Intracellular delivery and biodistribution study of CRISPR/Cas9 ribonucleoprotein loaded bioreducible lipidoid nanoparticles†
Yamin
Li
 a,
Justin
Bolinger
a,
Yingjie
Yu
a,
Zachary
Glass
a,
Nicola
Shi
a,
Justin
Bolinger
a,
Yingjie
Yu
a,
Zachary
Glass
a,
Nicola
Shi
 a,
Liu
Yang
a,
Ming
Wang
b and
Qiaobing
Xu
a,
Liu
Yang
a,
Ming
Wang
b and
Qiaobing
Xu
 *a
*a
aDepartment of Biomedical Engineering, Tufts University, Medford, MA 02155, USA. E-mail: Qiaobing.Xu@tufts.edu; Fax: +1 617-627-3231; Tel: +1 617-627-4322
bBeijing National Laboratory for Molecular Sciences, Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences, Beijing, China 100190
First published on 31st July 2018
Abstract
CRISPR/Cas9 ribonucleoprotein (RNP) complexes with transient therapeutic activity and minimum off-target effects have attracted tremendous attention in recent years for genome editing and have been successfully employed in diverse targets. One ongoing challenge is how to transport structurally and functionally intact Cas9 protein and guide RNA molecules into cells efficiently and safely. Here we report a combinatorial library of disulfide bond-containing cationic lipidoid nanoparticles (LNPs) as carrier systems for intracellular Cas9/sgRNA delivery and subsequent genome editing. Nanoparticles with high efficacies of targeted gene knockout as well as relatively low cytotoxicities have been identified through in vitro screening. The in vivo biodistribution profiles were studied utilizing fluorescent dye labeled and RNP complexed LNPs. Results from this study may shed some light on the design of effective cationic lipidoids for intracellular delivery of genome editing platforms, as well as optimizing the nanoparticle formulations for further disease modeling and therapeutic applications.
Introduction
Since 2013, as the microbial clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 9 (Cas9) adaptive immune system was successfully employed for genome editing in mammalian cells,1,2 increased attention has been paid to the CRISPR/Cas9 technology. This system uses a complementary guide RNA (gRNA) molecule to target a specific DNA site in the genome and introduce a double-strand break (DSB) precisely at that position.3,4 Gene knockout or knock-in could be achieved by subsequent repair of the DNA DSB through the nonhomologous end joining (NHEJ) or homology directed repair (HDR) processes.5 The CRISPR/Cas9 has dramatically changed the landscape of genome engineering over the last few years, while recent studies revealed some potential obstacles in translating CRISPR/Cas9 such as pre-existing adaptive immunity6 and p53-mediated damage response.7 Furthermore, great demands still exist for intracellular delivery systems that can introduce structurally and functionally intact Cas9 and gRNA into targeted cells and the nucleus with high efficiency and minimum safety issues. Successful gene editing has been achieved by delivering either DNA (plasmid DNA and viral genome that encoding Cas9), mRNA or protein, and the advantages as well as disadvantages of delivering each cargo format have been summarized in recent reviews.5,8–10 In general, delivering the Cas9:gRNA ribonucleoprotein (RNP) complex offers a most straightforward strategy and transient therapeutic activity with minimum off-target effects expected. In this context, physical methods including microinjection, electroporation, and acoustic-assisted transfection have been reported for CRISPR/Cas9 delivery,11–13 while in vivo applications are complicated by their invasive features and the need for direct physical access to the target cells and tissues. Covalent protein modification with targeting ligand (cell penetrating peptide and nuclear localization sequence)14,15 and carrier-based (lipid and lipid-like (lipidoid) nanoparticles (LNPs),16–18 biopolymers,19 gold nanoparticles,20–23 and zeolitic imidazole frameworks,24etc.) delivery strategies, on the other hand, have also been explored for CRISPR/Cas9 RNP complex delivery. Among these, successful local delivery, including inner ear,16 tumor,19 and muscle,21 of CRISPR/Cas9 RNP complexes and subsequent genome editing have been reported by using cationic lipid nanoparticles, DNA nanoclews, and polymer modified gold nanoparticles, respectively. While systemic delivery of CRISPR/Cas9 RNP complex is still challenging, viral vectors have already been proved to be efficient for both gene knock-in and knockout through systemic administrations.25–27 Combinatorial libraries of cationic lipidoid nanoparticles are promising candidates for intracellular biomacromolecule delivery, as demonstrated by us28–33 as well as others.34–38 In this study, we further expand our disulfide bond containing biodegradable lipidoid library,39 by introducing an amide linker between the hydrophilic amine heads and aliphatic tail groups. The reducing reagents triggered degradation and cargo release behaviours of the bioreducible lipidoid nanoparticles have been studied previously and enhanced intracellular delivery efficiencies were obtained comparing with the non-degradable LNPs counterparts.39 Furthermore, these lipidoids have showed effectiveness for genome editing proteins (Cre recombinase and Cas9:sgRNA complex) delivery both in vitro and in vivo (intracranial injection).17 LNPs were then fabricated through self-assembly procedures, and the intracellular delivery efficacy and cytotoxicity of CRISPR/Cas9 RNP complex loaded LNPs were examined. Top LNP candidates with best performances were identified and further utilized for in vivo biodistribution study through the systemic administration to Balb/c mice. The in vitro screening results as well as biodistribution profiles observed in this study were expected to shed some light on RNP/LNPs formulation optimization and further therapeutic applications.Experimental
