Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Lanthanides compete with calcium for binding to cadherins and inhibit cadherin-mediated cell adhesion†
Lewis L.
Brayshaw
 a,
Rosanna C. G.
Smith
a,
Rosanna C. G.
Smith
 ab,
Magd
Badaoui
c,
James A.
Irving
c and
Stephen R.
Price
ab,
Magd
Badaoui
c,
James A.
Irving
c and
Stephen R.
Price
 *a
*a
aResearch Department of Cell and Developmental Biology, UCL, Gower Street, WC1E 6BT, London, UK. E-mail: Stephen.price@ucl.ac.uk
bCentre for Human Development, Stem Cells, and Regeneration, University of Southampton, Southampton, SO17 1BJ, UK
cResearch Department of Respiratory Medicine, UCL, Gower Street, WC1E 6BT, London, UK
First published on 8th March 2019
Abstract
Lanthanides are rare-earth metals with a broad range of applications in biological research and medicine. In addition to their unique magnetic and spectroscopic properties, lanthanides are also effective mimics of calcium and can stimulate or inhibit the function of calcium-binding proteins. Cadherins are a large family of calcium-binding proteins that facilitate cell adhesion and play key roles in embryo development, tissue homeostasis and tumour metastasis. However, whether lanthanides can bind cadherins and functionally replace calcium binding has not been comprehensively explored. In this study, we investigated the effect of lanthanide binding on cadherin structure and function using terbium, which is a commonly used lanthanide for protein spectroscopy and a proposed anti-cancer agent. We demonstrate that terbium can compete with calcium for binding to calcium-binding sites in cadherins. Terbium binding to cadherins abolished their cell adhesive activity and rendered cadherins sensitive to proteolysis by trypsin. Molecular dynamics simulations indicate that replacement of calcium by terbium results in structural rearrangements and increases the flexibility of the cadherin ectodomain. These changes in structure and dynamics are likely to underlie the inability of lanthanide-bound cadherins to support cell adhesion. Taken together, our findings further knowledge on lanthanide interactions with calcium-binding proteins and provide new insight into the influence of metal chemistry on cadherin structure, dynamics and function.
Significance to metallomicsLanthanides are widely used in medicine and biological research. A unique feature of lanthanides is their ability to mimic calcium in biological processes and the binding of lanthanides is known to induce a range of effects in calcium-binding proteins. This study investigated the effect of lanthanides binding to cadherins, which are calcium-dependent cell adhesion molecules that have key roles in embryo development and tumour metastasis. Our findings expand knowledge on the bioactivity of lanthanides and provide new insight into the sensitivity of cadherin function to the chemistry of binding metals. |
Introduction
Lanthanides are a group of metals with extensive use in biological research and medicine.1–4 They are frequently used in clinical magnetic resonance imaging and, due to their spectroscopic properties, are powerful tools for probing physiological processes. For example, lanthanides have been used to monitor E- and N-cadherin dynamics in the membranes of cells undergoing epithelial-to-mesenchymal transition.5 Additionally, lanthanides are highly bioactive and there is significant interest in their use as therapeutics.6 Lanthanum carbonate for instance, is a potent binder of phosphate and is an approved treatment for hyperphosphatemia in patients with chronic kidney disease.7 Furthermore, considerable research exists to support the use of lanthanides (in particular Tb3+ and Gd3+) as anti-cancer agent, given their ability to induce apoptosis and inhibit cell motility in cancer cells.8–11A significant characteristic of lanthanide ions is their chemical similarity to Ca2+ ions, which is a result of their comparable ionic radii and metal coordination chemistry.12 For example, Tb3+ has an ionic radius of 0.98 Å and Ca2+ has an ionic radius of 1.06 Å, and both are typically coordinated by 6–8 ligands.13–15 Lanthanides binding within a number of Ca2+-binding proteins has been demonstrated, and due to their greater electro-positivity, lanthanides often have a stronger binding affinity to proteins than Ca2+.16,17 However, the effect that lanthanide binding has on the function of proteins is hard to predict. For certain proteins, lanthanide binding has been shown to effectively substitute for Ca2+ in terms of protein function.18,19 For example, Tb3+ binding to β1-bungarotoxin stimulates the protein's phospholipase A2 activity.20 For others, such as Gd3+ binding to stretch activated Ca2+ channels, lanthanide ions have a potent inhibitory effect.21–23 Given their wide-ranging application in biology, it is important to understand the bioactivity of lanthanides on Ca2+-binding proteins.
Cadherins are a large family of calcium-dependent membrane proteins with over 100 members. Cadherins have vital roles in embryo development and tissue homeostasis, and a number of human diseases, such as cancer, are associated with alterations in cadherin function.24–27 Cadherins facilitate cell–cell adhesion through trans-binding of their extracellular regions, which are known as ectodomains.28 The ectodomains of classical cadherins are composed of five extracellular domains (EC1–5), which are connected by flexible linker regions. X-ray crystal structures indicate that three Ca2+ ions can bind at each of the four linker regions and that the cadherin protein gradually attains its functional conformation as Ca2+ ions bind to the 12 binding sites.29–31 Successive Ca2+-binding results in the precise relative positioning of EC domains and imparts rigidity and curvature onto the cadherin molecule which is an essential feature for cadherin function.32–34 Furthermore, as Ca2+ ions bind to a cadherin molecule, the binding affinity for successive Ca2+ ions increases due to cooperativity in binding and conformational changes which are transmitted distally along the cadherin.35,36 Whether lanthanides can bind cadherins and effectively replace calcium in supporting cadherin function has not been comprehensively investigated.
In this study, we investigated the binding of the lanthanide Tb3+ to two classical cadherins with fundamental functions in embryo development and tumour metastasis, E-cadherin and N-cadherin. Cell-based assays, protein biophysics and molecular dynamics have been used to demonstrate that Tb3+ competes with Ca2+ for binding to cadherin Ca2+-binding sites, and Tb3+-bound cadherins have enhanced flexibility and do not support cell–cell adhesion.
Methods
Cell culture
Chinese hamster ovary cells were stably transfected with human N-cadherin (N-CHO cells) or human E-cadherin (E-CHO cells) using Lipofectamine 2000 (Thermo), and cells were selected for expression using G418 (Gibco). Parental CHO, N-CHO and E-CHO cells were cultured in DMEM/F12 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Gibco) containing 10% FBS (Gibco). Hs578t and MCF-7 cells were obtained from ATCC and were cultured in DMEM containing 10% FBS. Culture media for Hs578t additionally contained 10 μg mL−1 bovine insulin (Sigma).
1 (Gibco) containing 10% FBS (Gibco). Hs578t and MCF-7 cells were obtained from ATCC and were cultured in DMEM containing 10% FBS. Culture media for Hs578t additionally contained 10 μg mL−1 bovine insulin (Sigma).
