Open Access Article
Open Access ArticleRecent advances in biosensing approaches for point-of-care breast cancer diagnostics: challenges and future prospects
Anju
Joshi
a,
Anil
Vishnu G. K.
ab,
Tushar
Sakorikar
a,
Arif M.
Kamal
a,
Jayant S.
Vaidya
 c and
Hardik J.
Pandya
c and
Hardik J.
Pandya
 *a
*a
aDepartment of Electronic Systems Engineering, Division of EECS, Indian Institute of Science, Bangalore, India. E-mail: hjpandya@iisc.ac.in
bCentre for BioSystems Science and Engineering, Indian Institute of Science, Bangalore, India
cDivision of Surgery and Interventional Science, University College London, 4919 London, UK
First published on 12th August 2021
Abstract
Timely and accurate diagnosis of breast cancer is essential for efficient treatment and the best possible survival rates. Biosensors have emerged as a smart diagnostic platform for the detection of biomarkers specific to the onset, recurrence, and therapeutic drug monitoring of breast cancer. There have been exciting recent developments, including significant improvements in the validation, sensitivity, specificity, and integration of sample processing steps to develop point-of-care (POC) integrated micro-total analysis systems for clinical settings. The present review highlights various biosensing modalities (electrical, optical, piezoelectric, mass, and acoustic sensing). It provides deep insights into their design principles, signal amplification strategies, and comparative performance analysis. Finally, this review emphasizes the status of existing integrated micro-total analysis systems (μ-TAS) for personalized breast cancer therapeutics and associated challenges and outlines the approach required to realize their successful translation into clinical settings.
1. Introduction
Breast cancer is presently the most common cancer in women,1 with two million new cases and 626![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 deaths in 2020.2 Timely diagnosis and optimal treatment have reduced mortality to 50% in the last few decades. For effective treatment, reliable and efficient diagnosis is necessary. In resource-limited health systems, reducing the cost of diagnosis is crucial as it could enable many more women to be diagnosed in a timely manner so that they can be treated effectively.
700 deaths in 2020.2 Timely diagnosis and optimal treatment have reduced mortality to 50% in the last few decades. For effective treatment, reliable and efficient diagnosis is necessary. In resource-limited health systems, reducing the cost of diagnosis is crucial as it could enable many more women to be diagnosed in a timely manner so that they can be treated effectively.
The existing state-of-the-art diagnostic process includes clinical examination, mammography, ultrasonography, and biopsy. Magnetic resonance imaging (MRI) and positron emission tomography (PET) are needed in some cases.3,4 The necessity of sophisticated instrumentation and expert personnel, particularly for the assessment of tissue biopsy, reduces their wider application for routine investigative consultation for early signs of cancer.5 The bottlenecks associated with the existing diagnostic techniques and continuous demand for early diagnosis have led to the identification of novel potential molecular signatures, diverse genetic and epigenetic patterns and molecular alterations (abnormal expression levels of miRNA and protein), and the validation of existing FDA approved biomarkers for the diagnosis of breast cancer at the earliest stage.6 Therefore, there is a need for developing novel and innovative biosensing technologies to detect such breast cancer-specific entities at clinically relevant concentrations.
Recent years have witnessed a rapid explosion in innovative biosensing assays for breast cancer through different transducers that use the differential between cancer and normal tissue with respect to several different physical properties, viz., electrochemical, optical, piezoelectric, quartz crystal microbalance, acoustic, colorimetric, and electrical impedance. In addition, signal amplification strategies and bio-receptors have improved significantly.7 The primary emphasis remains on improvising the analytical performance towards breast cancer-specific biomarkers in biological fluids to open the possibilities of performing non-invasive liquid biopsy.8
Despite such technological advancements, only a few lab-based biosensing technologies are in popular use and used at the point-of-care (POC).9 The existing knowledge in this field has been described previously with significant emphasis on biomarkers, bio-receptors, bio-transducers, nanostructure mediated signal amplification strategies, and nanobiosensing modalities associated with breast cancer detection.7,10–12
The present review is aimed at complementing these reviews by highlighting various biosensing modalities, technological prerequisites, detailed performance analysis, and how necessary scientific details and engineering aspects are required for translating these lab-based biosensing modalities to clinically significant POC devices for the assessment of patients with breast problems. Compared to existing articles, this review summarizes the latest advancements in the field of microfluidic biochips for the detection of breast cancer-specific biomarkers (proteinaceous, DNA base change, and miRNAs). Additionally, integrated micro-total analysis systems capable of separation, preconcentration, and detection of breast cancer-specific biomarkers (peptides, miRNAs, and exosomes) are also discussed. The present review also critically discusses the performance analysis of different sensing modalities (in terms of amplification strategies, affinity towards the bioreceptor, limit of detection, dynamic range, and specificity achieved towards breast cancer-specific biomarkers) and challenges faced, and emphasizes the requirement of self-competent diagnostic systems capable of processing complex biological matrices and detection on a single platform. Finally, the recent advances in integrated micro-total analysis systems for breast cancer diagnosis are summarized.
2. Biosensors and nano-biosensors for breast cancer diagnosis
Biosensors have emerged as potential self-contained integrated analytical platforms for the ultrasensitive monitoring of breast cancer biomarkers. The accurate quantification of over/under-expressed biomarkers allows the identification of the tumor and its type (benign/cancerous). Nanomaterials with a tuneable morphology, large surface-to-volume ratio, and critical size are integrated to enhance their analytical performance (limit of detection (LOD), detection range, sensitivity, and specificity) towards breast cancer-specific biomarkers. Their exceptionally high surface area and numerous catalytic sites facilitate the biorecognition process (at the electrode surface) as a direct consequence of the increased active transducer area of the biosensor.7,13 The next section discusses different biosensing modalities for breast cancer detection (Fig. 1).2.1. Biosensing modalities for the detection of breast cancer
The activation and overexpression of various biomolecules (receptor proteins on cell surfaces, gene mutations, and microRNAs) are considered promising signatures to predict breast cancer.16 In this regard, different proteinaceous (carcinoembryonic antigen (CEA)),17 human epidermal growth factor receptor 2 (HER2),18 cytokeratin, mucin (MUC1),19 mammaglobin,20 DNA (BRCA1 and BRCA2),21 and miRNA (miR-21, 16, 27a, 150, and 191)7 based biomarkers have been identified as promising candidates for the detection of breast cancer.
Abnormal levels of HER2 (overexpression, which occurs in 15–20% of breast cancer cases) have also been associated with the faster growth and progression of breast cancer (20–30%).22 Therefore, an immunosensing platform was developed by functionalizing 3-aminopropyltrimethoxysilane (APTES) with magnetite nanoparticles (APTMS-Fe3O4) as a platform bioconjugate (PB) for further modification with magnetic gold nanoparticles.23 An innovative RecJf exonuclease-assisted amplification strategy was introduced to develop a high-performance electrochemical aptasensor towards HER2.24 The detection procedure relied on the selective recognition of HER2 on an Apt HER2 (HER2 specific aptamer) conjugated ferrocene-labeled DNA/Au nanosphere (FcNS) modified Au electrode. The highly specific interaction between HER2 and its complementary Apt HER2 spontaneously releases FcNS to hybridize with DNA/Au nanospheres functionalized with HRP (HRPNS). Here, RecJf exonuclease-assisted amplification benefitted the assay by degrading the HER2 aptamer to generate enormous FcNS/HRPNS. These FcNS/HRPNS were electrochemically detectable at the gold electrode surface. The proposed strategy enabled an ultra-low DL (4.9 ng mL−1) with excellent specificity towards HER2, indicating its suitability for the early detection of breast cancer.
The exceptionally high affinity of polycytosine DNA towards inorganic materials (molybdate) was utilized to develop a versatile electrochemical platform against HER2.25 Here, a gold (Au) nanoparticle decorated gold working electrode served as an efficient sensing interface for the immobilization of a polycytosine DNA sequence (dC20) and HER2 specific antibodies (anti-HER2). The presence of HER2 leads to the formation of a sandwich immunocomplex electrochemically detectable due to the formation of redox-active molybdophosphate precipitates (through interaction between dC20 phosphate backbones with molybdate). The assay was found promising to detect an ultra-low concentration of HER2 (0.5 pg mL−1) without any potential interference from biological co-interferents (human IgG, human IgA, p53, carcinoembryonic antigen (CEA), and protein kinase (PKA)).
Likewise, a significant reduction in the detection limit was observed using the HER2 specific aptamer (Apt HER2) and phosphate decorated MnO2 nanosheets to develop electrochemically active molybdophosphate as a consequence of an efficient sandwich reaction (between phosphate, aptamer, and HER2 specific aptamer).26 The sensor demonstrated a DL of 0.05 pg mL−1 towards HER2 with suitability for clinical samples as well.
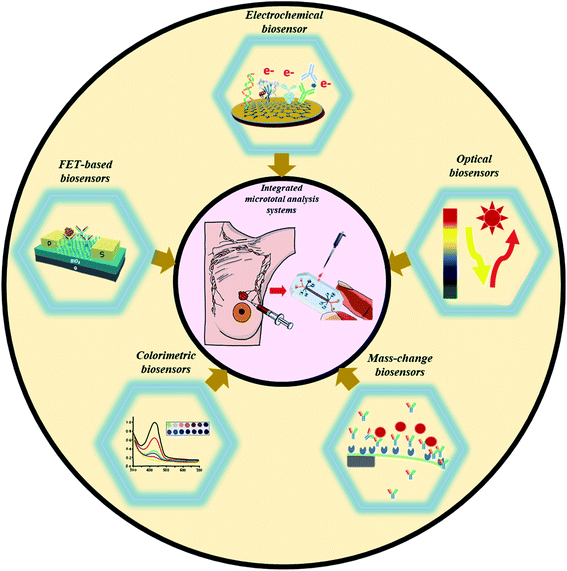
Furthermore, gold nanorod-based bioconjugates (GNR@Pd SSs—Apt—HRP) (as an amplification probe) were explored to develop a DNA tetrahedron biosensor for HER2(Fig. 2I).27 As compared with ss-DNA-based aptamers, the presence of DNA tetrahedrons significantly improved the recognition capability towards HER2. Furthermore, the HRP conjugated GNR@Pd allowed a DL of 0.15 ng mL−1 through hydrogen peroxide facilitated oxidation of hydroquinone.
 | ||
| Fig. 2 Different examples of electrochemical biosensing approaches for breast cancer detection. (I) Schematic representation of GNR@Pd SSs—Apt—HRP: formation and its application for HER2 detection. Reproduced with permission.27 Copyright 2019, Elsevier. (II) Schematics of the steps involved in the electrochemical detection of mi-RNA 155. Reproduced with permission.35 Copyright 2016, Elsevier. (III) Detailed process of graphene facilitated BRCA 1 detection using gold-thiolate chemistry. Reproduced with permission.41 Copyright 2014, Elsevier. | ||
Human mammaglobin (MG) (a highly specific biomarker against all breast cancer cell lines) was electrochemically targeted using 25-base synthetic oligonucleotides to diagnose the hematogenous spread of breast cancer.28 After detailed optimization studies (in terms of the time taken for the immobilization process, rinse process, and hybridization event), the hybridization process was monitored using impedance spectroscopy (frequency range: 1 Hz to 105 Hz). As a result, the biosensor demonstrated a linear dependence of charge transfer resistance with complementary oligonucleotides in the concentration range of 1.0 × 10−9 to 2.0 × 10−8 M with a DL of 5.0 × 10−10 M. Recently, a competitive interaction between MUC1 and cDNA-ferrocene/MXene was explored to perform electrochemical aptasensing of MUC1.29 The presence of high surface area MXene (Ti3C2) nanosheets facilitated the conjugation of ferrocene functionalized complementary DNA (cDNA-Fc) to produce amplified signals. The significant electrical change arising from the competition reaction of MUC1 enabled a DL of 0.33 pM. The efforts towards MUC1 detection were further advanced using a dopamine/MUC1 functionalized carbon nanotube-based electroactive interface.30 The sensor employed EDC-NHS mediated chemistry to establish gelatin (GE) for avoiding the biofouling effects caused due to non-specific biological materials. The synergistic effects of the electroactive functionalized carbon interface and gelatin allowed the detection of MUC1 (0.01 U mL−1) in clinical samples.
