Engineering human islet organoids from iPSCs using an organ-on-chip platform†
Tingting
Tao
ad,
Yaqing
Wang
ad,
Wenwen
Chen
ad,
Zhongyu
Li
a,
Wentao
Su
a,
Yaqiong
Guo
ad,
Pengwei
Deng
ad and
Jianhua
Qin
 *abcd
*abcd
aDivision of Biotechnology, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, 457 Zhongshan Road, Dalian 116023, China. E-mail: jhqin@dicp.ac.cn
bInstitute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing, China
cCAS Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, Shanghai, China
dUniversity of Chinese Academy of Sciences, Beijing, China
First published on 23rd January 2019
Abstract
Human pluripotent stem cell (hPSC)-derived islet cells provide promising resources for diabetes studies, cell replacement treatment and drug screening. Recently, hPSC-derived organoids have represented a new class of in vitro organ models for disease modeling and regenerative medicine. However, rebuilding biomimetic human islet organoids from hPSCs remains challenging. Here, we present a new strategy to engineer human islet organoids derived from human induced pluripotent stem cells (hiPSCs) using an organ-on-a-chip platform combined with stem cell developmental principles. The microsystem contains a multi-layer microfluidic device that allows controllable aggregation of embryoid bodies (EBs), in situ pancreatic differentiation and generation of heterogeneous islet organoids in parallel under perfused 3D culture in a single device. The generated islet organoids contain heterogeneous islet-specific α and β-like cells that exhibit favorable growth and cell viability. They also show enhanced expression of pancreatic β-cell specific genes and proteins (PDX1 and NKX6.1) and increased β-cell hormone specific INS gene and C-peptide protein expressions under perfused 3D culture conditions compared to static cultures. In addition, the islet organoids exhibit more sensitive glucose-stimulated insulin secretion (GSIS) and higher Ca2+ flux, indicating the role of biomimetic mechanical flow in promoting endocrine cell differentiation and maturation of islet organoids. This islet-on-a-chip system is robust and amenable to real-time imaging and in situ tracking of islet organoid growth, which may provide a promising platform for organoid engineering, disease modeling, drug testing and regenerative medicine.
Introduction
Diabetes mellitus caused by damaged pancreatic islets leads to an increase in morbidity and mortality. It is characterized by hyperglycemia due to insufficient insulin secretion resulting from the destruction of pancreatic islet cells.1,2 Human pluripotent stem cells (hPSCs) make it possible to derive an unlimited source of insulin-producing pancreatic β cells for disease studies and cellular replacement therapy due to their self-renewal and differentiation capability.3–6 Recent studies have demonstrated the feasibility of in vitro generation of functional islet β-cells with insulin secretion from hPSCs.7 Most of the existing approaches for the differentiation of hPSCs into pancreatic endocrine β-cells are based on a series of sequential chemicals or factors in a monolayer culture system;7,8 however, it is unlikely that these β-cells will represent the multicellular components and complex biology of pancreatic islets involved in human islet function.Recently, hPSC-derived organoids have represented a new class of in vitro organ models for development biology studies, disease modeling and regenerative medicine. Breakthroughs in stem cell organoids have facilitated high-fidelity modeling of the original tissue or organ via self-renewal and self-organization, albeit on a much smaller scale compared to human organs in vivo.9 These organoids can recapitulate the organ-specific functions, cellular architecture and morphologies.10–12 The remarkable advances in organoid technology have led to the successful generation of diverse human organoids in 3D culture, such as the brain,13 liver14 and intestines.15 Presently, several studies have attempted to generate islet-like organoids from hPSCs in the forms of spheroid, microcapsule and scaffold cultures;16–18 however, pancreatic islet organogenesis in vivo is tightly regulated by complex and dynamic cellular niche signals involving blood flow, transcription factors, and multicellular interactions, thereby yielding functional islets responsible for glucose homeostasis.19 Moreover, blood flow as a source of developmental signals plays an important role in pancreatic differentiation and functional maturation.20–22 As such, there is a critical need for a 3D dynamic culture and biomimetic platforms that allow the integration of supporting heterogeneous cell populations and facilitate islet function maturation.
Advances in organ-on-a-chip technologies have offered unprecedented opportunities to rebuild in vivo-like human tissues or organs (such as the lungs, kidneys and intestines) by mimicking the multicellular architecture, key tissue–tissue interfaces and relevant physical microenvironment of the functional unit of living organs.23–25 Compared to the conventional monolayer culture, organ-on-chip platforms have significant advantages in providing dynamic 3D culture and offering biomimetic cellular or tissue microenvironments by accurate control over the mechanical fluid flow, biochemical signals, cell–cell interactions, etc. Recently, the integration of organ-on-a-chip and microfabricated approaches with hPSC technologies has emerged as an attractive and promising 3D culture platform for synergistic engineering of organ-specific stem cell organoids, such as brain organoids and liver organoids, for various applications.26–30
Here, we present a new and robust strategy for creating heterogeneous human islet organoids derived from hiPSCs using an organ-on-chip platform combined with stem cell developmental principles. The perfusable microfluidic chip on this platform consists of a multilayer configuration, including parallel 3D culture chambers, a porous membrane and fluid channel network layers, which enables the formation of controllable EBs from hiPSCs, in situ pancreatic differentiation and generation of 3D islet organoids in parallel on a single device. The generated islet organoids exhibited heterogeneous islet-like components and functionalities. The expressions of pancreatic islet-specific genes and proteins in islet organoids under static or perfused culture conditions were identified and evaluated. The glucose-stimulated insulin secretion and Ca2+ flux in islet organoids were also examined.