General
All chemicals used for lipidoids synthesis were purchased from Sigma-Aldrich and Oakwood Chemical without further purification unless otherwise noted. Fluorescent (−30)GFP-Cre recombinase, S. pyogenes Cas9 (spCas9) and single guide RNA (sgRNA) targeting GFP gene were produced according to our previously reported protocols.17 ATTO 550 labeled guide RNA was purchased from IDT and used for determination of RNP loading efficiency. 1H and 13C NMR spectra were recorded on a Bruker AVIII 500 MHz NMR spectrometer operated in the Fourier transform mode. Hydrodynamic sizes and polydispersity indexes were measured by Zeta-PALS particle size analyzer (Brookhaven Instruments). TEM images were taken on a FEI Technai Transmission Electron Microscope. HeLa-DsRed and GFP-HEK cells were cultured in Dulbecco's modified eagle's medium (DMEM, Sigma-Aldrich) with 10% fetal bovine serum (FBS, Sigma-Aldrich) and 1% penicillin–streptomycin (Gibco). Fluorescence profiles of HeLa-DsRed and GFP-HEK cells were analyzed using flow cytometer (BD FACS Calibur, BD Science, CA). Fluorescence biodistribution images of whole mice body and collected organs were obtained using a PerkinElmer IVIS Spectrum CT Biophotonic Imager.Synthesis of lipidoids
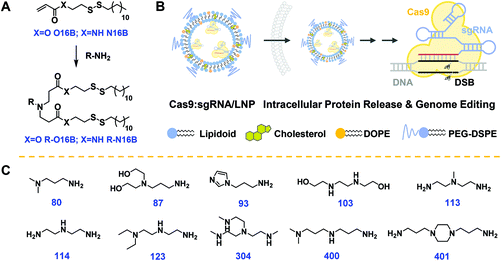
The lipidoid tails, O16B and N16B, were synthesized using the routes outlined in Fig. S1.† The chemical structures of O16B and N16B were confirmed by 1H and 13C NMR spectra and the results are shown in Fig. S2 and S3.† Then, the acrylate tails were reacted with amine head groups (R groups; 80, 87, 93, etc.) at 2.5/1 molar ratio in Teflon-lined glass screw-top vials for 48 h at 70 °C. The crude products were purified using a Teledyne Isco Chromatography system.Preparation of LNPs
Following our previously reported protocol,17 lipidoid nanoparticles were fabricated for protein complexation and intracellular delivery. Briefly, lipidoid was combined in a clean vial with precalculated amount of cholesterol (Sigma-Aldrich), DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Avanti Polar Lipids), DSPE-PEG2k (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000], Avanti Polar Lipids) at a 16/4/1/1 weight ratio. Lipid films were formed after evaporation of the organic solvent and 100 μL of ethanol was added. The mixture was then briefly sonicated to completely dissolve all materials. The ethanol solution was then added dropwise into 800 μL of sodium acetate buffer (25 mM, pH 5.2) with continuous stirring. Finally, the mixture was transferred into a dialysis cassette (MWCO 3.5 kDa, Slide-A-Lyzer, ThermoFisher Scientific) and dialyzed against pure water to remove the ethanol. The fabricated LNPs were then stored at 4 °C for future use.Intracellular delivery of (−30)GFP-Cre/LNP
A 48-well plate was seeded with HeLa-DsRed cells at an initial concentration of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well dispersed in 250 μL of DMEM cell culture media and incubated for 24 h. The fabricated LNPs were mixed with (−30)GFP-Cre protein in PBS buffer and incubated for another 30 min under room temperature. The (−30)GFP-Cre/LNPs were then added into each well. The final concentration of (−30)GFP-Cre is 50 nM and lipidoid is 6.6 μg mL−1. The cells were incubated for another 8 h and then analyzed by flow cytometry.
000 cells per well dispersed in 250 μL of DMEM cell culture media and incubated for 24 h. The fabricated LNPs were mixed with (−30)GFP-Cre protein in PBS buffer and incubated for another 30 min under room temperature. The (−30)GFP-Cre/LNPs were then added into each well. The final concentration of (−30)GFP-Cre is 50 nM and lipidoid is 6.6 μg mL−1. The cells were incubated for another 8 h and then analyzed by flow cytometry.
Intracellular delivery of Cas9:sgRNA/LNP
A 48-well plate was seeded with GFP-HEK cells at an initial concentration of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well dispersed in 250 μL of DMEM media and incubated for 24 h. Cas9 and sgRNA were mixed in PBS at 1/1 molar ratio and left undisturbed for 20 min. LNPs were then added and the mixture was incubated for another 30 min under room temperature. The Cas9:sgRNA/LNP solution was then added to the plate and the plate was incubated for 48 h before flow cytometry analysis. The final concentrations of Cas9:sgRNA complex is 25 nM and lipidoid is 3.3 μg mL−1.
000 cells per well dispersed in 250 μL of DMEM media and incubated for 24 h. Cas9 and sgRNA were mixed in PBS at 1/1 molar ratio and left undisturbed for 20 min. LNPs were then added and the mixture was incubated for another 30 min under room temperature. The Cas9:sgRNA/LNP solution was then added to the plate and the plate was incubated for 48 h before flow cytometry analysis. The final concentrations of Cas9:sgRNA complex is 25 nM and lipidoid is 3.3 μg mL−1.