Enlightened by abnormal and tissue-specific expressions, miRNAs are considered potential tumor markers for breast cancer diagnosis.31 In this regard, an ultra-sensitive label-free strategy was proposed to detect miRNA-21 using duplex specific nuclease (DSN) and mutual enhancement through the recycling effects of the target and 2′O-methyl modified DNAzyme.32 The application of guanine nanowire amplified tetrahedral DNA nanostructures enabled the detection of miRNA-21 at a concentration of 176 fM.33 However, the proposed assay required further improvements in the linear range and automation for better clinical applications.
Furthermore, specific bipedal DNA walkers were designed to monitor the presence of miR-21 through conformational changes and an increased signal ratio generated using target-respond and target-independent reporters.34 The proposed signal cascade-based amplification strategy (using DNA walkers and reference reporters) allowed the proposed biosensor to demonstrate an ultra-low DL of 67 aM towards exosomal miRNA extracted from breast cancer cells.
A simple and convenient design was executed to detect miRNA-155 in patient's serum samples.35 The proposed sensor involved a synthetic complementary oligonucleotide modified with dimercaptosuccinic acid for targeted interaction with miRNA-155 (Fig. 2II). Based on the simple design principle, the sensor was claimed to be promising for clinical settings by detecting the attomolar concentration of miRNA-155.
The identification of more than 100 mutations in the human caretaker gene (BRCA1) (corresponding to 40% of breast cancer cases) suggests its potential applicability for probing the onset of breast cancer. After a few pioneering studies by the Wang group in early 2000,36–38 a single-walled carbon nanotube modified disposable electrochemical biosensor was developed for BRCA1. The detection procedure involved a simple hybridization event between the target inosine-BRCA1 and its complementary DNA at the electrode surface.39 The hybridization event was further monitored using a guanine-assisted oxidation signal detectable through electrochemical impedance spectroscopy.
The sensor exhibited a DL of 378.52 nM towards the target BRCA1 gene, which was further improved to a value of 0.05 nM using a novel lambda exonuclease-assisted target recycling approach proposed by Xu and coworkers.40 A graphene–DNA decorated electrochemical sensor was developed to perform a hybridization event between a reporter probe (DNA-R) and target probe (DNA-T) to monitor electrochemical signals arising from the oxidation of gold nanoparticles conjugated to DNA-R (Fig. 2III).41 The sensor showed a DL of 1 fM, indicating its potential applicability for performing early cancer diagnosis. In recent years, exosomal miRNAs have gained considerable attention as a remarkable biomarker for the reliable detection of breast cancer.42 Therefore, a target-driven cascade primer exchange reaction (PER) was carried out to generate a single strand through the synergistic assembly of a primer, DNA polymerase, and gated hairpin. The detection process was amplified through the catalysis of hydrogen peroxide using multi-layered nanozymes (MIL-88 @Pt@MIL-88) to obtain a DL of 0.29 fM.
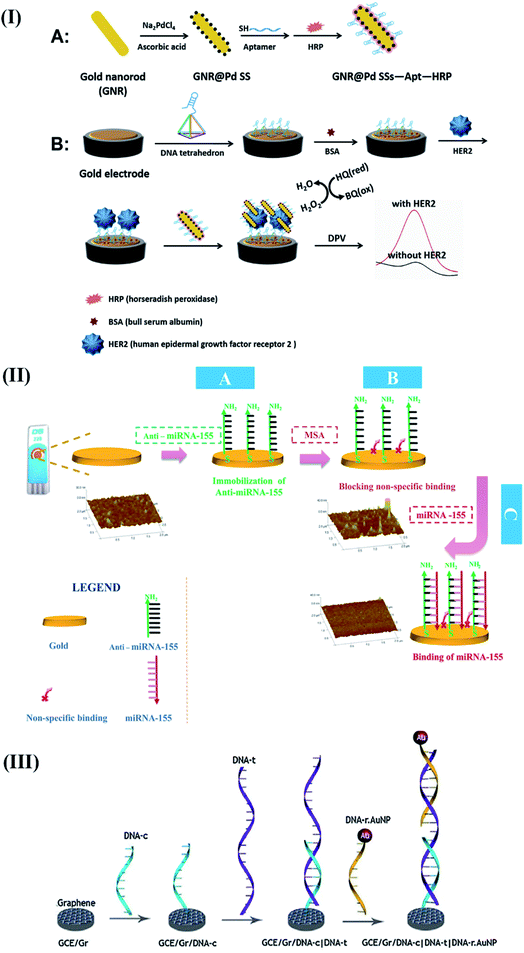
Fluorescence-based optical biosensors. The steady advancement of virtuous, photostable, and biocompatible fluorescent labels,50 QDs,51,52 dye-doped silica nanoparticles,53,54 and diverse nanostructures has led to the development of novel optical sensing platforms for the quantification of breast cancer-specific biomarkers (for, e.g., MUC1,55 HER2,56 c-erbB,57 and Erα).58 In this regard, an innovative strategy involving Exo III nuclease facilitated recycling of the target followed by quadruplex formation was proposed to develop a label-free, fluorescent biosensor to detect c-erbB-2 in the saliva of humans.57 The sensor detected 20 fM of DNA without any potential interference from mismatched DNA sequences. Furthermore, the high affinity of the HER2 binding aptamer (HApt) was synergistically combined with the amplification effects of silver nanocrystals (AgNCs) to detect 0.0904 fM of HER2.59 The proposed turn-on sensor involved a hybridization event between the HApt, G-rich DNA, AgNCs, and complementary base sequences in forming an Ag NC decorated ds-DNA template for the detection process (Fig. 3I). The target HER2 was found to increase the proximity between AgNCs and G-rich DNA sequences to enhance the fluorescence signal. The authors claimed that the proposed assay was a sensitive, convenient, and universal approach for the clinical detection of HER2.
 | ||
| Fig. 3 Different optical biosensors for breast cancer specific biomarkers. (I) AgNC facilitated fluorescence aptasensing of HER2. Reproduced with permission.59 Copyright 2019, Elsevier. (II) Optical Magnetoactuated immunoassay for pre-concentration, and detection of breast cancer cell derived exosome. Adapted with permission.61 Copyright 2020, Elsevier. (III) An all-in-one optical DNA biosensor for multiplexed detection of miRNAs (exosomal origin) using the competitive strand displacement strategy. Reproduced with permission.64 Copyright 2020, Royal Society of Chemistry. | ||
Later, the same group designed a dual-signal nanoprobe to detect microRNA-21 and HER2 together.56 The nanoprobe included a DNA sequence labeled with Black Hole Quencher (BHQ2), fluorescein (FAM)-decorated HER2 specific aptamers, and DNA 2 sequence labeled with sulfo-cyanine5 carboxylic acid (Cy5). The presence of target analytes (microRNA-21 and HER2) was found to increase the fluorescence intensity with a subsequent increase in the distance between Cy5 and FAM (present within the nanoprobe).
The sensor demonstrated a wide detection range (HER2: 0.5 to 3.5 ng mL−1; microRNA-21: 0.1 to 20 pM) and a DL of 0.042 ng mL−1 and 0.048 pM towards HER2 and microRNA-21, respectively.
Furthermore, the analytical response was found to be unaffected in the presence of clinically relevant biomolecules (e.g., microRNA-96, microRNA-300, microRNA-382, human serum albumin, immunoglobulin G, insulin, and MUC1).
Additionally, the energy donor–acceptor pair of a La(III)-metal–organic framework (MOF) and silver nanoparticles (Ag NPs) was utilized to detect miRNA-155 in a sensitive, convenient, and label-free manner.60 Here, sandwich-type architectures were generated through glutaraldehyde-assisted conjugation of the MOF and Ag NPs using 5′-amino-labeled aptamers. The proposed turn-off FRET biosensor enabled a DL of 0.04 ppb (ng mL−1) or 5.5 fM after detailed optimization studies carried out in terms of the pH, miRNA-155 concentration, contact time, and temperature of the miRNA-155 biomarker.
Recently, the application of magneto-actuated immunoassay was demonstrated for the preconcentration and quantification of exosomes from MCF-7, MDA-MB-231, and SKBR3 breast cancer cells (Fig. 3II).61 The extracted exosomes were further modified with a HRP labeled secondary antibody to detect 105 exosomes per μL directly in human serum. The proposed assay was claimed as a successful alternative of flow cytometry to perform multiplexed detection of breast cancer-specific exosomes in resource-limited laboratories.
After a few attempts,62,63 the diagnostic applications of exosomal miRNAs were further strengthened by implementing a competitive strand displacement process to perform DNA-based biosensing of miRNAs.64 The biosensor included three Y-type scaffold oligonucleotides (Y1, Y2, and Y3) extended at their 5′ through single-stranded capture sequences against target miRNAs (miR-21, miR-27a, and miR-375) with the help of quenchers (BHQ1, BHQ2, and BHQ3) respectively (Fig. 3III).
The extended capture sequences were further modified with fluorophores (FAM, Cy3, and Cy5) and labeled reporter sequences (RP-21, RP-27a, and RP-375) to develop a self-quenched fluorescence-based DNA biosensor. As compared to existing reports, the proposed work highlighted the biosensor's capability to enter into exosomes and perform duplex formation (hybridization process with target miRNAs) with the concomitant release of reporter sequences to produce fluorescence signals. The treatment of MCF-7 exosomes with the biosensor enabled the detection of miR-21, miR-27a and miR-375 at a concentration of 0.116 μg mL−1, 0.125 μg mL−1 and 0.287 μg mL−1 respectively. Moreover, the sensor response remains unaltered in the presence of MCF-10A cancer cell lines to prove its practical applicability for routine multiplexed detection of exosomal miRNAs for breast cancer diagnosis.
Surface plasmon resonance (SPR) based biosensors. Liedberg and Nylander first introduced the practical application of surface plasmon resonance for biosensing applications in 1983.65 Since then, SPR biosensors have been extensively used for investigating antigen–antibody interactions, ligand-receptor kinetics, enzyme–substrate reactions, and epitope mapping.66 In 2010, the combined application of microfluidics and optofluidic ring resonator (OFRR) technology was realized to develop a convenient and sensitive biosensor to detect the HER2 biomarker.67 The detection process of HER2 was further improved using an optical fiber-based sensor designed with a taper interferometer cascaded with a Fiber Bragg Grating (FBG).68 Here, the taper interferometer was modified with anti-HER2 using glutaraldehyde-APTES functionalization chemistry to serve as a sensing interface for HER2.
Furthermore, the insensitive response of the FBG towards the refractive index (RI) allowed its application as a temperature thermometer to probe the temperature perturbations arising as a result of the highly specific interaction of anti-HER2 with the HER2 biomarker. The compatible surface functionalization strategies and innovative concepts enabled the sensor to detect ultra-low concentrations (2 ng mL−1) of HER2. However, a significant analytical response towards HER2 was observed by applying a gold nanoparticle (GNP) amplified sandwich assay.69
The authors used SPR chips modified with a carboxymethylated dextran hydrogel to develop an immunocomplex in the presence of target HER2. The sensor exhibited excellent sensitivity towards HER2 (LOD: 180 pg mL−1) in serum samples of human and cell lysate samples from SK-BR-3, MCF-7, and MDA-MB-436 cancer cell lines. Although being sensitive, SPR based biosensing remains challenging for biosensing events taking place beyond 200 nM. Therefore, Bloch surface wave supported 1D photonic crystals were proposed as a novel optical transducer towards label-free, fluorescence detection of ERBB2 (Neu, or HER2).70 The detection procedure relied on sandwich immunocomplex (in response to the target ERBB2 biomarker) formation to tailor the photonic crystals supported with Bloch surface waves. The proposed design successfully accommodated LF or FLUO modes and detected 1.7 ng mL−1 of ERBB2 without any change within the optical system.
