, Xiamen, China. E-mail: llyang@xmu.edu.cn
First published on 6th June 2023
Abstract
A generic strategy to construct virus protein-based hybrid nanomaterials is reported by using a macromolecular glue inspired by mussel adhesion. Commercially available poly(isobutylene-alt-maleic anhydride) (PiBMA) modified with dopamine (PiBMAD) is designed as this macromolecular glue, which serves as a universal adhesive material for the construction of multicomponent hybrid nanomaterials. As a proof of concept, gold nanorods (AuNRs) and single-walled carbon nanotubes (SWCNTs) are initially coated with PiBMAD. Subsequently, viral capsid proteins from the Cowpea Chlorotic Mottle Virus (CCMV) assemble around the nano-objects templated by the negative charges of the glue. With virtually unchanged properties of the rods and tubes, the hybrid materials might show improved biocompatibility and can be used in future studies toward cell uptake and delivery.
Introduction
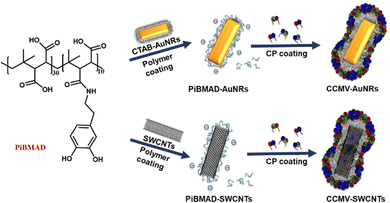
Nanomaterials have found numerous applications in catalysis,1,2 biosensors,3 drug delivery4 and bioimaging5,6 due to their controlled chemical compositions and structures, large surface-to-volume ratios, various functional groups, and surfaces. Among them, protein cages, such as virus-based hybrid nanomaterials, show interesting properties in electronics,7 catalysis,8 drug/gene delivery,9,10 imaging and immune-therapy,11 that are largely based on their reversible assembly, uniform size, facile modification, and capability of functional cargo encapsulation.12,13These functional protein-organic/inorganic hybrid nanomaterials are prepared via self-assembly of proteins,12,14–17 chemical conjugation,5,18 and protein immobilization.19 Protein cages derived from the Cowpea Chlorotic Mottle Virus (CCMV) have been widely used as nanocarriers with various cargos such as polystyrene sulfonate (PSS),20 oligonucleotides,21 functionalized enzymes,22 gold nanoparticles,23 and luminescent materials.14 These hybrid materials combine efficiently the biocompatibility of the protein coating with the functionality of the cargo, as exemplified by our recent study on nanodiamonds for intracellular trajectory analysis.24 It is, however, crucial in the formation of these hybrid materials to maintain the functional properties of the nanomaterials. The capsid protein of CCMV is capable of self-assembly into empty capsids at pH 5 with high salt concentration, when the main driving force is the interaction between capsid proteins. While for efficient loading, negatively charged cargo surfaces are generally required to guarantee strong electrostatic interactions between the capsid proteins and the desired cargos at neutral pH. Therefore, for diverse nanometer-sized cargo candidates, such as silicon quantum dots, gold nano-objects and carbon nanotubes with neutral surfaces, a surface modification either by coupling oligonucleotides,6 PSS or thiol-based ligands is a prerequisite.7 However, this surface modification usually involves tedious organic synthesis, meanwhile, it suffers from the aggregation of these nano-objects that lead to instability and the loss of intrinsic properties. To tackle this problem, we propose a generic adhesive material that can serve as an interfacial molecular glue to facilitate the coating of nanometer-sized cargo with CCMV capsid proteins, to further expand the toolbox for the colloidal stabilization, biocompatibility and eventual cellular uptake of functional nanomaterials.24
The structural design and core capabilities of this molecular glue should meet two requirements: (i) as an adhesive material, it can intimately interact with a wide range of nanomaterial surfaces; and (ii) as a template to induce the coating of capsid proteins on nanomaterials, it should incorporate negatively charged functional groups (i.e, carboxylic acids, sulfates, phosphates). Catechol functionalized polymers inspired by natural mussels have been developed as tissue adhesives for bio-applications25–27 Since the catechol group is capable of having strong non-covalent interactions with metal ions, polarized organic groups, or covalent crosslinking via oxidation.26,28–30 Hence, the catechol group is an ideal building block to meet the first requirement for constructing the molecular glue. Meanwhile, commercially available poly(isobutylene-alt-maleic anhydride) that inherently contains multiple carboxyl groups can serve as the template building block to meet the second requirement. Therefore, we prepared catechol modified poly(isobutylene-alt-maleic anhydride) as the molecular glue for constructing the desired nanomaterials.