MTT assay
The cytotoxicity of the LNPs against GFP-HEK cells were measured via MTT assay. A 96-well plate was seeded with GFP-HEK cells at an initial concentration of 5000 cells per well dispersed in 100 μL of DMEM media and incubated for 24 h. Cas9:sgRNA/LNP solution was added into the plate and the plate was incubated for another 48 h. MTT solution (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; 5 mg mL−1 in PBS) was added to each well and the cells were incubated for another 4 h at 37 °C. The media was then removed from each well, and 200 μL dimethyl sulfoxide (DMSO) was added to each well. The plate was gently agitated by an orbital shaker for 10 min and absorbance measurements were immediately read by a microplate reader (Molecular Devices, SpectraMax M2) at a wavelength of 570 nm.Hemolysis assay
Human red blood cells (hRBCs) were washed with PBS buffer three times and collected after centrifugation at 1000 rpm for 5 min. The resulting stock solution was diluted 3 fold in PBS buffer to give the assay solution (∼3% v/v hRBCs). Then, 90 μL of hRBCs assay solution was mixed with 10 μL of LNPs solutions (final concentration of lipidoids = 3.3 μg mL−1) and incubated at 37 °C for 1 h. Then the mixture was centrifuged again and supernatant was collected. 10 μL of the supernatant was further diluted into 90 μL of PBS buffer, and the absorbance at 405 nm (OD405) was recorded using a microplate reader (Molecular Devices, SpectraMax M2). The PBS buffer and Triton X-100 (1% v/v) were used as negative and positive controls, respectively.Biodistribution study
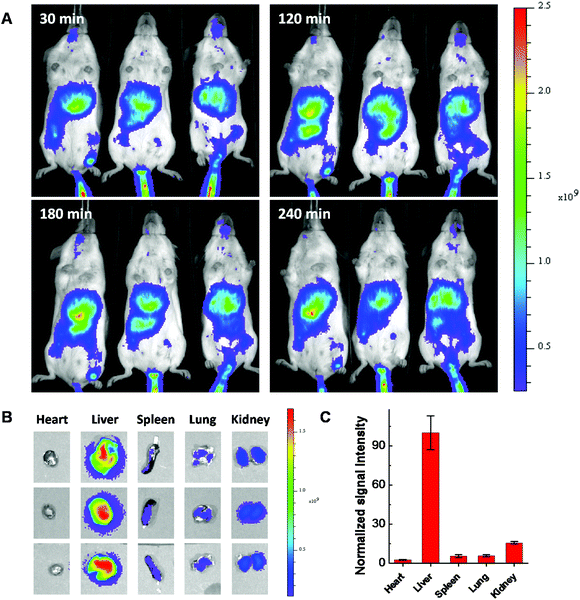
All experiments with animals were performed according to the NIH guidelines for the care and use of experimental animals and were approved by the Tufts University Institutional Animal Care and Use Committee (IACUC). Cy7.5 labeled LNPs (lipidoid/Cholesterol/DOPE/DSPE-PEG2k-Cy7.5 = 16/4/1/1, weight ratio) were prepared for biodistribution study. Balb/c mice were housed in a temperature and humidity controlled facility with a 12 h light/dark cycle. Three mice in each group were injected with Cas9:sgRNA/LNPs formulations through the tail vein, with 50 μg Cas9 protein for each injection. Whole mouse body and major organs including heart, liver, spleen, lung and kidney from all groups were imaged using a PerkinElmer IVIS Spectrum CT Biophotonic Imager at different time intervals after the injection and results were analyzed using the Living Image Software.Results and discussion
Lipidoids synthesis, LNPs fabrication and characterization
The hydrophobic tails O16B and N16B were synthesized at first via the routes outlined in Fig. S1.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17,39,40 The structures of the O16B and N16B tails were confirmed by 1H and 13C NMR spectra (Fig. S2 and S3†). The tails were then reacted with commercially available amine heads (R groups; 80, 87, 93 etc.; Fig. 1C) to create amphiphilic lipidoids, as shown in Fig. 1A. Formulated lipidoid nanoparticles were then prepared together with helper lipids including cholesterol, DOPE, and DSPE-PEG2k (lipidoid/cholesterol/DOPE/DSPE-PEG2k = 16/4/1/1, weight ratio), and complexed with (−30)GFP-Cre protein or Cas9:sgRNA RNP complex for further studies (Fig. 1B), following our previously reported procedures.17
17,39,40 The structures of the O16B and N16B tails were confirmed by 1H and 13C NMR spectra (Fig. S2 and S3†). The tails were then reacted with commercially available amine heads (R groups; 80, 87, 93 etc.; Fig. 1C) to create amphiphilic lipidoids, as shown in Fig. 1A. Formulated lipidoid nanoparticles were then prepared together with helper lipids including cholesterol, DOPE, and DSPE-PEG2k (lipidoid/cholesterol/DOPE/DSPE-PEG2k = 16/4/1/1, weight ratio), and complexed with (−30)GFP-Cre protein or Cas9:sgRNA RNP complex for further studies (Fig. 1B), following our previously reported procedures.17
The hydrodynamic diameters and polydispersity indexes of blank and Cas9:sgRNA loaded LNPs were determined by dynamic light scattering (DLS) analysis. As shown in Fig. 2A, RNP loaded LNPs (Cas9:sgRNA/LNP) showed slightly size variations comparing to the blank LNPs, as both increases (80-N16B, 87-O16B, 87-N16B, etc.) and decreases (80-O16B, 93-N16B, 103-O16B, etc.) in averaged hydrodynamic diameter (〈Dh〉) were recorded. Furthermore, a clear majority of the blank and Cas9:sgRNA loaded LNPs tested have 〈Dh〉 values between 100 and 350 nm, which are considered to be suitable for intracellular delivery applications, and polydispersity indexes between 0.1–0.3 (Fig. S4†), which further point to the uniformity and relative homogeneity of initial particle sizes as well as to a lack of evident aggregation of the blank and loaded LNPs. Typical diameter distribution profiles of blank (80-O16B, 〈Dh〉 = 202.8 nm, μ2/Γ2 = 0.28; 80-N16B, 〈Dh〉 = 278.8 nm, μ2/Γ2 = 0.33) and Cas9:sgRNA loaded LNPs (Cas9:sgRNA/80-O16B, 〈Dh〉 = 187.8 nm, μ2/Γ2 = 0.29; Cas9:sgRNA/80-N16B, 〈Dh〉 = 310.9 nm, μ2/Γ2 = 0.30) are shown in Fig. 2B.