Materials and methods
Design and fabrication of the microfluidic chip device
The microfluidic organ-on-a-chip device was designed and fabricated using standard soft lithography and micromolding methods.31 The device was made of PDMS (Sylgard 184, Dow Corning, U.S.) at a weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and was cured at 80 °C for 1 to 2 h in an oven. The microfluidic device with a multi-layer configuration consisted of four layers, including a top polydimethylsiloxane (PDMS) layer, through-hole PDMS layer (500 μm pore size), polycarbonate porous membrane (8 μm pore) and bottom PDMS layer, followed by specific binding between different layers. The top and bottom layers were fabricated with one chamber (1 mm height, 5 mm width and 10 mm length) and one inlet and outlet for medium/cell infusion, respectively. After fabrication of different layers, the top layer was adhered to the through-hole PDMS membrane using oxygen plasma treatment. The bottom layer was adhered to the polycarbonate porous membrane using oxygen plasma treatment as well. Then, the through-hole PDMS membrane and polycarbonate porous membrane were sealed with PDMS glue to form an array of micro-well structures for the cell culture. Before the experiments, the device was placed in water and sterilized in an autoclave.
1 and was cured at 80 °C for 1 to 2 h in an oven. The microfluidic device with a multi-layer configuration consisted of four layers, including a top polydimethylsiloxane (PDMS) layer, through-hole PDMS layer (500 μm pore size), polycarbonate porous membrane (8 μm pore) and bottom PDMS layer, followed by specific binding between different layers. The top and bottom layers were fabricated with one chamber (1 mm height, 5 mm width and 10 mm length) and one inlet and outlet for medium/cell infusion, respectively. After fabrication of different layers, the top layer was adhered to the through-hole PDMS membrane using oxygen plasma treatment. The bottom layer was adhered to the polycarbonate porous membrane using oxygen plasma treatment as well. Then, the through-hole PDMS membrane and polycarbonate porous membrane were sealed with PDMS glue to form an array of micro-well structures for the cell culture. Before the experiments, the device was placed in water and sterilized in an autoclave.
HiPSC culture
Undifferentiated iPSC lines (donated by Dr. Ning Sun) were kept on Matrigel-treated six-well plates (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 dilution, BD) in mTeSR1 medium (StemCell) at 37 °C in a humidified incubator with 5% CO2, with the fresh medium being changed daily. When about 80% to 90% confluence was reached, the iPSC colonies were passaged at a 1
50 dilution, BD) in mTeSR1 medium (StemCell) at 37 °C in a humidified incubator with 5% CO2, with the fresh medium being changed daily. When about 80% to 90% confluence was reached, the iPSC colonies were passaged at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 ratio by digestion with Accutase (Sigma) for experiments.
6 ratio by digestion with Accutase (Sigma) for experiments.
Embryoid body formation and differentiation of islet organoids
Islet organoids were generated by initial embryoid body (EB) formation followed by pancreatic induction differentiation. The differentiation protocols were followed by a previous report32 after modification. Dissociated hiPSCs resuspended in mTeSR1 medium with 10 μM Y27632 (Selleck) are infused into the microfluidic chip via inlets. EBs are spontaneously differentiated and formed by iPSCs in the device with microwell configurations after 12 h culture.After the EB formation, the medium was replaced with definitive endoderm (DE) induction medium containing basal DMEM/F12 (Invitrogen), 1% KSR (Invitrogen), 1% B27 supplement (Invitrogen), 50 ng ml−1 activin-A (PeproTech), 1% GlutaMAX (Invitrogen) and 1% penicillin–streptomycin (Sigma) for 5 days. For pancreatic endoderm (PE) differentiation, the medium containing DMEM (Invitrogen), 0.5% B27 supplement (Invitrogen), 2 μM dorsomorphin (Selleck), 2 μM retinoic acid (RA, Sigma), 10 μM SB431542 (Selleck), 5 ng ml−1 basic fibroblast growth factor (bFGF; R&D systems), and 250 nM SANT-1 (Selleck) was used to culture EBs for 6 days. Then, to facilitate endocrine progenitor (EP) cell differentiation, the medium was replaced with DMEM containing 0.5% B27 supplement (Invitrogen), 2 μM dorsomorphin (Selleck), 10 μM SB431542 (Selleck), 50 μg ml−1 ascorbic acid (Sigma), and 10 μM DAPT (Abcam Biochemicals). After day 15, the islet organoids were cultured in CMRL 1066 (Invitrogen) containing 25 mM glucose (Sigma), 0.5% B27 supplement (Invitrogen), 10 mM nicotinamide (Sigma), 10 μM SB431542 (Selleck), 50 μg ml−1 ascorbic acid (Sigma), 2 μM dorsomorphin (Selleck), and 0.5% penicillin–streptomycin (Invitrogen) for a long-term culture.
For the generation of islet organoids under the dynamic perfusable system, the culture medium was continuously injected to the medium channels in order to provide nutrients for the long-term culture. Different flow rates (50, 100 and 200 μl h−1) were tested in the experiment. The flow rate of 100 μl h−1 was selected as the optimized condition in the following experiment. In addition, the organoids were cultured on a Petri dish under static conditions using the same medium as a control group.
Immunohistochemistry
At the end of the differentiation period, islet organoids were collected and fixed in 4% paraformaldehyde (PFA) and then washed 3 times with PBS at room temperature (RT). Tissues were permeabilized in 0.2% Triton X-100 (Sigma) for 15 min and then washed three times with PBS. Next, islet spheroids were blocked in a blocking solution containing 10% goat serum for 2 h and then incubated with primary antibodies at 4 °C for 12 h. The antibodies used in the experiment were as follows: E-cadherin (mouse, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, CST 14472s), PDX1 (rabbit, 1
50, CST 14472s), PDX1 (rabbit, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, CST D59H3), NKX6.1 (rabbit, 1
1000, CST D59H3), NKX6.1 (rabbit, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250, CST D804R), insulin (rabbit, 1
250, CST D804R), insulin (rabbit, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250, CST C27C9), C-peptide (rabbit, 1
250, CST C27C9), C-peptide (rabbit, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800, Abcam ab14181), and GCG (rabbit, 1
800, Abcam ab14181), and GCG (rabbit, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400, Bioss bs-3796R). The spheroids were washed three times with PBS and then incubated with Alexa Fluor 488- or 594-conjugated secondary antibodies (1
400, Bioss bs-3796R). The spheroids were washed three times with PBS and then incubated with Alexa Fluor 488- or 594-conjugated secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) at RT for 1 h. DAPI (1
500) at RT for 1 h. DAPI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000, CST 4083) was used to stain the cell nuclei. Z-stack images of spheroids starting at the apex of each organoid were collected at 10 μm intervals using a confocal microscope.