SERS (surface-enhanced Raman scattering) based biosensors. Surface-Enhanced Raman Scattering (SERS) is a robust detection method for providing a reliable response towards breast cancer-specific biomarkers. The strength of the SERS method lies in excellent amplification effects (10–15 orders) exerted by the localized surface plasmon resonance (LSPR) generated in metallic nanostructures at the hotspots.71 In one such report, silver nanoparticle (Ag NP) decorated gold nanorod (Au NR) nanostructures were developed as a SERS probe for ultra-low detection of the MUC1 biomarker (4.3 aM).72 The detection process involved the formation of Ag NP and Au NR core-satellite nanostructures with high SERS signals as a consequence of the coupling process of MUC1 specific target DNA with a partial complementary sequence. Furthermore, the highly specific recognition of MUC1 with target DNA quenched the SERS signal due to the release of the core-satellite assemblies. The SERS-based detection of MUC1 was further improved by using a combinatorial approach of magnetic separation powered SERS and colorimetric visualization to obtain a DL of 0.1 U mL−1.73 The proposed biosensor involved magnetic nanobead decorated MUC1 specific aptamers as a capture probe and Raman reporter modified gold–silver–core–shell nanoparticles and MUC1 complementary sequences as a signal indicator to accomplish the dual-mode detection process. The proposed strategy was found competent to perform ultra-sensitive detection of MUC1 in real samples.
As a significant advancement over existing reports, the concept of inverse Molecular Sentinel (iMS) nanoprobes was proposed to perform turn-on SERS detection of breast cancer-specific miRNAs (miR-21 and miRNA-34a).74 It is worth mentioning that a unique target triggered strand displacement process and reorientation of hairpin probes enabled the simultaneous detection of selected miRNAs without any labeling process. The proposed plasmonic nano-star assisted assay was found promising to detect two breast cancer-specific miRNAs (miR-21 and miR-34a) with excellent potential for real sample analysis.
Recently, plasmonic substrates of head-flocked gold nanopillars were designed to generate multiple hotspots as a consequence of interactions of miRNAs and locked nucleic acid probes (LNA).75 The proposed nanopillars were fabricated on silicon wafers (diameter: 200 nm, height: 800 nm, and gap width: 100 to 200 nm) using mask-less reactive ion etching (RIE). The sensor's response relied on the sandwich hybridization process between the miRNAs and short LNA probe (complementary to miRNA) detectable through Cy3-labeled LNA detection probes to attain a DL of 1 aM. The sensor response was found to be consistent with conventional qRT-PCR. However, the highlight of the work was its ability to identify single base mismatch and investigate the subtypes of breast cancer through the expression patterns of exosomal miRNA.
Luminescence-based biosensors. Luminescence-based optical biosensors are well-recognized for analysis in a hazardous environment in a non-invasive way. Therefore, novel luminescent labels are continuously evolving to attain clinically relevant sensitivity and selectivity in real samples. In this regard, up-conversion nanoparticles (α-NaYF4:Yb3+, Er3+) have contributed significantly as luminescent probes for detecting breast cancer specific biomarkers due to their inherent attributes (tunable light emissions, stability against chemicals, biocompatibility, and resistance towards photobleaching).
Lan and coworkers developed a sensitive luminesce assay to detect the vascular endothelial growth factor (VEGF) using α-NaYF4:Yb3+, Er3+ nanoparticles with a diameter of 6–7 nM.76 Here, the synthesis procedure involved thermal decomposition of rare earth stearate for efficient tagging of VEGF specific aptamers. The recognition process of the target VEGF biomarker resulted in a linearly increasing upconversion luminescence intensity within a concentration range of 50 pM to 2000 pM. The assay demonstrated a detection limit of 6 pM. Furthermore, the acceptable recoveries ranging from 98–113% authenticated the clinical suitability of the assay for real sample analysis.
Furthermore, NaYF4:Yb3+/Er3+ luminescent labels were combined with the amplification effects of exonuclease III (Exo III) assisted target recycling and DNA concatemers for the detection of the c-erbB2 oncogene.77 Here, signal amplification was achieved as a consequence of hybridization, degradation, and further hybridization in the presence of Exo III. Furthermore, the presence of AP1 and AP2 (auxiliary probes) triggered the development of super sandwich DNA concatemers to generate an amplified up-conversion luminescence signal (excitation at 980 nm) towards c-erbB2 oncogene (detection limit of 40 aM).
Likewise, a DNA cascade-based amplification strategy was combined with NaYF4:Yb3+/Er3+ luminescent labels to obtain an amplified interference-free response towards circulating miRNA-21 in serum samples.78 Motivated by the enormous potential of up-conversion nanoparticles, quantum dots (QDs) of Cd/ZnS in a core/shell architecture were combined with HER2 specific antibodies to visualize the low levels of HER2 using confocal microscopy.79
Recently, the presence of a wide band-gap, 2-D structure, and resistance towards chemicals has facilitated the application of MoS2 as a luminescent probe.80 Therefore, MoS2 flakes were epitaxially grown to facilitate the hybridization of target miRNA-21 (using thiolated miRNA-21 probe). The photoluminescence-based detection process measured a significant red-shift through perturbation in the local dielectric permittivity due to the hybridization process.
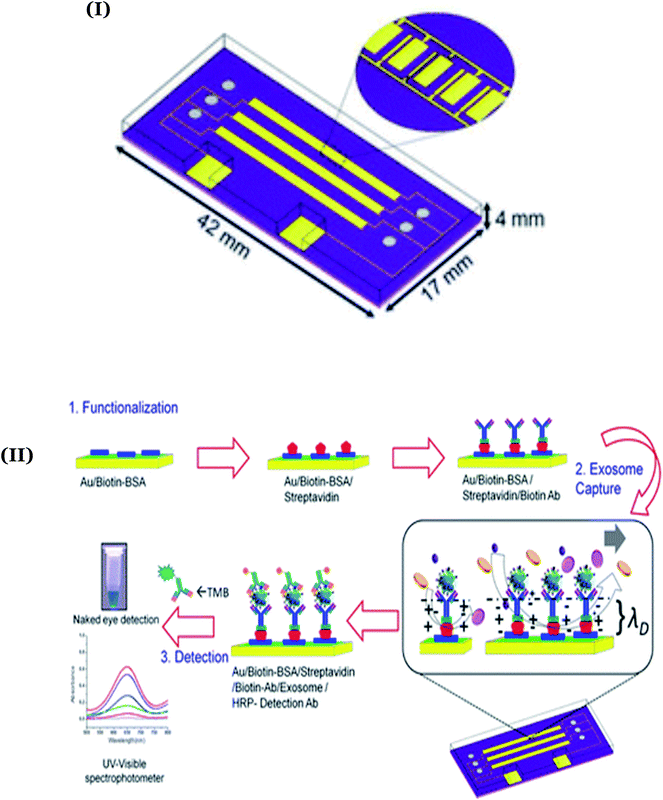
Chemiluminescence based biosensors. Chemiluminescence (CL) detection technologies are preferred for the fabrication of portable and disposable biosensors due to their inherent attributes like high sensitivity, selectivity, low detection limit, and convenient set-up. Therefore, Qiao and coworkers targeted the application of mercaptopropionic acid (MPA) functionalized Eu3+ doped CdS nanocrystals (MPA-CdS:Eu NCs) to detect exosomes derived from MCF-7 breast cancer cells.81 Here, the detection process relied on recognizing the target exosomes through the CD63 aptamer to form G-quadruplex/hemin DNAzymes to facilitate the decomposition of hydrogen peroxide. These events decreased the electro-chemiluminescence (ECL) signals of MPA-CdS:Eu NCs within a wide concentration range of 3.4 × 105–1.7 × 108 particles. The sensor exhibited a detection limit of 7.41 × 104 particles per mL with significant reproducibility and standard recoveries (96.97–106.6%) to authenticate its practical utility for human serum samples. Likewise, CD63 specific aptamer and MUC1 modified Ru(bpy)32+@SiO2 nanoparticles (Ru@SiO2 NPs) were explored to target MCF-7 breast cancer cells for the quantification of MUC1 and derived exosomes.82 Here, the high surface area of SiO2 nanoparticles promoted the immobilization of a large amount of Ru(bpy)32+ to amplify ECL intensity for the detection of MCF-7 cancer cell derived exosomes (detection range (3.22 × 10−4–156 μg mL−1) with a detection limit of 2.73 × 10−4 μg mL−1).
Recently, an innovative approach involving catalytic hairpin assembly (CHA) and hybridization chain reactions (HCRs) was combined with the aptamer-induced release of emitters to develop a versatile detection platform for miRNA-21 and MUC1.83 Here, the detection process of target microRNA-21 (miRNA-21) relied on the release of abundant double-stranded DNA (dsDNA) to intercalate with ([Ru(bpy)2dppz]Cl2) as an ECL indicator and attain a detection limit of 0.1 fM. Furthermore, the presence of MUC1 leads to a significant decrease in the ECL intensity corresponding to the removal of [Ru(bpy)2dppz]Cl2 from the sensing surface. The major highlight of this work was its ability to conveniently amplify and detect multiple biomarkers derived from MDA-MB-231 cells. Within recent years, nanostructures have been considered an integral part of the amplification strategies for various biomedical assays due to their high surface area, stability, reactivity, and small size.84 In this regard, Santa Barbara Amorphous-15 (SBA-15) was functionalized with Ag nanoparticles and Ru(bpy)32+ to covalently immobilize antibodies specific to CA 15-3.85 The proposed nanostructured immunosensor exhibited excellent biocompatibility and intense ECL signals to obtain a detection limit of 0.004 U mL−1 towards CA 15-3.
The ECL response towards CA 15-3 was further improved with the application of a graphene oxide-PEI-carbon quantum dot (CQD)-Au nanohybrid as a signal probe by Qin and coworkers.86 Here, the presence of a high surface area of Ag nanoparticles and polydopamine (AgNPs-PDA) based nanocomposites (developed through a redox reaction between Ag+ and dopamine) provided an efficient immobilization surface for primary antibodies specific to CA 15-3 (Ab1). Furthermore, gold nanostructure modified carbon quantum dot decorated polyethyleneimine–graphene oxide (CQD-decorated PEI-GO) substrates assisted in immobilizing secondary antibodies (Ab2). Here, the synergistic contribution of various nanomaterials facilitated the ECL-assisted electrochemical detection of CA 15-3. The fabricated immunosensor demonstrated a wide linear detection range (0.005 to 500 U mL−1) with a detection limit of 0.0017 U mL−1 (signal-to-noise ratio of 3). Although the authors were successful in achieving a clinically significant, reliable, and sensitive response towards CA 15-3, the fabrication process was considered cumbersome. Therefore, a convenient single step immunoassay was proposed for CA 15-3 through the modification of the high surface area UiO-66-NH2 metal–organic framework with the ECL indicator Ru(bpy)62+.87 The sensor demonstrated excellent sensitivity with an ultra-low detection limit of 1.7705 × 10−5 U mL−1 towards CA 15-3.
Colorimetric biosensors. Colorimetric biosensing techniques are well-recognized for providing a convenient detection process, straightforward interpretation, and rapid and visual read-out. However, their clinical application is strictly hampered by a lack of sensitivity and translation of color-less events into a colored read-out.88,89 The colorimetric detection of breast cancer-specific biomarkers gained momentum with the development of naphthoquinone as a potent inhibitor against an ER+ breast cancer-specific biomarker (arylamine N-acetyltransferases (hNAT1)).90 The same probe was further explored to develop an innovative non-covalent colorimetric approach for the detection of hNAT1(N-acetyltransferase 1) and mNat2 (murine homolog of hNAT1).91 The instantaneous color changes in response to the interaction between the naphthoquinone probe and target enzyme served as a detection mechanism for early estrogen-receptor-positive breast cancer.
Sang and coworkers took advantage of the deoxyribonuclease activity of T7 exonuclease (T7 Exo) to develop a sensitive signal-off colorimetric bioassay for the determination of microRNA-21.92 In the presence of target miRNA-21, the assay procedure relied upon the hybridization event between the miRNA and capture probe (Cp) to form RNA/DNA duplexes.
These duplexes were further subjected to exonuclease activity of T7 exonuclease for the digestion of Cps and subsequent release of miRNAs. These digested Cps remain inaccessible to form a G-quadruplex DNAzyme (GDNA) complex through hybridization with hairpin probes (Hp) in the presence of miRNA-21 to elicit a reduced output signal. In contrast, the absence of target miRNA-21 leads to G-quadruplex/hemin DNAzyme complex formation for the catalytic oxidation of the substrate ABTS by H2O2.