Aggregation assays
Monolayer cell cultures were treated with 0.01% trypsin + 1 mM CaCl2 in HBSS (Gibco) for 30 minutes at 37 °C. This trypsin treatment dissociates monolayers into single cells whilst leaving surface cadherins intact. Cells were resuspended in HBSS + 1% BSA (Sigma) + 10% FBS to inhibit further trypsin degradation and then resuspended in HBSS + 1% BSA to give a concentration of 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per 50 μL. 150
000 cells per 50 μL. 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded into 24-well ultra-low attachment plates (Corning Costar) in 0.5 mL HBSS + 1% BSA containing CaCl2, TbCl3 or combinations of both. Plates were rotated horizontally at 75 rpm at 37 °C on an orbital shaker Model 3500 (WR) for either 30 minutes (N-CHO) or 40 minutes (E-CHO, Hs578t, MCF-7). The aggregation potential was calculated by 1 minus the number of single cells at end of aggregation (Ne) divided by the number of single cells at the beginning of aggregation (Ns). Aggregation potential = 1 − Ne/Ns. The number of single cells was quantified using a 0.2 mm Improved Neubauer haemocytometer counting chamber (Hawksley).
000 cells were seeded into 24-well ultra-low attachment plates (Corning Costar) in 0.5 mL HBSS + 1% BSA containing CaCl2, TbCl3 or combinations of both. Plates were rotated horizontally at 75 rpm at 37 °C on an orbital shaker Model 3500 (WR) for either 30 minutes (N-CHO) or 40 minutes (E-CHO, Hs578t, MCF-7). The aggregation potential was calculated by 1 minus the number of single cells at end of aggregation (Ne) divided by the number of single cells at the beginning of aggregation (Ns). Aggregation potential = 1 − Ne/Ns. The number of single cells was quantified using a 0.2 mm Improved Neubauer haemocytometer counting chamber (Hawksley).
Trypsin protection assays
1.5 × 106 cells were seeded into 10 cm culture plates (Triple Red) with media 24 h prior to experimentation. The cells were washed twice with HBSS and then incubated with HBSS + 5 mM EDTA for 8 min to remove Ca2+ ions. Cells were washed with HBSS and then incubated with HBSS + required ions (Ca2+, EGTA, Tb3+) for 10 min. Trypsin (Sigma) was added to a final concentration of 0.04% and cells were incubated for 80 min at 25 °C. Cells were then pelleted, resuspended in HBSS + 0.5 mg trypsin inhibitor (Sigma) and placed on ice.Western blotting
Cells were lysed and protein collected using MEM-PER Plus membrane extraction kit (Thermo) with trypsin inhibitor and Halt protease inhibitor cocktail (Thermo). Protein concentration in samples was quantified using Pierce BCA Protein Assay Kit (Thermo). Samples were boiled in SDS-DTT sample buffer for 5 min, separated on an 8% SDS-polyacrylamide gel and then transferred to a nitrocellulose membrane (Biorad). The membrane was blocked with TBS-T (tris buffered saline plus 0.1% Tween) + 5% milk for 1 hour at room temperature and then incubated with primary antibody diluted in TBS-T + 5% milk overnight at 4 °C. Membranes were washed four times in TBS-T and incubated with secondary antibody for 1 hour at 25 °C. Following four further washes in TBS-T, membranes were developed with Luminata Western HRP substrate (Millipore) and exposed using a ChemiDoc MP system (Biorad). Image collection and analysis was performed with ImageLab software (Biorad).Primary antibodies used include mouse anti-N-cadherin (BD Biosciences, 610920) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2500), mouse anti-E-cadherin (BD Biosciences, 610181) (1
2500), mouse anti-E-cadherin (BD Biosciences, 610181) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2500), and rabbit anti-β-actin (Cell Signaling Technology, 4970) (1
2500), and rabbit anti-β-actin (Cell Signaling Technology, 4970) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000). Secondary antibodies used include anti-mouse HRP-conjugate (Promega, W4021) (1
1000). Secondary antibodies used include anti-mouse HRP-conjugate (Promega, W4021) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) and anti-rabbit HRP-conjugate (Promega, W4011) (1
2000) and anti-rabbit HRP-conjugate (Promega, W4011) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000).
2000).
Tb3+-FRET
Wild-type human E-cadherin recombinant protein fragment aa155-710 (Sigma) was dialysed within 2 × 5 L 10 mM Tris–HCl, 100 mM KCl, 120 mM NaCl (all Sigma) pH 7.4 at 4 °C using Slide-A-Lyzer Dialysis cassettes (Thermo). The protein sample was aliquoted and stored at −20 °C. E-cadherin titration and Tb3+ titration experiments were carried out on a SpectraMax-M5 (Molecular Devices). For the E-cadherin titration, a stock solution of E-cadherin was added to 100 μM TbCl3 in buffer (10 mM Tris–HCl, 100 mM KCl, 120 mM NaCl) or buffer alone and emission at 543 nm measured during excitation at 282 nm. For the Tb3+ titration, a stock solution of Tb3+ was added to 43.7 nM E-cadherin in buffer and emission at 543 nm measured during excitation at 282 nm. The fluorescence competition assay was carried out on a Cary Eclipse Fluorescence Spectrophotometer (Agilent). Stock concentrations of Ca2+ were added to 3 μM E-cadherin in 170 μM Tb3+ and buffer. Emission was measured at 542 nm during excitation at 280 nm. For all experiments, spectra were corrected for the background contribution of Tb3+ to 542–543 nm emissions by measuring and subtracting the 542–543 nm emissions of each Tb3+ concentration in the absence of protein. All samples were gently mixed and allowed to equilibrate for 15 minutes prior to data collection.Molecular dynamics
The LEaP software, part of AmberTools, was used to generate systems from the crystal structure of mouse E-cadherin (PDB code 3Q2V).31,37 Residues 504–507 were not resolved by crystallography and were modelled in using Pymol.38 Disulphide bridges found in physiological conditions were generated in all systems.31 The Ca-Ecad system consisted of the protein and the 12 crystallographically resolved Ca2+ ions found at the linker regions between EC domains. All 12 crystallographically resolved Ca2+ ions were replaced with Tb3+ ions for the Tb-Ecad system, and the 12 Ca2+ ions were removed for the Apo-Ecad system. All other crystallographically resolved non-protein molecules were removed in systems. Systems were neutralised through the addition of bulk Na+ ions and solvated explicitly in boxes of TIP3P water molecules with edges no closer than 10 Å from atoms in the protein.39Simulations were carried out using the SANDER package in Amber16 with the Parm99 parameter sets for Ca2+ and Tb3+ ions.37 For all simulations, 2 fs time steps were used and a potential cut-off of 8 Å was assumed for van der Waals interactions. Constant volume periodic boundary conditions were assumed. The systems were minimized with 500 steps of steepest descent minimization followed by 500 steps of conjugate gradient minimization using 2 kcal mol−1 Å−2 quadratic constraints. After minimisation, the temperature of the systems was increased to 300 K over 2500 steps using Langevin dynamics. Free molecular dynamics simulations were then carried out for 40 ns.
The coordinates of atoms in the systems were saved every 10 ps during simulation and trajectories were analysed in VMD.40,41 RMSDs were calculated using non-hydrogen atoms in VMD.