4000, CST 4083) was used to stain the cell nuclei. Z-stack images of spheroids starting at the apex of each organoid were collected at 10 μm intervals using a confocal microscope.
Real-time quantitative PCR
To detect the gene expression level, mRNA was extracted from the whole islet organoids using a Trizol reagent (TAKARA) and the final concentration was adjusted to 250 ng μl−1. The cDNA was then synthesized using reverse transcription polymerase chain reaction (RT-PCR) (TAKARA) according to the manufacturer's protocol. Real-time PCR was performed using SYBR Green (TAKARA) under the following reaction conditions (40 cycles): denaturation at 95 °C for 1 min, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. The primer pairs used were as follows: SOX17 forward: 5′-CAG AAT CCA GAC CTG CAC AA-3′, reverse: 5′-GCG GCC GGT ACT TGT AGT T-3′; FOXA2 forward: 5′-AAC AAG ATG CTG ACG CTG AG-3′, reverse: 5′-CAG GAA ACA GTC GTT GAA GG-3′; GATA4 forward: 5′-TCC AAA CCA GAA AAC GGA AG-3′, reverse: 5′-CTG TGC CCG TAG TGA GAT GA-3′; HNF1β forward: 5′-AGC CCA CCA ACA AGA AGA TG-3′, reverse: 5′-CAT TCT GCC CTG TTG CAT TC-3′; PDX1 forward: 5′-GTT CCG AGG TAG AGG CTG TG-3′, reverse: 5′-AAC ATA ACC CGA GCA CAA GG-3′; NKX2.2 forward: 5′-TGG CCA TGT AAA CGT TCT GA-3′, reverse: 5′-GGA AGA AAG CAG GGG AAA AC-3′; NKX6.1 forward: 5′-ATT CGT TGG GGA TGA CAG AG-3′, reverse: 5′-TGG GAT CCA GAG GCT TAT TG-3′; INS forward: 5′-TGT ACC AGC ATC TGC TCC CTC TA-3′, reverse: 5′-TGC TGG TTC AAG GGC TTT ATT CCA-3′; GCG forward: 5′-AGG CAG ACC CAC TCA GTG A-3′, reverse: 5′-AAC AAT GGC GAC CTC TTC TG-3′; E-cadherin forward: 5′-GAG AAC GCA TTG CCA CAT ACA-3′, reverse: 5′-ACC TTC CAT GAC AGA CCC CTT AA-3′; GAPDH forward: 5′-CTT CGC TCT CTG CTC CTC CT-3′, reverse: 5′-GTT AAA AGC AGC CCT GGT GA-3′. In the expression quantification, the GAPDH gene was used as the reference gene for each sample.Cell viability analysis
The islet cell viability was tested at days 23, 45, and 60 during the long-term procedure in the perfusable system using calcein-AM and ethidium homodimer-1 (Eth-D) (LIVE/DEAD viability/cytotoxicity assay kit, Gibco). The procedure was performed according to the manufacturer's instructions. The images were taken using a fluorescence microscope at different time points.Glucose-stimulated insulin secretion assay
Islet organoids cultured on the perfusable chip or Petri dish from day 23 to day 30 were sampled. Approximately 150 organoids were collected and washed with Krebs buffer (Krb) and then cultured in 2.5 mM glucose Krb for 2 h to remove residual insulin. Spheroids were then sequentially exposed to 2.5 mM and 25 mM glucose Krb (1 ml each) for 30 min. This sequence was performed twice. Fresh 2.5 mM glucose Krb was used to wash spheroids three times between each procedure. Finally, spheroids were incubated in 2.5 mM glucose Krb containing 30 mM KCl (depolarization challenge) for 30 min, and then the supernatant was collected. Supernatant samples containing secreted insulin were processed using the human ultrasensitive insulin enzyme-linked immunosorbent assay (ELISA; ATECE Diagnostics).Calcium imaging
Islet organoids derived from hiPSCs were collected at day 23 and encapsulated in Matrigel solution (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution; BD) for 12 h. Settled islet organoids were washed and incubated in Krb solution at 37 °C for 30 min. They were then stained with Fluo-4 AM (Invitrogen) according to the manufacturer's instructions. Time series images were acquired every 10 s for 20 min. Sequential exposure to 2.5 mM and 25 mM glucose Krb for 5 min was repeated twice, and then the organoids were incubated in 2.5 mM glucose Krb containing 30 mM KCl for 5 min. The fluorescence intensity of the islet organoids was recorded and analyzed using a confocal laser scanning system (Andor Technology, Revolution WD).
1 dilution; BD) for 12 h. Settled islet organoids were washed and incubated in Krb solution at 37 °C for 30 min. They were then stained with Fluo-4 AM (Invitrogen) according to the manufacturer's instructions. Time series images were acquired every 10 s for 20 min. Sequential exposure to 2.5 mM and 25 mM glucose Krb for 5 min was repeated twice, and then the organoids were incubated in 2.5 mM glucose Krb containing 30 mM KCl for 5 min. The fluorescence intensity of the islet organoids was recorded and analyzed using a confocal laser scanning system (Andor Technology, Revolution WD).
Scanning electron microscopy
The spheroids were collected on days 5, 11 and 23 and fixed in 4% PFA for 30 min at room temperature and then washed with PBS three times for 5 min each. Fixed spheroids were dehydrated with a graded series of ethanol concentrations (25%, 50%, 75%, 95% and 100% ethanol for 10 min each). Subsequently, the samples were air-dried on a clean bench for 1 h and sputter coated with a thin gold layer (∼10 nm). Finally, the spheroid morphology was monitored using SEM (Hitachi, S4800).Statistical analysis
The data are expressed as mean ± standard deviation (SD) for three independent experiments. p-Values were calculated using Student's t-test. Experimental groups showed significant differences when *p < 0.05, **p < 0.01 and ***p < 0.001.Results and discussion
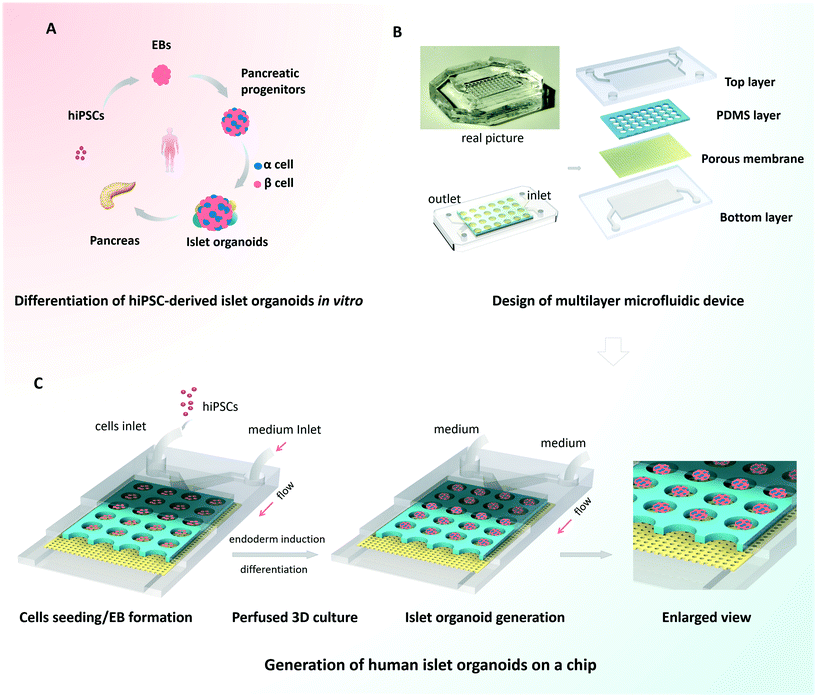
Design and operation of the islet-on-a-chip device
Organoids are characterized by their capacity for spontaneous organization into organ-like 3D tissues.10,12 To generate the islet organoids in vitro, EBs were initially formed from hiPSCs, followed by endoderm induction, islet differentiation and maturation [Fig. 1A]. Pancreas organogenesis during embryonic development in vivo is a complex and dynamic process, relying on the control of mechanical and biochemical cues. In this study, we designed and fabricated a multilayer microfluidic chip device to generate islet organoids in a dynamic cellular microenvironment [Fig. 1B]. The device consists of four parts: top and bottom PDMS layers separated by a through-hole PDMS membrane and a polycarbonate porous membrane. The microwell array of the upper layer was formed from the through-hole PDMS membrane and polycarbonate porous membrane, which allowed 3D culture of EBs and the perfusion of media flow. Notably, polycarbonate porous membranes were used to interconnect the medium flow in the upper and bottom channels of the device. The multilayer chip design permitted circulatory flow, which may facilitate sufficient medium exchange and uniform fluid stress to islet organoids.Prior to the assay, hiPSCs were dissociated into single cells and the suspension was injected into the upper layer of the device. The injected cells dispersed evenly throughout the channel and microwells, and then the EBs formed overnight. The cells outside the wells could be flushed out by a high rate of fluid flow. A microwell chip in a Petri dish was used to generate islet organoids from hiPSCs under static culture conditions, which served as the control group. Similarly, dissociated hiPSCs were seeded on the microwell chip in the Petri dish and cultured with sufficient media. As shown in Fig. 1C, the uniform EBs were restrained in the microwell of the chip and differentiated into 3D islet organoids in a stepwise process. After EB formation, fresh media with different chemical factors were pumped into the device via the upper and bottom layers during the whole differentiation process. The condition of the media flow was maintained at a rate of 100 μl h−1, which provided continuous media and nutrients to support the formation and long-term culture of the islet organoids.
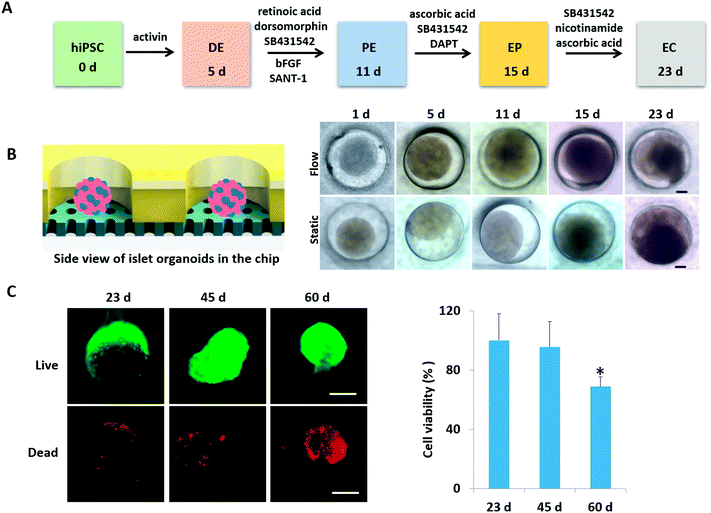
HiPSC-derived islet organoid formation and 3D culture on a chip
According to pancreas developmental principles in vivo, the generation of islet organoids in vitro involved endoderm induction, pancreatic progenitor differentiation and expansion, and endocrine cell maturation. To develop a one-stepwise protocol for in situ generation of islet organoids from hiPSCs on the chip, different chemical factors were sequentially added to the culture medium during the developmental process as shown in Fig. 2A. These factors can induce the efficient differentiation of pancreatic progenitor cells and insulin-producing cells, including activins (endoderm induction), dorsomorphin and retinoic acid (pancreatic definitive endoderm induction), and nicotinamide (pancreatic β-cell induction). The chemical factors are essential for efficient differentiation of endocrine cells, while the cellular microenvironment of derived cells and their fate in insulin-producing cells remain to be optimized.To evaluate the growth of islet organoids in the perfusable organ-on-a-chip system, we examined the tissue morphology during the organoid differentiation period. As the morphology of EBs is associated with the lineage-specific differentiation of stem cells and the cell viability of organoids,33,34 it is essential to ensure the integrity of their morphology to reduce the possible variability of islet organoids. In this device, the micro-well array can serve as permeable chambers for uniform EB formation and organoid growth with sufficient nutrients and oxygen exchange under perfusable 3D culture (Fig. 2B). It appears that the formed EBs and organoids could maintain their spherical shape with a smooth edge under perfusion culture. As a comparison, those with an irregular shape have a tendency to attach to the surface of the micro-well array under static culture conditions. Furthermore, cell debris tends to gather surrounding the organoids generated under static conditions, thus, leading to possible cell apoptosis. The development of islet organoids is a long-term process; we further examined the cell viability of organoids during the long-term culture on the device. As shown in Fig. 2C, the organoids showed high proportions of living cells on days 23 and 45 by the live–dead assay, indicating the capability of this microfluidic device to maintain the cell viability of islet organoids during the prolonged perfused 3D culture for more than one month.
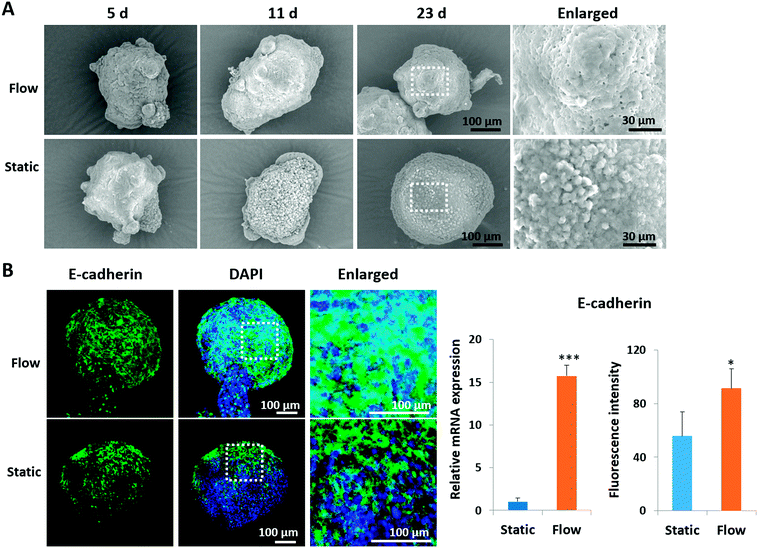
It is known that the intercellular junctions of pancreatic islet cells can regulate pancreatic cell aggregation, islet cell type segregation, architectural organization, the state of differentiation, and hormone secretion.35–37 To investigate the effects of perfused culture conditions on the growth of islet organoids, the ultrastructural morphology of the organoids during the dynamic 3D culture was identified using SEM. As shown in Fig. 3A, the organoids had smooth and tightly connected surfaces in perfused cultures, whereas they had rough surfaces with visible individual cells under static cultures. Moreover, the expression of E-cadherin, a type of cell junction protein, was assessed in the organoids by using immunofluorescence staining. The relevant mRNA expressions were quantified using real-time PCR. As shown in Fig. 3B, the expression levels of the E-cadherin protein and gene were significantly upregulated under perfused culture conditions, indicating the advantages of perfused cultures and fluid flow to facilitate the formation of cell–cell junctions during organoid formation in vitro.
Identification of the differentiation of islet organoids
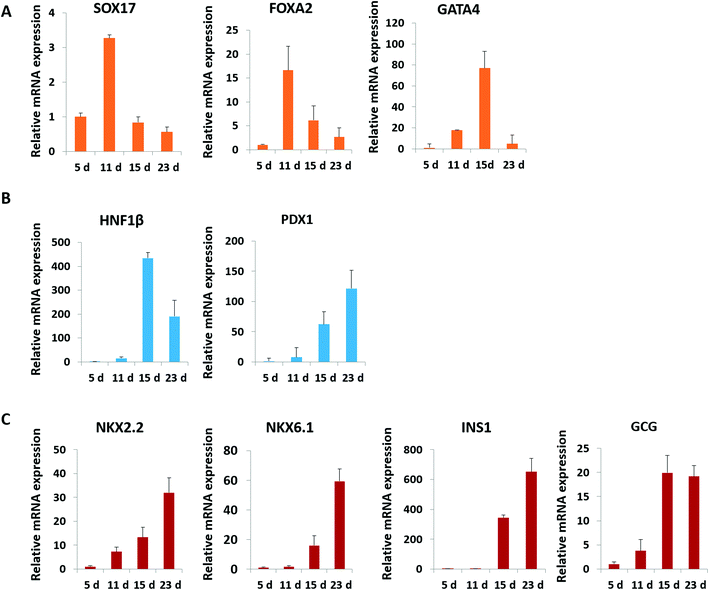
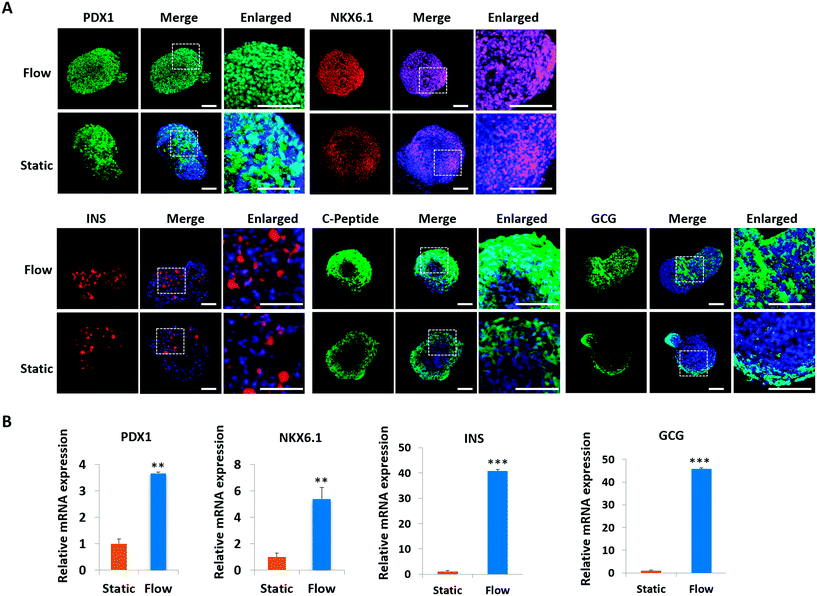
Pancreatic islet development is a complex and dynamic process. To assess the detailed characterization of islet organoids' differentiation under perfused culture conditions, the gene expression profiles of the pancreatic lineage were analyzed using real-time PCR [Fig. 4]. Desired spheroids were collected on days 5, 11, 15 and 23 during the differentiation process, which represented the differentiation of DE, PE, EP and endocrine cells, respectively. As shown in Fig. 4A, DE markers (SOX17 and FOXA2) were upregulated on days 5 and 11, and GATA4 was significantly upregulated on day 15. The pancreatic progenitor markers (HNF1β and PDX1) were detected and exhibited significantly high expressions in organoids on days 15 and 23 [Fig. 4B]. The expressions of β-cell associated transcriptional factors (NKX2.2, NKX6.1) were remarkably elevated on day 23 [Fig. 4C], especially the NKX6.1 gene relative to the maturation of the β-cell. Additionally, α-cell specific marker GCG and β-cell specific marker INS exhibited significantly high expression in organoids on days 20 and 30 indicating that the human islet organoids generated under flow conditions were composed of the two typical endocrine cells. These results demonstrate that the islet organoids generated using the perfusable islet-on-a-chip system are, to some extent, analogous to human pancreatic islets in terms of internal development.In vivo, pancreatic islets are multicellular tissues that can secrete insulin and maintain blood glucose homeostasis. To assess the degree of maturity and functionality of islet organoids under perfused culture conditions, protein markers related to insulin secretion and cell maturation in islet organoids were examined using immunostaining assay [Fig. 5A]. The organoids were collected on day 23 at the end of the differentiation process. The organoids generated on the chip with continuous media flow showed higher expression of the pancreatic progenitor marker PDX1 and mature β-cell marker NKX6.1 protein than those generated under static cultures. Recent studies have shown that differentiation protocols that generate high levels of NKX6.1 could improve the β-cell outcomes of the pancreatic progenitor.38 To assess the insulin secretion function of islet organoids, we examined the expression of INS and C-peptide proteins. The results showed that these proteins' expression of organoids in the perfusable system was significantly upregulated, indicating that the fluid flow could promote the differentiation of hiPSCs into insulin-producing cells. Furthermore, the islet organoids displayed more increased expression of α-cell specific marker GCG under perfused cultures, suggesting multicellular components in the islet organoids. Consistently, the real-time PCR results showed that the islet organoids generated on the chip with continuous media flow had higher expression levels of pancreatic β-cell genes (PDX1 and NKX6.1) and pancreatic hormone genes (INS and GCG; Fig. 5B). Overall, these results demonstrate the efficient differentiation and generation of islet organoids in the dynamic physiological microenvironments on the chip, providing favorable conditions for developing further maturity and functional maintenance of islet organoids in vitro.
In vitro pancreatic function of islet organoids from hiPSCs
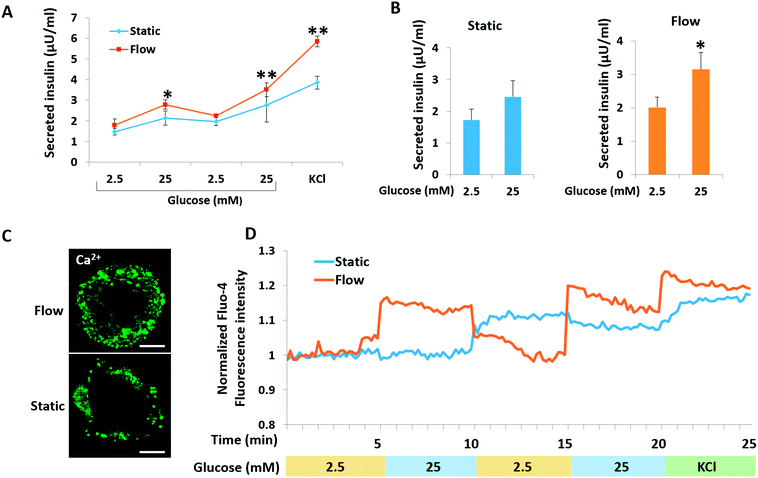
A key functional feature of β-cells is their ability to repeatedly perform glucose-stimulated insulin secretion (GSIS). However, it is a challenge to generated β-like cells from hiPSCs are response to series glucose challenge.6 We initially examined the insulin secretion of islet organoids generated on the chip in response to multiple sequential high-glucose challenges. About 150 islet organoids were collected from the perfusable chip to test their response to two more sequential high-glucose challenges. Similarly, the response of the islet organoids generated under static culture conditions to the sequential high-glucose challenges was examined. As shown in Fig. 6A, the islet organoids generated under perfused cultures showed a more sensitive response to high-glucose challenges. Additionally, the average value was calculated from insulin secreted in high glucose (25 mM) to low glucose (2.5 mM) for all sequential challenges indicating that the islet organoids generated under flow conditions performed higher levels of insulin secretion (Fig. 6B). What's more, KCl mediated cell depolarization in islet organoids with higher levels of insulin secretion. These results demonstrated that organoids cultured in the perfusable system improved the islet secretion function over the static cultures.Calcium flux plays an important role in glucose-stimulated insulin secretion from β cells. High blood glucose can induce β cell depolarization and extracellular calcium flow into the cell, which causes insulin exocytosis.39 To monitor the intracellular calcium influx, the generated islet organoids were stained with Fluo-4 AM, a cell-permeable fluorescent calcium indicator dye. As shown in Fig. 6C, the fluorescence images show that the islet organoids exhibited higher levels of cytosolic Ca2+ under perfused culture conditions than those under static culture conditions. Furthermore, calcium signaling in the islet organoids was analyzed under the different culture conditions. The results showed that the islet organoids generated under static and flow conditions both showed Ca2+ oscillation in the presence of high glucose. But the islet organoids generated on the chip showed more immediate Ca2+ influx after high glucose stimulation, indicating that the islet organoids generated under perfused culture conditions exhibit more sensitive Ca2+ oscillations than those generated under static cultures (Fig. 6D). Overall, these data indicate that the dynamic culture conditions could promote mature β cell-like properties in terms of both Ca2+ oscillation and insulin secretion in hiPSC-derived islet organoids.
Conclusions
In this work, we proposed a new and robust strategy to generate human islet organoids from hiPSCs in a perfusable organ-on-chip system. The generated islet organoids showed a suitable tissue morphology and multicellular complexity resembling human pancreatic islets in vivo. Moreover, they showed enhanced expression of mature β cell-associated genes and proteins, insulin secretion levels and Ca2+ flux release ability in response to glucose under perfused culture conditions, highlighting the role of biomimetic mechanical cues in enhancing the islet organoid function and maturation.The novelty of this work lies in: 1) offering a new and robust engineering approach to generate functional islet organoids derived from hiPSCs in a biomimetic microenvironment; 2) easy integration of controlled EB formation, in situ differentiation and generation of heterogeneous islet organoids in parallel on a single chip; 3) providing a continuous perfusion system that enables long-term 3D culture of islet organoids with improved cell viability and islet specific functions, such as glucose-stimulated insulin secretion and Ca2+ flux; 4) being amenable to real-time imaging and in situ tracking of the growth of islet organoids on a chip.
This human islet-on-a-chip provides a proof of concept for synergistic engineering of heterogeneous islet organoids from hiPSCs by combining organ-on-a-chip technology and stem cell development biology. We envision that this platform could be further integrated with additional microfluidic elements, advanced materials and biosensors to facilitate human islet maturation and functional monitoring in an in vitro setting, holding great promise for developmental biology studies, diabetes modeling and drug testing. It can also be extended to a wide range of applications in stem cell-based organoid engineering and regenerative medicine.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA16020900, XDB29050301), the National Key R&D Program of China (No. 2017YFB0405400), the Key Program of the Chinese Academy of Sciences (KFZD-SW-213), the National Natural Science Foundation of China (No. 91543121, 81573394, 81803492), the China Postdoctoral Science Foundation (No. 2018 M631836), and the Innovation Program of Science and Research from the DICP, CAS (DICP TMSR201601).References
- A. M. Shapiro, M. Pokrywczynska and C. Ricordi, Nat. Rev. Endocrinol., 2017, 13, 268–277 CrossRef CAS PubMed.
- D. A. Melton, Curr. Top. Dev. Biol., 2016, 117, 65–73 CrossRef PubMed.
- B. Blum, S. S. Hrvatin, C. Schuetz, C. Bonal, A. Rezania and D. A. Melton, Nat. Biotechnol., 2012, 30, 261–264 CrossRef CAS PubMed.
- J. V. Schiesser and J. M. Wells, Ann. N. Y. Acad. Sci., 2014, 1311, 124–137 CrossRef CAS PubMed.
- F. M. Ashcroft and P. Rorsman, Cell, 2012, 148, 1160–1171 CrossRef CAS PubMed.
- Y. Kondo, T. Toyoda, N. Inagaki and K. Osafune, J. Diabetes Invest., 2018, 9, 234–243 CrossRef PubMed.
- F. W. Pagliuca, J. R. Millman, M. Gurtler, M. Segel, A. Van Dervort, J. H. Ryu, Q. P. Peterson, D. Greiner and D. A. Melton, Cell, 2014, 159, 428–439 CrossRef CAS PubMed.
- Y. Kunisada, N. Tsubooka-Yamazoe, M. Shoji and M. Hosoya, Stem Cell Res., 2012, 8, 274–284 CrossRef CAS PubMed.
- T. Takebe, B. Zhang and M. Radisic, Cell Stem Cell, 2017, 21, 297–300 CrossRef CAS PubMed.
- M. A. Lancaster and J. A. Knoblich, Science, 2014, 345, 1247125 CrossRef PubMed.
- Y. Sasai, Cell Stem Cell, 2013, 12, 520–530 CrossRef CAS PubMed.
- A. L. Bredenoord, H. Clevers and J. A. Knoblich, Science, 2017, 355, eaaf9414 CrossRef PubMed.
- M. A. Lancaster, M. Renner, C. A. Martin, D. Wenzel, L. S. Bicknell, M. E. Hurles, T. Homfray, J. M. Penninger, A. P. Jackson and J. A. Knoblich, Nature, 2013, 501, 373–379 CrossRef CAS PubMed.
- T. Takebe, K. Sekine, M. Enomura, H. Koike, M. Kimura, T. Ogaeri, R. R. Zhang, Y. Ueno, Y. W. Zheng, N. Koike, S. Aoyama, Y. Adachi and H. Taniguchi, Nature, 2013, 499, 481–484 CrossRef CAS PubMed.
- M. Crespo, E. Vilar, S. Y. Tsai, K. Chang, S. Amin, T. Srinivasan, T. Zhang, N. H. Pipalia, H. J. Chen, M. Witherspoon, M. Gordillo, J. Z. Xiang, F. R. Maxfield, S. Lipkin, T. Evans and S. Chen, Nat. Med., 2017, 23, 878–884 CrossRef CAS PubMed.
- D. An, A. Chiu, J. A. Flanders, W. Song, D. Shou, Y. C. Lu, L. G. Grunnet, L. Winkel, C. Ingvorsen, N. S. Christophersen, J. J. Fels, F. W. Sand, Y. Ji, L. Qi, Y. Pardo, D. Luo, M. Silberstein, J. Fan and M. Ma, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E263–E272 CrossRef CAS PubMed.
- J. Candiello, T. S. P. Grandhi, S. K. Goh, V. Vaidya, M. Lemmon-Kishi, K. R. Eliato, R. Ros, P. N. Kumta, K. Rege and I. Banerjee, Biomaterials, 2018, 177, 27–39 CrossRef CAS PubMed.
- Y. C. Kuo, Y. C. Liu and R. Rajesh, Mater. Sci. Eng., C, 2017, 77, 384–393 CrossRef CAS PubMed.
- M. E. Cerf, Cell Biol. Int., 2011, 35, 1065–1078 CrossRef PubMed.
- E. Lammert, O. Cleaver and D. A. Melton, Science, 2001, 294, 564–567 CrossRef CAS PubMed.
- K. Wang, X. Wang, C. Han, W. Hou, J. Wang, L. Chen and Y. Luo, Adv. Mater., 2017, 29 DOI:10.1002/adma.201604600.
- J. A. Diez, E. D. R. Arrojo, X. Zheng, O. V. Stelmashenko, M. Chua, R. Rodriguez-Diaz, M. Fukuda, M. Kohler, I. Leibiger, S. B. B. Tun, Y. Ali, G. J. Augustine, V. A. Barathi and P. O. Berggren, Cell Rep., 2017, 20, 1490–1501 CrossRef CAS PubMed.
- K. H. Benam, R. Villenave, C. Lucchesi, A. Varone, C. Hubeau, H. H. Lee, S. E. Alves, M. Salmon, T. C. Ferrante, J. C. Weaver, A. Bahinski, G. A. Hamilton and D. E. Ingber, Nat. Methods, 2016, 13, 151–157 CrossRef CAS PubMed.
- L. Wang, T. Tao, W. Su, H. Yu, Y. Yu and J. Qin, Lab Chip, 2017, 17, 1749–1760 RSC.
- H. J. Kim and D. E. Ingber, Integr. Biol., 2013, 5, 1130–1140 CrossRef CAS PubMed.
- Y. Wang, H. Wang, P. Deng, W. Chen, Y. Guo, T. Tao and J. Qin, Lab Chip, 2018, 18, 3606–3616 RSC.
- Y. Wang, L. Wang, Y. Zhu and J. Qin, Lab Chip, 2018, 18, 851–860 RSC.
- Y. Zhu, L. Wang, F. Yin, Y. Yu, Y. Wang, H. Liu, H. Wang, N. Sun, H. Liu and J. Qin, Integr. Biol., 2017, 9, 774–781 CrossRef CAS PubMed.
- Y. Zhu, L. Wang, H. Yu, F. Yin, Y. Wang, H. Liu, L. Jiang and J. Qin, Lab Chip, 2017, 17, 2941–2950 RSC.
- F. Yin, Y. Zhu, Y. Wang and J. Qin, ACS Biomater. Sci. Eng., 2018 DOI:10.1021/acsbiomaterials.8b00160.
- Z. Li, Y. Guo, Y. Yu, C. Xu, H. Xu and J. Qin, Integr. Biol., 2016, 8, 1022–1029 CrossRef CAS PubMed.
- Y. Kim, H. Kim, U. H. Ko, Y. Oh, A. Lim, J. W. Sohn, J. H. Shin, H. Kim and Y. M. Han, Sci. Rep., 2016, 6, 35145 CrossRef CAS PubMed.
- A. W. Xie, B. Y. K. Binder, A. S. Khalil, S. K. Schmitt, H. J. Johnson, N. A. Zacharias and W. L. Murphy, Sci. Rep., 2017, 7, 14070 CrossRef PubMed.
- Z. Li, H. Sun, J. Zhang, H. Zhang, F. Meng and Z. Cui, PLoS One, 2013, 8, e72612 CrossRef CAS PubMed.
- V. Cirulli, Diabetes, 2015, 64, 709–711 CrossRef CAS PubMed.
- R. Jain and E. Lammert, Diabetes, Obes. Metab., 2009, 11(Suppl 4), 159–167 CrossRef CAS PubMed.
- Y. Jun, A. R. Kang, J. S. Lee, S. J. Park, D. Y. Lee, S. H. Moon and S. H. Lee, Biomaterials, 2014, 35, 4815–4826 CrossRef CAS PubMed.
- J. E. Bruin, A. Rezania, J. Xu, K. Narayan, J. K. Fox, J. J. O'Neil and T. J. Kieffer, Diabetologia, 2013, 56, 1987–1998 CrossRef PubMed.
- P. Gilon, H.-Y. Chae, G. A. Rutter and M. A. Ravier, Cell Calcium, 2014, 56, 340–361 CrossRef CAS PubMed.
Footnote |
| † Dedicated to the 70th anniversary of Dalian Institute of Chemical Physics, CAS. |
| This journal is © The Royal Society of Chemistry 2019 |