Open Access Article
Open Access ArticleA micro-spray-based high-throughput screening system for bioplastic-degrading microorganisms†
Giyoung
Shin
a,
Seul-A.
Park
 a,
Jun Mo
Koo
a,
Jun Mo
Koo
 a,
Minsun
Kim
a,
Minkyung
Lee
a,
Jonggeon
Jegal
a,
Sung Yeon
Hwang
a,
Minsun
Kim
a,
Minkyung
Lee
a,
Jonggeon
Jegal
a,
Sung Yeon
Hwang
 *ab,
Jeyoung
Park
*ab,
Jeyoung
Park
 *ab and
Dongyeop X.
Oh
*ab and
Dongyeop X.
Oh
 *ab
*ab
aResearch Center for Bio-based Chemistry, Korea Research Institute of Chemical Technology (KRICT), Ulsan 44429, Republic of Korea. E-mail: crew75@krict.re.kr; jypark@krict.re.kr; dongyeop@krict.re.kr
bAdvanced Materials and Chemical Engineering, University of Science and Technology (UST), Daejeon 34113, Republic of Korea
First published on 25th June 2021
Abstract
Designing efficient plastic-digesting microorganisms is necessary to accelerate the decomposition of biodegradable plastics leaking into outer environments. However, screening of microorganisms for hydrophobic plastics is labor-intensive and time-consuming. Herein, a high-throughput, straightforward micro-spray-based screening system is presented; the whole process only takes 1–5 days using a minimal amount of polymers. Bioplastic microparticles sprayed on an agar plate share a large interface with microorganisms, and therefore their discovery is readily recognizable. This method inspires finding new microorganisms that digest conventional plastics known as non-degradable plastics.
Over 360 million tons of plastics are produced annually worldwide.1 The annual generation of plastic waste worldwide is estimated at approximately 150 million tons.2 However, the recycling rate only accounts for less than 9% of the plastic produced.3 Plastic wastes are mainly managed by disposal in landfills or by incineration.4,5 A massive amount of non-degradable plastics is accumulating and intensifying the destruction of the ecosystem.6–9
As an alternative to non-degradable plastics, biodegradable plastics have been developed. Biodegradable plastics are mainly produced from renewable biomass raw materials or partially from fossil resources.10,11 Biodegradable plastics are utilized as a source of carbon and energy by microorganisms present in the environment and are completely decomposed into CO2, H2O, and biomass.12 The global production capacity of biodegradable plastics reached 1.23 million tons in 2020.13 It accounts for approximately 60% of the total 2.11 million tons of global bioplastic production including bio-based/non-biodegradable bioplastics. Polylactic acid (PLA),14 polybutylene succinate (PBS),15 poly(butylene adipate-co-terephthalate) (PBAT),16 and polyhydroxyalkanoate (PHA)17,18 make up a large portion of the total production of biodegradable plastics.13,19
The degradation of plastics depends not only on the chemical structure of the polymer, but also on environmental conditions.20,21 To date, studies on bioplastic decomposition have been limited in the sense that a majority of them aim to understand the degradability of representative biodegradable plastics such as PLA, PBS, PBAT, and PHA in particular environments, especially under composting conditions that have abundant organic matter and microorganisms.22–26 However, current studies are not focused on the fundamental mechanisms of biodegradation of biodegradable plastics when they are exposed to unfertile conditions or extreme environments such as deep sea, deserts, low- and high-temperature areas, and underground anaerobic zones. New biodegradable plastics with reinforced mechanical properties have been invented because traditional bioplastics have undesirable mechanical properties.27–29 To determine the biodegradation mechanisms of synthetic polymers and to estimate and control the by-products after the use of these polymers in the environment, studies on bioplastic-degrading microorganisms are essential. Therefore, it is important to determine microorganisms capable of plastic decomposition that are naturally present in the environment or by modifying existing organisms in order to successfully explore the end-of-life process of plastics. In addition, the discovery of novel microbes can provide a platform that can be used to produce value-added products from alternative feedstock through metabolic pathway redesign.30
The need for biodegradable plastics is expected to increase continuously, and their wastes are already being buried in landfills or treated in industrial composting plants.31 Currently, at the composting stage, there is a need to accelerate bioplastic degradation using high-concentration microbial solutions with excellent decomposition activities, which will contribute to efficient waste management.32,33
Microorganisms that degrade plastics can be discovered and isolated from the natural environment via a screening process.34,35 Innovative methods based on metagenomic libraries are currently being developed, but their applications are still limited.36,37 The conventional culture-based screening method consists of (i) collection of diverse natural substrata, (ii) preparation of a culture medium that contains plastic as the sole carbon source, (iii) inoculation of microbes on the medium, and (iv) colony isolation. Among other screening methods, this is the most fundamental and straightforward method for the discovery and isolation of novel microorganisms with specific bioplastic-degrading activities.38,39 However, the major challenge of this method is that it is labor intensive and time-consuming.
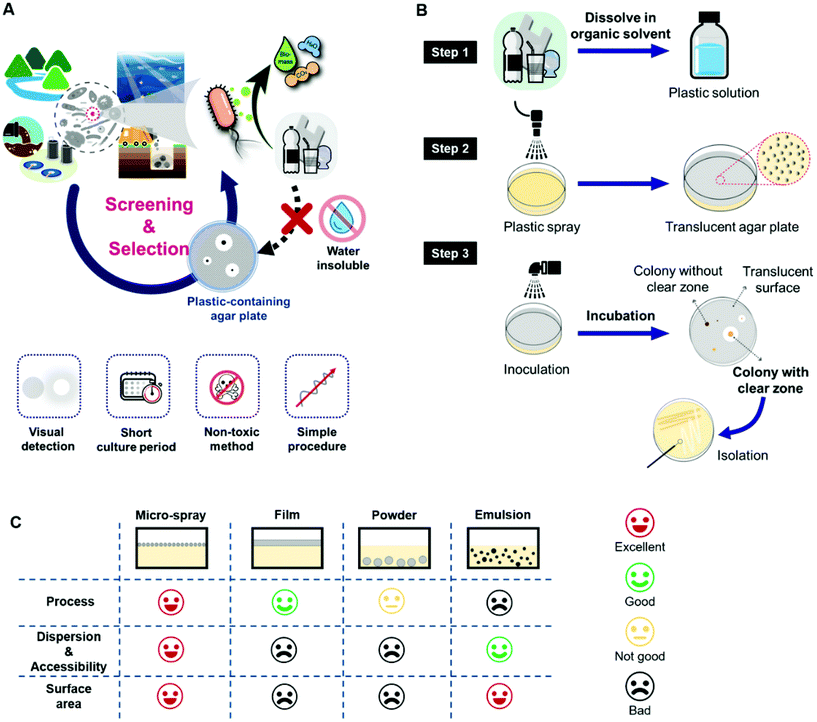
To improve the screening efficiency, especially in the ii–iv processes, the culture medium should satisfy the conditions given in Fig. 1A. Briefly, (1) the target carbon source, i.e. the plastic sample, should be evenly distributed throughout the medium with a large interface area so that the interaction with microorganisms and the decomposition efficiency are improved, (2) the medium should be capable of allowing clear visual detection to easily detect the degradation activity of microorganisms, (3) the medium should be non-toxic to prevent the inhibition of microbes, (4) the medium should be able to supply nutrients other than carbon sources appropriately, and (5) the medium should be economical in terms of time, cost, and its quantity used.
However, plastics, including bioplastics, are generally not soluble in water, which makes the preparation of plastic-containing media a fastidious, time-consuming, and labor-intensive task. It is difficult to homogeneously distribute plastics while ensuring a large surface area among water, nutrients, and microorganisms on solid media to produce an effective screening medium. The emulsion method is cumbersome to optimize as it is intricate and requires sophisticated procedures and skilled operators, and many plastics are not applicable for this method.40–42 It is also difficult to exclude solvents or surfactants that may have potential cytotoxicity. The emulsion particles are mainly inside the solid medium rather than on the surface. The bulk film method is simple to produce in contrast to the emulsion method but offers a limited surface area between microorganisms and the plastic sample, which decreases the biodegradation efficiency and increases the experimental duration.43 Due to the lack of preparation methods for selective media, studies on the screening of degrading microbes have been limited. Therefore, the production of efficient screening plates is important in reducing the experimental workload for the isolation of bioplastic-degrading microorganisms.
In this study, we propose a new approach for the efficient screening of bioplastic-degrading microorganisms by spraying a biodegradable plastic solution dissolved in an appropriate organic solvent on agar plates. The environmental inoculum, such as a soil suspension, seawater, or river sample, was also sprayed on the plastic microparticles and allowed to be evenly distributed over the agar plate. After incubation, the formation of a clear zone around each colony was easily detected and these colonies were selected as strains possessing plastic-decomposition activities. Microparticles of biodegradable plastics were thus introduced on a solidified agar medium through an air-blowing spray (Fig. 1B). PLA, PBS, PBAT, and PHA were dissolved in chloroform to form the solutions for spraying. The polymer solutions were sprayed using 25G core/8G sheath nozzles with compressed air at a pressure of 140 kPa. To totally inhibit any toxic effects on the microorganisms, chloroform was fully evaporated in a fume hood at room temperature (20–25 °C). Inoculation was done by spraying the inoculum on the plates, as conventional inoculation methods such as streaking or spreading are likely to peel off the plastic particles, since the plastic microparticles on the agar plate were not firmly fixed to the basal media. Conditions for the spray-based inoculation were optimized in terms of serial dilution and spray time. Then, the inoculated screening plates were cultured in an incubator, and colonies forming clear zones were selected as strains that produce plastic-degrading enzymes. The entire process for screening with the micro-spray system can be found in Movie S1.† Compared to other preparation methods, such as blending with powders or emulsion or applying plastic films, the micro-spray-based screening plate method facilitated easier preparation of plates and allowed the plastic particles to be homogeneously distributed throughout the plate surface (Fig. 1C).
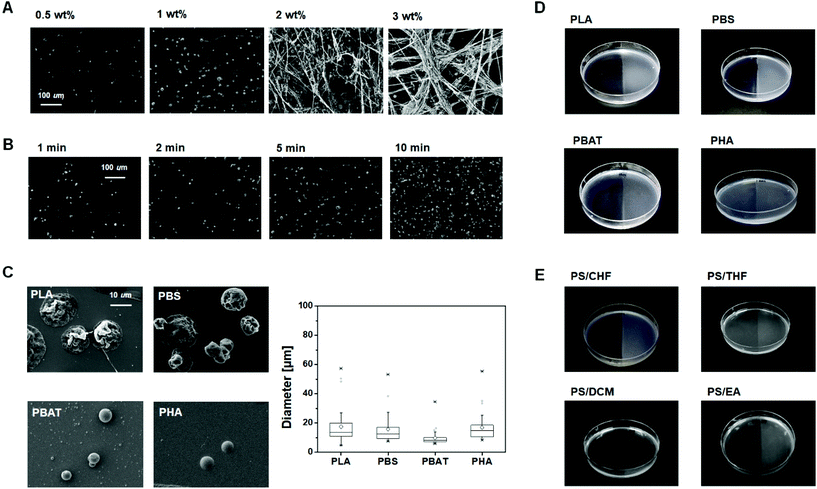
The morphology of the sprayed PLA microparticles on the agar plate was affected by the polymer concentration (0.5 to 3 wt%) and spraying time (1–10 min). As shown in the scanning electron microscopy (SEM) images (Fig. 2A), PLA solutions of 0.5 and 1 wt% formed spherical beads with a diameter of 7.31 ± 2.38 and 12.87 ± 5.03 μm, respectively, while 2 and 3 wt% solutions produced bundles consisting of 0.09 ± 0.02 and 0.28 ± 0.08 μm fibers in diameter, respectively (Fig. S1†). The viscosity values of the PLA solutions with 0.5, 1, 2, and 3 wt% were 0.065, 0.075, 0.131, and 0.161 Pa s, respectively. A polymer solution with a viscosity less than 0.100 Pa s is likely to produce spherical beads. In the high-concentration polymer spray, the nozzle was clogged frequently owing to solvent evaporation and viscosity. Therefore, spraying of a relatively low concentration solution in which fibers are rarely produced is suitable for the fabrication of the screening plate.
The number of spherical PLA microparticles placed on the surface increased with the spray time, while the size of the PLA microparticles did not significantly change (Fig. 2B). This means that the quantity of relatively regular-sized plastic particles on the medium plate can be controlled through the spray time. Furthermore, it is noteworthy that even if the particles overlap with each other, they do not merge into a single lump but are piled up, one by one, owing to fast solvent evaporation (Fig. 2C, left). This phenomenon increases the surface area to volume ratio of the particles.
The micro-sprayed particles of PLA, PBS, PBAT, and PHA from the 1 wt% polymer solution had a mean diameter of 17.6 ± 12.8, 15.8 ± 9.9, 9.8 ± 5.8, and 17.1 ± 9.8 μm, respectively (Fig. 2C, right). The diameter of the micro-sprayed particles was one order of magnitude smaller than those of the ground PLA powder, which was found to be 662 ± 157 μm. Plastic microparticles with large surface areas are more likely to be utilized more efficiently by microorganisms than bulk films or powders. In addition, it takes only 15 mg of the plastic sample to fabricate a screening plate. Therefore, microorganism screening can be accomplished using a minimal amount of a newly synthesized polymer.
After plastic microparticle spraying, the transparent surface of the solid medium changed to translucent (Fig. 2D). The plastic-sprayed and unprocessed surfaces of the agar plate could thus be readily distinguished with the naked eye. This is advantageous in isolating plastic decomposition strains, as a clear visual change reveals if the plastic present on the surface is getting degraded by bioplastic-degrading strains. As shown in Fig. 2E, the polystyrene screening plate was also successfully formed by applying the spray method using chloroform solution. Furthermore, it was confirmed that a screening plate could be made by spraying polystyrene dissolved in not only chloroform but also tetrahydrofuran, dichloromethane and ethyl acetate. With these results, the method proposed here is a versatile and simple method that can be used for microbial screening of a wide range of plastics, even petroleum-based conventional plastics.
To discover and isolate bioplastic-degrading microorganisms using a plastic micro-sprayed screening plate, an appropriate method is required for applying the inoculum onto the plate. This is because hydrophobic plastic microparticles have relatively weak adhesion towards the hydrophilic agar surface. Thus, general inoculation methods such as spreading or streaking were not suitable. To avoid damaging the plastic microparticle layer, microorganisms derived from the environment were inoculated on the plastic screening plates by the micro-spray method. The microbial inoculum spray process utilized environmental samples or microbial culture media which was sprayed evenly on the surface of solid media by utilizing an air-brush with an air pressure of 70 kPa. Through this process, the bacterial cells were placed on or around plastic particles. The SEM image shows that the bacterial cells were placed on or around the PLA particles by the spray inoculation method (Fig. S2†).
It is necessary to investigate whether the plastic microparticle-sprayed plate is a suitable environment for living microorganisms by assessing if the residual organic solvent and the pressure during micro-spray have a negative effect on bacterial cells. Serially diluted Escherichia coli was inoculated and cultured on PLA particle-sprayed screening plates were made from a nutrient-rich Luria–Bertani (LB) agar plate, while unprocessed LB agar plates with E. coli were used as a negative control. All the plates were incubated at 37 °C overnight. The residual solvents and pressure during spraying did not induce a negative effect on the growth of microorganisms, as the number of colonies between the negative control and plastic micro-sprayed plates were roughly the same. The number of colonies from 100 μL of 104 diluted E. coli culture was 623 and 703 in the non-treated LB plate and PLA screening plate, respectively. Colony counting was performed using the ImageJ software. From 100 μL of 106 diluted culture, 16 ± 4.6 and 14 ± 2 colonies were detected in the non-treated and PLA screening plates, respectively (Fig. S3†). This was done by manually counting three replicates. There were no significant differences between the two types of solid agar media plates in terms of the number of colonies. The effects of air pressure and organic solvent during spraying on microorganisms were also evaluated against Gram-positive bacteria (Staphylococcus aureus) and yeast (Saccharomyces cerevisiae). As shown in Fig. S4,† there was no significant difference in colony formation between the plates tested for each strain. Therefore, it was confirmed that there is no inhibitory effect on the growth of microorganisms from the plastic particles or residual solvents, and bacteria could efficiently proliferate due to proper nutrient supplementation on plastic micro-sprayed solid media. In addition, since intervention is minimized, the risk of contamination can be avoided, and the possibility that any toxic emulsifiers or organic solvents that may have a negative effect on the growth and reproduction of microorganisms in the media is avoided. In our proposed method, the organic solvent is immediately evaporated during the spraying of the plastic solution.44 Even if trace amounts of the organic solvent remain, the residual solvent can be removed through an additional evaporation process.
Before screening wild microorganisms from the environment, we evaluated the screening performance of the plastic-particle-sprayed screening plate using a previously reported PLA-degrading strain, Amycolatopsis orientalis spp. orientalis.45,46 The reference strain (KCTC 9412) was obtained from the Korean Collection for Type Cultures (KCTC) (Jeongeup, Jeollabuk-do, Republic of Korea). The strain was streaked on a carbon-free agar plate, followed by spraying with the PLA solution. Here, A. orientalis spp. was directly exposed to chloroform gas. The tested plate was incubated at 30 °C until the colonies and clear zones were formed. The degradation of PLA by A. orientalis spp. was observed as clear zones after 4 days of incubation (Fig. 3A). These results show that the developed screening plate can screen microorganisms capable of biodegradation. This also indicates that if microbial cells are directly exposed to chloroform gas during spraying, it would be at a sub-toxic level. Although, in the actual screening experiments, the chloroform is completely dried after spraying, and then the inoculum is sprayed.
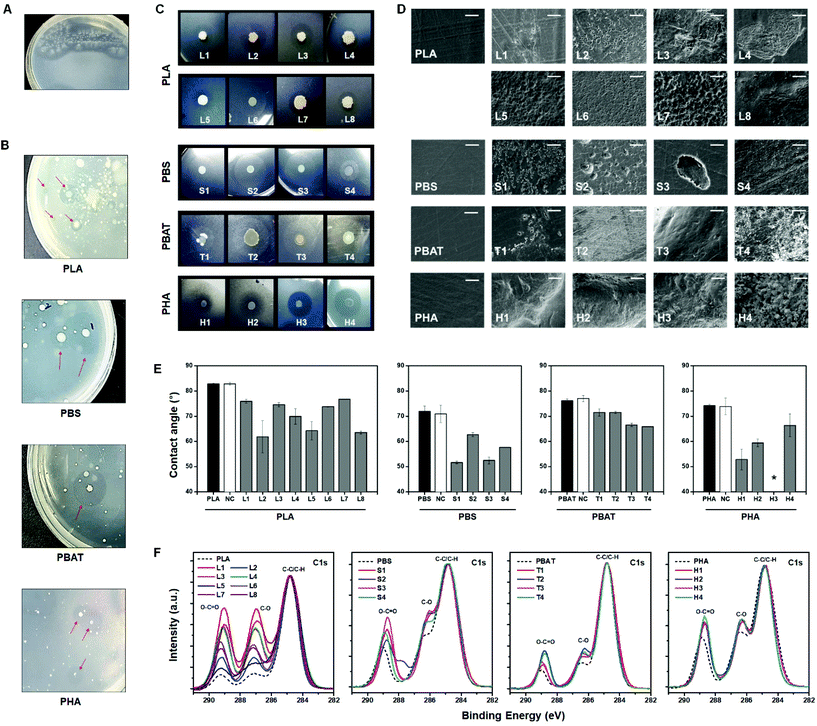 | ||
| Fig. 3 Plastic particle-coated screening plates. (A) PLA degradation activities of A. orientalis on a PLA-sprayed agar plate. (B) Clear zone formation by bioplastic-degrading microorganisms on plastic-sprayed screening plates from environmental samples. (C) Selected bacteria cultivation. Isolated single bacterial species were grown on plastic-sprayed agar plates. The bacteria used are listed in Table S1.† (D) The SEM images of plastic films degraded by isolated species. Scale bars, 10 μm. (E) Water contact angle values of the plastic films before incubation (black), incubated films without bacteria (white), and the biodegraded plastic films by the selected bacteria (grey). Error bars indicate mean ± standard deviation (n = 3). *, Not measurable. (F) XPS narrow-scan spectra for the C 1s of the PLA, PBS, PBAT, and PHA before (dotted line) and after (solid line) incubation with screened bacteria. | ||
To discover bioplastic-degrading microorganisms from wild environmental samples, activated sludge samples were inoculated into the PLA, PBS, PBAT, and PHA screening plates through spraying. The plates were incubated at 30 °C until a clear zone appeared. After 1 to 5 days, the microorganisms generally formed colonies, which created neighboring clear zones due to the bioplastic-degrading activity. It was possible to visually detect these clear zones by the naked eye (Fig. 3B). The plates containing a mixture of plastic powder or film were cultured for the same period under the same conditions. However, it was difficult to observe colony formation or morphological changes of bioplastic samples in both the bulk film and powder plates with the naked eye (Fig. S5†). In the powder system, the plastic powders with a diameter of <1 mm did not provide an interfacial area as large as the microparticles, and they settled down during the agarose solidification process. In the bulk film system, it took a long duration for the microorganisms to significantly decompose the film. These results suggest why the discovery of novel bioplastic-degrading microorganisms is limited. Applying the micro-spray screening method has the advantages of reducing the experimental workload, time, and cost of screening. Therefore, our approach will significantly improve the throughput for the discovery of bioplastic-degrading microorganisms.
Microorganisms that were isolated from environmental samples using the PLA screening plates were: Bacillus velezensis, Bacillus subtilis, Bacillus sp., Bacillus altitudinis, Bacillus amyloliquefaciens, and Bacillus stratosphericus. The isolates from the PBS screening plate were: Marinomonas sp., Marinomonas primoryensis, and Pseudomonas migulae. The isolates from the PBAT screening plate were Marinomonas sp., Bacillus sp., Aeromonas media, and Pseudomonas sp. The isolates from the PHA screening plate were: Pseudomonas stutzeri, Pseudomonas sp., Acidovorax facilis, and Vibrio sp. A clear zone was formed around the colonies of the isolates from the screening plates when these isolates were cultured again on corresponding plastic micro-sprayed plates. For this, a drop of the isolated bacterial suspension (5 μL) was spotted on the screening plate and incubated at 30 °C, and a clear zone was observed within 3 to 5 days of cultivation (Fig. 3C). It has been previously reported that species belonging to Pseudomonas are the prominent microbes associated with polymer degradation.47 Besides this, the genus Bacillus specializes in adsorbing and colonizing the polymer film; for example, Bacillus licheniformis is involved in the degradation of PLA.43,48 Moreover, extracellular lipase activity was detected in members of the genus Marinomonas.49 Screening plate tests against Gram-negative(E. coli), Gram-positive (S. aureus) bacteria and yeast (S. cerevisiae) are shown in Fig. S6.† After 2 weeks of incubation, there was no significant formation of recognizable to clear-zone in the PLA, PBS, PBAT, and PHA screening plates. Furthermore, it was confirmed that clear-zones were not observed after the selected strains that degrade PLA, PBS, PBAT, and PHA were inoculated on a PS screening plate and incubated (Fig. S7†). These results are indicative of plastic depolymerization activity by bacteria but not by the abiotic process such as diffusion of sprayed plastic microparticles.
We also further investigated the biodegradation of plastic films by the isolated microorganisms mentioned above. Isolated bacterial cultures were spread on a carbon-free solid media, and the corresponding plastic films were placed on the surface of the inoculated solid media. All the plates were incubated at 30 °C for 2 weeks except for PHA. The PHA films were retrieved from plates within 1 week. As shown in Fig. 3D, pits and crevices were observed on the surface of all the retrieved films. The PHA films were extensively damaged within 1 week. The changes in the surface hydrophobicity of the incubated films were analysed by measuring the water contact angle. The surface hydrophobicity of the plastic films that were inoculated corresponding to the bacteria was notably decreased compared to films before incubation and incubated films without bacterial inoculation (Fig. 3E). To further elucidate the biodegrading abilities of screened bacteria, narrow X-ray photoelectron spectroscopy (XPS) scans of plastic films before and after incubation with screened bacteria were performed. As displayed in Fig. 3F, the XPS results provided evidence of biodegradation. After incubation with bacteria, the peak proportions of C–O and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O normalized by C–C and C–H were increased compared to the pristine plastic film. The enzymatic cleavage of ester linkages from screened bacteria formed the hydroxyl- and carboxyl-terminated chemicals on the surface. The results of SEM observation, water contact angle measurement, and XPS analysis indicate that the bacteria isolated from the screening plates have biodegradation activity against the corresponding bioplastics.
O normalized by C–C and C–H were increased compared to the pristine plastic film. The enzymatic cleavage of ester linkages from screened bacteria formed the hydroxyl- and carboxyl-terminated chemicals on the surface. The results of SEM observation, water contact angle measurement, and XPS analysis indicate that the bacteria isolated from the screening plates have biodegradation activity against the corresponding bioplastics.
Our data suggest that the strains that form a clear zone in the screening plate are potential candidates for plastic biodegradation. The formation of a clear zone is a result of the hydrolysis of water-insoluble polymers to low-molecular-weight intermediates by microbial enzyme activities. In the biological degradation process, the step involving the hydrolysis of water-insoluble polymers to the formation of intermediates that microorganisms can uptake into their metabolic pathway is considered the rate-limiting step.50 Thus, the generation time, growth rate, size, and degree of clearness of the clear zone are all closely related to the hydrolysis activity of bioplastic-degrading microorganisms. Our screening strategy, utilizing the plastic micro-spray method, allows a faster clear zone formation and higher clarity by producing plastic particles with small sizes and high surface areas.
We emphasize that this spraying-based method can also be applied to studies on the decomposition of non-degradable plastics. Studies on the decomposition of non-degradable plastics, such as polyethylene, polystyrene, and polyethylene terephthalate, have been largely limited due to their slow degradation rates.51,52 If the surface area of the non-decomposable plastic is maximized by applying our proposed screening method, we hope that research on microorganisms that degrade non-degradable plastics can also be potentially accelerated. Encouragingly, methods to coat non-degradable plastic microparticles through spraying have already been well established.53,54 In the near future, we would like to study the screening of microorganisms that degrade conventional plastics known as non-degradable using the spray-based screening plate system.
Conclusions
The objective of this study was to develop a simple, straightforward and efficient method to isolate plastic degrading microorganisms. To achieve this purpose, we developed and optimized a method of spraying biodegradable microplastics onto the surface of the solid media using a plastic solution. Through this method, it was possible to obtain translucent solid media containing plastic particles of variable diameters on the surface within a few minutes. This method has the advantages of being easy to reproduce and time efficient. Microbial screening to isolate bioplastic-degrading activities was performed using a wild environmental inoculum for the biodegradation of PLA, PBS, PBAT, and PHA. Biodegradable plastic-degrading bacteria such as Pseudomonas and Bacillus were successfully isolated by utilizing our screening method. Our micro-spray screening method has the advantage of being applied for a wide range of applications, regardless of the type of basal media used.The plastic decomposition mechanisms and rates differ depending on biotic and abiotic factors, such as degrading microorganisms, temperature, and humidity. Therefore, bioplastic-degrading microorganisms and the degrading enzymes discovered under various environmental conditions, along with the information accumulated through the identification of their mechanisms can be used to develop newer plastic materials, and this is also important for the production of biodegradable plastics with a controllable biodegradation rate. These plastics can therefore be more suitable for the environment as well.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Bio-Industrial Technology Development Program (20008628) funded by the Ministry of Trade, Industry & Energy (MI, Korea), the Korea Research Institute of Chemical Technology (KRICT) core project (SS2142-10), and the Korea Institute for Advancement of Technology (KIAT) through the System Industrial Base Institution Support Program (P0001939). Dr Sejin Choi at KRICT gave an advice to optimize the spray conditions.Notes and references
- Plastics - the Facts 2020, https://www.plasticseurope.org/application/files/5716/0752/4286/AF_Plastics_the_facts-WEB-2020-ING_FINAL.pdf, (accessed January 2021).
- K. Beydoun and J. Klankermayer, ChemSusChem, 2020, 13, 488–492 CrossRef CAS PubMed
.
- R. Geyer, J. R. Jambeck and K. L. Law, Sci. Adv., 2017, 3, e1700782 CrossRef PubMed
.
- A. K. Mohanty, S. Vivekanandhan, J.-M. Pin and M. Misra, Science, 2018, 362, 536–542 CrossRef CAS PubMed
.
- A. Alassali, S. Fiore and K. Kuchta, Waste Manag., 2018, 82, 71–81 CrossRef CAS PubMed
.
- S. B. Borrelle, J. Ringma, K. L. Law, C. C. Monnahan, L. Lebreton, A. McGivern, E. Murphy, J. Jambeck, G. H. Leonard, M. A. Hilleary, M. Eriksen, H. P. Possingham, H. De Frond, L. R. Gerber, B. Polidoro, A. Tahir, M. Bernard, N. Mallos, M. Barnes and C. M. Rochman, Science, 2020, 369, 1515–1518 CrossRef CAS PubMed
.
- C. M. Rochman, M. A. Browne, B. S. Halpern, B. T. Hentschel, E. Hoh, H. K. Karapanagioti, L. M. Rios-Mendoza, H. Takada, S. Teh and R. C. Thompson, Nature, 2013, 494, 169–171 CrossRef CAS PubMed
.
- M. A. Browne, A. Dissanayake, T. S. Galloway, D. M. Lowe and R. C. Thompson, Environ. Sci. Technol., 2008, 42, 5026–5031 CrossRef CAS PubMed
.
- K. L. Law, N. Starr, T. R. Siegler, J. R. Jambeck, N. J. Mallos and G. H. Leonard, Sci. Adv., 2020, 6, eabd0288 CrossRef PubMed
.
- H. Kim, H. Jeon, G. Shin, M. Lee, J. Jegal, S. Y. Hwang, D. X. Oh, J. M. Koo, Y. Eom and J. Park, Green Chem., 2021, 23, 2293–2299 RSC
.
- T. Iwata, Angew. Chem., Int. Ed., 2015, 54, 3210–3215 CrossRef CAS PubMed
.
- S. Chinaglia, M. Tosin and F. Degli-Innocenti, Polym. Degrad. Stab., 2018, 147, 237–244 CrossRef CAS
.
- Bioplastics market development update 2020, https://docs.european-bioplastics.org/conference/Report_Bioplastics_Market_Data_2020_short_version.pdf, (accessed January, 2021).
- F. Carrasco, P. Pagès, J. Gámez-Pérez, O. O. Santana and M. L. Maspoch, Polym. Degrad. Stab., 2010, 95, 116–125 CrossRef CAS
.
- J. Xu and B.-H. Guo, Biotechnol. J., 2010, 5, 1149–1163 CrossRef CAS PubMed
.
- B. V. M. Rodrigues, A. S. Silva, G. F. S. Melo, L. M. R. Vasconscellos, F. R. Marciano and A. O. Lobo, Mater. Sci. Eng., C, 2016, 59, 782–791 CrossRef CAS PubMed
.
- M. A. M. Reis, L. S. Serafim, P. C. Lemos, A. M. Ramos, F. R. Aguiar and M. C. M. Van Loosdrecht, Bioprocess Biosyst. Eng., 2003, 25, 377–385 CrossRef CAS PubMed
.
- D. Cha, H. S. Ha and S. K. Lee, Bioresour. Technol., 2020, 309, 123332 CrossRef CAS PubMed
.
- T. P. Haider, C. Völker, J. Kramm, K. Landfester and F. R. Wurm, Angew. Chem., Int. Ed., 2019, 58, 50–62 CrossRef CAS PubMed
.
- S. Bi, B. Tan, J. L. Soule and M. J. Sobkowicz, Polym.
Degrad. Stab., 2018, 155, 9–14 CrossRef CAS
.
- E. Castro-Aguirre, R. Auras, S. Selke, M. Rubino and T. Marsh, Polym. Degrad. Stab., 2017, 137, 251–271 CrossRef CAS
.
- K. Yamamoto-Tamura, S. Hiradate, T. Watanabe, M. Koitabashi, Y. Sameshima-Yamashita, T. Yarimizu and H. Kitamoto, AMB Express, 2015, 5, 10 CrossRef PubMed
.
- L. S. Dilkes-Hoffman, P. A. Lant, B. Laycock and S. Pratt, Mar. Pollut. Bull., 2019, 142, 15–24 CrossRef CAS PubMed
.
- A. S. Al Hosni, J. K. Pittman and G. D. Robson, Waste Manag., 2019, 97, 105–114 CrossRef CAS PubMed
.
- M. J. Krause and T. G. Townsend, Environ. Sci. Technol. Lett., 2016, 3, 166–169 CrossRef CAS
.
- M. T. Zumstein, A. Schintlmeister, T. F. Nelson, R. Baumgartner, D. Woebken, M. Wagner, H.-P. E. Kohler, K. McNeill and M. Sander, Sci. Adv., 2018, 4, eaas9024 CrossRef CAS PubMed
.
- T. Kim, H. Jeon, J. Jegal, J. H. Kim, H. Yang, J. Park, D. X. Oh and S. Y. Hwang, RSC Adv., 2018, 8, 15389–15398 RSC
.
- H. Kim, T. Kim, S. Choi, H. Jeon, D. X. Oh, J. Park, Y. Eom, S. Y. Hwang and J. M. Koo, Green Chem., 2020, 22, 7778–7787 RSC
.
- L. Botta, V. Fiore, T. Scalici, A. Valenza and R. Scaffaro, Materials, 2015, 8, 7770–7779 CrossRef CAS
.
- H. G. Lim, D. H. Kwak, S. Park, S. Woo, J. S. Yang, C. W. Kang, B. Kim, M. H. Noh, S. W. Seo and G. Y. Jung, Nat. Commun., 2019, 10, 2486 CrossRef PubMed
.
- R. A. Gross and B. Kalra, Science, 2002, 297, 803–807 CrossRef CAS PubMed
.
- Q. N. M. Tran, H. Mimoto and K. Nakasaki, Int. Biodeterior. Biodegrad., 2015, 104, 377–383 CrossRef CAS
.
- R. Barrena, E. Pagans, G. Faltys and A. Sánchez, J. Chem. Technol. Biotechnol., 2006, 81, 420–425 CrossRef CAS
.
- S. Yoshida, K. Hiraga, T. Takehana, I. Taniguchi, H. Yamaji, Y. Maeda, K. Toyohara, K. Miyamoto, Y. Kimura and K. Oda, Science, 2016, 351, 1196–1199 CrossRef CAS PubMed
.
- Z. Wang, X. Xin, X. Shi and Y. Zhang, Sci. Total Environ., 2020, 726, 138564 CrossRef CAS PubMed
.
- A. Popovic, T. Hai, A. Tchigvintsev, M. Hajighasemi, B. Nocek, A. N. Khusnutdinova, G. Brown, J. Glinos, R. Flick, T. Skarina, T. N. Chernikova, V. Yim, T. Brüls, D. L. Paslier, M. M. Yakimov, A. Joachimiak, M. Ferrer, O. V. Golyshina, A. Savchenko, P. N. Golyshin and A. F. Yakunin, Sci. Rep., 2017, 7, 44103 CrossRef PubMed
.
- D. Danso, J. Chow and W. R. Streit, Appl. Environ. Microbiol., 2019, 85, e01095–19 CrossRef PubMed
.
- B. K. Singh, Trends Biotechnol., 2010, 28, 111–116 CrossRef CAS PubMed
.
- R. Molitor, A. Bollinger, S. Kubicki, A. Loeschcke, K.-E. Jaeger and S. Thies, Microb. Biotechnol., 2020, 13, 274–284 CrossRef CAS PubMed
.
- T. Teeraphatpornchai, T. Nakajima-Kambe, Y. Shigeno-Akutsu, M. Nakayama, N. Nomura, T. Nakahara and H. Uchiyama, Biotechnol. Lett., 2003, 25, 23–28 CrossRef CAS PubMed
.
- A. Jarerat, H. Pranamuda and Y. Tokiwa, Macromol. Biosci., 2002, 2, 420–428 CrossRef CAS
.
- R. H. Staff, D. Schaeffel, A. Turshatov, D. Donadio, H.-J. Butt, K. Landfester, K. Koynov and D. Crespy, Small, 2013, 9, 3514–3522 CrossRef CAS PubMed
.
- M. Y. Kim, C. Kim, J. Moon, J. Heo, S. P. Jung and J. R. Kim, J. Microbiol. Biotechnol., 2017, 27, 342–349 CrossRef CAS PubMed
.
- E. S. Medeiros, G. M. Glenn, A. P. Klamczynski, W. J. Orts and L. H. C. Mattoso, J. Appl. Polym. Sci., 2009, 113, 2322–2330 CrossRef CAS
.
- F. Li, S. Wang, W. Liu and G. Chen, FEMS Microbiol. Lett., 2008, 282, 52–58 CrossRef CAS PubMed
.
- A. Jarerat, Y. Tokiwa and H. Tanaka, Appl. Microbiol. Biotechnol., 2006, 72, 726–731 CrossRef CAS PubMed
.
- V. M. Pathak and Navneet, Bioresour. Bioprocess., 2017, 4, 15 CrossRef
.
- B. Nowak, J. Pająk, M. Drozd-Bratkowicz and G. Rymarz, Int. Biodeterior. Biodegrad., 2011, 65, 757–767 CrossRef CAS
.
- Y. Yu, H. Li, Y. Zeng and B. Chen, Polar Biol., 2009, 32, 1539–1547 CrossRef
.
- R.-J. Mueller, Process Biochem., 2006, 41, 2124–2128 CrossRef CAS
.
- I. Taniguchi, S. Yoshida, K. Hiraga, K. Miyamoto, Y. Kimura and K. Oda, ACS Catal., 2019, 9, 4089–4105 CrossRef CAS
.
- B. T. Ho, T. K. Roberts and S. Lucas, Crit. Rev. Biotechnol., 2018, 38, 308–320 CrossRef CAS PubMed
.
- Y.-S. Yu, M.-C. Wang and X. Huang, Sci. Rep., 2017, 7, 14118 CrossRef PubMed
.
- L. T. Duarte, E. M. Paula e Silva, J. R. Branco and V. F. Lins, Surf. Coat. Technol., 2004, 182, 261–267 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section and additional experimental data. See DOI: 10.1039/d1gc01916c |
| This journal is © The Royal Society of Chemistry 2021 |