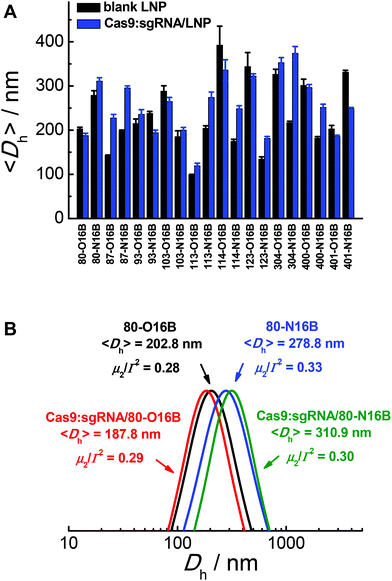 | ||
| Fig. 2 (A) Averaged hydrodynamic sizes and (B) typical size distribution profiles of blank and Cas9:sgRNA RNP loaded LNPs. | ||
Then, the morphologies of LNPs were examined by transmission electron microscopy (TEM) and Fig. 3 and S5† show typical TEM images of blank and Cas9:sgRNA loaded 80-O16B and 80-N16B LNPs. Spherical nanoparticles most likely vesicles/liposomes with lamellar phase bilayer structures were observed for both 80-O16B and 80-N16B LNPs, and their estimated mean sizes are 158.6 nm and 198.4 nm, respectively. The lipidoid vesicles which are also typical morphologies obtained from our previously synthesized cationic LNP libraries,17,33 are hollow spheres with hydrophobic bilayer walls sandwiched by hydrophilic internal and external coronas.41 Negatively charged cargos such as (−30)GFP-Cre fusion protein or Cas9:sgRNA can form nanocomplexes with the LNPs mainly through electrostatic interaction. As shown in Fig. 3 and S5,† the morphologies of Cas9:sgRNA/80-O16B and Cas9:sgRNA/80-N16B are similar to their blank counterparts and the estimated diameters are 151.8 nm and 232.2 nm, respectively. The RNP loading efficiencies of 80-O16B and 80-N16B LNPs were determined to be 95.3% and 97.4%, respectively, indicating most of the RNP molecules are complexed with the LNPs (Fig. S6†). Furthermore, the averaged sizes of both blank and Cas9:sgRNA loaded LNPs determined by TEM observations are smaller than the hydrodynamic diameters obtained from DLS measurements, which is reasonable considering these TEM images are taken under dehydrated conditions. Meanwhile, the aggregations of LNPs shown in TEM images (Fig. 3 and S5†) are considered to be resulted from the drying/staining procedures during the sample preparation, which is also observed in previous studies.33
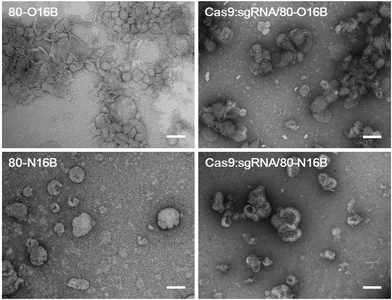 | ||
| Fig. 3 Typical TEM images of blank and Cas9:sgRNA loaded 80-O16B and 80-N16B nanoparticles. Scale bar = 100 nm. | ||
Stability of blank and Cas9:sgRNA loaded LNPs
The stability of blank and Cas9:sgRNA loaded LNPs were characterized by DLS and fluorescence measurements.42Fig. 4A shows a representative example of the relative 〈Dh〉 value variations exhibited by 80-O16B, Cas9:sgRNA/80-O16B, 80-N16B and Cas9:sgRNA/80-N16B over 48 h of storage at room temperature. All the nanoparticles tested exhibit a change in 〈Dh〉 value of no greater than 25% at any of the 8 h intervals over the span of the study, indicating high stability with statistically insignificant aggregation over two days of storage at room temperature. Furthermore, the time-dependent DLS measurement results of the stability study for other Cas9:sgRNA/LNPs are shown in Fig. S7,† from which, except for one particular condition Cas9:sgRNA/93-O16B, all the Cas9:sgRNA/LNPs showed good stability over 48 h of storage. Extended storage stability of nanoparticles were studied by using the fluorescence resonance energy transfer (FRET) pair, DiO (3,3′-Dioctadecyloxacarbocyanine perchlorate) and DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbo-cyanine perchlorate), loaded 80-O16B, Cas9:sgRNA/80-O16B, 80-N16B and Cas9:sgRNA/80-N16B nanoparticles, following previously reported procedures.42 The FRET pair loaded LNPs were excited at the wavelength of DiO absorption (450 nm), fluorescence emissions of FRET donor (DiO) and acceptor (DiI) at 505 nm and 575 nm were recorded, and FRET ratios, i.e. I575/(I575 + I505) were calculated accordingly.43 Minor changes in the FRET ratio indicate the high kinetic stability of the self-assemblies and the degradation or dissociation processes of the supramolecular structures could induce significant variations of the FRET ratios. As shown in Fig. 4B, both the blank (80-O16B and 80-N16B) and Cas9:sgRNA loaded nanoparticles (Cas9:sgRNA/80-O16B and Cas9:sgRNA/80-N16B) showed negligible FRET ratio (I575/(I575 + I505)) variations, as <±5% variations after three days and <±12.5% variations after seven days of storage were observed, indicating the structure integrity and long-term storage stability of these blank and Cas9:sgRNA RNP complex loaded nanoparticles.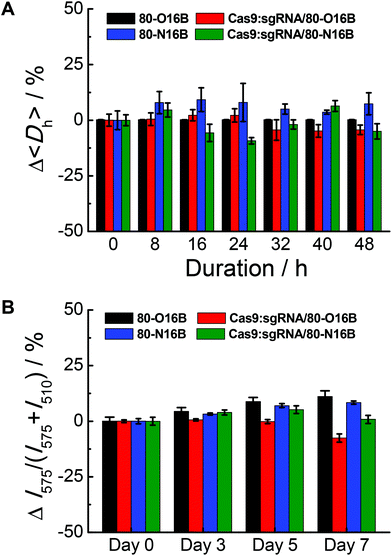 | ||
| Fig. 4 Stability tests of blank and Cas9:sgRNA loaded 80-O16B and 80-N16B nanoparticles using (A) DLS and (B) fluorescence measurement. | ||
Internalization study using (−30)GFP-Cre protein model
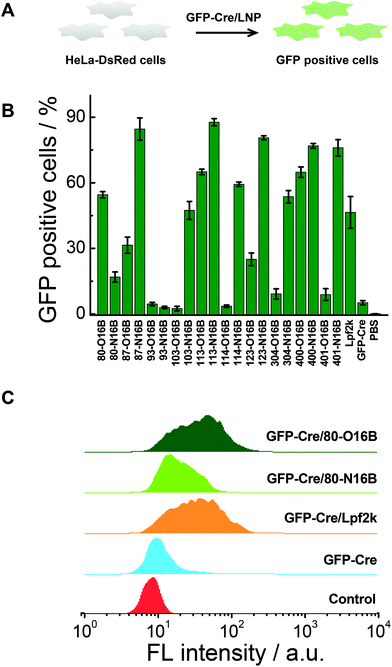
Negatively charged fluorescent recombinant protein (−30)GFP-Cre was used to complex with LNPs for the internalization study. (−30)GFP-Cre/LNPs were prepared at first following our previously reported procedures and the (−30)GFP-Cre protein was found to be loaded quantitatively into 80-O16B and 80-N16B LNPs (Fig. S6†). Then the (−30)GFP-Cre/LNPs were incubated with HeLa-DsRed cells (Fig. 5A). The cells were harvested after 8 h of exposure (50 nM of (−30)GFP-Cre) and analyzed via flow cytometry to evaluate the internalization efficiencies as measured by GFP-positive cell percentages.17,33 Lpf2k (Lipofectamine 2000, commercial available transfection reagent), naked (−30)GFP-Cre protein, and PBS treated samples were used as controls. As shown in Fig. 5B, negative controls ((−30)GFP-Cre and PBS) induced negligible GFP-positive cell portions, as less than 5.5% GFP-positive cells were recorded for both groups, while (−30)GFP-Cre/Lpf2k treated cells showed 46.5% of GFP-positive portion. As to the LNPs synthesized in this study, most of the (−30)GFP-Cre/LNPs could induce 15–88% of GFP-positive cells, and 55% of the LNPs (11 out of 20) performed better than or at a statistically similar level (≥∼45%) to the positive control, (−30)GFP-Cre/Lpf2k. LNPs with the highest transfection efficacies were identified as 87-N16B and 113-N16B, as these nanoparticles were able to readily facilitate the uptake of (−30)GFP-Cre, and the GFP-positive cells are determined as 84.6 and 87.7%, respectively. 123-N16B, 400-N16B, and 401-N16B also performed well, with 80.6%, 76.9%, and 76.0% of the cells being identified as GFP-positive, respectively. The typical flow cytometry profiles of (−30)GFP-Cre/LNPs and control groups are shown in Fig. 5C. Both the untreated and naked (−30)GFP-Cre treated cells showed low fluorescence intensities, while enhanced green fluorescence emission intensities were recorded from (−30)GFP-Cre/LNPs and (−30)GFP-Cre/Lpf2k treated cells. Furthermore, (−30)GFP-Cre/80-O16B and (−30)GFP-Cre/Lpf2k treated cells possessed similar fluorescence profiles and the mean fluorescence intensities are higher than that of (−30)GFP-Cre/80-O16B treated cells, which are all consistent with the results shown in Fig. 5B. Above all, using (−30)GFP-Cre fluorescent protein model, the LNPs were demonstrated to be efficient for intracellular protein delivery and top candidates could be identified as with comparable or even higher transfection efficacy as Lpf2k.Intracellular delivery of Cas9:sgRNA RNP complex
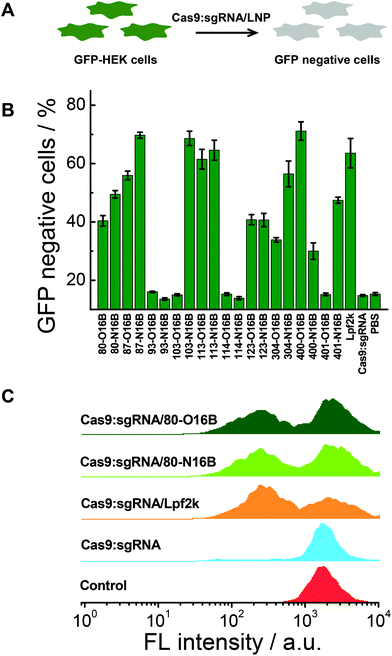
The gene knockout study of the protein/LNPs nanocomplexes were performed using Cas9:sgRNA RNP complex targeting GFP gene as the cargo and GFP expressing HEK cells (GFP-HEK) as the cell model. Cas9:sgRNA/LNPs were fabricated following our previously reported procedures and incubated with GFP-HEK cells.17,33 The fluorescence properties of the GFP-HEK cells were analyzed by flow cytometry after 48 h of exposure (25 nM of Cas9:sgRNA). Naked Cas9:sgRNA RNP complex, Cas9:sgRNA/Lpf2k and PBS treated cells were used as controls. In this study, LNPs yielding higher percentages of GFP negative cells are considered more effective, and therefore more desirable, at transfecting RNP complexes and editing GFP-HEK cells (Fig. 6A). As shown in Fig. 6B, naked Cas9:sgRNA RNP complex induced the same minimum level of GFP knockout as the other negative control, PBS, which is consistent with our previously reported results,17 as Cas9:sgRNA alone is not able to enter the cells efficiently. Lpf2k, on the other hand, could efficiently facilitate Cas9:sgRNA RNP complex enter into the GFP-HEK cells and 63.6% of GFP negative cells were observed. As to the O16B and N16B tailed LNPs, some LNPs (e.g. 93-O16B, 93-N16B, 103-O16B) exhibited little to no transfection into the GFP-HEK cells, while 35% of the LNPs (7 out of 20) tested performed better than or at the comparable level as the Lpf2k. Most of the inefficient LNPs for Cas9:sgRNA RNP delivery (i.e. 93-O16B, 93-N16B, 103-O16B, 114-O16B, 401-O16B) also showed negligible efficacy for fluorescent (−30)GFP-Cre protein delivery in the internalization study (Fig. 5A and B), indicating these LNPs cannot be efficiently internalized into either GFP-HEK or HeLa-DsRed cell lines regardless the cargo proteins. On the other hand, LNPs with the best performance for intracellular Cas9:sgRNA RNP complex delivery were identified as 87-N16B, 103-N16B, and 400-O16B, as these