Results and discussion
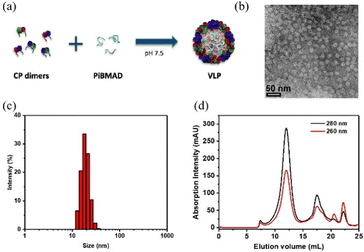
The molecular glue was synthesized by amination of poly(isobutylene-alt-maleic anhydride) with dopamine (PiBMAD).31 Dopamine was used to introduce the catechol group for the adhesiveness of this generic molecular glue, as it is a well-known molecular mimic of mussel foot protein.32,33 As a proof of concept, two representative nanomaterials, gold nanorods (AuNRs) and single-walled carbon nanotubes (SWCNTs), have been selected as cargo candidates to verify the applicability of this molecular glue (Fig. 1).PiBMAD was synthesized as described in the ESI† and Fig. S1a (ESI†). Commercially available PiBMA with an average molechular weight (Mw) of 6 kDa was used (Fig. S1b, ESI†). To maintain water solubility of the polymer and keep sufficient negative charges, around 12.5% of the available carboxylate groups were equipped with dopamine according to 1H-NMR analysis (Fig. S1c, ESI†). The capability of PiBMAD to induce the assembly of capsid proteins was first checked by incubating the proteins in a buffer with the polymer at pH 7.2 (see ESI† for details). The formation of virus-like particles was evident and purified from fast protein liquid chromatography (FPLC) equipped with a UV-vis detector (Fig. 2d). The absorption data showed an overlapping signal at two detection wavelengths (λ = 260 nm and 280 nm) at an elution volume of V ≈ 11 mL, whereas excess capsid protein eluted out at a larger elution volume (Fig. S8, ESI†) indicating the formation of virus-like particles. The fraction was isolated and analyzed by transmission electron microscopy (TEM) and dynamic light scattering (DLS) (Fig. 2b and c). TEM analysis revealed the formation of spherical nanoparticles with an average diameter of 19 nm. DLS showed that the isolated fraction contained nanoparticles with a diameter of 18.0 ± 3.6 nm, which is consistent with the TEM results. These data are in line with the previously confirmed T = 1 icosahedral symmetry for CCMV based virus-like particles formed at neutral pH on a polyanionic template.34
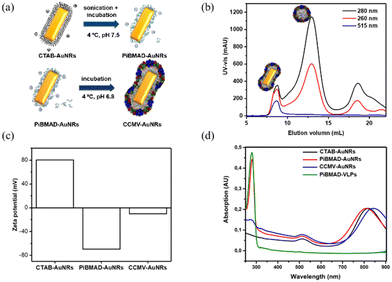
Gold nanorods (AuNRs) belong to an interesting class of nanosized objects for a plethora of biomedical and biotechnological applications such as sensing, imaging and others. However, these applications rely on their stability in biological fluids, avoiding aggregation and precipitation. Surface coating of gold nanorods with biocompatible proteins may facilitate the bioapplications of these nanomaterials. To this end, surface coating of AuNRs by CCMV proteins was successfully realized by PiBMAD scaffolding (Fig. 3a), detailed experimental methods are described in the ESI.† The coated AuNRs were analyzed by FPLC (Fig. 3b) monitoring the elution at λ = 260, 280 and 515 nm. The elution volume of the first fraction around 7.5 mL, of all three coinciding wavelengths, is smaller than the typical elution volume of virus-like particles (11 mL), suggesting the formation of larger particles. Other than that, the transverse plasmon resonance peak of AuNRs at λ = 515 nm was also and only detected in the first fraction, confirming that this fraction contains coated gold nanorods (CCMV-AuNRs).35
Zeta potential measurements were carried out to check the change of surface charge of AuNRs during coating (Fig. 3c). When the AuNRs were first coated by PiBMAD, the zeta potential of AuNRs reversed from positive to negative, due to the abundant carboxyl groups of polymer chains. When finally coated by capsid proteins, as expected, the zeta potential of CCMV-AuNRs increased from −75 mV to −10 mV. The longitudinal plasmon resonance peak (LPRW) of the original AuNRs was located at λ = 808 nm, after coating by PiBMAD, the LPRW of PiBMAD-AuNRs was slightly shifted to λ = 805 nm, while after coating by capsid proteins, a 50 nm red shift of the longitudinal plasmon resonance (Fig. 3d) was observed.
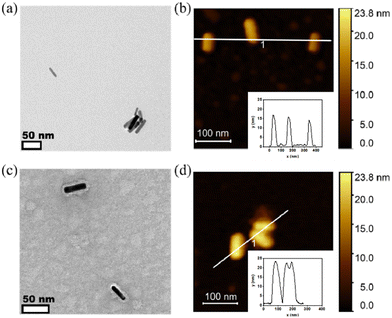
The morphology of PiBMAD-AuNRs and CCMV-AuNRs were studied by transmission electronic microscopy (TEM) and atomic force microscopy (AFM). Both TEM and AFM analysis showed that the morphology of AuNRs was unchanged after coating with PiBMAD (Fig. 4a and b). The original height of AuNRs was 10 nm, while the overall height of PiBMAD-AuNRs was around 15 nm (Fig. 4b insert), suggesting a ∼2.5 nm thickness of polymer layer. The thickness of capsid proteins layer on CCMV-AuNRs is estimated to be ∼5 nm (Fig. 4c and d), which is consistent with the thickness of the native CCMV capsid shell, suggesting a monolayer coating of capsid proteins on the AuNRs surface. AFM micrograph and height analysis on single PiBMAD-AuNRs and CCMV-AuNRs further supports this conclusion (Fig. S5, ESI†).36,37 According to the literature37 and our investigation, we conclude that capsid proteins formed subunits such as heximers and pentamers while coating on the surface of AuNRs, and defects can be seen from Fig. 4c.
To avoid the aggregation of AuNRs during the coating process, an optimized concentration of PiBMAD is required (Fig. S2, ESI†). The strongest LPRW absorption peak of PiBAMD-AuNRs suggests minimal aggregation when the PiBMAD concentration is 3 mg mL−1. This is explained by less catechol being attached to the surface of AuNRs at lower concentrations of PiBMAD and when the concentration of PiBMAD is too high, catechol groups of PiBMAD might cause inter-nanoparticle crosslinking.38,39 It should be further noted that the buffer solution (i.e, salt concentrations, pH) also plays an important role in coating capsid proteins onto the AuNRs (Fig. S3, ESI†). FPLC data (Fig. S3a, ESI†) indicates a thicker layer of capsid proteins is coated on AuNRs at pH 6.8 compared to pH 7.2, which is in line with a higher UV absorption at λ = 280 nm by capsid proteins (Fig. S3b, ESI†).
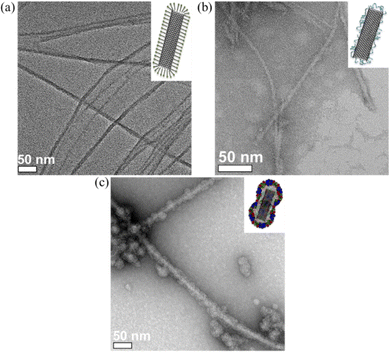
Because of the large surface area, excellent chemical stability and rich polyaromatic structure, carbon nanotubes (CNTs) are an emerging nanomaterial as scaffolds for biosensors40–42 and anticancer drug delivery cargos.43 The main challenge of CNTs as biomaterials, is their poor aqueous solubility due to their high hydrophobicity. To improve the aqueous solubility, the functionalization of CNTs with biocompatible and hydrophilic functional organic species are the general approach.44–46 Herein, we demonstrate a facile protein coating strategy on single-walled carbon nanotubes (SWCNTs) by employing the molecular glue PiBMAD without modification of SWCNTs. The detailed coating process is described in the ESI.†
SWCNTs dispersed with sodium dodecyl sulfate (SDS) showed few bundles (Fig. 5a). After ligand exchange with PiBMAD, the size and distribution of SWCNTs were almost unchanged (Fig. 5b). While after coating with capsid proteins, the average diameter of CCMV-SWCNTs was around 20 nm (Fig. 5c, Fig. S7, ESI†), suggesting a protein monolayer with ∼5 nm thickness on the surface of SWCNTs. Raman spectroscopy (Fig. S4, ESI†) also confirmed the presence of SWCNT before and after coating with PiBMAD, based on the unchanged characteristic peaks associated with CNT.
In control experiments, other ligands such as bis(p-sulfonatophenyl)phenylphosphine dihydrate dipotassium salt (BSPP) and poly-dopa were used as templates for CCMV capsid proteins coating (Fig. S6, ESI†). For the AuNRs, both BSPP and PiBMAD templated the formation of CCMV-AuNRs and an apparent single layer of capsid proteins coating was observed, while poly-dopa templated coating induced aggregation of AuNRs which might be caused by crosslinking of AuNRs by the poly-dopa (Fig. S6a–c, ESI†). With respect to SWCNTs, only PiBMAD induced the formation of well-dispersed CCMV-SWCNTs (Fig. S6d–f, ESI†). These results are in line with the concept of using PiBMAD as a generic molecular glue for the construction of viral protein coated hybrid nanomaterials.
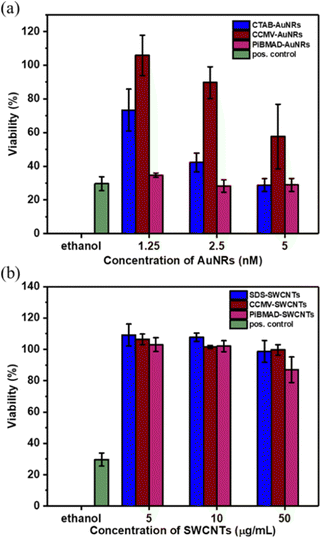
Increasing the colloidal stability of functional nanomaterials can be achieved by a variety of chemical means. We chose, however, the biorelevant coating of virus capsid proteins to potentially improve biocompatibility and eventually direct cell uptake.10,24 To demonstrate the compatibility of CCMV-AuNRs and CCMV-SWCNTs for potential bio-applications, cytotoxicity studies were carried out at various concentrations of the biohybrid materials in HeLa cells. As shown in Fig. 6a, the cells incubated with CCMV-AuNRs revealed significantly higher viability compared to CTAB-AuNRs and PiBMAD-AuNRs over the tested concentration range (from 1.25 to 5 nmol L−1), although a concentration dependent cytotoxicity was shown for all three samples. The SWCNTs with different surface coatings demonstrate similar cell viability at the various concentrations studied (range from 5–50 μg mL−1) (Fig. 6b). Both AuNRs47 and CCMV48 based nanostructures are presumed to be taken up in HeLa cells by clathrin mediated endocytose. The different surface chemistry of the three samples in Fig. 6a, with CTAB-AuNRs being positive and CCMV-AuNRs only slightly negatively charged, likely accounts for the differences in cytotoxicity. In the case of the SWCNT based samples the marginal difference in cell viability might be attributed to limited uptake of these samples by the cells. For both classes of nanomaterials, further and more detailed studies are needed to understand the interactions with cells. It is clear, however, that the successful coating with capsids proteins allows for targeted modification of the surface, for example, with proteins equipped with cell penetrating peptides.49
Conclusions
In summary, we have demonstrated a generic strategy to prepare viral protein-based hybrid biomaterials mediated by a macromolecular glue that is inspired by mussel adhesives. AuNRs and SWCNTs were selected as typical nanomaterials to verify the concept of this macromolecular glue. Catechol modified negatively charged polymers show the capability to mediate the co-assembly of these nanometer-sized cargos and CCMV capsid proteins. This strategy provides a facile and general coating process compared with traditional methods.In contrast to other commonly used ligands, such as, thiolate polymers and single-strand short DNA fragments, the feasibility of preparing PiBMAD with a diversity of other functional moieties makes this polymer a promising interfacial molecular glue for the construction of viral protein-based hybrid nanomaterials.
Experimental
1. Material
All chemicals were purchased from Sigma-Aldrich and used as received unless stated. Deionized water used for buffers, reactions and dialysis media was of ultrapure quality (Milli-Q, 18.2 MΩ cm). Membrane filters (Spectra/Por® regenerated cellulose tubing) used for dialysis were purchased from Spectrum Laboratories, Inc., USA.2. Methods
3. Sample preparation
100 mg PiBSA was dissolved in 5 mL dimethylformamide (DMF) containing 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC·HCl, 0.1 M) and N hydroxysuccinimide (NHS, 0.1 mol L−1). This mixture was stirred for 20 min. Meanwhile, a second solution was prepared of 47.4 mg dopamine·HCl (0.5 mol L−1) in 0.5 mL aqueous NaHCO3 (0.5 mol L−1) and Na2S2O3 (reducing agent, 0.1 mol L−1). The mixture was then added to the organic PiBSA solution and left to react overnight under mild magnetic stirring. The mixture was diluted 10× using deionized water and purified by dialysis against deionized water (1 kD MWCO dialysis tubing), refreshing the medium every 3 h for at least five times. The solution was then freeze-dried (Labconco FreeZone 4.5) for 48 h, after which the PiBMAD powder is collected.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 10 min. This is repeated once more at 14
000 g for 10 min. This is repeated once more at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 10 min, resulting in a total of four pellets after one round of centrifugation. All the centrifugation of AuNRs was performed in this manner, unless stated differently.
000 g for 10 min, resulting in a total of four pellets after one round of centrifugation. All the centrifugation of AuNRs was performed in this manner, unless stated differently.
The ligand exchange of CTAB-coated gold nanorods with PiBMAD was based on general methods found in literature,50 where (poly)dopamine coatings are applied to gold nanorods. However, since oxidation of dopamine is undesired in our case, pH was lowered to 7.5 and a small amount of reducing agent (Na2S2O3) was included. Briefly, 1 mL CTAB-AuNRs (optical density = 0.95) from stock solution was centrifuged at 20 °C. The pellet was suspended in 2 mL Tris buffer (10 mmol L−1, pH 7.5) containing PiBMAD (0.5–4.0 mg mL−1 or 50–400 equiv. by weight conc.) and 5 mmol L−1 Na2S2O3. The nanorod suspension was then ultrasonicated for 30 min to accelerate displacement of the CTAB bilayer from the surface and subsequently incubated overnight at 4 °C. PiBMAD coated AuNRs were pelleted by centrifugation at 20 °C and resuspended in 1 mL deionized water. UV-vis-NIR absorption spectra and zeta potentials were recorded to confirm the presence of gold (SPR peaks) and polymer respectively.
Subsequently 1 mL PiBMAD-AuNRs were centrifuged once at 20 °C. The pellets were suspended and concentrated to OD ≈ 2–2.5 in 200 μL pH 6.8 cold encapsulation buffer (50 mmol L−1 Tris, 200 mmol L−1 NaCl and 5 mmol L−1 MgCl2). CP solution from storage, was freshly dialyzed to cold pH 6.8 encapsulation buffer, and diluted to a concentration of 6.0 mg mL−1. The PiBMAD-AuNRs and CP solution were then mixed in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio and gently stirred overnight at 4 °C. Note that due to the difficult purification of the nanorods after ligand exchange, there is still free PiBMAD in the nanorod solution, which is able to induce the formation of spherical VLPs. Hence an excess amount of CP is applied. After encapsulation CCMV-AuNRs were purified by FPLC. The collected fractions could be further purified by centrifugation at 6000 g for 10 min and were analyzed using UV-vis-NIR and TEM.
1 volume ratio and gently stirred overnight at 4 °C. Note that due to the difficult purification of the nanorods after ligand exchange, there is still free PiBMAD in the nanorod solution, which is able to induce the formation of spherical VLPs. Hence an excess amount of CP is applied. After encapsulation CCMV-AuNRs were purified by FPLC. The collected fractions could be further purified by centrifugation at 6000 g for 10 min and were analyzed using UV-vis-NIR and TEM.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 1 h, precipitating all undispersed and amorphous carbon. The supernatant, containing SDS-SWCNTs was collected, and the presence of carbon nanotubes was investigated using Raman spectroscopy and TEM. Then 2 mg mL−1 PiBMAD solution in pH 7.5 Tris (20 mmol L−1, 10 mmol L−1 Na2S2O3) was prepared and added to aqueous SDS-SWCNT in a 1
000 g for 1 h, precipitating all undispersed and amorphous carbon. The supernatant, containing SDS-SWCNTs was collected, and the presence of carbon nanotubes was investigated using Raman spectroscopy and TEM. Then 2 mg mL−1 PiBMAD solution in pH 7.5 Tris (20 mmol L−1, 10 mmol L−1 Na2S2O3) was prepared and added to aqueous SDS-SWCNT in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 vol. ratio. The mixture was then ultrasonicated for 2–3 h and centrifuged at 16
1 vol. ratio. The mixture was then ultrasonicated for 2–3 h and centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 30 min to remove aggregated SWCNT. The supernatant was collected and dialyzed (12–14 kD MWCO) against two to five volumes of 300 mL pH 6.8 encapsulation buffer (50 mmol L−1 Tris, 200 mmol L−1 NaCl, and 5 mmol L−1 MgCl2) to remove free polymer. After dialysis, aggregated carbon nanotubes were sonicated for 30 min for potential redispersion and then centrifuged for 30 min at 16
000 g for 30 min to remove aggregated SWCNT. The supernatant was collected and dialyzed (12–14 kD MWCO) against two to five volumes of 300 mL pH 6.8 encapsulation buffer (50 mmol L−1 Tris, 200 mmol L−1 NaCl, and 5 mmol L−1 MgCl2) to remove free polymer. After dialysis, aggregated carbon nanotubes were sonicated for 30 min for potential redispersion and then centrifuged for 30 min at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g. Due to insufficient removal of the polymer during dialysis, the solution was washed at least six times using a 100 kD spin filter with pH 6.8 encapsulation buffer. The carbon nanotubes, mostly sticking to the filter (around 90%), are entered back in the solution in aggregated form by thoroughly flushing the filter with 100 μL encapsulation buffer using a pipette and could be redispersed by 5–20 min ultrasonication. Then PiBMAD-SWCNTs (100 μL pH 6.8 encapsulation buffer) were added to 10 μL CP (6 mg mL−1, pH 6.8 encapsulation buffer) and gently stirred overnight at 4 °C. Excess CP was removed by washing at least three times with deionized water using a 100 kD spin filter.
000 g. Due to insufficient removal of the polymer during dialysis, the solution was washed at least six times using a 100 kD spin filter with pH 6.8 encapsulation buffer. The carbon nanotubes, mostly sticking to the filter (around 90%), are entered back in the solution in aggregated form by thoroughly flushing the filter with 100 μL encapsulation buffer using a pipette and could be redispersed by 5–20 min ultrasonication. Then PiBMAD-SWCNTs (100 μL pH 6.8 encapsulation buffer) were added to 10 μL CP (6 mg mL−1, pH 6.8 encapsulation buffer) and gently stirred overnight at 4 °C. Excess CP was removed by washing at least three times with deionized water using a 100 kD spin filter.
3.5.1. PolyDOPA. For the growth of PDA films on gold nanorods, 1 mL CTAB-AuNRs from stock solution was centrifuged and suspended in 2 mL Tris pH 8.5 buffer containing 0.5 mg mL−1L-DOPA. The solution was ultrasonicated for 30 min to allow for film growth, where the thickness depends on the sonication time and concentration of L-DOPA. After sonication, the PDA-AuNRs could not be precipitated into pellet anymore, unless the pH of the solution was significantly lower than the pKa of the carboxylic group (≈ 2.3) of L-DOPA. Centrifugation at pH = 1.8 resulted in a pellet which was suspended into 300 μL pH 6.8 encapsulation buffer (50 mmol L−1 Tris, 200 mmol L−1 NaCl, 5 mmol L−1 MgCl2). The PDA-AuNRs were then added (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 vol.) to 6.0 mg mL−1 CP (pH 6.8 enc. buffer) and mildly stirred overnight at 4 °C. Samples were inspected using TEM.
1 vol.) to 6.0 mg mL−1 CP (pH 6.8 enc. buffer) and mildly stirred overnight at 4 °C. Samples were inspected using TEM.
3.5.2. Bis(p-sulfonatophenyl)phenylphosphine (BSPP). The capping of AuNRs by BSPP was performed according to the method of capping spherical gold nanoparticles (AuNPs) described by A. Liu et al. Briefly,23 1 mL AuNRs from stock solution was centrifuged and the pellet was suspended into 2 mL aqueous BSPP (0.5–3.0 mg mL−1). The solution was mildly stirred overnight at room temperature. BSPP-AuNRs were then centrifuged and concentrated in 200 μL pH 6.8 encapsulation buffer (50 mmol L−1 Tris, 200 mmol L−1 NaCl, 5 mmol L−1 MgCl2). A solution of freshly dialyzed CCMV CP into pH 6.8 encapsulation buffer was then diluted to 6 mg mL−1 and added to the BSPP-AuNR solution in a 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio. The solution was then mildly stirred overnight at 4 °C. CCMV-BSPP-AuNRs were then purified by size exclusion chromatography (FPLC), the collected gold fractions were analyzed with TEM and UV-vis-NIR. The CCMV coated nanorods could be concentrated by centrifugation at 6000 g for 10 min at 4 °C and resuspension of the pellet.
1 volume ratio. The solution was then mildly stirred overnight at 4 °C. CCMV-BSPP-AuNRs were then purified by size exclusion chromatography (FPLC), the collected gold fractions were analyzed with TEM and UV-vis-NIR. The CCMV coated nanorods could be concentrated by centrifugation at 6000 g for 10 min at 4 °C and resuspension of the pellet.
Attempts to coat SWCNTs with BSPP and PDA were made. The coating of SWCNTs with BSPP and PDA were performed nearly identical to the method described previously for AuNRs. But BSPP was added in deionized water instead of tris buffer. For L-DOPA the following modifications were applied: a slightly elevated pH of 8.5 and the exclusion of Na2S2O3 to facilitate oxidation and a shorter ultrasonic treatment (1 h instead of 3 h) to limit film growth.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge financial support from the ERC Consolidator Grant (Protcage #616907) to J.J.L.M.C. The authors are grateful to Dr E. G. Keim and Dr Jun Wang (MESA+ Institute for Nanotechnology, University of Twente) for assistance with TEM and AFM respectively.Notes and references
- I. J. Minten, V. I. Claessen, K. Blank, A. E. Rowan, R. J. M. Nolte and J. J. L. M. Cornelissen, Catalytic capsids: the art of confinement, Chem. Sci., 2011, 2(2), 358–362 RSC.
- A. Liu, M. V. de Ruiter, W. Zhu, S. J. Maassen, L. Yang and J. J. L. M. Cornelissen, Compartmentalized Thin Films with Customized Functionality via Interfacial Cross-linking of Protein Cages, Adv. Funct. Mater., 2018, 28(34), 1801574 CrossRef.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection, ACS Nano, 2020, 14(5), 5268–5277 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing, Nat. Nanotechnol., 2020, 15(4), 313–320 CrossRef CAS PubMed.
- H. Li, X. Wang, T. Y. Ohulchanskyy and G. Chen, Lanthanide-Doped Near-Infrared Nanoparticles for Biophotonics, Adv. Mater., 2021, 33(6), e2000678 CrossRef PubMed.
- F. Tajoli, N. Dengo, M. Mognato, P. Dolcet, G. Lucchini, A. Faresin, J. D. Grunwaldt, X. Huang, D. Badocco, M. Maggini, C. Kübel, A. Speghini, T. Carofiglio and S. Gross, Microfluidic Crystallization of Surfactant-Free Doped Zinc Sulfide Nanoparticles for Optical Bioimaging Applications, ACS Appl. Mater. Interfaces, 2020, 12(39), 44074–44087 CrossRef CAS PubMed.
- R. Zinelli, S. Soni, J. J. L. M. Cornelissen, S. Michel-Souzy and C. A. Nijhuis, Charge transport across proteins inside proteins: Tunneling across encapsulin protein cages and the effect of cargo proteins, Biomolecules, 2023, 13, 174–189 CrossRef CAS PubMed.
- A. Liu, C. H. H. Traulsen and J. J. L. M. Cornelissen, Nitroarene Reduction by a Virus Protein Cage Based Nanoreactor, ACS Catal., 2016, 6(5), 3084–3091 CrossRef CAS.
- J. A. Tejeda-Rodriguez, A. Nunez, F. Soto, V. Garcia-Gradilla, R. Cadena-Nava, J. Wang and R. Vazquez-Duhalt, Virus-Based Nanomotors for Cargo Delivery, ChemNanoMat, 2019, 5(2), 194–200 CrossRef CAS.
- F. Xue, J. J. L. M. Cornelissen, Q. Yuan and S. Cao, Delivery of MicroRNAs by plant virus-based nanoparticles to functionally alter the osteogenic differentiation of human mesenchymal stem cells, Chin. Chem. Lett., 2023, 34(1), 107448 CrossRef CAS.
- Y. Wu, H. Yang, Y.-J. Jeon, M.-Y. Lee, J. Li and H.-J. Shin, Surface modification of cowpea chlorotic mottle virus capsids via a copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) reaction and their adhesion behavior with HeLa cells, Biotechnol. Bioprocess Eng., 2014, 19(4), 747–753 CrossRef CAS.
- A. D. Malay, N. Miyazaki, A. Biela, S. Chakraborti, K. Majsterkiewicz, I. Stupka, C. S. Kaplan, A. Kowalczyk, B. M. A. G. Piette, G. K. A. Hochberg, D. Wu, T. P. Wrobel, A. Fineberg, M. S. Kushwah, M. Kelemen, P. Vavpetič, P. Pelicon, P. Kukura, J. L. P. Benesch, K. Iwasaki and J. G. Heddle, An ultra-stable gold-coordinated protein cage displaying reversible assembly, Nature, 2019, 569(7756), 438–442 CrossRef CAS PubMed.
- L. Schoonen and J. C. M. van Hest, Modification of CCMV nanocages for enzyme encapsulation, Methods Mol. Biol., 2018, 1798, 69–83 CrossRef CAS PubMed.
- S. Sinn, L. Yang, F. Biedermann, D. Wang, C. Kübel, J. J. L. M. Cornelissen and L. De Cola, Templated Formation of Luminescent Virus-like Particles by Tailor-Made Pt(II) Amphiphiles, J. Am. Chem. Soc., 2018, 140(6), 2355–2362 CrossRef CAS PubMed.
- H. Sun, Q. Luo, C. Hou and J. Liu, Nanostructures based on protein self-assembly: From hierarchical construction to bioinspired materials, Nano Today, 2017, 14, 16–41 CrossRef CAS.
- Q. Liu, A. Shaukat, D. Kyllönen and M. A. Kostiainen, Polyelectrolyte Encapsulation and Confinement within Protein Cage-Inspired Nanocompartments, Pharmaceutics, 2021, 13, 1551 CrossRef CAS PubMed.
- S. B. P. E. Timmermans, A. Ramezani, T. Montalvo, M. Nguyen, P. van der Schoot, J. C. M. van Hest and R. Zandi, The Dynamics of Viruslike Capsid Assembly and Disassembly, J. Am. Chem. Soc., 2022, 144(28), 12608–12612 CrossRef CAS PubMed.
- S. F. Oliveira, G. Bisker, N. A. Bakh, S. L. Gibbs, M. P. Landry and M. S. Strano, Protein functionalized carbon nanomaterials for biomedical applications, Carbon, 2015, 95, 767–779 CrossRef CAS.
- W. Zhang, K. Patel, A. Schexnider, S. Banu and A. D. Radadia, Nanostructuring of Biosensing Electrodes with Nanodiamonds for Antibody Immobilization, ACS Nano, 2014, 8(2), 1419–1428 CrossRef CAS PubMed.
- M. Comellas-Aragonès, A. de la Escosura, A. J. Dirks, A. van der Ham, A. Fusté-Cuñé, J. J. L. M. Cornelissen and R. J. M. Nolte, Controlled Integration of Polymers into Viral Capsids, Biomacromolecules, 2009, 10(11), 3141–3147 CrossRef PubMed.
- M. V. de Ruiter, N. J. Overeem, G. Singhai and J. J. L. M. Cornelissen, Induced Förster resonance energy transfer by encapsulation of DNA-scaffold based probes inside a plant virus based protein cage, J. Phys.: Condens. Matter, 2018, 30(18), 184002 CrossRef PubMed.
- L. Schoonen, S. Maassen, R. J. M. Nolte and J. C. M. van Hest, Stabilization of a Virus-Like Particle and Its Application as a Nanoreactor at Physiological Conditions, Biomacromolecules, 2017, 18(11), 3492–3497 CrossRef CAS PubMed.
- A. Liu, M. Verwegen, M. V. de Ruiter, S. J. Maassen, C. H. H. Traulsen and J. J. L. M. Cornelissen, Protein Cages as Containers for Gold Nanoparticles. The, J. Phys. Chem. B, 2016, 120(26), 6352–6357 CrossRef CAS PubMed.
- Y. Wu, S. Cao, M. N. A. Alam, M. Raabe, S. Michel-Souzy, Z. Wang, M. Wagner, A. Ermarkova, J. J. L. M. Cornelissen and T. Weil, Fluorescent nanodiamonds encapsulated by Cowpea Chlorotic Mottle Virus (CCMV) proteins for intracellular 3D-trajectory analysis, J. Mater. Chem. B, 2010, 9, 5621–5627 RSC.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Mussel-Inspired Surface Chemistry for Multifunctional Coatings, Science, 2007, 318(5849), 426–430 CrossRef CAS PubMed.
- W. Zhu, Y. Peck, J. Iqbal and D. A. Wang, A novel DOPA-albumin based tissue adhesive for internal medical applications, Biomaterials, 2017, 147, 99–115 CrossRef CAS PubMed.
- J. Xu, G. M. Soliman, J. Barralet and M. Cerruti, Mollusk glue inspired mucoadhesives for biomedical applications, Langmuir, 2012, 28(39), 14010–14017 CrossRef CAS PubMed.
- D. E. Fullenkamp, L. He, D. G. Barrett, W. R. Burghardt and P. B. Messersmith, Mussel-inspired histidine-based transient network metal coordination hydrogels, Macromolecules, 2013, 46(3), 1167–1174 CrossRef CAS PubMed.
- L. Yang, J. Kong, D. Zhou, J. M. Ang, S. L. Phua, W. A. Yee, H. Liu, Y. Huang and X. Lu, Transition-metal-ion-mediated polymerization of dopamine: mussel-inspired approach for the facile synthesis of robust transition-metal nanoparticle-graphene hybrids, Chemistry, 2014, 20(25), 7776–7783 CrossRef CAS PubMed.
- B. Wang, Y. Jeon, S. H. Bhang and J. Kim, Bioinspired dopamine-conjugated polyaspartamide as a novel and versatile adhesive material, eXPRESS Polym. Lett., 2017, 11, 601–610 CrossRef CAS.
- L. Graña Suárez, W. Verboom and J. Huskens, Cyclodextrin-based supramolecular nanoparticles stabilized by balancing attractive host–guest and repulsive electrostatic interactions, Chem. Commun., 2014, 50(55), 7280–7282 RSC.
- Q. Ye, F. Zhou and W. Liu, Bioinspired catecholic chemistry for surface modification, Chem. Soc. Rev., 2011, 40(7), 4244–4258 RSC.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Mussel-inspired surface chemistry for multifunctional coatings, Science, 2007, 318(5849), 426–430 CrossRef CAS PubMed.
- I. J. Minten, Y. Ma, M. A. Hempenius, G. J. Vancso, R. J. M. Nolte and J. J. L. M. Cornelissen, CCMV capsid formation induced by a functional negatively charged polymer, Org. Biomol. Chem., 2009, 7(22), 4685–4688 RSC.
- T. Zhou, M. Yu, B. Zhang, L. Wang, X. Wu, H. Zhou, Y. Du, J. Hao, Y. Tu, C. Chen and T. Wei, Inhibition of Cancer Cell Migration by Gold Nanorods: Molecular Mechanisms and Implications for Cancer Therapy, Adv. Funct. Mater., 2014, 24(44), 6922–6932 CrossRef CAS.
- C. Zeng, G. Rodriguez Lázaro, I. B. Tsvetkova, M. F. Hagan and B. Dragnea, Defects and Chirality in the Nanoparticle-Directed Assembly of Spherocylindrical Shells of Virus Coat Proteins, ACS Nano, 2018, 12(6), 5323–5332 CrossRef CAS PubMed.
- A. L. Durán-Meza, M. I. Escamilla-Ruiz, X. F. Segovia-González, M. V. Villagrana-Escareño, J. R. Vega-Acosta and J. Ruiz-Garcia, Encapsidation of Different Plasmonic Gold Nanoparticles by the CCMV capsid proteins, Molecules, 2020, 25(11), 2628 CrossRef PubMed.
- H. S. Jung, K. J. Cho, Y. Seol, Y. Takagi, A. Dittmore, P. A. Roche and K. C. Neuman, Polydopamine encapsulation of fluorescent nanodiamonds for biomedical applications, Adv. Funct. Mater., 2018, 28, 33 Search PubMed.
- A. Trapaidze, M. D'Antuono, P. Fratzl and M. J. Harrington, Exploring mussel byssus fabrication with peptide-polymer hybrids: Role of pH and metal coordination in self-assembly and mechanics of histidine-rich domains, Eur. Polym. J., 2018, 109, 229–236 CrossRef CAS.
- P. Hu, J. Zhang, Z. Wen and C. Zhang, Network single-walled carbon nanotube biosensors for fast and highly sensitive detection of proteins, Nanotechnology, 2011, 22(33), 335502 CrossRef PubMed.
- P. W. Barone; E. S. Jeng; D. A. Heller and M. S. Strano, Biosensors based on single-walled carbon nanotube near-infrared fluorescence, John Wiley & Sons Ltd., 2007, pp. 843–854 Search PubMed.
- C. C. Cid, J. Riu, A. Maroto and F. X. Rius, Biosensors based on carbon nanotube-network field-effect transistors, Methods Mol. Biol., 2010, 625, 213–225 CrossRef CAS PubMed.
- C. L. Lay, J. Liu and Y. Liu, Functionalized carbon nanotubes for anticancer drug delivery, Expert Rev. Med. Devices, 2011, 8(5), 561–566 CrossRef CAS PubMed.
- G. R. Dieckmann, A. B. Dalton, P. A. Johnson, J. Razal, J. Chen, G. M. Giordano, E. Muñoz, I. H. Musselman, R. H. Baughman and R. K. Draper, Controlled Assembly of Carbon Nanotubes by Designed Amphiphilic Peptide Helices, J. Am. Chem. Soc., 2003, 125(7), 1770–1777 CrossRef CAS PubMed.
- A. Ortiz-Acevedo, H. Xie, V. Zorbas, W. M. Sampson, A. B. Dalton, R. H. Baughman, R. K. Draper, I. H. Musselman and G. R. Dieckmann, Diameter-Selective Solubilization of Single-Walled Carbon Nanotubes by Reversible Cyclic Peptides, J. Am. Chem. Soc., 2005, 127(26), 9512–9517 CrossRef CAS PubMed.
- M. Zheng, A. Jagota, E. D. Semke, B. A. Diner, R. S. McLean, S. R. Lustig, R. E. Richardson and N. G. Tassi, DNA-assisted dispersion and separation of carbon nanotubes, Nat. Mater., 2003, 2(5), 338–342 CrossRef CAS PubMed.
- M. Kus-Liśkiewicz, P. Fickers and I. Ben Tahar, Int. J. Mol. Sci., 2021, 22, 10952–10973 CrossRef PubMed.
- M. de Ruiter, R. M. van der Hee, A. J. M. Driesen, E. M. Keurhorst, M. Hamid and J. J. L. M. Cornelissen, J. Controlled Rel., 2019, 307, 342–354 CrossRef CAS PubMed.
- A. Hassani-Mehraban, S. Creutzburg, L. van Heereveld and R. Kormelink, BMC Biotechnol., 2015, 15, 80 CrossRef PubMed.
- K. C. Black, J. Yi, J. G. Rivera, D. C. Zelasko-Leon and P. B. Messersmith, Polydopamine-enabled surface functionalization of gold nanorods for cancer cell-targeted imaging and photothermal therapy, Nanomedicine, 2013, 8(1), 17–28 CrossRef CAS PubMed.
- Z. Sun, V. Nicolosi, D. Rickard, S. D. Bergin, D. Aherne and J. N. Coleman, Quantitative Evaluation of Surfactant-stabilized Single-walled Carbon Nanotubes: Dispersion Quality and Its Correlation with Zeta Potential. The, J. Phys. Chem. C, 2008, 112(29), 10692–10699 CrossRef CAS.
- E. Ramos, W. A. Pardo, M. Mir and J. Samitier, Dependence of carbon nanotubes dispersion kinetics on surfactants, Nanotechnology, 2017, 28(13), 135702 CrossRef PubMed.
- W. H. Duan, Q. Wang and F. Collins, Dispersion of carbon nanotubes with SDS surfactants: a study from a binding energy perspective, Chem. Sci., 2011, 2(7), 1407–1413 RSC.
- Y. H. Ding, M. Floren and W. Tan, Mussel-inspired polydopamine for bio-surface functionalization, Biosurf. Biotribology, 2016, 2(4), 121–136 CrossRef CAS PubMed.
- M. Sajitha, A. Vindhyasarumi, A. Gopi and K. Yoosaf, Shape controlled synthesis of multi-branched gold nanocrystals through a facile one-pot bifunctional biomolecular approach, RSC Adv., 2015, 5(119), 98318–98324 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb02688k |
| This journal is © The Royal Society of Chemistry 2023 |