The sensor provided a convenient, efficient visual detection of miRNA 21 with a DL of 0.69 pM (S/N: 3).
The color transition properties of gold NPs were explored to perform an ultra-sensitive real-time detection of estrogen receptor alpha (ERα) protein in breast cancer.93 The coating of the ERα-RNA aptamer (GGGGUCAAGGUGACCCC) on AuNPs was found to prevent salt-induced aggregation of AuNPs, which is further overcome in the presence of ERα with an evident change in the color (wine red to deep blue). This color transition-based phenomenon enabled the sensitive detection of ERα within a detection range of 10 ng mL−1 to 5 μg mL−1 and a DL of 0.64 ng mL−1. The proposed well-planned strategy has a promising application for the POC analysis of ERα in clinical samples.
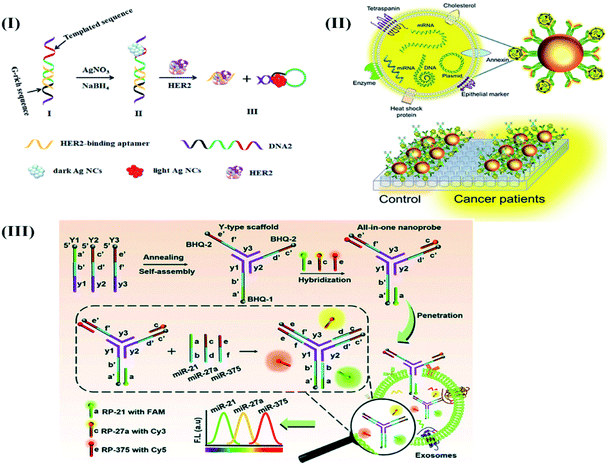
Likewise, the application of DNAzyme assisted recycling was accompanied by rolling circle amplification (RCA) to develop G-quadruplex/hemin complexes as a read-out for the colorimetric detection of BRCA1.94 The binding event of target gene BRCA1 was found to trigger Pb2+ assisted DNA recycling, which generates methylene blue (MB) fragment-padlock template probes to initiate rolling circle amplification (RCA) (Fig. 4). The cascade amplification was found to impart excellent sensitivity to the assay procedure for promising early breast cancer diagnosis.
 | ||
| Fig. 4 DNAzyme assisted DNA recycling and rolling circle amplification for the colorimetric detection of the BRCA1 genetic biomarker for breast cancer. Reproduced with permission.94 Copyright 2015, Royal Society of Chemistry. | ||
3. Performance evaluation of different biosensing modalities for the detection of breast cancer
The application of biosensors holds enormous potential to revolutionize existing breast cancer diagnostics. The performance significantly relies on the choice of the biomarker, robust recognition elements, and signal amplification strategies employed to enhance the biorecognition process. An in-depth literature review (2010–2020) on various transduction methodologies (Section 2.1) revealed step-wise progression towards improvisation in the assay protocol, novel arrangements, and nanomaterial induced signal amplification strategies for excellent sensitivity and specificity against diverse breast cancer-specific biomarkers.Therefore, this section discusses a comparative performance analysis between different biosensing modalities to identify the potential pitfalls (associated with different biosensing modalities) and how the necessary modifications can be proposed to enhance their clinical applications to realize the POC settings for early breast cancer diagnosis.
Different serum-based biomarkers (HER2, MUC1, and human mammaglobin), CTCs, and genetic molecular signatures have been routinely targeted to probe the onset, progression, and metastases stages of breast cancer. Electrochemical biosensors detected an ultra-low concentration of HER2 (0.05 pg mL−1 or 0.2 fM) using a sandwich assay between a HER2 specific peptide and phosphate group and HER2 aptamer functionalized MnO2 nanosheets as an electrochemical probe (Table 1).
| S. no. | Sensing interface | Electrochemical technique | Electrode | Target analyte | Real samples | Strategy | Detection limit | Linear range | Advantages | Limitations | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GCE/Gr/DNA-C|DNA-t|DNA-r AuNP | CV and chronoamperometry | Glassy carbon electrode | BRCA1 | NS | Gold nps | 1 fM | 1 fM to 1 nM | Ultra-sensitive detection of BRCA1 | Gold nanoparticle-based labelled approach adds to the cost and is affected by the instability of the labels | 41 |
| Real sample investigations are not performed | |||||||||||
| 2 | dsDNA/MCH | EIS and DPV | Gold electrode | BRCA1 | NS | Label-free | 0.05 nM | 0.1–10 nM | Label-free sensitive detection of BRCA1 | Immobilized exonucleases adds to the cost and affects the reproducibility of the sensor | 40 |
| 3 | ss-DNA | EIS | GCE | Mammaglobin | Clinical breast cancer samples | Label-free | 5.0 × 10−10 M | 1.0 × 10−9 to 2.0 × 10−8 M | Convenient, sensitive, and time-saving approach | Sensor's response should be extended to other biomarkers to ensure reliable detection | 28 |
| 4 | SPCE-AuNP/HER2-ECD anti-HER2-ECD-AP | LSV | Screen printed carbon electrode | HER2-ECD | Human serum | Alkaline phosphatase | 5 U mL−1 | 0–70 U mL−1 | Simultaneous detection of HER2 and CA 15-3 for accurate breast cancer prediction | Immobilized alkaline phosphatase can add to the cost and affects the reproducibility of the sensor | 100 |
| 5 | AuNPs/Apt/Ab-EGFR | DPV | Magnet | EGFR | Human serum | Gold nps | 50 pg mL | 1–40 ng mL−1 | First approach of Apt/Ab sandwich assay of EGFR | Proposed approach is still in the proof-of-concept stage | 101 |
| 6 | Polycytosine DNA/gold electrode | SWV | Gold electrode | HER2 | Human serum | Gold nps | 0.5 pg mL−1 | 1 pg mL−1 to 1 ng mL−1 | Versatile and sensitive approach for the detection of HER2 | Sensor's response should be extended to other biomarkers to ensure reliable detection | 25 |
| 7 | cDNA/Fc/MXene/Apt/Au/GCE | SWV | Glassy carbon electrode | MUC1 | Human serum | Fc labelled cDNA | 0.33 pM | 1.0 pM to 10 μM | Sensitive and stable approach with clinical potential | Ferrocene-cDNA labelling process can add to the cost and processing steps | 29 |
| 8 | GCE/APTMS-Fe3O4 NPs/Ab/HER2/hyd@AuNPs-APTMS-Fe3O4 labeled Ab-AgNO3 | DPV | Glassy carbon electrode | HER2 | Human serum | Fe3O4 labeled Ag NO3 | 2 × 10−5 ng mL | 5.0 × 10−4 to 50.0 ng mL | AuNP-promoted silver enhancement-based amplification strategy | Ag nanoparticle-based labelled approach | 23 |
| 9 | Au NPs/β-CD/HT/FcNS/HRPNS | DPV | Gold electrode | HER2 | Human serum | HRPNS | 4.9 ng mL−1 | 10–150 ng mL−1 | RecJf exonuclease and peroxidase-based signal amplification towards HER2 | Immobilization of multiple enzymes may add to the cost and affect reproducibility due to random orientation. | 24 |
| 10 | GNR@Pd SSs—Apt—HRP/DNA tetrahedron | DPV | Gold electrode | HER2 | Human serum | HRP | 0.15 ng mL | 10–200 ng mL | Combined application of a DNA tetrahedron and GNR@Pd SSs—Apt—HRP for amplification towards HER2 | Time consuming labelling process and instability due to the presence of the HRP enzyme | 27 |
| 11 | Tetrahedral DNA nanostructure and G-wire | Chronoamperometry | Gold electrode | miRNA 21 | Human serum | Label-free | 176 fM | 0.5–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pM 000 pM |
Tetrahedral DNA nanostructure probes and guanine nanowire amplification assisted detection of miRNA-21 | Regeneration of the biosensor is not feasible and improvisation in the detection range is required | 33 |
| 12 | SWNT-SPE | EIS and DPV | Screen printed electrode | BRCA1 | NS | Label-free | 378.52 nM | 10–120 μg mL−1 | Nano-structuring and screen printing facilitated disposable and sensitive platform for the detection of BRCA1 | Complex DNA based chemistry can lead to false signals as a consequence of varying DNA structures. | 39 |
| 13 | Au-SPR/anti-miRNA-155 | EIS and SWV | Gold–SPE | miRNA −155 | Human serum | Label-free | 5.7 aM | 10 aM to 1.0 nM | Simple and sensitive approach for multiplexed detection of miRNA-155 | Sensor's response should be extended to multiplexed miRNA detection in human serum | 35 |
| 14 | HER2-peptide/Apt HER2/phosphate ion | SWV | Gold electrode | HER2 | Human serum samples | Label-free | 0.05 pg mL−1 | 0.1–500 pg mL−1 | Dual signal amplification based sensitive approach for HER2 detection | Adsorption based immobilization of phosphate ions onto MnO2 nanosheets can affect the reproducibility of sensor response | 26 |
| 15 | 2′-O-Methyl modified G-quadruplex-hemin DNAzyme/DSN | Amperometry (current–time) | Gold Electrode | miR-21 | Human serum | Label-free | 8 aM | 50–500 aM | 2′-O-methyl modified G-quadruplex-hemin DNAzyme and DSN assisted target recycling-based amplification of miR-21 detection | Instability of DNAzymes and high cost can affect sensor performance | 32 |
| 16 | Bipedal DNA walkers/LNA | DPV | Gold electrode | miR-21 | Breast cancer patient serum | LNA labelled captured probe | 67 aM | 0.1–100 fM | Cascade signal amplification assisted by two footed DNA walkers and excellent specificity due to LNA and TMSDR approaches | Further improvisation in the performance (extension of the walking duration of DNA walkers) is required and integration of components to execute exosomal capture, release, crush and detection on a single platform is required | 34 |
| 17 | MOF@Pt@MOF | DPV | Gold electrode | miR-21 | Breast cancer patient serum | PER (primer exchange reaction) | 0.29 fM | 1 fM to 1 nM | Ultrasensitive response due to cascade PER and multiple layered nanozymes | Time-consuming and high cost of PER constituents (primer, gated hairpin, and DNA polymerase) | 42 |
The proposed dual signal amplification using peptide targeted capture of HER2 followed by the formation of redox-active phosphomolybdate was claimed to be facile, cost-effective, and capable of reaching clinical cut-off values. Likewise, enzyme-assisted amplification (e.g., HRP and alkaline phosphatase) strategies were also found promising to achieve a low detection limit towards other breast cancer-specific serum biomarkers (EGFR, MUC1, and CEA) using electrochemical transduction methods. However, the denaturation of antibodies and enzyme labels remains a critical issue that can severely affect their electrochemical performance.
Therefore, de novo synthesized antibodies, aptamers (developed through the SELEX procedure), and molecularly imprinted polymers have emerged as suitable alternatives to antibodies for providing stability to the sensing platform. The concern of enzyme instability has been efficiently addressed with the use of functional nanomaterials CuO and silver nanoflake supported Pt NPs (Pt@AgNFs) as enzyme mimics without compromising their performance. Furthermore, the employment of innovative signal strategies like DSN, T7 Exo, and rolling circle amplification provided 102–105 fold improvements in DLs to detect genetic aberrations related to breast cancer.