nanoparticles were able to transfect the GFP-HEK cells and showed GFP knockout efficiencies ∼70%. Additionally, 113-O16B and 113-N16B also performed well, as 61.5% and 64.6% GFP knockout efficiencies were determined, respectively. Typical flow cytometry profiles of GFP-HEK cells treated by Cas9:sgRNA/80-O16B, Cas9:sgRNA/80-N16B, Cas9:sgRNA/Lpf2k, naked Cas9:sgRNA RNP complex and PBS are shown in Fig. 6C. Additionally, it was found that most of the LNPs with high Cas9:sgRNA RNP delivery efficacies (e.g. 87-N16B, 103-N16B, 400-O16B, 113-O16B and 113-N16B; Fig. 6) are also proved efficacious for (−30)GFP-Cre delivery (Fig. 5), which demonstrated the possibility of using negatively charged fluorescent Cre recombinase as a potential cargo model for quick and high-throughput in vitro screening of lipidoid nanoparticles for intracellular delivery purposes.Cytotoxicity test and structure–activity relationship analysis
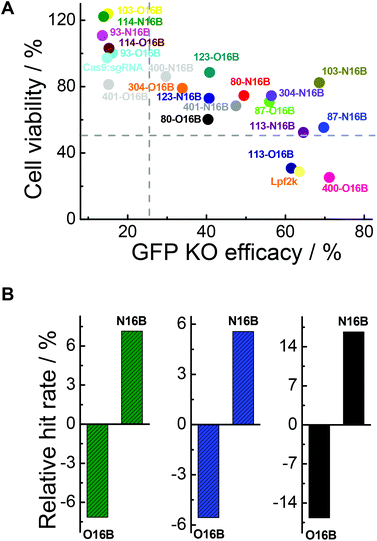
In addition to intracellular transfection efficiency, cell compatibility should also be evaluated when developing bioactive cargo delivery systems.44–47 In this context, the MTT assay was employed for cytotoxicity test of the Cas9:sgRNA/LNPs against GFP-HEK cells. As shown in Fig. S8,† after 48 h of exposure, notably, 45% of the Cas9:sgRNA/LNPs (9 out of 20) exhibited minimal cytotoxicity (cell viability >80%), with 93-O16B, 93-N16B, 103-O16B, 114-O16B and 114-N16B having the most favorable performance with respect to percentages of cells staying alive after 48 h of incubation. This is in dramatic contrast to the commercial Lpf2k, which is relatively toxic (cell viability 28.8% after 48 h of exposure). This indicates that many of the tested LNPs have cytotoxicities low enough to be considered potentially viable for genome editing protein delivering therapeutics with extended exposure time.As mentioned above, two important factors for determining which LNPs might be suitable for future protein-based therapies involving intracellular delivery are their ability to efficiently transfect cells to affect desired functional changes, and their ability to do so without harming or adversely affecting the cells’ normal functionalities. Fig. 7A plots cell viabilities against GFP knockout efficacies of all 20 LNPs studied here, with dotted lines denoting 50% and 25% values for cell viability and GFP knockout efficacy, respectively. In general, it is obvious that cell viability is negatively correlated with GFP knockout efficacy, which could potentially be expected because higher transfection efficiency probably stands for stronger interactions between the cells and foreign entities, i.e. LNPs, which could interfere with the homeostasis of the cell. Further analysis revealed that, among all the O16B and N16B tailed LNPs, 0% of the LNPs (0 out of 20) had poor cell viability and poor GFP knockout efficacy (lower left quadrant); this indicates that none of the LNPs studied here were purely toxic to the cells. 30% of LNPs (6 out of 20) had high cell viability and poor GFP knockout efficacy (upper left quadrant, naked Cas9:sgRNA RNP complex was found in this region), indicating these LNPs did minimal to harm the cells when compared with some of their counterparts, but also did not deliver the Cas9:sgRNA RNP complex to the targets effectively. 10% of the LNPs (2 out of 20) had low cell viability and high GFP knockout efficacy (lower right quadrant, Lpf2k was found in this region), meaning these LNPs delivered the complex to the cells effectively while also being fairly toxic. 60% of the LNPs (12 out of 20) showed relatively low cytotoxicity while high transfection efficacies (upper right quadrant), which demonstrates the effectiveness of the combinatorial methods in building lipidoids library for intracellular delivery applications. Detailing the upper right region further, 87-O16B (70.9% cell viability, 55.9% GFP knockout efficacy) and 304-N16B (74.5%, 56.5%) were LNPs with high cell viability and adequate knockout efficacy, while 113-N16B (52.4%, 64.6%) and 87-N16B (55.4%, 69.7%) had adequate cell viability and high knockout efficacy. The best performing LNP identified was 103-N16B (82.4%, 68.6%), exhibiting a combination of both high cell viability and knockout efficacy. These LNPs found in the upper right quadrant of Fig. 7A are considered to be strong candidates for future studies surrounding the delivery of Cas9:sgRNA RNP complexes based therapies.
The structure–activity relationship of the lipidoids was further explored. According to previous studies, the intracellular delivery efficiencies of LNPs are considered to be related to the chemical structures of amine heads (R groups), hydrophobic tails, substitution numbers, tail numbers and apparent pKa values, etc.33,36,38 In this study, the relationship between tail structures (O16B and N16B) and delivery performances (delivery efficacy and cytotoxicity) were investigated. For this analysis, LNPs with >25% of GFP knockout efficiencies and >50% cell viabilities after 48 h exposure were considered to be efficacious and less toxic LNPs, respectively, as shown in Fig. 7A. The lipidoid library was categorized into two groups based on their hydrophobic tail structures (O16B and N16B), with each tail group making up 50% of the overall library. As there are 14 efficacious LNPs in total and 57.1% of them (8 out of 14) are with N16B tails, the relative hit rates of O16B and N16B LNPs for being efficacious LNPs are determined to be −7.14% and 7.14%, as shown in Fig. 7B (green bars), meaning that lipidoids with O16B tails are underrepresented among LNPs with GFP knockout efficacy >25%, while lipidoids with N16B tails are overrepresented in this group. This result suggests that comparing to O16B tails, N16B tails are more associated with efficacious LNPs. As to the cytotoxicity of LNPs, 18 LNPs showed >50% cells viabilities, in which 55.6% are N16B tailed LNPs. Therefore, the relative hit rates for being less toxic LNPs for O16B and N16B tails are −5.56% and 5.56% (Fig. 7B, blue bars), respectively, indicating N16B tailed LNPs are also overrepresented among the less toxic LNPs. Furthermore, LNPs with best performances are defined as LNPs that can both induce >25% GFP knockout efficiency and possess >50% cell viability, which are also represented in the upper right quadrant of Fig. 7A. Further analysis revealed that 12 LNPs are located in the best performance LNPs region and 66.7% are N16B tailed LNPs (8 out of 12). Accordingly, −16.67% and 16.67% are the relative hit rates determined for O16B and N16B tailed LNPs regarding being the best performance LNPs (Fig. 7B, black bars). Above all, these results indicate that, to all the lipidoids studied here as intracellular Cas9:sgRNA RNP delivery nanocarriers targeting GFP-HEK cell line, N16B tailed LNPs generally perform better than the O16B tailed LNPs at both GFP gene knockout efficiency and cell compatibility. This phenomenon may carry over to other genome-editing protein platforms and cell types, and may be taken into consideration when fabricating new LNPs for intracellular delivery purposes. Our future research would assess more amine head groups with combinations of O16B and N16B tails across other cell types and cargos to further test this hypothesis, exploit the structure–activity relationship regarding amine head groups, and broaden these LNPs’ applications as carrier systems for bioactive molecules.
Hemolysis test and biodistribution study
Considering using lipid or lipidoid nanoparticles for CRISPR/Cas9 RNP delivery through a systemic administration has not been reported yet,26,27 the biodistribution of Cas9:sgRNA RNP complex loaded LNPs was further studied using fluorescent dye Cy7.5 labeled 103-N16B LNPs (103-N16B/Cholesterol/DOPE/DSPE-PEG2k-Cy7.5 = 16/4/1/1, weight ratio). A better understanding of the biodistribution of LNPs after systemic delivery is expected to be beneficial to future in vivo therapeutic applications.48–50 The hemolysis test was conducted at first to verify the compatibility of the LNPs with blood cells.51 Human red blood cells were used for this study, in which Triton X-100 and PBS are used as positive and negative controls, respectively. Fig. S9† shows that although certain LNPs like 113-O16B and 113-N16B appeared to induce a minimal amount of hemolysis (3.79% and 6.80%, respectively), the majority of LNPs synthesized in this study were shown to induce a negligible amount of hemolysis (<4% of hemolysis effect) which indicating their good compatibility with red blood cells. After that, Balb/c mice (n = 3) were injected with 50 μg of Cas9:sgRNA RNP loaded Cy7.5 labeled 103-N16B LNPs through the tail vein and fluorescence images were collected using a IVIS small animal imager. As shown in Fig. 8A, strong fluorescence signals were observed after the i.v. injections from similar locations at 30, 120, 180 and 240 min time intervals for all 3 mice. After 240 min, mice were sacrificed and major organs were collected and imaged. As shown in Fig. 8B and S10,† positive signals were recorded from lung, liver, kidney and spleen, while the liver showed strongest signals (Fig. 8C and S10†), indicating efficient accumulation of nanoparticles in the liver, which is a common phenomenon observed for drug delivery nanosystems.52,53 From a therapeutic perspective, it is expected that the LNPs reported here could be used for liver delivery of CRISPR/Cas9 platforms by designing and utilizing guide RNA molecules that target functional genes in liver, e.g. PCSK9.54 And it is also possible that by further in vivo screening and formulation optimization, LNPs that could deliver RNPs into other organs with high specificity could be developed.Conclusions
In summary, a lipidoids library containing O16B and N16B tails with disulfide bonds was synthesized and nanoparticles were fabricated. The hydrodynamic size, morphology, and stability of these nanoparticles were examined by DLS, TEM and fluorescence measurements. The typical LNPs showed spherical shapes with the size between 100–350 nm before and after Cas9:sgRNA RNP complex loading, as well as good stabilities during storage. Then fluorescent protein (−30)GFP-Cre was used as macromolecular cargo model and complexed with LNPs for internalization efficiency study. It was found that many of the reported LNPs were able to induce efficient protein transfection into the cells at a rate equal or greater than an effective and commercially available transfect reagent, Lpf2k. Then Cas9:sgRNA RNP complex targeting GFP gene was complexed with LNPs and the gene knockout efficiency and cytotoxicity were studied. Efficacious LNPs with >25% GFP knockout efficiencies as determined by flow cytometry and less toxic LNPs with >50% cell viabilities as determined by MTT assay were identified and further analysis showed that LNPs with N16B tails are overrepresented in both groups. Furthermore, the relative hit rate of LNPs with N16B tails in the LNPs with best performance (with both >25% GFP knockout efficiencies and >50% cell viabilities) group were also found to be positive indicating these lipidoids are good candidates for further biomedical applications.55–59 Hemolysis tests showed that these O16B and N16B tailed LNPs possess good compatibility with red blood cells and the in vivo biodistribution study revealed that the Cas9:sgRNA RNP complex loaded LNPs could efficiently accumulate into liver after the systemic administration (intravenous injection). Applications of gene editing in various areas,19,60–62 such as genetic disorders, are undergoing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by National Science Foundation Grant DMR 1452122 and NIH (1R21EB024041-01).References
- L. Cong, F. A. Ran, D. Cox, S. L. Lin, R. Barretto, N. Habib, P. D. Hsu, X. B. Wu, W. Y. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS PubMed
.
- P. Mali, L. H. Yang, K. M. Esvelt, J. Aach, M. Guell, J. E. DiCarlo, J. E. Norville and G. M. Church, Science, 2013, 339, 823–826 CrossRef CAS PubMed
.
- P. Horvath and R. Barrangou, Science, 2010, 327, 167–170 CrossRef CAS PubMed
.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed
.
- H. X. Wang, M. Li, C. M. Lee, S. Chakraborty, H. W. Kim, G. Bao and K. W. Leong, Chem. Rev., 2017, 117, 9874–9906 CrossRef CAS PubMed
.
- C. T. Charlesworth, P. S. Deshpande, D. P. Dever, B. Dejene, N. Gomez-Ospina, S. Mantri, M. Pavel-Dinu, J. Camarena, K. I. Weinberg and M. H. Porteus, bioRxiv, 2018 DOI:10.1101/243345
.
- E. Haapaniemi, S. Botla, J. Persson, B. Schmierer and J. Taipale, Nat. Med., 2018, 24, 927–930 CrossRef CAS PubMed
.
- Z. Glass, M. Lee, Y. M. Li and Q. B. Xu, Trends Biotechnol., 2018, 36, 173–185 CrossRef CAS PubMed
.
- L. Li, Z. Y. He, X. W. Wei, G. P. Gao and Y. Q. Wei, Hum. Gene Ther., 2015, 26, 452–462 CrossRef CAS PubMed
.
- M. Wang, Z. A. Glass and Q. Xu, Gene Ther., 2017, 24, 144–150 CrossRef CAS PubMed
.
- M. Hansen-Bruhn, B. E. F. de Avila, M. Beltran-Gastelum, J. Zhao, D. E. Ramirez-Herrera, P. Angsantikul, K. V. Gothelf, L. F. Zhang and J. Wang, Angew. Chem., Int. Ed., 2018, 57, 2657–2661 CrossRef CAS PubMed
.
- S. Kim, D. Kim, S. W. Cho, J. Kim and J. S. Kim, Genome Res., 2014, 24, 1012–1019 CrossRef CAS PubMed
.
- L. C. Tang, Y. T. Zeng, H. Z. Du, M. M. Gong, J. Peng, B. X. Zhang, M. Lei, F. Zhao, W. H. Wang, X. W. Li and J. Q. Liu, Mol. Genet. Genomics, 2017, 292, 525–533 CrossRef CAS PubMed
.
- S. Ramakrishna, A. K. Dad, J. Beloor, R. Gopalappa, S. K. Lee and H. Kim, Genome Res., 2014, 24, 1020–1027 CrossRef CAS PubMed
.
- B. T. Staahl, M. Benekareddy, C. Coulon-Bainier, A. A. Banfal, S. N. Floor, J. K. Sabo, C. Urnes, G. A. Munares, A. Ghosh and J. A. Doudna, Nat. Biotechnol., 2017, 35, 431–434 CrossRef CAS PubMed
.
- X. Gao, Y. Tao, V. Lamas, M. Q. Huang, W. H. Yeh, B. F. Pan, Y. J. Hu, J. H. Hu, D. B. Thompson, Y. L. Shu, Y. M. Li, H. Y. Wang, S. M. Yang, Q. B. Xu, D. B. Polley, M. C. Liberman, W. J. Kong, J. R. Holt, Z. Y. Chen and D. R. Liu, Nature, 2018, 553, 217–221 CrossRef CAS PubMed
.
- M. Wang, J. A. Zuris, F. T. Meng, H. Rees, S. Sun, P. Deng, Y. Han, X. Gao, D. Pouli, Q. Wu, I. Georgakoudi, D. R. Liu and Q. B. Xu, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2868–2873 CrossRef CAS PubMed
.
- J. A. Zuris, D. B. Thompson, Y. Shu, J. P. Guilinger, J. L. Bessen, J. H. Hu, M. L. Maeder, J. K. Joung, Z. Y. Chen and D. R. Liu, Nat. Biotechnol., 2015, 33, 73–80 CrossRef CAS PubMed
.
- W. J. Sun, W. Y. Ji, J. M. Hall, Q. Y. Hu, C. Wang, C. L. Beisel and Z. Gu, Angew. Chem., Int. Ed., 2015, 54, 12029–12033 CrossRef CAS PubMed
.
- Z. Glass, Y. M. Li and Q. B. Xu, Nat. Biomed. Eng., 2017, 1, 854–855 CrossRef PubMed
.
- K. Lee, M. Conboy, H. M. Park, F. G. Jiang, H. J. Kim, M. A. Dewitt, V. A. Mackley, K. Chang, A. Rao, C. Skinner, T. Shobha, M. Mehdipour, H. Liu, W. C. Huang, F. Lan, N. L. Bray, S. Li, J. E. Corn, K. Kataoka, J. A. Doudna, I. Conboy and N. Murthy, Nat. Biomed. Eng., 2017, 1, 889–901 CrossRef PubMed
.
- R. