Concentrated polymer brush-modified cellulose nanofibers promote chondrogenic differentiation of human mesenchymal stem cells by controlling self-assembly†
Punnida
Nonsuwan‡
a,
Nanami
Nishijima‡
ab,
Keita
Sakakibara
 *c,
Tadashi
Nakaji-Hirabayashi
*c,
Tadashi
Nakaji-Hirabayashi
 *abd and
Chiaki
Yoshikawa
*abd and
Chiaki
Yoshikawa
 *a
*a
aResearch Center for Functional Materials, National Institute for Materials Science (NIMS), Tsukuba, Ibaraki 305-0047, Japan. E-mail: yoshikawa.chiaki@nims.go.jp
bFaculty of Engineering, Academic Assembly, University of Toyama, 3190 Gofuku, Toyama, Toyama 930-8555, Japan
cResearch Institute for Sustainable Chemistry, National Institute of Advanced Industrial Science and Technology (AIST), 3-11-32 Kagamiyama, Higashi-hiroshima, Hiroshima 739-0046, Japan
dGraduate School of Innovative Life Science, University of Toyama, 2630 Sugitani, Toyama, Toyama 930-0194, Japan
First published on 19th January 2022
Abstract
In order to develop new three-dimensional (3D) cell culture systems for articular cartilage regeneration, concentrated poly(styrene sulfonate sodium salt) brush-modified cellulose nanofibers were employed as building blocks for the self-assembly of human mesenchymal stem cells (hMSCs). Unique 3D cellular structures, such as giant spheres and sheets, were formed by controlling hMSC self-assembly.
Introduction
The human body is formed through organized self-assembly. Sophisticated and hierarchical structures of our tissues and organs are built from smaller individual components such as DNA, peptides, collagens, and cells.1 Inspired by nature's design principles, we have developed a cellular flocculation system using cellulose nanofibers modified with concentrated polymer brushes (CNF–CPBs).2 We demonstrated that human hepatocyte cells (HepG2) spontaneously formed flocs with CNF–CPBs, affording the ability to control their size and shape, and enhance cellular functions. Considering the potential of CNF–CPBs as a new 3D cell culture tool, particularly as a novel cellular self-assembling system, we aimed to extend this system to regenerate articular cartilage using human bone marrow-derived mesenchymal stem cells (hMSCs).Articular cartilage is a connective tissue found in our joints, which has a low frictional coefficient to facilitate smooth movement, and a high compression modulus to transmit loads at the joint.3 It is comprised of three major components: chondrocytes, extracellular matrix (ECM) (mainly, collagen fibers), and 70–80% water. However, as articular cartilage is devoid of blood vessels, lymphatics, and nerves, it is limited in terms of self-repair.3 Therefore, many scientists have extensively studied the repair or regeneration of articular cartilage using cells or materials individually, or in combination; however, many challenges still remain.3
Cellulose is the most abundant, sustainable, and environmentally safe biopolymer, and has attracted significant attention as a naturally occurring nanofiber, called cellulose nanofiber (CNF) or nano-fibrillated cellulose (NFC), for many applications including nanocomposite fillers, (opto)electronic substrates, templates, biomedicals, and pharmaceuticals.4–7 CNF gel networks produced from bacteria, known as bacterial cellulose, have been applied to tissue engineering.5 More recently, CNF dispersion, isolated from wood and plant biomass via mechanical or chemical disintegration, has emerged as an interesting cellular scaffold due to its biocompatibility, cost-effectiveness, abundance, and accessibility to surface modification.2,8–16 Importantly, the dimension of CNF, typically 3–20 nm in diameter and several micrometers in length, is analogous to that of collagen, which is the main component of ECM, resulting in the enhancement of cell proliferation.
In this study, CPBs obtained via surface-initiated living radical polymerization (SI-LRPs) is the key principle for success.17–26 It is known that CPBs exhibit unique structures and properties, in comparison to the corresponding semi-dilute polymer brushes (SDPBs).19,21–26 For example, swollen CPBs in good solvents can extend very close to their full length. Owing to such highly extended structures, CPBs exhibit high-modulus, super lubrication, and size-exclusion effect (bioinert property). These so-called CPB effects have never been realized in the corresponding SDPBs.19,21–26 Herein, we prepared concentrated poly(styrene sulfonate sodium salt) (PSSNa) on CNFs (CNF–CPBs) as an artificial ECM. The CPB of PSSNa can provide CNFs with stable dispersion in water and exhibit specific charge interactions with cells, which is the driving force for flocculation. In addition, we previously demonstrated that the combination of a bacterial CNF network and CPB exhibited synergistic properties including high mechanical strength, ultralow friction, and bioinertness, which are comparable to those of articular cartilage.27 Hence, the CNF–CPBs developed in this study are capable of satisfying the mechanical properties required for articular cartilage.