Like electrochemical transduction methods, FET-based biosensors are also considered promising to perform ultra-sensitive detection of diverse breast cancer-specific biomarkers (mutated genes, protein, and DNA) without any fluorescence/electrochemical labels (Table 2).
| S. no. | Target biomarker | Nanomaterials | Detection limit | Advantages | Limitations | Reference |
|---|---|---|---|---|---|---|
| 1 | BRCA1 | Graphene oxide | 0.2 nM | Convenient strategy with potential for lab-on-chip application to detect exon 20 of BRCA1 | PCR assisted amplification is required for the specific DNA region, 157 bp | 44 |
| 2 | DNA and cancer cells (universal approach) | Graphene oxide ink | 0.1 pM | Versatile approach with excellent selectivity with 0.1 pM of the target DNA | Needs association of integrated circuits for wireless POC applications | 45 |
| 3 | HER3 | Pt modified graphene | 300 fg mL−1 | Scalable fabrication of arrays of graphene field effect transistors (GFETs). A novel approach for scalable fabrication of 52 arrays of graphene field effect transistors (GFETs) for sensitive detection of HER3 | Real sample analysis needs to be done in the presence of HER1, HER2, and HER4 | 46 |
| 4 | HER2 and EGFR | r-GO-silicon oxide NPs | 1 pM and 100 pM | Convenient, sensitive, and selective approach | Sensor's response should be extended to multiple breast cancer specific biomarkers on a single platform | 47 |
| 5 | miRNA-155 | MoS2 | 0.03 fM | Convenient and sensitive approach | Sensor's response should be extended to multiplexed miRNA detection | 43 |
A detection limit of 1 pM was obtained towards HER2 using 3D hierarchical nanostructures of a graphene oxide-based FET. As compared with both electrochemical and electrical modalities, a slight improvement in the DL was observed for HER2 using fluorescence-assisted optical transduction of HER2 and Apt HER2 (HER2 specific aptamer) interactions. The AgNCs facilitated the hybridization process to obtain a concentration-dependent fluorescence signal with a DL of 0.0904 fM towards HER2 (Table 3).
| S. no. | Type of sensor | Target analyte | Matrices | Detection limit | Linear range | Advantages | Limitations | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | Fluorescence | c-erbB-2 oncogene | Human saliva | 20 fM | 100 fM to 1 pM | Sensitive detection of c-erbB-2 in saliva using thioflavin-T as the fluorescent indicator | Loss of fluorescence due to photobleaching | 57 |
| 2 | Fluorescence | HER 2 | Human serum | 0.0904 fM | 8.5–255 fM | Convenient and sensitive turn-on approach for the detection of HER2 | Temperature and pH-dependent fluorescence response | 59 |
| 3 | Fluorescence | HER2 and microRNA-21 | Human serum | 0.042 ng mL−1; 0.048 pM | 0.5 to 3.5 ng mL−1; 0.1 to 20 pM | Convenient approach for sensitive multiplexed detection of HER2 and microRNA-21 | Loss of fluorescence due to photobleaching | 56 |
| 4 | Fluorescence | miRNA-155 | Human serum samples | 5.5 fM | 2.70 fM to 0.01 pM | Ultra-sensitive FRET approach for the detection of miRNA-155 using a La(III)-metal−organic framework (MOF) and silver nanoparticles (Ag NPs) as an energy-donor couple | Approach should be extended for multiplexed detection of miRNAs | 60 |
| 5 | Fluorescence | HER2 | Cell lysates | 1.5 pM | NA | Sensitive assay for the detection of HER2 within 30 min | Future studies to be directed to perform the detection of HER2 in human plasma samples | 70 |
| Application of two different quantification techniques (LF or FLUO modes) without any change in optical systems was proposed | ||||||||
| 6 | Fluorescence | miR-21, miR-27a, miR-375 | Human serum samples | 0.116 μg mL−1, 0.125 μg mL−1, 0.287 μg mL−1 | 5–140 nM, 2–100 nM, 7–70 nM | Self-competent biosensor with a capability to enter into exosomes and perform multiplexed detection of miRNAs (miR-21, miR-27a and miR-375) | Collection of exosomes from cell lysates and extraction is time consuming (48 h). | 64 |
| Attached fluorophores are subject to photo-bleaching | ||||||||
| 7 | SPR | HER2 | Serum | 180 pg mL | 0.23–55 ng/mL | Gold nanoparticle assisted amplification leads to a 180 pg mL−1 detection limit towards HER2 | Detection process should be extendable to other biomarkers for accuracy in the detection process | 69 |
| 8 | SPR | HER2 | NA | 2 ng mL | NA | Compact, disposable, and sensitive (2 ng mL−1) approach towards the detection of HER2 | Sensor's response should be extended to multiple breast cancer biomarkers | 68 |
| 9 | SPR | HER2 | Serum and cell lysates | 10 ng mL | 13 to 100 ng mL | Convenient and efficient approach | Streptavidin-avidin antibody modifiers are required for better and oriented interaction with the target | 67 |
| 10 | SERS | miR-21, miR-34a | NA | NA | NA | Strategy holds potential for multiplexing capabilities | Detailed studies are required for implementation in clinical practice | 74 |
| 11 | SERS | MUC1 | Human serum samples | 4.3 aM | 0.005–1 fM | Sensitive technique for the detection of MUC1 (4.3 aM) using core–shell nanostructures | Multiplexed detection of breast cancer biomarkers is required | 72 |
| 12 | SERS-colorimetric | MUC1 | Human serum samples | 0.1 U mL−1 | 0.1–100 U mL−1 | Visual naked eye based sensitive detection of MUC1 | Time consuming assay procedure (MUC1 capture: 90 min and magnetic separation: 75 min) | 73 |
| 13 | SERS | Exosomal miRNAs | Human serum samples | 1 aM | 1 aM to 100 nm | Versatile approach for sensitive detection of mi-RNA's | Incorporation of sample extraction, processing, and detection capabilities on a single platform is required | 75 |
In recent years, SERS-based optical biosensors have emerged as a sensitive and non-destructive method for trace-level detection of breast cancer-specific biomarkers. With the application of silver nanoparticle (Ag NP) decorated gold nanorods (Au NRs) as a SERS probe, a detection limit of 4.3 aM towards MUC1 (Table 3) was observed. Recently, the potential of the SERS technique has been extended to detect eight breast cancer-specific biomarkers in human tears for the diagnosis of asymptotic breast cancer patients using multivariate statistical analysis and it has been found promising.102 The analytical response of colorimetric and mass change-based transducers incorporated cost-effectiveness and ease of visual detection of target biomarkers. However, the performance was severely restricted with respect to sensitivity and selectivity.
As sensitivity, portability, and performance have been concerned, electrochemical, FET, and optical transduction are best suited for detecting a wide range of breast cancer-specific biomarkers. However, the poor refractive index and turbidity of complex biological samples remain a major bottleneck to perform optical biosensing of breast cancer-specific biomarkers. Although SERS-based biosensors have enabled optical biosensing techniques to show excellent sensitivity in a non-invasive way, their potential as a standalone device remains doubtful due to complications in the integration of SERS and Raman components together along with sample handling techniques. Therefore, considering the present status, electrical and optical biosensors hold enormous potential to establish themselves under POC clinical settings. However, an integration of electrical/optical transducers with multiple components (isolation process of biomarkers from complex biological fluids, concentration, and detection) on a single platform is still in infancy to revolutionize this field for ensuring POC breast cancer detection. The next section discusses the recent advancements in the field of integrated micro-total analysis systems for breast cancer diagnosis.
4. Integrated micro-total analysis systems for breast cancer diagnosis
Biosensors have demonstrated enormous potential to revolutionize the existing breast cancer diagnostics. Few commercially available solutions like CELLSEARCH® and Videssa Breast® have been attempted to realize POC settings for breast cancer diagnosis. However, the clinical translation of such lab-driven biosensing platforms requires an integration of multiple bioanalytical functions into a miniaturized portable instrument with functionalities of full-sized laboratories (pre-treatment of complex biological specimens, isolation of the required analyte of interest, and then quantification through a suitable read-out).9 With remarkable innovations in this field, a few micro-total analysis systems (μTASs) were developed for the detection of breast cancer (Table 4) and have been discussed in the following section.| S. no. | Sensing modalities | Technique | Target entity | Detection limit | Detection range | Cost | Real samples | Advantages | Limitations | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Electrochemical | Electrochemical impedance spectroscopy | ErbB2 | 1 fM | 1.0 fM to 0.1 pM | NA | NA | Clinically relevant high sensitivity towards ErbB2. | Device's response should be extended for other potential breast cancer specific biomarkers. | 103 |
| 0.1 pM to 0.1 μM | Efficient discrimination between ErbB2, ErbB3, and ErbB4 biomarkers | Sample processing steps needs to be incorporated for lab-on-chip settings | ||||||||
| 2 | Electrochemical | Amperometry | ERα | 10.0 fg mL−1 | 16.6–513.3 fg mL | $0.20 | Calf serum | Inexpensive approach, quick and sensitive detection (2 h) | Device response should be extended to other breast cancer specific biomarkers for reliable prediction | 104 |
| 3 | Electrochemical | Amperometry | HER2 | 12 pg mL−1 | 2 pg mL−1 to 12.5 ng mL−1 | $0.25 | Serum | Inexpensive, ultra-fast assay process (15 min) | Multiplexing is required for other breast cancer specific biomarkers for reliable prediction, and sample processing steps needs to be incorporated into the device | 105 |
| 4 | Electrochemical | Capacitance electrochemical impedance | HER2 | 1 pM | 1 pM to 100 nM | NA | Serum | Capacitance-based sensitive detection of HER2 in undiluted serum. | Response of the device should be extended to other breast cancer specific biomarkers for reliable prediction | 106 |
| 5 | Optical | FRET | let-7a and miRNA-195 | 0.061 nM; 0.064 nM | 0–10 nM; 0–10 nM | NA | Serum | Sensitive response towards multiple breast cancer specific biomarkers | Minimization of the noise and background interference is required for human serum samples | 108 |
| 6 | Optical | Fluorescence microfluidics | miR-21 | NA | NA | NA | 51 (30 healthy and 21 breast cancer positive patients) blood samples | Efficient and low-cost approach for discriminating breast cancer patients from healthy individuals. | Multiplexing capabilities need to be incorporated for enhancing the performance of the device. | 109 |
| 7 | Optical | Fluorescence | miR-4732, miR-3646, miR-4484, miR-K12-5 | 1 pM | 10−7 to 10−12 M | NA | Serum | Sensitive multiplexed detection of miRNAs (∼30 min) | Sample processing steps need to be integrated to avoid interference in the detection process | 110 |
| 8 | Optical | Fluorescence microfluidics | HER2 positive exosomes | NA | NA | NA | Plasma sample from breast cancer patients | Device capable of performing on-chip capture and quantification of circulating HER2 positive exosomes | Analytical performance of the device (concentration responses and interference studies) should be investigated in detail | 111 |
| 9 | Optical | Fluorescence microfluidics | EpCAM positive exosomes | NA | NA | NA | Blood of breast cancer patients | Device capable of performing on-chip isolation and detection of circulating exosomes | Requirement of the secondary antibody labelled approach for exosome enrichment | 112 |
| 10 | Optical | SERS | Peptides from BRCA1 | 0.1 ng μL−1 | NA | NA | NA | Device competent in extracting breast cancer specific peptides and performing their detection | Multiplexed separation and SERS-based detection of breast cancer specific peptides are required | 113 |
Ali and coworkers emphasized the ultra-sensitive detection of the ErbB2 gene (located on the 17q chromosome), relying on its close association with aggressive primary breast cancer expression.103 Here, the authors developed an electrochemical microfluidic biochip with a 3-D immunoelectrode comprising graphene foam (GF) and an n-TiO2 composite (GF-TiO2) for the efficient immobilization of anti-ErbB2. The required three-electrode setup on the micro-fluidic chip was acquired through the conventional soft-lithographic fabrication process. Owing to the synergistic combination of the high reaction kinetics and biocompatibility of n-TiO2 with porous GF and promising EDC-NHS chemistry, the proposed electrochemical microfluidic device demonstrated an extraordinary analytical response towards ErbB2 (detection range: 1.0 fM to 0.1 μM; sensitivity: 123.5 kΩ pM−1). Moreover, the device's potential was also extendable to detect ErbB1, ErbB3, and ErbB4 together in biological fluids.
Likewise, a disposable microfluidic electrochemical array device (μFED) was proposed for the detection of estrogen receptor alpha (ERα) by Uliana and coworkers.104 The low cost (US$ 0.20), inexpensive printing, and convenient fabrication procedure were a few attributes that made the proposed device attractive for the mainstream research/commercial market.
The proposed μFED was fabricated by sandwiching two polystyrene sheets with screen-printed counter electrodes (CEs), reference electrodes (REs), and a microarray of 8 working electrodes (WEs) using adhesive polystyrene cards (Fig. 5I). The surface of the WEs was modified with estrogen response elements (DNA-ERE) to bind target ERα. However, to ensure an amplified electrochemical response, the target ERα was magnetically enriched using anti ERα, and a HRP labeled magnetic particle bioconjugate (MP-Ab-HRP). The resulting bioconjugate was further introduced into the detection chamber of μFED (microarrays of WEs modified with DNA-ERE) to perform electrochemical detection of ERα through the formation of a sandwich assay after suitable incubation of 30 min. The proposed μFED detected ERα amperometrically at −0.2 V bias voltage using 100 μL of 1.0 × 10−3 M hydrogen peroxide and 1.0 × 10−4 M hydroquinone (HQ). Moreover, the device demonstrated a DL of 10.0 fg mL−1 with excellent recoveries ranging between 94.7 and 108% to perform the detection of ERα in MCF-7 cancer cell lysates. The proposed design was found affordable for delivering breast cancer diagnostics in developing countries.
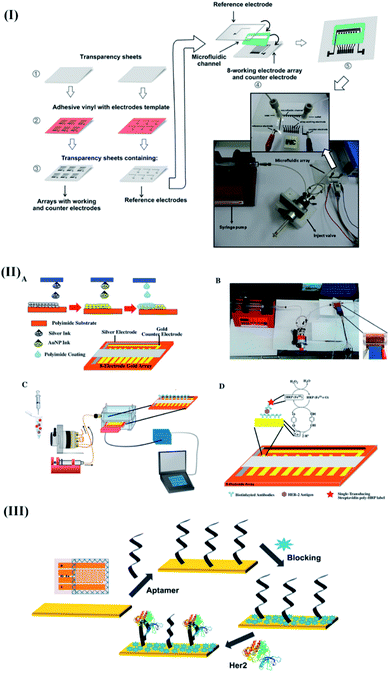 | ||
| Fig. 5 Recent advancements in electrochemical transducer based integrated microfluidic devices for breast cancer specific biomarker detection. (I) Detailed steps involved during the construction of the μFED device for estrogen receptor alpha (ERα) detection. Reproduced with permission.104 Copyright 2018, Elsevier. (II) Electrochemical fully inkjet printed array for sandwich immunoassay-based detection of HER2. Reproduced with permission.105 Copyright 2018, Elsevier. (III) Capacitive electrochemical device for the detection of HER2. Reproduced with permission.106 Copyright 2018, Elsevier. | ||
In another report, Carvajal and coworkers implemented a rapid, inexpensive (<$0.25), and simple immunoassay to improvise the clinical utility of POC devices towards HER2.105 The proposed device included an electrochemical platform with eight inkjet-printed gold electrode arrays (WEA)s, a CE, and an Ag/AgCl electrode. The device was designed to implement a sandwich immunoassay in the presence of the target analyte, biotin functionalized antibody, and HRP labels (Fig. 5II). The low-cost device (0.25 $) exhibited a quick response (assay time: 15 min) and a DL of 12 pg mL−1 towards HER2. Moreover, the device's analytical performance was reproducible, sensitive, and stable, with standard recoveries ranging from 76% to 103%. Later, the potential of the DNA aptasensor (fabricated through the immobilization of a thiolated HER2 specific aptamer on gold interdigitated microelectrodes (IDμEs)) (Fig. 5III) was combined with capacitive measurements to improvise the analytical response of the biosensor towards HER2.106 The proposed non-faradaic mode of impedance spectroscopy predominantly relied on the charging currents (rather than faradaic current arising from the oxidation/reduction of the redox couple (Fe2+/Fe3+)) originating from the perturbations in the surface dielectric, conductance, and charge distribution in the presence of HER2. As compared with the faradaic impedance measurements, the proposed single frequency capacitive measurement exhibited enhanced e-transfer kinetics, high sensitivity, and a better signal-to-noise ratio to acquire excellent sensitivity (0.035 μF/log([HER2] pM)) towards HER2 without any pre-addition of redox labels.
Genetic anomalies are considered promising for investigating the onset and metastases of breast cancer. Therefore, Wee and coworkers microfabricated an electrochemical device for monitoring single DNA base changes to predict the onset of breast cancer.107 The micropatterned electrodes present within the device were fabricated using photolithographic techniques to measure amplified long and short knife motifs (through the ligase chain reaction (LCR)) corresponding to the change of DNA bases in a target sequence.
As compared with short knife motifs, DNA sequences with long knife motifs were intentionally labeled with HRP (using streptavidin-biotin chemistry) to perform the reduction of HRP (Fe3+) to HRP (Fe2+). The resulting (Fe2+) further extends the catalytic cycle to produce a detectable electrochemical response corresponding to a single DNA base change (example: C to T base change). The proposed strategy successfully distinguished methylated and non-methylated cytosines in DNA samples taken from breast cancer cell lines and serum samples.
In addition to electrochemical transducers, various optical-based μTASs have witnessed success in detecting different breast cancer-specific biomarkers. Especially, miRNAs remain the target of interest due to their peculiar relevance for probing the onset of breast cancer. In this regard, Panesar and coworkers developed a microchip targeted against miRNAs to ensure minimally invasive breast cancer detection.108 The authors integrated quantum dots with an in-house developed microfluidic box with Arduino control to monitor the TR-FRET measurements.
As a consequence of a stable sandwich sequence formed between the complementary sequence and supporter sequence (in the presence of target microRNAs), strong FRET signals were achieved. The sensor demonstrated its applicability as an efficient and field-deployable tool for rapidly detecting breast cancer signatures with excellent sensitivity (DL: 0.061 nM and 0.064 nM for let-7a and miRNA-195, respectively). Later, a portable and sensitive miRNA beacon probe-based microfluidic device was devised to perform a convenient blood test to detect miRNA-21.109 The proposed micromachined polymethyl methacrylate (PMMA) based device involved a hybridization interaction of the beacon probe with the target overexpression of miRNA-21 in the patients' serum samples within 30 minutes. The microfluidic platform was further integrated with a fluorescence reader to provide portability and cost-effectiveness to the detection procedure. The group evaluated the performance of the device for clinical samples (51 (30 healthy and 21 breast cancer positive patients) blood samples)) using a voltage-dependent expression of miRNA-21 to authenticate the potential of the device for effective discrimination of breast cancer patients from healthy individuals.
Recently, quantitative and simultaneous detection of four different breast cancer-specific miRNAs (miR-4732, miR-3646, miR-4484, and miR-K12-5) was performed using high-throughput microfluidic chips supported by self-assembled poly-L-lysine (PLL) slides.110 The device explored complementary base pairing between the capture probe and target miRNAs at one end of the segmented hybridization system. The target miRNA's other end remains accessible for pairing with a fluorescence-labeled detection probe to produce fluorescence signals corresponding to the presence of target miRNAs even at a concentration of 1 pM within 30 minutes. Fang and coworkers developed an integrated microfluidic chip to perform immunocapture-based analysis of overexpressed circulating exosomes in patient's serum samples using immunofluorescent labeling techniques.111 The proposed device provided significant opportunities to detect circulating exosomes in breast cancer patients in a convenient, efficient, and cost-efficient manner. Likewise, a self-competent device capable of performing isolation and in situ detection of exosomes within 1.5 h was developed.112
The device's exceptional performance relied on high surface area immunomagnetic beads (biotinylated EpCAM antibodies) for on-chip enrichment of exosomes, and well-directed microvalve assisted fluid control (Fig. 6II). The captured exosomes were further subjected to immunofluorescence assay for quantification and downstream analysis (western blot analysis and electron microscopy imaging). The device demonstrated high prediction accuracy (sensitivity: 90%; specificity: >95%) for circulating exosomes.
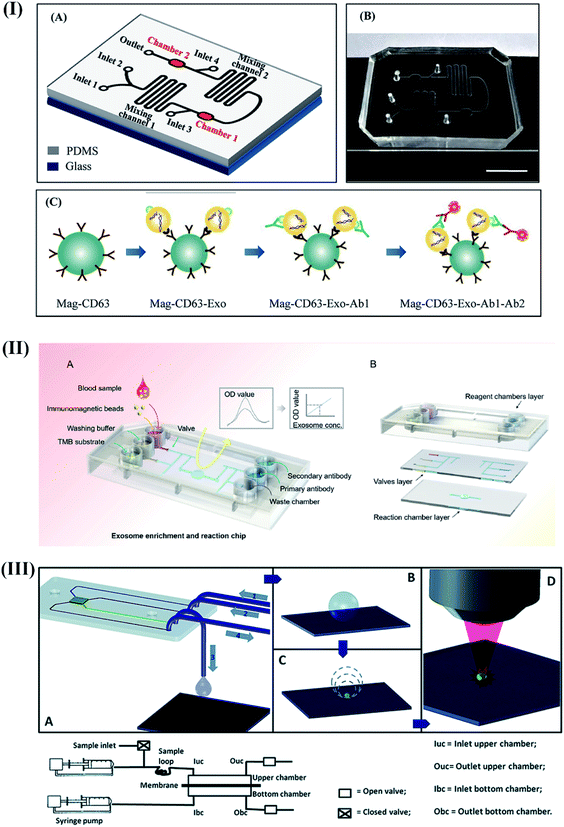 | ||
| Fig. 6 Different integrated optical microfluidic devices for the detection of breast cancer specific biomarkers. (I) Microfluidic device for the detection of circulating CD63 positive exosomes. Reproduced with permission.111 Copyright 2017, PLOS ONE. (II) Immunofluorescence based isolation and detection of the EpCAM expressing exosome. Reproduced with permission.112 Copyright 2019, American Institute of Physics (AIP). (III) Self-competent SERS based sensing platform for the detection of BRCA1 specific peptides. Reproduced with permission.113 Copyright 2014, Royal Society of Chemistry. | ||
A self-competent modular SERS-based sensing platform capable of separation, preconcentration, and detection was proposed to detect peptides derived from BRCA1 protein.113 The device was designed to filter peptides using a porous membrane (cut-off of 12–14 kD) to get concentrated for quantification on a silver nanograin decorated micropillar based superhydrophobic surface (Fig. 6III). These nanograins served as analysis sites for SERS-based sensing of the sample after evaporation. The excellent synergy between the nano-component of the sensor and microfluidic techniques enabled a detection resolution of 0.1 ng μL−1 with enormous potential for early breast cancer diagnosis without any pre-treatment or labeling procedures.
In addition to electrochemical and optical transducers, colorimetric transducers have also been explored for straightforward visual detection of breast cancer-specific biomarkers. In this regard, Vaidyanathan and coworkers have emphasized the application of shear forces generated through alternating current (ac) electrohydrodynamics (ac-EHD) for naked-eye detection of exosomes derived from HER2 positive cells.114 The authors witnessed a significant improvement in the DL as compare to the conventional hydrodynamic flow assays. The proposed device included a pair of gold microelectrodes and three channels fabricated using two-step lithography processes (Fig. 7I). The asymmetric gold microelectrodes were functionalized with anti-HER2, anti-CD9, and anti-PSA using well-established avidin-biotin chemistry for the enrichment and detection of breast cancer cells' exosomes with HER2 expression (Fig. 7II). A similar concept was further extended for the detection of HER2 in serum samples.115 Here, owing to the nano-shearing phenomenon, the device attained a convenient visual detection of HER2 (100 fg mL−1) through minimizing the non-specific adherence of co-interfering biological species present in the blood.
 | ||
| Fig. 7 (I) Design of the ac-EHD based multiplexed device for naked-eye detection of exosomes derived from HER2 positive cells. Adapted with permission.114 Copyright 2014, American Chemical Society. (II) Detailed steps involved during the avidin-biotin assisted functionalization of the device with anti-HER2 and anti-CD9/anti-PSA. Reproduced with permission.114 Copyright 2014, American Chemical Society. | ||
5. Conclusion
Nanotechnology has significantly revolutionized the existing biosensing technologies to ensure an affordable and convenient diagnostic platform for breast cancer using molecular-level analysis. The present review highlights the recent advancements in the sensitivity, selectivity, and detection range of nano-biosensing technologies for breast cancer diagnosis. A detailed comparative analysis elucidated the significant relevance of electrochemical and optical transducers (compared with other biosensing modalities) to achieve clinically relevant sensitivity against breast cancer-specific biomarkers. However, to replace the existing gold standard diagnostics and commercialize emerging biosensing technologies, the major hurdles in this field such as (i) the validation of target biomarkers, (ii) the necessity to validate in a large number of samples, (iii) clinically relevant sensitivity and specificity, (iv) sample processing, purification, and amplification, (v) cost-effectiveness, (vi) automation of complex bioanalytical processes, and (vii) minimal interference and false positives from environmental contamination need to be addressed. These challenges can be resolved by integrating microfabrication techniques and novel nanomaterial-assisted signal amplification strategies to develop μ-TAS based biosensors for breast cancer diagnosis. These integrated systems hold enormous potential to overcome the hurdles associated with existing lab-based diagnostics and facilitate personalized POC testing for breast cancer.In this regard, novel amplification strategies (enzyme-based, DNA cascade, DNA concatemers, exonuclease III, rolling circle amplification (RCA), and nanozymes), microfluidics, sample collection, and pre-concentration steps need to be integrated on a single platform. Furthermore, efforts should be targeted to develop a versatile, convenient approach to perform simultaneous detection of multiple breast cancer biomarkers to deal with the heterogeneous nature of breast cancer.
To realize the concept of point-of-care, priority should be given to provide convenience to end-users (patient compliance, minimal operational steps and sample collection, processing, and user-friendly read-out) to ensure reliable detection. To execute nucleic acid-based tests (with the requirement of a heater for the amplification process), efficient power sources (e.g., batteries) need to be included to advance the functionality further. Apart from design features, reliable data quantification, storage, and further sharing are crucial through the inclusion of smartphone apps and the internet of things (IoT).
Author contributions
Anju Joshi contributed to the data curation, methodology, and preparation of the manuscript. Anil Vishnu G K, Tushar Sakorikar, Arif M. Kamal, and Jayant S. Vaidya contributed to the preparation of the manuscript. Hardik J. Pandya contributed to the methodology, preparation of the manuscript, and supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Anju Joshi acknowledges the Science and Engineering Research Board, Government of India for a National Postdoctoral Fellowship (PDF/2019/002621). Anil Vishnu G. K. acknowledges the award funding from BIRAC under the SITARE-GYTI scheme. Hardik J. Pandya acknowledges the Core Research Grant support (CRG/2019/004963) from the Science and Engineering Research Board (SERB).References
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed
.
- I. Abrao Nemeir, J. Saab, W. Hleihel, A. Errachid, N. Jafferzic-Renault and N. Zine, Sensors, 2019, 19, 2373 CrossRef PubMed
.
- J. Michaelson, S. Satija, R. Moore, G. Weber, E. Halpern, A. Garland, D. Puri and D. B. Kopans, Cancer, 2002, 94, 37–43 CrossRef PubMed
.
- G. Yahalom, D. Weiss, I. Novikov, T. B. Bevers, L. G. Radvanyi, M. Liu, B. Piura, S. Iacobelli, M. T. Sandri and E. Cassano, Biomarkers Cancer, 2013, 5, S13236 CrossRef PubMed
.
- R. Ranjan, E. N. Esimbekova and V. A. Kratasyuk, Biosens. Bioelectron., 2017, 87, 918–930 CrossRef CAS PubMed
.
- S. A. Soper, K. Brown, A. Ellington, B. Frazier, G. Garcia-Manero, V. Gau, S. I. Gutman, D. F. Hayes, B. Korte and J. L. Landers, Biosens. Bioelectron., 2006, 21, 1932–1942 CrossRef CAS PubMed
.
- S. Mittal, H. Kaur, N. Gautam and A. K. Mantha, Biosens. Bioelectron., 2017, 88, 217–231 CrossRef CAS PubMed
.
- M. Labib, R. M. Mohamadi, M. Poudineh, S. U. Ahmed, I. Ivanov, C.-L. Huang, M. Moosavi, E. H. Sargent and S. O. Kelley, Nat. Chem., 2018, 10, 489–495 CrossRef CAS PubMed
.
- B. Hayes, C. Murphy, A. Crawley and R. O'Kennedy, Diagnostics, 2018, 8, 39 CrossRef PubMed
.
- M. Sharifi, A. Hasan, F. Attar, A. Taghizadeh and M. Falahati, Talanta, 2020, 121091 CrossRef CAS PubMed
.
- L. Wang, Sensors, 2017, 17, 1572 CrossRef PubMed
.
- P. Ranjan, A. Parihar, S. Jain, N. Kumar, C. Dhand, S. Murali, D. Mishra, S. K. Sanghi, J. Chaurasia and A. K. Srivastava, Anal. Biochem., 2020, 113996 CrossRef CAS PubMed
.
- V. Gajdosova, L. Lorencova, P. Kasak and J. Tkac, Sensors, 2020, 20, 4022 CrossRef CAS PubMed
.
- N. J. Ronkainen, H. B. Halsall and W. R. Heineman, Chem. Soc. Rev., 2010, 39, 1747–1763 RSC
.
- C. Zhu, G. Yang, H. Li, D. Du and Y. Lin, Anal. Chem., 2015, 87, 230–249 CrossRef CAS PubMed
.
- S. Lal, A. E. M. Reed, X. M. de Luca and P. T. Simpson, Methods, 2017, 131, 135–146 CrossRef CAS PubMed
.
- S. Tang, F. Zhou, Y. Sun, L. Wei, S. Zhu, R. Yang, Y. Huang and J. Yang, Breast Cancer, 2016, 23, 813–819 CrossRef PubMed
.
- R. Salahandish, A. Ghaffarinejad, S. M. Naghib, K. Majidzadeh-A, H. Zargartalebi and A. Sanati-Nezhad, Biosens. Bioelectron., 2018, 117, 104–111 CrossRef CAS PubMed
.
- A. Nicolini, P. Ferrari and G. Rossi, Advances in Cancer Biomarkers, 2015, 197–225 CAS
.
- B. K. Zehentner, D. H. Persing, A. Deme, P. Toure, S. E. Hawes, L. Brooks, Q. Feng, D. C. Hayes, C. W. Critichlow and R. L. Houghton, Clin. Chem., 2004, 50, 2069–2076 CrossRef CAS PubMed
.
- P. J. O'Donovan and D. M. Livingston, Carcinogenesis, 2010, 31, 961–967 CrossRef PubMed
.
- Z. Mitri, T. Constantine and R. O'Regan, Chemother. Res. Pract., 2012, 2012 Search PubMed
.
- M. Shamsipur, M. Emami, L. Farzin and R. Saber, Biosens. Bioelectron., 2018, 103, 54–61 CrossRef CAS PubMed
.
- S. Yang, M. You, F. Zhang, Q. Wang and P. He, Sens. Actuators, B, 2018, 258, 796–802 CrossRef CAS
.
- X. Li, C. Shen, M. Yang and A. Rasooly, Anal. Chem., 2018, 90, 4764–4769 CrossRef CAS PubMed
.
- Y. Chai, X. Li and M. Yang, Microchim. Acta, 2019, 186, 1–6 CrossRef CAS PubMed
.
- D. Chen, D. Wang, X. Hu, G. Long, Y. Zhang and L. Zhou, Sens. Actuators, B, 2019, 296, 126650 CrossRef CAS
.
- X. Xu, X. Weng, A. Liu, Q. Lin, C. Wang, W. Chen and X. Lin, Anal. Bioanal. Chem., 2013, 405, 3097–3103 CrossRef CAS PubMed
.
- H. Wang, J. Sun, L. Lu, X. Yang, J. Xia, F. Zhang and Z. Wang, Anal. Chim. Acta, 2020, 1094, 18–25 CrossRef CAS PubMed
.
- S. Rashid, M. H. Nawaz, I. u. Rehman, A. Hayat and J. L. Marty, Sens. Actuators, B, 2021, 330, 129351 CrossRef CAS
.
- J. Lu, G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak and A. A. Ferrando, nature, 2005, 435, 834–838 CrossRef CAS PubMed
.
- X. Zhang, D. Wu, Z. Liu, S. Cai, Y. Zhao, M. Chen, Y. Xia, C. Li, J. Zhang and J. Chen, Chem. Commun., 2014, 50, 12375–12377 RSC
.
- Y. L. Huang, S. Mo, Z. F. Gao, J. R. Chen, J. L. Lei, H. Q. Luo and N. B. Li, Microchim. Acta, 2017, 184, 2597–2604 CrossRef CAS
.
- J. Zhang, L.-L. Wang, M.-F. Hou, Y.-K. Xia, W.-H. He, A. Yan, Y.-P. Weng, L.-P. Zeng and J.-H. Chen, Biosens. Bioelectron., 2018, 102, 33–40 CrossRef CAS PubMed
.
- A. R. Cardoso, F. T. Moreira, R. Fernandes and M. G. F. Sales, Biosens. Bioelectron., 2016, 80, 621–630 CrossRef CAS PubMed
.
- J. Wang, A.-N. Kawde, A. Erdem and M. Salazar, Analyst, 2001, 126, 2020–2024 RSC
.
- J. Wang, A.-N. Kawde and M. Musameh, Analyst, 2003, 128, 912–916 RSC
.
- J. Wang, D. Xu, A. Erdem, R. Polsky and M. A. Salazar, Talanta, 2002, 56, 931–938 CrossRef CAS PubMed
.
- C.-z. Li, H. Karadeniz, E. Canavar and A. Erdem, Electrochim. Acta, 2012, 82, 137–142 CrossRef CAS
.
- H. Xu, L. Wang, H. Ye, L. Yu, X. Zhu, Z. Lin, G. Wu, X. Li, X. Liu and G. Chen, Chem. Commun., 2012, 48, 6390–6392 RSC
.
- P. A. Rasheed and N. Sandhyarani, Sens. Actuators, B, 2014, 2014, 777–782 CrossRef
.
- X. Li, X. Li, D. Li, M. Zhao, H. Wu, B. Shen, P. Liu and S. Ding, Biosens. Bioelectron., 2020, 168, 112554 CrossRef CAS PubMed
.
- S. M. Majd, A. Salimi and F. Ghasemi, Biosens. Bioelectron., 2018, 105, 6–13 CrossRef CAS PubMed
.
- M. Filippidou, C. M. Loukas, G. Kaprou, E. Tegou, P. Petrou, S. Kakabakos, A. Tserepi and S. Chatzandroulis, Microelectron. Eng., 2019, 216, 111093 CrossRef CAS
.
- D.-H. Lee, H.-S. Cho, D. Han, R. Chand, T.-J. Yoon and Y.-S. Kim, J. Mater. Chem. B, 2017, 5, 3580–3585 RSC
.
- Rajesh, Z. Gao, R. Vishnubhotla, P. Ducos, M. D. Serrano, J. Ping, M. K. Robinson and A. T. C. Johnson, Adv. Mater. Interfaces, 2016, 3, 1600124 CrossRef
.
- S. Myung, A. Solanki, C. Kim, J. Park, K. S. Kim and K. B. Lee, Adv. Mater., 2011, 23, 2221–2225 CrossRef CAS PubMed
.
- F. S. Ligler and J. J. Gooding, Anal. Chem., 2019, 91, 8732–8738 CrossRef CAS PubMed
.
- D. Dey and T. Goswami, J. Biomed. Biotechnol., 2011, 2011 Search PubMed
.
- X. Hun and Z. Zhang, Spectrochim. Acta, Part A, 2009, 74, 410–414 CrossRef PubMed
.
- X. Hua, Z. Zhou, L. Yuan and S. Liu, Anal. Chim. Acta, 2013, 788, 135–140 CrossRef CAS PubMed
.
- T. Y. Rakovich, O. K. Mahfoud, B. M. Mohamed, A. Prina-Mello, K. Crosbie-Staunton, T. Van Den Broeck, L. De Kimpe, A. Sukhanova, D. Baty and A. Rakovich, ACS Nano, 2014, 8, 5682–5695 CrossRef CAS PubMed
.
- L. Cai, Z.-Z. Chen, M.-Y. Chen, H.-W. Tang and D.-W. Pang, Biomaterials, 2013, 34, 371–381 CrossRef CAS PubMed
.
- M.-Y. Chen, Z.-Z. Chen, L.-L. Wu, H.-W. Tang and D.-W. Pang, Analyst, 2013, 138, 7411–7416 RSC
.
- Y. Ding, J. Ling, H. Wang, J. Zou, K. Wang, X. Xiao and M. Yang, Anal. Methods, 2015, 7, 7792–7798 RSC
.
- M. Zhang, X. Li, G. Gao and C. Deng, Anal. Lett., 2020, 53, 371–384 CrossRef CAS
.
- J. Chen, J. Lin, X. Zhang, S. Cai, D. Wu, C. Li, S. Yang and J. Zhang, Anal. Chim. Acta, 2014, 817, 42–47 CrossRef CAS PubMed
.
- P. Kumar, S. C. Sharma and A. Deep, Sens. Actuators, B, 2014, 202, 404–409 CrossRef CAS
.
- M. Zhang, G. Gao, Y. Ding, C. Deng, J. Xiang and H. Wu, Talanta, 2019, 199, 238–243 CrossRef CAS PubMed
.
- A. Afzalinia and M. Mirzaee, ACS Appl. Mater. Interfaces, 2020, 12, 16076–16087 CrossRef CAS PubMed
.
- S. L. Moura, C. G. Martín, M. Martí and M. I. Pividori, Talanta, 2020, 211, 120657 CrossRef CAS PubMed
.
- J. H. Lee, J. A. Kim, S. Jeong and W. J. Rhee, Biosens. Bioelectron., 2016, 86, 202–210 CrossRef CAS PubMed
.
- Y. Xia, L. Wang, J. Li, X. Chen, J. Lan, A. Yan, Y. Lei, S. Yang, H. Yang and J. Chen, Anal. Chem., 2018, 90, 8969–8976 CrossRef CAS PubMed
.
- H. Wang, D. He, K. Wan, X. Sheng, H. Cheng, J. Huang, X. Zhou, X. He and K. Wang, Analyst, 2020, 145, 3289–3296 RSC
.
- B. Liedberg, C. Nylander and I. Lunström, Sens. Actuators, 1983, 4, 299–304 CrossRef CAS
.
- N. Bellassai, R. D'agata, V. Jungbluth and G. Spoto, Front. Chem., 2019, 7, 570 CrossRef CAS PubMed
.
- J. T. Gohring, P. S. Dale and X. Fan, Sens. Actuators, B, 2010, 146, 226–230 CrossRef CAS
.
- D. Sun, Y. Ran and G. Wang, Sensors, 2017, 17, 2559 CrossRef PubMed
.
- U. Eletxigerra, J. Martinez-Perdiguero, R. Barderas, J. M. Pingarrón, S. Campuzano and S. Merino, Anal. Chim. Acta, 2016, 905, 156–162 CrossRef CAS PubMed
.
- A. Sinibaldi, C. Sampaoli, N. Danz, P. Munzert, L. Sibilio, F. Sonntag, A. Occhicone, E. Falvo, E. Tremante and P. Giacomini, Biosens. Bioelectron., 2017, 92, 125–130 CrossRef CAS PubMed
.
- P. L. Truong, C. Cao, S. Park, M. Kim and S. J. Sim, Lab Chip, 2011, 11, 2591–2597 RSC
.
- J. Feng, X. Wu, W. Ma, H. Kuang, L. Xu and C. Xu, Chem. Commun., 2015, 51, 14761–14763 RSC
.
- N. Li, S. Zong, Y. Zhang, Z. Wang, Y. Wang, K. Zhu, K. Yang, Z. Wang, B. Chen and Y. Cui, Anal. Bioanal. Chem., 2020, 412, 5707–5718 CrossRef CAS PubMed
.
- H.-N. Wang, B. M. Crawford, A. M. Fales, M. L. Bowie, V. L. Seewaldt and T. Vo-Dinh, J. Phys. Chem. C, 2016, 120, 21047–21055 CrossRef CAS PubMed
.
- J. U. Lee, W. H. Kim, H. S. Lee, K. H. Park and S. J. Sim, Small, 2019, 15, 1804968 CrossRef PubMed
.
- J. Lan, L. Li, Y. Liu, L. Yan, C. Li, J. Chen and X. Chen, Microchim. Acta, 2016, 183, 3201–3208 CrossRef CAS
.
- J. Lan, Y. Liu, L. Li, F. Wen, F. Wu, Z. Han, W. Sun, C. Li and J. Chen, Sci. Rep., 2016, 6, 1–8 CrossRef PubMed
.
- J. Lan, F. Wen, F. Fu, X. Zhang, S. Cai, Z. Liu, D. Wu, C. Li, J. Chen and C. Wang, RSC Adv., 2015, 5, 18008–18012 RSC
.
- T. Y. Rakovich, O. K. Mahfoud, B. M. Mohamed, A. Prina-Mello, K. Crosbie-Staunton, T. Van Den Broeck, L. De Kimpe, A. Sukhanova, D. Baty, A. Rakovich and S. A. Maier, ACS Nano, 2014, 8, 5682–5695 CrossRef CAS PubMed
.
- S. Catalán-Gómez, M. Briones, S. Cortijo-Campos, T. García-Mendiola, A. de Andrés, S. Garg, P. Kung, E. Lorenzo, J. L. Pau and A. Redondo-Cubero, Sci. Rep., 2020, 10, 1–9 CrossRef PubMed
.
- B. Qiao, Q. Guo, J. Jiang, Y. Qi, H. Zhang, B. He, C. Cai and J. Shen, Analyst, 2019, 144, 3668–3675 RSC
.
- R. Li, Y. An, T. Jin, F. Zhang and P. He, J. Electroanal. Chem., 2021, 882, 115011 CrossRef CAS
.
- Y. Nie, X. Yuan, P. Zhang, Y. Q. Chai and R. Yuan, Anal. Chem., 2019, 91, 3452–3458 CrossRef CAS PubMed
.
- C. Hu, W. Yue and M. Yang, Analyst, 2013, 138, 6709–6720 RSC
.
- X. Li, Y. Li, R. Feng, D. Wu, Y. Zhang, H. Li, B. Du and Q. Wei, Sens. Actuators, B, 2013, 188, 462–468 CrossRef CAS
.
- D. Qin, X. Jiang, G. Mo, J. Feng, C. Yu and B. Deng, ACS Sens., 2019, 4, 504–512 CrossRef CAS PubMed
.
- X. Xiong, Y. Zhang, Y. Wang, H. Sha and N. Jia, Sens. Actuators, B, 2019, 297, 126812 CrossRef CAS
.
- J. Li, H.-E. Fu, L.-J. Wu, A.-X. Zheng, G.-N. Chen and H.-H. Yang, Anal. Chem., 2012, 84, 5309–5315 CrossRef CAS PubMed
.
- Y. Song, K. Qu, C. Zhao, J. Ren and X. Qu, Adv. Mater., 2010, 22, 2206–2210 CrossRef CAS PubMed
.
- N. Laurieri, M. H. Crawford, A. Kawamura, I. M. Westwood, J. Robinson, A. M. Fletcher, S. G. Davies, E. Sim and A. J. Russell, J. Am. Chem. Soc., 2010, 132, 3238–3239 CrossRef CAS PubMed
.
- N. Laurieri, J. E. Egleton, A. Varney, C. C. Thinnes, C. E. Quevedo, P. T. Seden, S. Thompson, F. Rodrigues-Lima, J. Dairou and J.-M. Dupret, PLoS One, 2013, 8, e70600 CrossRef CAS PubMed
.
- Y. Sang, Y. Xu, L. Xu, W. Cheng, X. Li, J. Wu and S. Ding, Microchim. Acta, 2017, 184, 2465–2471 CrossRef CAS
.
- R. Ahirwar and P. Nahar, Anal. Bioanal. Chem., 2016, 408, 327–332 CrossRef CAS PubMed
.
- D. Liang, W. You, Y. Yu, Y. Geng, F. Lv and B. Zhang, RSC Adv., 2015, 5, 27571–27575 RSC
.
- B. E. Rapp, F. J. Gruhl and K. Länge, Anal. Bioanal. Chem., 2010, 398, 2403–2412 CrossRef CAS PubMed
.
- R. Fogel, J. Limson and A. A. Seshia, Essays Biochem., 2016, 60, 101–110 CrossRef PubMed
.
- F. J. Gruhl and K. Länge, Anal. Biochem., 2012, 420, 188–190 CrossRef CAS PubMed
.
- O. Tigli, L. Bivona, P. Berg and M. E. Zaghloul, IEEE Transactions on Biomedical Circuits and Systems, 2010, 4, 62–73 CAS
.
- A. E. Murillo, L. Melo-Maximo, B. Garcia-Farrera, O. S. Martínez, D. V. Melo-Máximo, J. Oliva-Ramirez, K. Garcia, L. Huerta and J. Oseguera, J. Mater. Res. Technol., 2019, 8, 350–358 CrossRef CAS
.
- R. C. B. Marques, E. Costa-Rama, S. Viswanathan, H. P. A. Nouws, A. Costa-García, C. Delerue-Matos and M. B. González-García, Sens. Actuators, B, 2018, 255, 918–925 CrossRef CAS
.
- H. Ilkhani, M. Sarparast, A. Noori, S. Zahra Bathaie and M. F. Mousavi, Biosens. Bioelectron., 2015, 74, 491–497 CrossRef CAS PubMed
.
- S. Kim, T. G. Kim, S. H. Lee, W. Kim, A. Bang, S. W. Moon, J. Song, J.-H. Shin, J. S. Yu and S. Choi, ACS Appl. Mater. Interfaces, 2020, 12, 7897–7904 CrossRef CAS PubMed
.
- M. A. Ali, K. Mondal, Y. Jiao, S. Oren, Z. Xu, A. Sharma and L. Dong, ACS Appl. Mater. Interfaces, 2016, 8, 20570–20582 CrossRef CAS PubMed
.
- C. V. Uliana, C. R. Peverari, A. S. Afonso, M. R. Cominetti and R. C. Faria, Biosens. Bioelectron., 2018, 99, 156–162 CrossRef CAS PubMed
.
- S. Carvajal, S. N. Fera, A. L. Jones, T. A. Baldo, I. M. Mosa, J. F. Rusling and C. E. Krause, Biosens. Bioelectron., 2018, 104, 158–162 CrossRef CAS PubMed
.
- S. K. Arya, P. Zhurauski, P. Jolly, M. R. Batistuti, M. Mulato and P. Estrela, Biosens. Bioelectron., 2018, 102, 106–112 CrossRef CAS PubMed
.
- E. J. H. Wee, S. Rauf, K. M. Koo, M. J. A. Shiddiky and M. Trau, Lab Chip, 2013, 13, 4385–4391 RSC
.
- S. Panesar, X. Weng and S. Neethirajan, IEEE Sensors Letters, 2017, 1, 1–4 Search PubMed
.
- B. Salim, M. Athira, A. Kandaswamy, M. Vijayakumar, T. Saravanan and T. Sairam, Biomed. Microdevices, 2017, 19, 1–11 CrossRef CAS PubMed
.
- Y. Gao, L. Qiang, Y. Chu, Y. Han, Y. Zhang and L. Han, AIP Adv., 2020, 10, 045022 CrossRef CAS
.
- S. Fang, H. Tian, X. Li, D. Jin, X. Li, J. Kong, C. Yang, X. Yang, Y. Lu and Y. Luo, PLoS One, 2017, 12, e0175050 CrossRef PubMed
.
- W. Chen, H. Li, W. Su and J. Qin, Biomicrofluidics, 2019, 13, 054113 CrossRef PubMed
.
- G. Perozziello, P. Candeloro, F. Gentile, A. Nicastri, A. Perri, M. L. Coluccio, A. Adamo, F. Pardeo, R. Catalano and E. Parrotta, RSC Adv., 2014, 4, 55590–55598 RSC
.
- R. Vaidyanathan, M. Naghibosadat, S. Rauf, D. Korbie, L. G. Carrascosa, M. J. Shiddiky and M. Trau, Anal. Chem., 2014, 86, 11125–11132 CrossRef CAS PubMed
.
- R. Vaidyanathan, L. M. Van Leeuwen, S. Rauf, M. J. Shiddiky and M. Trau, Sci. Rep., 2015, 5, 1–7 Search PubMed
.
| This journal is © The Royal Society of Chemistry 2021 |