Results
Tb3+ inhibits E-cadherin- and N-cadherin-mediated cell adhesion
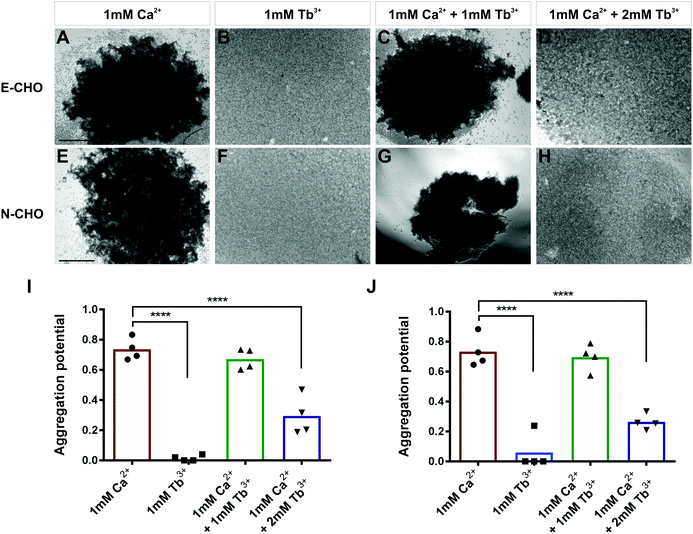
To investigate whether Tb3+ affects cadherin-mediated cell adhesion, cell aggregation assays were used which assess the ability of single cells to form cadherin-dependent aggregates. Cadherin-expressing cells were dissociated into single cells using 0.01% trypsin + 1 mM Ca2+ treatment, which cleaves proteins on the cell surface but does not affect cadherins. This treatment produces a single-cell population which can only form adhesions and aggregates via cadherin-mediated cell adhesion.30,42,43 To demonstrate that this method only permits cadherin-mediated cell aggregation, parental CHO cells were treated with 0.01% trypsin + 1 mM Ca2+ and assayed for 40 min in the presence of 1 mM Ca2+. Parental CHO cells have no endogenous cadherin expression and thus no aggregation was observed (Fig. S1, ESI†).In contrast, CHO cells stably expressing E-cadherin (E-CHO) and CHO cells stably expressing N-cadherin (N-CHO) formed dense aggregates within 40 minutes and 30 minutes respectively in the presence of 1 mM Ca2+ (Fig. 1A and E). However, no aggregation was visible in the presence of only 1 mM Tb3+ (Fig. 1B and F). Quantification of aggregation through the counting of unaggregated single cells confirmed that cadherin-mediated aggregation of E-CHO and N-CHO cells is significantly reduced in 1 mM Tb3+ compared with aggregation in 1 mM Ca2+ (Fig. 1I and J). Interestingly, cell aggregation in 1 mM Ca2+ + 1 mM Tb3+ was not significantly different from that in 1 mM Ca2+, but cell aggregation was significantly inhibited for both E-CHO and N-CHO cells in the presence of 1 mM Ca2+ + 2 mM Tb3+ (Fig. 1C, D, G, H, I and J). Similar results were obtained in cell aggregation assays in the presence of another lanthanide, Gd3+, suggesting that cadherin-mediated cell adhesion is inhibited by Ca2+-mimicking lanthanides (Fig. S2, ESI†).
In order to determine if Tb3+ had the same effect on cells expressing cadherins endogenously, aggregation assays using two human cancer cell lines were carried out. MCF-7 is an E-cadherin expressing breast cancer cell line and, similarly to E-CHO, MCF-7 E-cadherin mediated cell aggregation was significantly reduced in the presence of 1 mM Tb3+ and 1 mM Ca2+ + 2 mM Tb3+ compared with aggregation in the presence of 1 mM Ca2+ (Fig. S3A–D, I, ESI†).44 In the N-cadherin expressing breast cancer cell line Hs578t, cell aggregation was also significantly impaired in 1 mM Tb3+ and 1 mM Ca2+ + 2 mM Tb3+ (Fig. S3E–H, J, ESI†). Taken together, the lack of aggregation in 1 mM Tb3+ for all cell lines indicates Tb3+ cannot substitute for Ca2+ in mediating E- and N-cadherin adhesion. Additionally, Tb3+ is able to inhibit Ca2+-dependent cadherin cell adhesion when at a higher concentration than Ca2+, suggesting competitive binding between the two metal ions for cadherins.
Tb3+ does not protect E-cadherin or N-cadherin from trypsin degradation
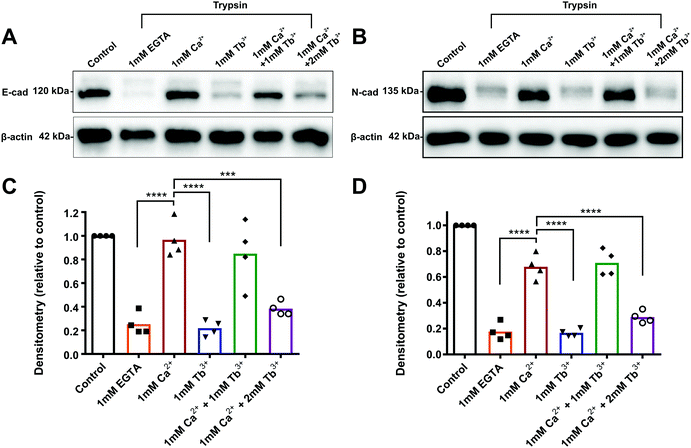
To determine how Tb3+ inhibits cadherin-mediated adhesion and whether Tb3+ competes with Ca2+ for binding at Ca2+ binding sites, trypsin resistance assays were carried out. When cadherins are fully bound to Ca2+, they are resistant to trypsin-mediated proteolysis when using a low concentration of trypsin.30,42,43 This is because Ca2+-bound cadherins adopt a rigid structural conformational that has no accessible trypsin recognition sites. In the absence of Ca2+ binding, cadherins adopt a flexible conformation that is sensitive to degradation as the trypsin recognition sites can become exposed. Thus, by assessing the resistance of cadherins to trypsin, we can demonstrate whether Tb3+-bound cadherins differ in structure or rigidity to Ca2+-bound cadherins and determine if Tb3+ can compete with Ca2+ for binding. Monolayers of E-CHO and N-CHO cells were first stripped of Ca2+via EDTA treatment and then treated with a low concentration of trypsin (0.04%) in the presence of different combinations of Ca2+ and Tb3+ for 80 min. Proteins were then extracted and western blots were carried out to analyse E-cadherin and N-cadherin proteins. To measure how Ca2+ and Tb3+ protects E-cadherin and N-cadherin from trypsin degradation, the densitometry of the protein bands corresponding to full-length cadherin proteins (120 kD for E-cadherin and 135 kD for N-cadherin) at each condition was quantified after normalisation to a β-actin protein loading control. The densitometry of the full-length band in each treatment was expressed as relative to measurement of the full-length band in the no trypsin control.As expected, when E-CHO cells were treated with trypsin + 1 mM Ca2+, there was significantly more detectable full-length E-cadherin than when E-CHO cells were treated with trypsin + 1 mM EGTA (Ca2+ chelator), demonstrating that Ca2+ is able to protect E-cadherin from trypsin degradation (Fig. 2). However, when cells were treated with trypsin + 1 mM Tb3+, significant loss of full-length E-cadherin protein occurred, indicating Tb3+ offers no trypsin protection to E-cadherin. Interestingly, trypsin treatment of E-CHO cells in 1 mM Ca2+ + 1 mM Tb3+ resulted in no significant difference in E-cadherin degradation compared with trypsin + 1 mM Ca2+. However, when the concentration of Tb3+ was greater than that of Ca2+, significant trypsin degradation of E-cadherin protein was observed. Trypsin resistance assays revealed similar results for N-CHO cells. Following treatment of N-CHO cells with trypsin, N-cadherin was significantly degraded when in the presence of 1 mM Tb3+ or 1 mM Ca2+ + 2 mM Tb3+ compared with N-cadherin in the presence of 1 mM Ca2+. Importantly, when E-CHO and N-CHO cells were incubated with the same combinations of Ca2+ and Tb3+ in the absence of trypsin, no significant differences in the amount of full-length cadherin were observed among the samples (Fig. S4, ESI†). This demonstrates that the different ion conditions have no effect on cadherin expression levels and only effect how sensitive cadherins are to trypsin degradation.
These results show that cadherins are degraded by trypsin when in the presence of Tb3+. Given that Ca2+-bound cadherin are resistant to trypsin, this indicates that Tb3+-bound cadherins adopt a different and/or more flexible structure that has exposed trypsin recognition sites. Additionally, when Tb3+ is present at a higher concentration than Ca2+, cadherins become sensitive to trypsin degradation. This suggests that Tb3+ has the ability to displace Ca2+ from cadherins and compete with Ca2+ for binding at cadherin Ca2+-binding sites.
Binding of Tb3+ to E-cadherin
The previous experiments suggest that Tb3+ binds cadherins and competes with Ca2+ for binding at Ca2+-binding sites, however direct biophysical evidence of this is required. In order to gather this evidence, we measured the fluorescence resonance energy transfer (FRET) between Tb3+ ions and human E-cadherin EC1–5 recombinant protein. Tb3+ ions are naturally fluorescent and their excitation is within the range of tryptophan and tyrosine emissions (348 nm and 303 nm respectively). Thus, when tryptophan and tyrosine residues in E-cadherin are excited (280–282 nm), one can quantify the amount of bound Tb3+ ions by the FRET to Tb3+ ions and the resulting Tb3+ emission (542–543 nm).There are 12 Ca2+-binding sites that Tb3+ ions might bind to, but FRET can only take place at sites within 10 Å of a tryptophan or tyrosine residue. There is no structure available for human E-cadherin EC1–5, thus the crystal structure for mouse E-cadherin EC1–5 (PDB 3Q2V) was used to identify binding sites potentially capable of FRET. The human and mouse homologues share an 81% sequence identity (450/553), and tryptophan (6/6) and tyrosine (13/13) residues are found in identical positions (Fig. S5B, ESI†).31,45 In the mouse E-cadherin crystal structure, there are tryptophan and/or tyrosine residues within 10 Å to binding sites 3 (EC1-2), 5 (EC2-3), 6 (EC2-3), 8 (EC3-4), and 10 (EC4-5) (Fig S5A, ESI†). If Tb3+ ions were to bind to these sites in E-cadherin, one would expect FRET to take place.
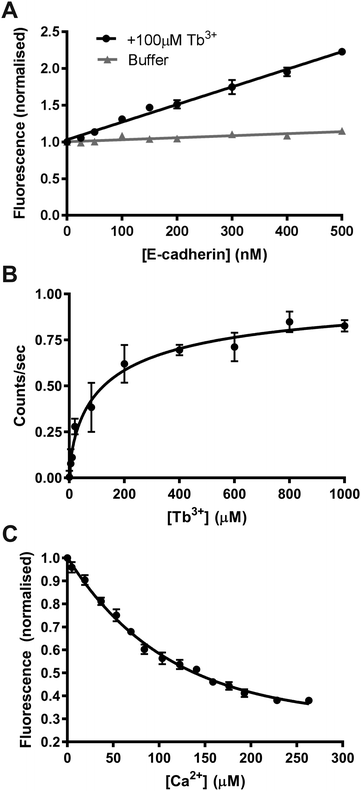
We first determined if FRET could be measured between tryptophan and tyrosine residues in E-cadherin and Tb3+ ions in order to confirm if Tb3+ binds to E-cadherin. E-cadherin was either titrated into buffer (10 mM Tris–HCl, 100 mM KCl, 120 mM NaCl) or into buffer + 100 μM Tb3+, and Tb3+ fluorescence at 543 nm was measured following excitation at 282 nm. In the absence of 100 μM Tb3+, the addition of E-cadherin protein resulted in no change in 543 nm emission (Fig. 3A). However, addition of E-cadherin protein to 100 μM Tb3+ resulted in a linear increase of Tb3+ fluorescence emission at 543 nm. This result indicates that FRET can occur between tryptophan and tyrosine residues in E-cadherin and bound Tb3+ ions, and thus suggests that Tb3+ can directly bind E-cadherin molecules.
In order to quantify the binding affinity of Tb3+ to E-cadherin, Tb3+ was titrated into a solution of E-cadherin and 543 nm emission was measured during 282 nm excitation. Data was fit to a Hill model using non-linear regression and the KD measured by this method was 143 μM (Fig. 3B). A Hill coefficient of 0.69 was obtained for the data, indicating that binding of Tb3+ to the E-cadherin protein had negative cooperativity.
A fluorescence competition assay was then carried out to determine if Tb3+ and Ca2+ compete for the same sites on E-cadherin. Ca2+ was titrated into a solution of E-cadherin in 170 μM Tb3+ and the change in Tb3+ fluorescence (542 nm) was measured during excitation of tryptophan and tyrosine residues (280 nm). Increasing the Ca2+ concentration reduced the level of Tb3+ fluorescence, suggesting displacement of Tb3+ ions at Ca2+-binding sites on E-cadherin by Ca2+ ions. Addition of Ca2+ was unable to completely remove Tb3+ fluorescence (∼60% total reduction with a half-maximal effect at ∼60 μM).
Differences in the molecular dynamics of Ca2+- and Tb3+-bound E-cadherin
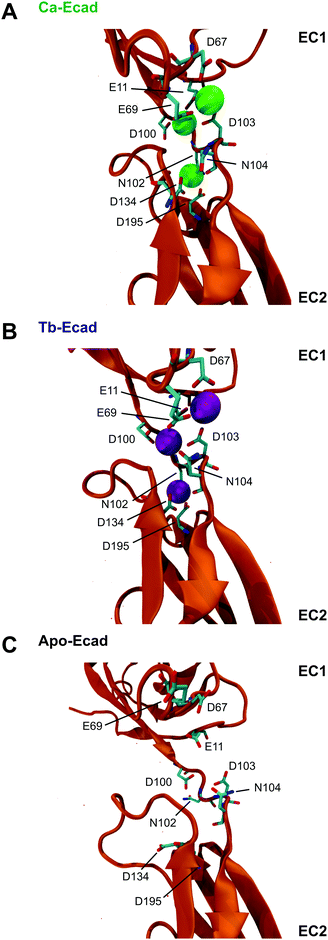
Our results indicate that Tb3+ can bind cadherins, but that Tb3+-bound cadherins are non-functional. In order to investigate the molecular basis of this, computational molecular dynamics simulations were performed to predict the differences in structure and dynamics of a cadherin protein bound to Ca2+ or Tb3+. Simulations were carried out using the crystal structure of the full ectodomain of E-cadherin (PDB 3Q2V) resolved to 3.4 Å.31 Three scenarios based on this E-cadherin were simulated: one with the 12 crystallographically resolved Ca2+ ions (Ca-Ecad), one with the 12 Ca2+ ions replaced with Tb3+ ions (Tb-Ecad) and one with the 12 Ca2+ ions removed (Apo-Ecad).The replacement of Ca2+ with Tb3+ resulted in a re-structuring of the cadherin linker regions, particularly in the charged residues in proximity to the metal ions (Fig. 4A and B). This is unsurprising given the greater electropositivity of Tb3+ ions and that Tb3+ ions typically exhibit higher coordination numbers within proteins.16,29,31 However, due to the resolution of the molecular dynamics simulations, it is not possible to comment on the exact coordination geometry and coordinating ligands of the metal ions. As expected, removal of Ca2+ in Apo-Ecad caused significant structural changes in the linker region as negative residues presumably undergo electrostatic repulsion (Fig. 4C).
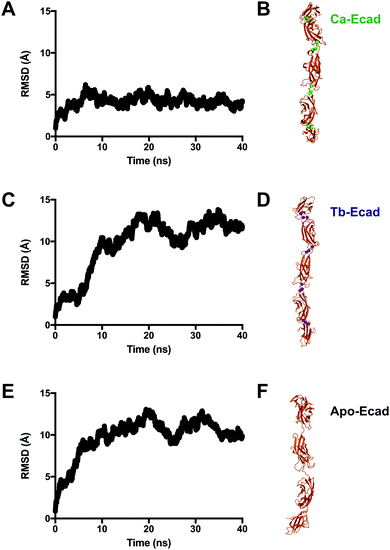
Root mean-square deviations (RMSD) were used to determine how much the theoretical E-cadherin tertiary structure changed over a simulation and the RMSD was calculated for Ca-Ecad, Tb-Ecad and Apo-Ecad (Fig. 5). As observed in previous studies with other cadherin ectodomains, the E-cadherin ectodomain experienced a higher RMSD, and thus higher structural fluctuation, when in the absence of Ca2+ (Apo-Ecad, RMSD average = 9.82 Å) compared when in the presence of Ca2+ (Ca-Ecad, RMSD average = 4.19 Å).33,34,46 These results are also consistent with structural studies which demonstrate the rigidification of cadherin ectodomains upon Ca2+ binding.29,30 Interestingly, the E-cadherin ectodomain in the presence of Tb3+ (Tb-Ecad, RMSD average = 9.74 Å) had a high overall RMSD that was similar to Apo-Ecad ectodomains, indicating a high level of structural flexibility. It is important to note that, like Ca2+ ions, all Tb3+ ions remained bound within the Ca2+-binding sites over the course of the simulation.
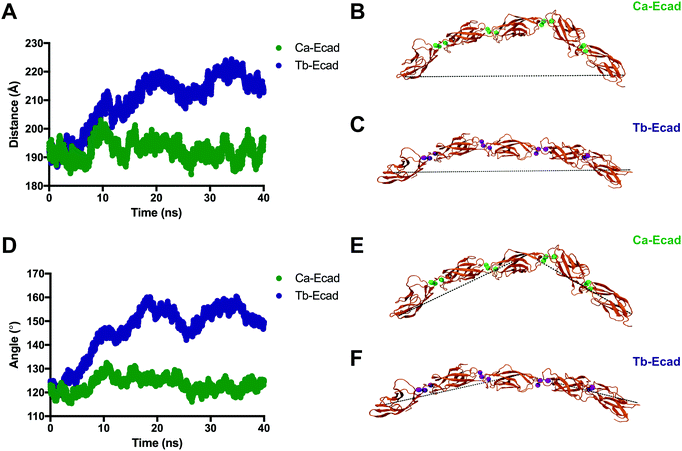
In order to compare how the structure and dynamics of the E-cadherin ectodomain differ in the presence of Ca2+ and Tb3+, the end-to-end distance and curvature of the ectodomains were measured (Fig. 6). The distance between α-Carbon atoms of two distal residues, Glu31 and Arg451, was measured for both scenarios over the 40 ns simulation. It was found that the E-cadherin ectodomain had a longer average end-to-end distance when in presence of Tb3+ compared with Ca2+ (209.6 Å vs. 192.6 Å). The curvatures of the ectodomains were compared by measuring the angle made between α-Carbon atoms of distal residues Glu31 and Arg451, and the residue that was closest to the geometric centre of EC3, Leu282. The curvature angle of the E-cadherin ectodomain with Tb3+ was also found to be larger than that for the ectodomain with Ca2+ (146.1° vs. 123.4°). Together these results suggest that the binding of Tb3+ leads to significant structural changes in the E-cadherin ectodomain and results in elongated structures with more shallow curvature compared with Ca2+-bound E-cadherin ectodomains. Furthermore, the Tb-Ecad ectodomain had greater fluctuation in both the end-to-end distance (Tb-Ecad, SD = 9.32 Å vs. Ca-Ecad, SD = 3.30 Å) and curvature angle (Tb-Ecad, SD = 10.66° vs. Ca-Ecad, SD = 3.18°), which indicates greater structural flexibility in the Tb3+-bound ectodomain. The outcomes from the simulations agree well with our experimental data, which demonstrated that Tb3+-bound cadherins cannot mimic the functional conformation of Ca2+-bound cadherins.
Our molecular dynamics simulations were used here to explore the impact of metal ion chemistry on cadherin structure and dynamics and provide atomistic explanations for our experimental findings. Further experimental work, such as crystallography or NMR, would be necessary to validate these computational predictions.
Discussion
The present study shows that Tb3+ inhibits cadherin-mediated cell adhesion and our evidence suggests that this is a result of competitive binding at cadherin Ca2+-binding sites. Cadherins require Ca2+-binding at all 12 sites in order to achieve a functional conformation and loss of Ca2+-binding at a single site is sufficient to disrupt cadherin-mediated cell adhesion.36,47 Here, we demonstrate that Tb3+ functions as a competitive antagonist in Ca2+-dependent cadherin-mediated cell aggregation assays and our trypsin resistance assays suggest that Tb3+ competes with Ca2+ for binding to cadherins. Furthermore, Tb3+ is a well-documented mimic of Ca2+ in biological processes and our molecular dynamics simulations show that Tb3+ remained bound within Ca2+-binding sites throughout the simulations. This is in contrast to Na+ and K+, which have no experimental evidence of binding to cadherins and these ions become unbound from cadherin Ca2+-binding sites during simulations.33 The Tb3+ FRET experiments additionally provide biophysical evidence for Tb3+ binding to cadherins and suggest that Tb3+ ions compete for the same sites as Ca2+ ions.Results from the fluorescence competition assay demonstrate that not all the observed Tb3+ fluorescence can be attributed to Tb3+ binding at Ca2+-binding sites. This might be an artefact of Tb3+ autofluorescence or potentially the result of Tb3+ binding to E-cadherin at sites other than Ca2+-binding sites. Whether Tb3+ binding at non-Ca2+-binding sites is possible and what effect this might have on cadherin structure remains to be determined.
It is not possible to determine at how many or at which Ca2+-binding sites Tb3+ ions can bind to. Only 5 out of the 12 Ca2+-binding sites in human E-cadherin are potentially reportable by FRET, and of these 5 sites one cannot accurately calculate how binding at each site would contribute to the Tb3+ fluorescence signal. For the same reasons, it is unknown how many Tb3+ ions could bind each E-cadherin molecule at non-Ca2+-binding sites. There might only be a very small amount of Tb3+ binding to non-Ca2+-binding sites but this binding might contribute to a large amount of the observed Tb3+ fluorescence, which is dependent on the distances between Tb3+ ions and tryptophan and tyrosine residues. Structural determination studies would be required to conclude exactly where and how Tb3+ binds to cadherins.
The ratio of [Tb3+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [Ca2+] required to inhibit cell adhesion and cadherin trypsin protection suggests that Ca2+ has a higher binding affinity than Tb3+ to cadherins. The competition fluorescence assay additionally suggests this as ∼60 μM Ca2+ had a half-maximal effect in reducing Tb3+ fluorescence in E-cadherin when in the presence of 170 μM Tb3+. A difference in binding affinity does not, however, fully explain why a steep dose–response relationship was observed for the cellular assays: no effect was observed at a [Tb3+]
[Ca2+] required to inhibit cell adhesion and cadherin trypsin protection suggests that Ca2+ has a higher binding affinity than Tb3+ to cadherins. The competition fluorescence assay additionally suggests this as ∼60 μM Ca2+ had a half-maximal effect in reducing Tb3+ fluorescence in E-cadherin when in the presence of 170 μM Tb3+. A difference in binding affinity does not, however, fully explain why a steep dose–response relationship was observed for the cellular assays: no effect was observed at a [Tb3+]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [Ca2+] ratio of 1
[Ca2+] ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 but effective inhibition was seen at a ratio of 2
1 but effective inhibition was seen at a ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. One explanation for these results is the potential binding of Tb3+ to both Ca2+- and non-Ca2+-binding sites on cadherins. At a ratio of 1
1. One explanation for these results is the potential binding of Tb3+ to both Ca2+- and non-Ca2+-binding sites on cadherins. At a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Tb3+ might bind elsewhere on cadherins as Ca2+ occupies the Ca2+-binding sites, but at a ratio of 2
1, Tb3+ might bind elsewhere on cadherins as Ca2+ occupies the Ca2+-binding sites, but at a ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Tb3+ may be at a sufficiently high concentration to outcompete Ca2+ for the Ca2+-binding sites and thus inhibit the Ca2+-dependent cadherin processes.
1, Tb3+ may be at a sufficiently high concentration to outcompete Ca2+ for the Ca2+-binding sites and thus inhibit the Ca2+-dependent cadherin processes.
It would be unusual if Tb3+ had a weaker binding affinity than Ca2+ to cadherins. Tb3+ typically binds more strongly to Ca2+-binding proteins, such as calmodulin, due to the greater electropositivity of Tb3+.16,17 One possible explanation for the apparent higher binding affinity of Ca2+ to cadherins is that Ca2+ binds to the 12 sites on cadherins with a high level of coorperativity.35,36,48 Binding of Ca2+ to one Ca2+-binding site induces specific structural changes that enhances the binding affinity of Ca2+ to the other sites on a cadherin molecule. Given that the extensive cooperativity of Ca2+ binding requires a precise network of molecular interactions across 12 binding sites, it is plausible that slight differences in metal chemistry result in Tb3+ being unable to mimic the cooperative binding of Ca2+ to cadherins. Results from the Tb3+ FRET assay suggest that Tb3+ binds to E-cadherin with negative cooperativity (KD = 143 μM, hill coefficient = 0.69), however values from this experiment should be treated with caution, as they are not specific for Tb3+ binding to only Ca2+-binding sites. Overall, it is possible that Ca2+-cooperative binding may be essential for cadherin's preferential binding of Ca2+ over other ions. This may be one mechanism by which cadherins maintain robust function in stochastic biological environments.
Upon Tb3+ binding to cadherins, data from trypsin protection assays indicate that cadherins differ in structure and/or conformational flexibility to Ca2+-bound cadherins. Molecular dynamics simulations support these findings and suggest that Tb3+-bound cadherins have a more elongated, less curved structure that exhibits enhanced structural fluctuations. These changes in tertiary structure are likely responsible for the inability of Tb3+-bound cadherins to support cell adhesion. Interestingly, a recent study found that binding of Tb3+ to another Ca2+-dependent protein, calmodulin, resulted in enhanced structural flexibility within Ca2+-binding sites and this was caused by the more electropositive Tb3+ adopting an alternative and more disordered coordination geometry.49 In the case of cadherins, changes in coordination geometry and flexibility within binding sites may also be the chemical driving forces behind global differences in Tb3+-bound and Ca2+-bound molecules. However, unlike in calmodulin, cadherins are non-functional upon binding to Tb3+, which provides further insight into the high level of sensitivity of cadherins to Ca2+ chemistry.50,51 Overall, it is fascinating that seemingly slight differences in the chemistry between Tb3+ and Ca2+ can significantly influence the structure, dynamics and function of cadherin proteins. Follow-on structural determination studies would be invaluable in providing conclusive evidence for how substitution of Ca2+ with Tb3+ affects the tertiary structure of cadherins and interactions within the binding sites.
We also show that Gd3+ inhibits cadherin adhesion in a similar Ca2+-competing manner and there is some evidence of La3+ competing with Ca2+ for binding to cadherins.52 Many lanthanide ions are known to chemically mimic Ca2+ and the Ca2+-binding pockets of classical cadherins are highly conserved, thus one would predict that the interaction elucidated here is universal between lanthanides and classical cadherins.16,53
Conclusion
Cadherin adhesions have widespread roles in physiology and there is a growing appreciation for how the interaction of cadherins with non-Ca2+ ions can regulate cadherin function. For example, Zn2+ ions have been shown to bind and inhibit N-cadherin binding and this interaction is believed to play a regulatory role in synaptic plasticity.54,55 Here, the lanthanide Tb3+, which has applications in protein spectroscopy and cancer therapy, was found to compete with Ca2+ for binding to cadherins. We show that Tb3+-bound cadherins are unable to support cell adhesion and demonstrate this is a result of differences in the structure and dynamics of cadherins following Ca2+- and Tb3+-binding. Unlike in other Ca2+-dependent proteins, Tb3+ cannot functionally-replace Ca2+ in cadherins and our work provides new insight into the sensitivity of cadherin structure, dynamics and function to the chemistry of Ca2+. Furthermore, our characterisation of the interaction between lanthanides and cadherins should be of value as lanthanides continue to be used in biology and medicine.Abbreviations
| Ca2+ | Calcium |
| Tb3+ | Terbium |
| Gd3+ | Gadolinium |
| EC | Extracellular domain |
| FRET | Fluorescence resonance energy transfer |
| N-CHO | N-Cadherin expressing CHO cell |
| E-CHO | E-Cadherin expressing CHO cell |
| aa | Amino acids |
| RMSD | Root mean square deviation |
| SEM | Standard error of the mean |
Financial support
This work was funded by the BBSRC, EPSRC and Wellcome Trust of the UK.Conflicts of interest
The authors indicate no potential conflicts of interest.Acknowledgements
We thank Dr Anwen Brown, Dr Shaline Fazal and Dr Tina Daviter for technical assistance.References
- E. Blumfield, M. M. Moore, M. K. Drake, T. R. Goodman, K. N. Lewis, L. T. Meyer, T. D. Ngo, C. Sammet, A. L. Stanescu, D. W. Swenson, T. L. Slovis and R. S. Iyer, Survey of gadolinium-based contrast agent utilization among the members of the Society for Pediatric Radiology: a Quality and Safety Committee report, Pediatr. Radiol., 2017, 47, 665–673 CrossRef PubMed.
- M. Bottrill, L. Kwok and N. J. Long, Lanthanides in magnetic resonance imaging, Chem. Soc. Rev., 2006, 35, 557–571 RSC.
- P. Caravan, J. J. Ellison, T. J. McMurry and R. B. Lauffer, Gadolinium(III) Chelates as MRI Contrast Agents: Structure, Dynamics, and Applications, Chem. Rev., 1999, 99, 2293–2352 CrossRef CAS PubMed.
- Z. Zhou and Z.-R. Lu, Gadolinium-Based Contrast Agents for MR Cancer Imaging, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5, 1–18 CrossRef CAS.
- S. Linden, M. K. Singh, K. D. Wegner, M. Regairaz, F. Dautry, F. Treussart and N. Hildebrandt, Terbium-based time-gated Förster resonance energy transfer imaging for evaluating protein-protein interactions on cell membranes, Dalton Trans., 2015, 44, 4994–5003 RSC.
- S. P. Fricker, The therapeutic application of lanthanides, Chem. Soc. Rev., 2006, 35, 524–533 RSC.
- A. J. Hutchison, R. J. Wilson, S. Garafola and J. B. Copley, Lanthanum carbonate: safety data after 10 years, Nephrology, 2016, 21, 987–994 CrossRef CAS PubMed.
- I. Kostova, Lanthanides as anticancer agents, Curr. Med. Chem.: Anti-Cancer Agents, 2005, 5, 591–602 CrossRef CAS PubMed.
- J. L. Cox, T. Lancaster and C. G. Carlson, Changes in the motility of B16F10 melanoma cells induced by alterations in resting calcium influx, Melanoma Res., 2002, 12, 211–219 CrossRef CAS PubMed.
- Y. Dai, J. Li, J. Li, L. Yu, G. Dai, A. Hu, L. Yuan and Z. Wen, Effects of rare earth compounds on growth and apoptosis of leukemic cell lines, In Vitro Cell. Dev. Biol.: Anim., 2002, 38, 373–375 CrossRef CAS.
- P. Shi and Z. Huang, Proteomic detection of changes in protein synthesis induced by lanthanum in BGC-823 human gastric cancer cells, Biometals, 2005, 18, 89–95 CrossRef CAS PubMed.
- W. D. Horrocks and D. R. Sudnick, Lanthanide Ion Probes of Structure in Biology. Laser- Induced Luminescence Decay Constants Provide a Direct Measure of the Number of Metal-Coordinated Water Molecules, J. Am. Chem. Soc., 1979, 101, 334–340 CrossRef CAS.
- A. K. Katz, J. P. Glusker, S. A. Beebe and C. W. Bock, Calcium ion coordination: A comparison with that of beryllium, magnesium, and zinc, J. Am. Chem. Soc., 1996, 118, 5752–5763 CrossRef CAS.
- R. D. Shannon, Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides, Acta Crystallogr., Sect. A: Cryst. Phys., Diffr., Theor. Gen. Crystallogr., 1976, 32, 751–767 CrossRef.
- E. E. Snyder, B. W. Buoscio and J. J. Falke, Calcium(II) site specificity: effect of size and charge on metal ion binding to an EF-hand-like site, Biochemistry, 1990, 29, 3937–3943 CrossRef CAS PubMed.
- E. Pidcock and G. R. Moore, Structural characteristics of protein binding sites for calcium and lanthanide ions, J. Biol. Inorg. Chem., 2001, 6, 479–489 CrossRef CAS PubMed.
- S. M. Mustafi, S. Mukherjee, K. V. R. Chary, C. Del Bianco and C. Luchinat, Energetics and Mechanism of Ca2+ Displacement by Lanthanides in a Calcium Binding Protein, Biochemistry, 2004, 43, 9320–9331 CrossRef CAS PubMed.
- B. C. Furie, K. G. Mann and B. Furie, Substitution of lanthanide ions for calcium in the activation of bovine prothrombin by activated factor X: high affinity metal binding sites, J. Biol. Chem., 1976, 251, 3235–3241 CAS.
- A. D. Sherry, A. D. Newman and C. G. Gutz, The activation of concanavalin A by lanthanide ions, Biochemistry, 1975, 14, 2191–2196 CrossRef CAS PubMed.
- S. T. Chu and Y. H. Chen, The intrinsic tryptophan fluorescence of beta 1-bungarotoxin and the Ca2+ -binding domains of the toxin as probed with Tb3+ luminescence., Biochem. J., 1989, 262, 773–779 CrossRef CAS PubMed.
- L. C. Adding, G. L. Bannenberg and L. E. Gustafsson, Basic experimental studies and clinical aspects of gadolinium salts and chelates, Cardiovasc. Drug Rev., 2001, 19, 41–56 CrossRef CAS PubMed.
- C. R. Halaszovich, C. Zitt, E. Jungling and A. Luckoff, Inhibition of TRP3 channels by lanthanides. Block from the cytosolic side of the plasma membrane, J. Biol. Chem., 2000, 275, 37423–37428 CrossRef CAS PubMed.
- Y. Wu, Lanthanide ions inhibit the activity of dihydrofolate reductase from chicken liver, Biometals, 2000, 13, 195–201 CrossRef CAS PubMed.
- M. Astick, K. Tubby, W. M. Mubarak, S. Guthrie and S. R. Price, Central topography of cranial motor nuclei controlled by differential cadherin expression, Curr. Biol., 2014, 24, 2541–2547 CrossRef CAS PubMed.
- L. L. Brayshaw and S. R. Price, in The Cadherin Superfamily: Key Regulators of Animal Development and Physiology, ed. T. S. Suzuki and S. Hirano, Springer, Japan, Tokyo, 2016, pp. 315–340 Search PubMed.
- H. G. Munshi, S. Ghosh, S. Mukhopadhyay, Y. I. Wu, R. Sen, K. J. Green and M. S. Stack, Proteinase suppression by E-cadherin-mediated cell-cell attachment in premalignant oral keratinocytes, J. Biol. Chem., 2002, 277, 38159–38167 CrossRef CAS PubMed.
- A. Jeanes, C. Gottardi and A. Yap, Cadherins and cancer: how does cadherin dysfunction promote tumor progression?, Oncogene, 2008, 27, 6920–6929 CrossRef CAS PubMed.
- D. Leckband and A. Prakasam, Mechanism and dynamics of cadherin adhesion, Annu. Rev. Biomed. Eng., 2006, 8, 259–287 CrossRef CAS PubMed.
- O. Pertz, D. Bozic, A. W. Koch, C. Fauser, A. Brancaccio and J. Engel, A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation, EMBO J., 1999, 18, 1738–1747 CrossRef CAS PubMed.
- S. Pokutta, K. Herrenknecht, R. Kemler and J. Engel, Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding, Eur. J. Biochem., 1994, 223, 1019–1026 CrossRef CAS PubMed.
- O. J. Harrison, X. Jin, S. Hong, F. Bahna, G. Ahlsen, J. Brasch, Y. Wu, J. Vendome, K. Felsovalyi, C. M. Hampton, R. B. Troyanovsky, A. Ben-Shaul, J. Frank, S. M. Troyanovsky, L. Shapiro and B. Honig, The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins, Structure, 2011, 19, 244–256 CrossRef CAS PubMed.
- T. J. Boggon, J. Murray, S. Chappuis-Flament, E. Wong, B. M. Gumbiner and L. Shapiro, Science, 2002, 296, 1308–1313 CrossRef CAS PubMed.
- M. Sotomayor and K. Schulten, The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin, Biophys. J., 2008, 94, 4621–4633 CrossRef CAS PubMed.
- J. Oroz, A. Valbuena, A. M. Vera, J. Mendieta, P. Gomez-Puertas and M. Carrion-Vazquez, Nanomechanics of the cadherin ectodomain: ‘Canalization’ by Ca2+ binding results in a new mechanical element, J. Biol. Chem., 2011, 286, 9405–9418 CrossRef CAS PubMed.
- O. Courjean, G. Chevreux, E. Perret, A. Morel, S. Sanglier, N. Potier, J. Engel, A. Van Dorsselaer and H. Feracci, Modulation of E-cadherin monomer folding by cooperative binding of calcium ions, Biochemistry, 2008, 47, 2339–2349 CrossRef CAS PubMed.
- A. Prakasam, Y. H. Chien, V. Maruthamuthu and D. E. Leckband, Calcium site mutations in cadherin: impact on adhesion and evidence of cooperativity, Biochemistry, 2006, 45, 6930–6939 CrossRef CAS PubMed.
- D. A. Case, R. M. Betz, W. Botello-Smith, D. S. Cerutti, T. E. Cheatham, III, T. A. Darden, R. E. Duke, T. J. Giese, H. Gohlke, A. W. Goetz, N. Homeyer, S. Izadi, P. Janowski, J. Kaus, A. Kovalenko, T. S. Lee, S. LeGrand, P. Li, C. Lin, T. Luchko, R. Luo and B. Madej, AMBER 2016, University of California, San Francisco, 2016 Search PubMed.
- L. Schrödinger, The PyMOL Molecular Graphics System, Version 1.8, 2015 Search PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79, 926 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, VMD-Visual Molecular Dynamics, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- J. Eargle, D. Wright and Z. Luthey-Schulten, Multiple Alignment of protein structures and sequences for VMD, Bioinformatics, 2006, 22, 504–506 CrossRef CAS PubMed.
- M. Takeichi, Functional correlation between cell adhesive properties and some cell surface proteins, J. Cell Biol., 1977, 75, 464–474 CrossRef CAS PubMed.
- M. Takeichi, Cadherins: a molecular family important in selective cell-cell adhesion, Annu. Rev. Biochem., 1990, 59, 237–252 CrossRef CAS PubMed.
- L. Hughes, C. Malone, S. Chumsri, A. M. Burger and S. McDonnell, Characterisation of breast cancer cell lines and establishment of a novel isogenic subclone to study migration, invasion and tumourigenicity, Clin. Exp. Metastasis, 2008, 25, 549–557 CrossRef PubMed.
- S. F. Altschul, T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res., 1997, 25, 3389–3402 CrossRef CAS PubMed.
- F. Cailliez and R. Lavery, Cadherin mechanics and complexation: the importance of calcium binding, Biophys. J., 2005, 89, 3895–3903 CrossRef CAS PubMed.
- M. Ozawa, J. Engel and R. Kemler, Single amino acid substitutions in one Ca2+ binding site of uvomorulin abolish the adhesive function, Cell, 1990, 63, 1033–1038 CrossRef CAS PubMed.
- J. R. Alattia, J. B. Ames, T. Porumb, K. I. Tong, Y. M. Heng, P. Ottensmeyer, C. M. Kay and M. Ikura, Lateral self-assembly of E-cadherin directed by cooperative calcium binding, FEBS Lett., 1997, 417, 405–408 CrossRef CAS PubMed.
- S. C. Edington, A. Gonzalez, T. R. Middendorf, D. B. Halling, R. W. Aldrich and C. R. Baiz, Coordination to lanthanide ions distorts binding site conformation in calmodulin, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 201722042 CrossRef PubMed.
- R. W. Wallace, E. A. Tallant, M. E. Dockter and W. Y. Cheung, Calcium Binding Domains of Calmodulin, J. Biol. Chem., 1982, 257, 1845–1854 CAS.
- W. Li and R. W. Aldrich, Activation of the SK potassium channel-calmodulin complex by nanomolar concentrations of terbium, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 1075–1080 CrossRef CAS PubMed.
- F. Hyafil, C. Babinet and F. Jacob, Cell-cell interactions in early embryogenesis: a molecular approach to the role of calcium., Cell, 1981, 26, 447–454 CrossRef CAS PubMed.
- L. Shapiro and W. I. Weis, Structure and biochemistry of cadherins and catenins, Cold Spring Harbor Perspect. Biol., 2009, 1, a003053 Search PubMed.
- E. Heiliger, A. Osmanagic, H. Haase, N. Golenhofen, A. M. Grabrucker, A. Weth and W. Baumgartner, N-cadherin-mediated cell adhesion is regulated by extracellular Zn2+, Metallomics, 2015, 7, 355–362 RSC.
- C. Wolf, A. Weth, S. Walcher, C. Lax and W. Baumgartner, Modeling of Zinc Dynamics in the Synaptic Cleft: Implications for Cadherin Mediated Adhesion and Synaptic Plasticity, Front. Mol. Neurosci., 2018, 11, 1–13 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8mt00317c |
| This journal is © The Royal Society of Chemistry 2019 |