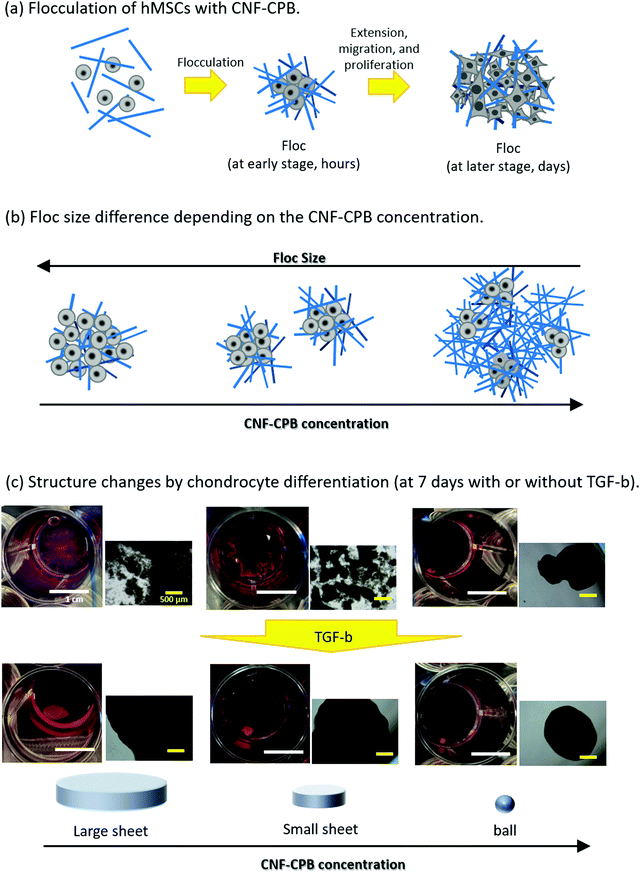
Scheme 1 shows representative results of the approach used here, supporting our proof-of-concept studies. hMSCs were mixed with CNF–CPBs and cultured in chondrogenic differentiated medium as well as in basal medium. hMSCs spontaneously flocculated with CNF–CPBs to form small flocs in basal medium and exhibited a change in their size (hundreds of micrometers), depending on the cultivation time and concentration of CNF–CPB used (Scheme 1a and b). Surprisingly, hMSCs cultured in chondrogenic differentiated medium self-assembled with CNF–CPBs to form a single unique structure, a giant sheet on a millimeter scale (Scheme 1c). The degree of chondrogenesis was evaluated by gene expression analysis, confirming that CNF–CPBs remarkably enhanced chondrogenic differentiation, in comparison to cell pellets, using a standard method. The degree of chondrogenesis was found to be dependent on the self-assembled 3D structures. These results strongly indicate that even in vitro, tissues or organs can be produced by self-assembly of cells and artificial ECM such as CNF–CPBs. To our knowledge, this is the first report to demonstrate cellular self-assembly on a macro scale. We believe that CNF–CPBs would be a powerful tool for 3D cell culture, not only for regeneration of articular cartilage, but also for other tissues.
Experimental
Materials
CNF with an ATRP initiator (CNF–Br) was prepared by surface 2-bromoisobutyryl esterification for mechanically disintegrated CNF from Radiata pine (kindly provided by Dr. K. Abe, Research Institute for Sustainable Humanosphere, Kyoto University) as described in our previous study.2 Cu(I)Br (99.99%, Wako), Cu(II)Br2 (99.99%, Wako Pure Chemicals, Japan), styrene sulfonate sodium salt (SSNa) (99.9%, Sigma-Aldrich, Japan), 2,2′-bipyridine (99.9%, Nacalai Tesque, Japan), and poly(ethylene glycol) methyl ether 2-bromoisobutyrate (Sigma-Aldrich) were used as received.Synthesis of CNF–CPB
SI-ATRP of SSNa from CNF–Br was carried out according to our previous report.2 The number-average molecular weight (Mn) and dispersity (Mw/Mn) of the free polymers produced simultaneously during SI-ATRP were determined using gel permeation chromatography (GPC). The procedure to determine the conversion of SSNa and graft density of PSSNa were detailed in our previous report.2 After polymerization, the CNF-CPBs were washed with Milli-Q water, and the concentration was adjusted to ca. 3 wt%. Samples were stored at 4 °C until use, and the solvent was exchanged with the cell culture medium before cell seeding.Characterization of PSSNa
GPC analysis of PSSNa was performed on a Shodex GPC-101 (Showa Denko K.K., Japan) equipped with two Shodex gel columns at a flow rate of 0.8 mL min−1 using a water/acetonitrile (6/4) mixture with 10 mM LiCl as the eluent (40 °C). The column system was calibrated using standard PSSNa.hMSC culture
Normal human bone marrow-derived mesenchymal stem cells (hMSCs) (multiple donors) were purchased from LONZA (Switzerland). hMSCs were maintained in basal growth medium (MSCGM BulletKit™) (LONZA PT-3001) at 37 °C in humidified air containing 5% CO2. At sub-confluence, the cells were harvested from the flasks using trypsin treatment. In this study, the cells were used at the fifth passage.hMSC culture with CNF–CPB and chondrogenic induction
A 0.5 mL aliquot of hMSC cells (1 × 106 cells per mL) were mixed with 0.5 mL of CNF–CPBs (0.2, 0.1, or 0.01 wt%). One milliliter of cell suspension (5 × 105 cells per mL) along with CNF–PSSNa (0.1, 0.05, and 0.005 wt%) was placed in a low-attachment 24-well multi-plate (PrimeSurface™ Plate 24F) (Sumitomo Bakelite Co., Ltd, Japan) and cultured for the prescribed duration. For chondrogenic induction, chondrogenic induction medium (hMSC BulletKit™) (LONZA PT-3003) with rhTGF-β3 was used. The basal and chondrogenic differentiation medium were changed every 2 or 3 days following the instruction manuals provided by LONZA.Cell pellet culture (control) without CNF–CPBs
Cell pellet culture (hMSCs only) was carried out using a low-attachment 96-well round-bottom multi-plate (PrimeSurface™ Plate 96U; Sumitomo Bakelite Co., Ltd). One milliliter of cell suspension (5 × 105 cells per mL) was placed in each well of the multi-plate.Cryosectioning and TUNEL assay
After the prescribed time, flocs (hMSCs/CNF–CPBs) were gently washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. Samples were then washed with PBS three times, embedded in water-soluble embedding medium (Tissue-Tek O.C.T. Compound; Sakura Finetek USA, Torrance, CA), and frozen by placing them in liquid nitrogen. The frozen samples were sectioned using a cryostat microtome (Leica CM1850, Germany). Sections of 3 μm thickness were allowed to adhere to the glass slide (FRONTIER FRC-01, Matsunami Glass Ind., Ltd), prior to use in the TUNEL assay.Cell viability was determined via the TUNEL assay according to the manufacturer's instructions (Takara Bio Inc., Japan). Cryosections (3 μm thickness) were washed with Milli-Q water, followed by PBS. Then, the sections were immersed in permeabilization buffer (100 μL) and left on ice for 2–5 min. The slides were washed with PBS, followed by application of the labelling reaction mixture (consisting of TdT Enzyme 5 μL + Labelling Safe Buffer 45 μL). The samples were incubated at 37 °C for 60–90 min. The reaction was terminated by washing the slides three times in PBS for 5 min. The nuclei were then stained with 4′,6-diamidino-2-phenylindole (DAPI) for 30 min at room temperature, washed with PBS, and mounted with SlowFade Diamond Antifade Mountant (Invitrogen, USA). Images were collected using a fluorescence microscope (IX71, Olympus, Japan).
Immunohistochemistry staining
To investigate chondrogenic differentiation, immunohistochemistry was performed for aggrecan. Sectioned samples were washed thrice with MilliQ water and subsequently blocked using a protein-blocking solution (Agilent Inc., USA) at room temperature for 1 h. Thereafter, the sectioned samples were incubated overnight at 4 °C with monoclonal mouse anti-aggrecan (Goat-Poly) (RD systems, Inc., USA) (0.2 mg mL−1) as a primary antibody. Subsequently, the secondary antibody, Alexa-Fluor anti-goat 546 (Life Technologies, USA) (×500 dilution), was added to the sectioned samples and incubated for 3 h at room temperature. The sections were then washed thrice with PBS. The nuclei were stained using DAPI, and the sections were observed using a fluorescence microscope (Olympus IX71).Gene expression analysis
hMSCs with CNF–CPBs in a 24-well multiplate were washed with PBS, followed by addition of 1 mL of ISOGEN reagent (Nippon Gene Company, Ltd, Tokyo, Japan) into each well and freezing at −80 °C. The frozen ISOGEN was thawed and mixed vigorously using a pipette to crush the cells. The freeze/thaw cycles were repeated thrice. Following the manufacturer's protocol, total RNA was isolated and stored at −80 °C. DNA in the total RNA isolates was digested using RNase-free recombinant DNase I (Takara Bio Inc.), and the isolates were purified with RNAClean® XP (Takara Bio Inc.). First-strand cDNA from each suspension was synthesized from purified total RNA (0.5 μg) using the PrimerScript RT reagent kit (Takara Bio Inc.). Quantitative real-time PCR (qPCR) was performed using a LightCycler 480 system (Roche, Germany). The primers used for qPCR are shown in Table S1 of the ESI.† Expression of the target genes was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The experimental run number was 1 for the preliminary test (Fig. S6, ESI†). For 3-week samples, experiments were performed in triplicate.Statistical analysis
Statistical analysis was performed using Prism 8.2 software package (GraphPad, San Diego, CA). Gene expression for the 3-week samples is presented as the mean ± standard deviation (SD) of three samples with one-way ANOVA and Tukey pairwise comparisons to analyze the statistical differences between data. Significance was determined at *p < 0.05, **p < 0.01, *** p < 0.001, and ****p < 0.0001.Results and discussion
Preparation of CNF–CPBs
The CNF–CPBs were prepared by SI-ATRP following our previously reported procedure.2 First, 2-bromoisobutyryloxy groups, used as ATRP initiators, were introduced to CNF in anhydrous N-methylpyrrolidone (Fig. S1, ESI†), enabling high esterification to obtain concentrated PSSNa brushes on the CNF surface. Then, PSSNa brushes were grafted from CNF–Br by SI-ATRP (Fig. S2, ESI†). The characteristics of the CNF-PSSNa obtained are summarized in Table S2 (ESI†). Using the criteria of CPB (σ* > 0.1) based on scaling theory,19 the two brushes were categorized as CPBs.Flocculation of hMSCs with CNF–CPBs in basal medium
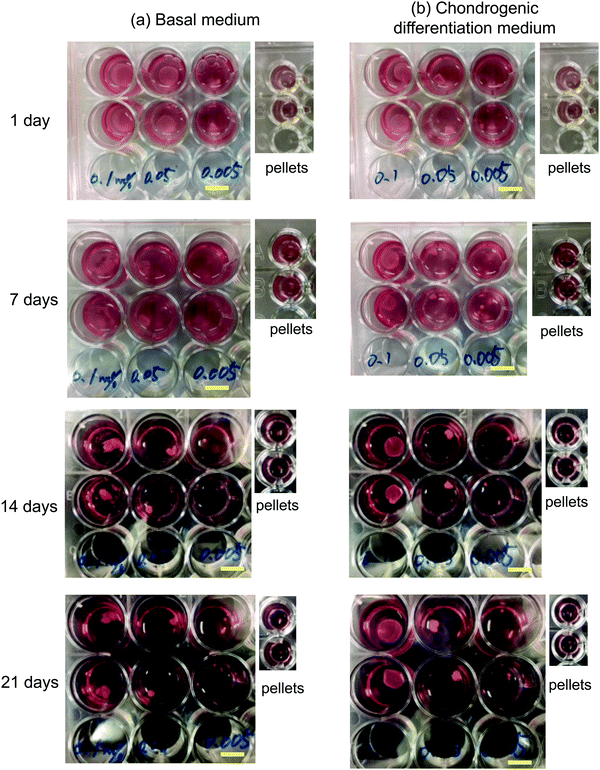
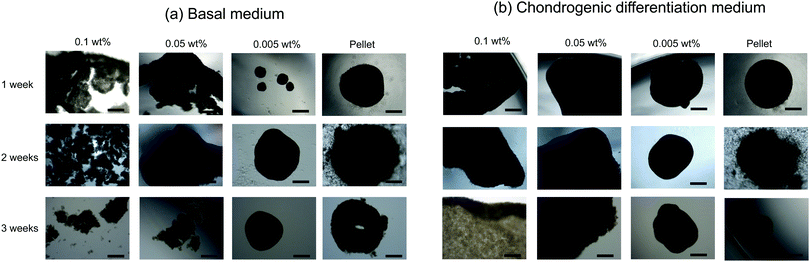
hMSCs were cultured with CNF–CPBs in a basal medium. Cell pellets (hMSCs without CNF–CPBs) were prepared as a control, which is the standard method for chondrogenic differentiation. The concentration of hMSCs was fixed at 5 × 105 cells per mL, whereas that of the CNF–CPBs varied from 0.005 to 0.1 wt%. The cell mixtures were placed in a multi-well plate and incubated for three weeks. Fig. 1a shows a photograph of the flocs obtained. The hMSCs formed flocs of different sizes, depending on the concentrations of the CNF–CPBs, and were found to increase in size with incubation time (Scheme 1a). The flocs were then observed by phase contrast microscopy. As shown in Fig. 2a, the flocs decreased in size with increasing CNF–CPB concentration (Scheme 1b). This was because excess CNF–CPBs prevented small flocs from aggregating and forming larger flocs. To further confirm this, we also co-cultured hMSCs with 0.5 wt% CNF–CPBs, higher concentration. The incubation time was 7 days. As shown in Fig. S3a (ESI†), the excess CNF–CPBs, which did not contribute to floc formation, sank to the bottom of the reaction vessel, and appeared as a white precipitate. In addition, the phase contrast micrograms showed that the floc sizes were smaller than those at lower concentrations (0.1, 0.05, and 0.005 wt%), and the excess fibers filled the gap among flocs, which appeared foggy (Fig. S4a, ESI†). Next, we measured the size of the flocs using phase-contrast micrograms and photos (e.g., Fig. 1). For oval flocs, the long axis was adopted as the size of the flocs (Fig. S5, ESI†). Fig. 3a shows the average size of the flocs obtained. Since cell proliferation occurred during the co-culture period and increased the cell density within a floc over time, the floc grew bigger under these conditions (Scheme 1a). At the same time, the flocs tend to gather to form a bigger floc, as observed in Fig. 2a and 3a. When comparing floc size over the same incubation period, we observed that the floc size decreased with increasing CNF–CPB concentration. As mentioned above, this was due to the excess CNF–CPBs, which prevented the aggregation of cells and/or flocs and prevented the formation of large flocs (Scheme 1b). Taken together, our results indicate that fiber concentration is an important factor in controlling floc size. This was consistent with the results obtained previously using HepG2 cells.2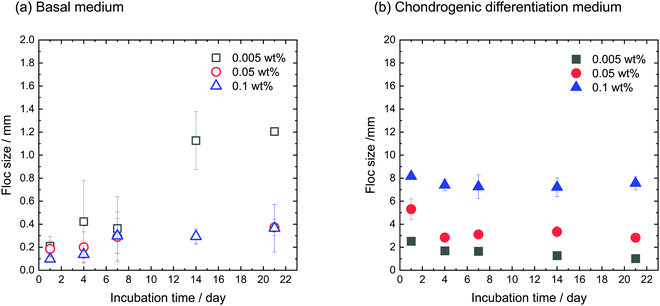 | ||
| Fig. 3 Average size of flocs cultivated using (a) basal medium and (b) chondrogenic differentiation medium. [CNF-CPB]0 = 0.1, 0.05, and 0.005 wt%. [hMSC]0 = 5 × 105 cells/well. | ||
Self-assembly of hMSCs with CNF–CPBs in chondrogenic induction medium
To differentiate hMSCs into chondrocytes, hMSCs (5 × 105 cells per mL) and CNF–CPBs (0.1, 0.05, and 0.005 wt%) were co-cultured in chondrogenic induction medium. Fig. 1b shows images of the flocs obtained. Surprisingly, in contrast to the co-culture in basal medium, the cells and CNF–CPBs self-assembled to form a giant aggregate on the millimeter scale after only 24 h. The giant flocs formed sheets when the CNF–CPB concentration was 0.1 and 0.05 wt%, while a ball shape was obtained at 0.005 wt%. Fig. 2b shows the phase-contrast images. In contrast to the co-culture in basal medium, no small flocs on the micrometer scale were observed when hMSCs were cultured in chondrogenic induction medium with CNF–CPB concentrations of 0.1 and 0.05 wt%. This was likely because hMSCs changed their nature in chondrogenic induction medium. It is known that hMSCs express specific surface proteins such as CD44, CD73, and CD90.28 However, these surface proteins disappear from the cell surface during differentiation. Therefore, hMSCs cultured in chondrogenic medium gradually changed their surface protein profiles, resulting in a charge interaction with the CNF–CPB. In addition, it is known that hMSCs cultured on a low-attachment dish aggregate to form a cell pellet via a cell–cell interaction.29 However, interactions of the cell–cell junction would also change during differentiation of hMSCs into chondrocytes. These may be the reasons why cells/CNF–CPBs did not form small flocs at the micro scale but did form one at the macroscale with a unique structure similar to a sheet.Fig. 3b shows the average size of the flocs. Floc sizes were found to increase with increasing CNF–CPB concentration. This trend was opposite to that observed when hMSCs were cultured in basal medium. As mentioned above, this is probably because the cells changed their surface properties in the chondrogenic medium. In addition, the flocs had little change in size over the 3-week incubation, which was different from the results obtained in basal medium. We attribute this to the gradual cessation of hMSC proliferation once chondrogenesis started. It should be noted that the cells/CNF–CPBs formed a single sheet with a diameter of approximately 8 mm, which is extremely large, in comparison to typical cell pellets (hundreds of micrometers). Thus, our cellular self-assembly system using CNF–CPBs may be able to automatically attain the 3D structure of articular cartilage.
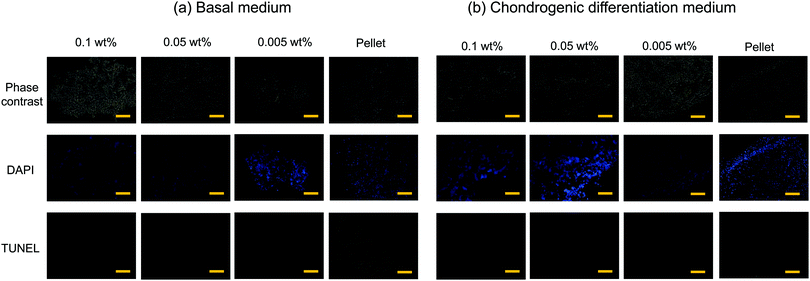
Cell viability in the flocs
Typical 3D cell cultures such as cell pellets (spheroids), or cells entrapped in hydrogels, often suffer from insufficient nutrient and oxygen supply, and hence necrosis, especially when they are relatively large in size and culture time is long. In contrast, cells inside the flocs would thrive because the CNF–CPBs provide significant gaps between cells. To examine this hypothesis, we assessed the viability of flocs and pellets cultured for 3 weeks by TUNEL staining. In order to observe cells inside the flocs and cell pellets, the samples were embedded in optimal cutting temperature (OCT) compound and cut into sections of 3 μm thickness using a cryostat. Next, the cryosections were transferred to glass slides, and apoptotic cells and nuclei were stained in green and blue, respectively. Fig. 4 shows the phase contrast micrograms and fluorescent images of the flocs and pellets that were cultured in (a) basal and (b) chondrogenic differentiation medium. The results confirmed that the cells existed homogenously in all aggregations, with or without CNF–CPBs. While few apoptotic cells were observed in the flocs with CNF–CPBs, cells in the pellets without CNF–CPBs died independently of the cell culture medium. Table 1 summarizes the number of dead cells in the images. Most cells in the pellets (about 90%) died independent of the culture medium used. In contrast, very few cells died in the flocs. This result strongly suggests that the presence of CNF–CPBs provide appropriate gaps for nutrient and oxygen supply, even in macroscale flocs, where nearly all cells survived after 3 weeks.| Medium | Concentration of CNF–PSSNa | Dead cell (%) |
|---|---|---|
| Basal medium | 0.1 wt% | 0 |
| 0.05 wt% | 0 | |
| 0.005 wt% | 7.1 | |
| Pellet | 95.0 | |
| Chondrogenic differentiation medium | 0.1 wt% | 0 |
| 0.05 wt% | 0 | |
| 0.005 wt% | 0 | |
| Pellet | 88.3 | |
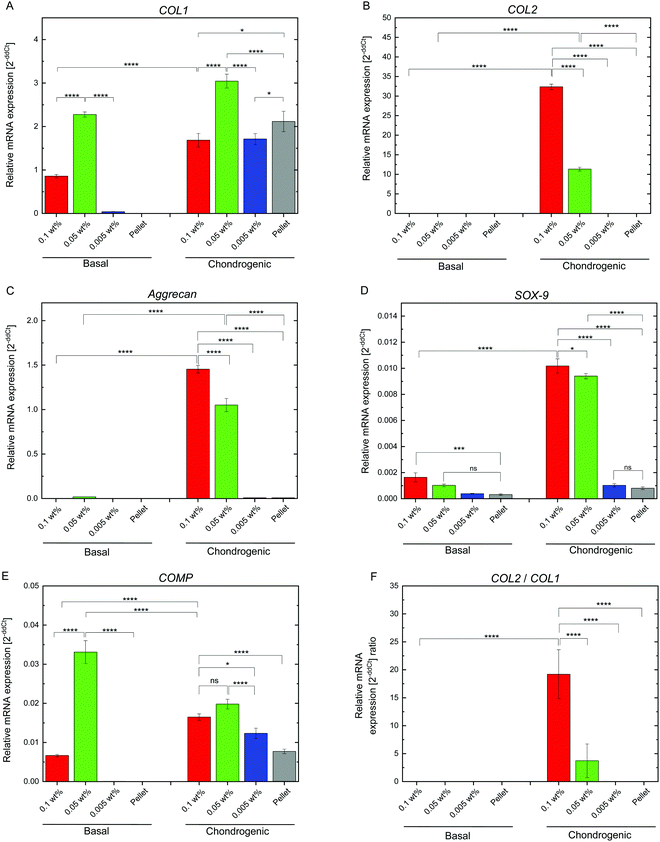
hMSC chondrogenic gene expression
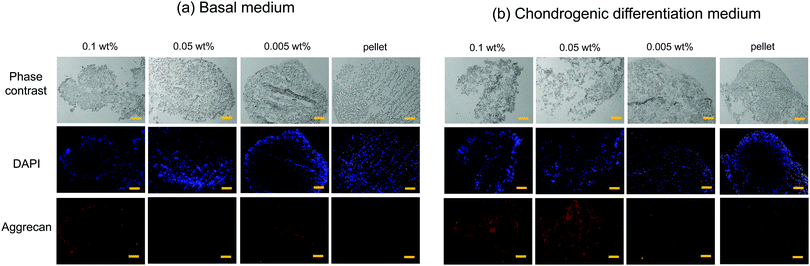
Chondrogenesis of the flocs and cell pellets was evaluated by RT-qPCR of type I collagen (COL1), type II collagen (COL2), aggrecan (aggrecan), SRY-box 9 (SOX-9), and cartilage oligomeric matrix protein (COMP) (Fig. S6, ESI†). All genes were normalized to GAPDH, a ubiquitous housekeeping gene. COL2, aggrecan, SOX-9, and COMP are important for cartilage, while COL1 is typically observed in undifferentiated hMSCs.30–33 To determine the minimal cultivation time necessary for chondrogenesis, as a preliminary test, gene expression analysis was conducted only once using the cellular flocs as well as pellets that were cultured for 1, 2, and 3 weeks. The results are shown in Fig. S6 (ESI†). COL1 (Fig. S6A, ESI†) was upregulated at 2 weeks, but downregulated at 3 weeks, in almost all samples. COL2 (Fig. S6B, ESI†) and aggrecan (Fig. S6C, ESI†) exhibited significant differences between the cultures in basal and chondrogenic media. In particular, the upregulation at 3 weeks was distinct in hMSCs cultured with 0.1 wt% and 0.05 wt% CNF–CPB. SOX-9 (Fig. S6D, ESI†) was significantly upregulated in the 0.1 and 0.05 wt% samples cultured in chondrogenic medium with gene expression dropping off at 2 weeks, followed by a spike at the 3 week point. This was because SOX-9 expression is parallel to COL2.34 In fact, COL2 expression was also found to suddenly spike at 3 weeks in the 0.1 and 0.05 wt% samples. COMP (Fig. S6E, ESI†) was upregulated in samples cultured in chondrogenic medium for 1 week but decreased over time. Since COMP is a marker for cartilage remodeling,33 the results obtained are reasonable because the remodeling associated with differentiation into chondrocytes is complete at some point. Overall, 3 weeks seemed to be sufficient to demonstrate chondrogenesis in cellular flocs. Then, we repeated three more experiments using the 3 week samples and performed gene expression analysis (Fig. 5). The results were almost consistent with those of the preliminary test. COL1 was upregulated in chondrogenic media (Fig. 5A). COL2, aggrecan, COMP, and SOX-9, which are important components of cartilage, were drastically upregulated in 0.1 wt% and 0.05 wt% samples cultured in chondrogenic medium (Fig. 5B–E). Interestingly, COMP was highly expressed on the CNF–CPBs (0.1 and 0.005 wt%), even in basal medium. Although the reason underlying this observation is unclear, it may be hypothesized that this observation is due to the CNF–CPB itself (chemical or physical stimulus) or the structural integrity of hMSCs/CNF–CPBs; we are investigating this further. In addition, the pellets little upregulated chondrogenic markers such as COL2 and aggrecan, even though the pellet culture is known to express these chondrogenic markers at around 3 weeks. The reason for this is unclear; however, under the examined culture condition, it is possible that the pellets need more than 3 weeks to differentiate. Then, we investigated the expression ratio of COL2 to COL1 (Fig. 5F), because the ratio is known to be relatively high when hMSCs differentiate into hyaline cartilage.35 The 0.1 and 0.05 wt% samples exhibited dramatically high ratios of COL2 to COL1, in comparison with the 0.005 wt% samples and the pellets. To further confirm hMSC chondrogenic differentiation in the flocs and pellets, immunohistochemistry was performed for aggrecan using the cross-sections. Fig. 6 shows immunostaining images of the flocs and pellets cultured for 3 weeks in basal and chondrogenic differentiation media. Aggrecan was barely observed on the sections cultured in basal medium, but showed distinct differences with respect to the sections cultured in chondrogenic medium: aggrecan was significantly exhibited on flocs with 0.1 and 0.05 wt% CNF–CPBs compared with flocs with 0.005 wt% and pellets. This result was consistent with the result of gene expression analysis (Fig. 5C), confirming that hMSCs in these flocs were more likely to differentiate into chondrocytes and produce the cartilaginous matrix protein. Overall, the 0.1 wt% CNF–CPBs exhibited the highest gene expression level among the samples, that is, it attained a much more hyaline cartilage. It should be noted that the self-assembly of hMSCs with CNF–CPBs (0.1 and 0.05 wt%), exhibited high cell viability and remarkably high enhancement of chondrogenesis, in comparison with the cell pellet culture, which is commonly used. Thus, this cellular self-assembly system using CNF–CPBs allows easy handling and provides controllable macroscopic 3D structures, homogeneous cell dispersion, high cell viability in the 3D structures, and enhancement of cell functions (chondrogenesis), which are advantages over typical 3D scaffolds such as hydrogel and electrospun fiber. In addition, CNF is renewable and cost-effective. Therefore, we expect that our self-assembly system will pave the way for a new avenue in tissue engineering.Conclusions
Here, we present a new 3D cell culture system for articular cartilage regeneration. The self-assembly of hMSCs and CNF–CPBs afforded macroscopic ball- or sheet-type cellular structures, depending on the CNF–CPB concentration in the presence of chondrogenic induction medium. The highest concentration of CNF–CPB (0.1 wt%) remarkably enhanced chondrogenic differentiation, in comparison to lower concentrations and a cell pellet. The system developed here could be useful in clinical therapy for the regeneration of articular cartilage, as well as other applications in regenerative medicine.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers JP18K05249 and JP19KK0368 (C. Y.), and partially supported by JP18K19907 (T. N.) and the NIMS Joint Hub Program. This work was performed in part on the NIMS Molecular and Material Synthesis Platform. We thank Ms Takako Honda for her technical support on qPCR.References
- A. C. Mendes, E. T. Baran, R. L. Reis and H. S. Azevedo, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5, 582–612 CAS.
- C. Yoshikawa, T. Hoshiba, K. Sakakibara and Y. Tsujii, ACS Appl. Nano Mater., 2018, 1, 1450–1455 CrossRef CAS.
- There are many good reviews. For example; (a) J. Malda, J. Groll and P. R. van Weeren, Nat. Rev. Rheumatol., 2019, 15, 571–572 CrossRef CAS PubMed; (b) H. Kwon, W. E. Brown, C. A. Lee, D. Wang, N. Paschos, J. C. Hu and K. A. Athanasiou, Nat. Rev. Rheumatol., 2019, 15, 550–570 CrossRef PubMed; (c) A. R. Armiento, M. J. Stoddart, M. Alini and D. Eglin, Acta Biomater., 2018, 65, 1–20 CrossRef CAS PubMed.
- D. Klemm, F. Kramer, S. Moritz, T. Lindström, M. Ankerfors, D. Gray and A. Dorris, Angew. Chem., Int. Ed., 2011, 50, 5438–5466 CrossRef CAS PubMed.
- D. Klemm, B. Heublein, H. P. Fink and A. Bohn, Angew. Chem., Int. Ed., 2005, 44, 3358–3393 CrossRef CAS PubMed.
- R. J. Moon, A. Martini, J. Nairn, J. Simonsen and J. Youngblood, Chem. Soc. Rev., 2011, 40, 3941–3994 RSC.
- K. Heise, E. Kontturi, Y. Allahverdiyeva, T. Tammelin, M. B. Linder, I. Nonappa and O. Ikkala, Adv. Mater., 2021, 33, 2004349 CrossRef CAS PubMed.
- M. Bhattacharya, M. M. Malinen, P. Lauren, Y. R. Lou, S. W. Kuisma, L. Kanninen, M. Lille, A. Corlu, C. Guguen-Guillouzo, O. Ikkala, A. Laukkanen, A. Urtti and M. Yliperttula, J. Controlled Release, 2012, 164, 291–298 CrossRef CAS PubMed.
- L. Geurds, J. Lauko, A. E. Rowan and N. Amiralian, J. Mater. Chem. A, 2021, 9, 17173–17188 RSC.
- M. M. Malinen, L. K. Kanninen, A. Corlu, H. M. Isoniemi, Y. R. Lou, M. L. Yliperttula and A. O. Urtti, Biomaterials, 2014, 35, 5110–5121 CrossRef CAS PubMed.
- Y. R. Lou, L. Kanninen, T. Kuisma, J. Niklander, L. A. Noon, D. Burks, A. Urtti and M. Yliperttula, Stem Cells Dev., 2014, 23, 380–392 CrossRef CAS PubMed.
- J. Liu, F. Cheng, H. Grénman, S. Spoljaric, J. Seppälä, J. E. E. Eriksson, S. Willför and C. Xu, Carbohydr. Polym., 2016, 148, 259–271 CrossRef CAS PubMed.
- H. J. Kim, D. X. Oh, S. Choy, H. Nguyen, H. J. Cha and D. S. Hwang, Cellulose, 2018, 25, 7299–7314 CrossRef CAS.
- R. Curvello, V. S. Raghuwanshi and G. Garnier, Adv. Colloid Interface Sci., 2019, 267, 47–61 CrossRef CAS PubMed.
- J. C. Courtenay, J. G. Filgueiras, E. R. deAzevedo, Y. Jin, K. J. Edler, R. I. Sharma and J. L. Scott, J. Mater. Chem. B, 2019, 7, 53–64 RSC.
- J. Athinarayanan, A. A. Alshatwi and V. S. Periasamy, Carbohydr. Polym., 2020, 235, 115961 CrossRef CAS PubMed.
- S. Edmondson, V. L. Osborne and W. T. S. Huck, Chem. Soc. Rev., 2004, 33, 14–22 RSC.
- Polymer Brushes: Synthesis, Characterization, Application , ed. R. C. Advincula, W. J. Brittain, K. C. Caster and J. Ruhe, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 2004 Search PubMed.
- Y. Tsujii, K. Ohno, S. Yamamoto, A. Goto and T. Fukuda, Adv. Polym. Sci., 2006, 197, 1–45 CrossRef CAS.
- R. Barbey, L. Lavanant, D. Paripovic, N. Schüwer, C. Sugnaux, S. Tugulu and H. A. Klok, Chem. Rev., 2009, 109, 5437–5527 CrossRef CAS PubMed.
- S. Yamamoto, M. Ejaz, Y. Tsujii, M. Matsumoto and T. Fukuda, Macromolecules, 2000, 33, 5602–5607 CrossRef CAS.
- S. Yamamoto, M. Ejaz, Y. Tsujii and T. Fukuda, Macromolecules, 2000, 33, 5608–5612 CrossRef CAS.
- Y. Tsujii, A. Nomura, K. Okayasu, W. Gao, K. Ohno and T. Fukuda, J. Phys.: Conf. Ser., 2009, 184, 012031 CrossRef.
- A. Nomura, K. Okayasu, K. Ohno, T. Fukuda and Y. Tsujii, Macromolecules, 2011, 44, 5013–5019 CrossRef CAS.
- C. Yoshikawa, A. Goto, Y. Tsujii, T. Fukuda, T. Kimura, K. Yamamoto and A. Kishida, Macromolecules, 2006, 39, 2284–2290 CrossRef CAS.
- C. Yoshikawa, A. Goto, Y. Tsujii, N. Ishizuka, K. Nakanishi and T. Fukuda, J. Polym. Sci., Part A: Polym. Chem., 2007, 45, 4795–4803 CrossRef CAS.
- K. Sakakibara, K. Maeda, C. Yoshikawa and Y. Tsujii, ACS Appl. Nano Mater., 2021, 4, 1503–1511 CrossRef CAS.
- E. M. Horwitz, K. Le Blanc, M. Dominici, I. Mueller, I. Slaper-Cortenbach, F. C. Marini, R. J. Deans, D. S. Krause and A. Keating, Cytotherapy, 2005, 7, 393–395 CrossRef CAS PubMed.
- A. Abbott, Nature, 2003, 424, 870–872 CrossRef CAS PubMed.
- D. S. Benoit, M. P. Schwartz, A. R. Durney and K. S. Anseth, Nat. Mater., 2008, 7, 816–823 CrossRef CAS PubMed.
- H. Haleem-Smith, R. Calderon, Y. Song, R. S. Tuan and F. H. Chen, J. Cell. Biochem., 2012, 113, 1245–1252 CrossRef CAS PubMed.
- C. Kiani, L. Chen, Y. J. Wu, A. J. Yee and B. B. Yang, Cell Res., 2002, 12, 19–32 CrossRef PubMed.
- S. Tseng, A. H. Reddi and P. E. Di Cesare, Biomarker Insights, 2009, 4, 33–44 CAS.
- W. Bi, J. M. Deng, Z. Zhang, R. R. Behringer and B. de Crombrugghe, Nat. Genet., 1999, 22, 85–89 CrossRef CAS PubMed.
- S. Marlovits, M. Hombauer, M. Truppe, V. Vècsei and W. Schlegel, J. Bone Jt. Surg., Br. Vol., 2004, 86, 286–295 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1tb02307a |
| ‡ The authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |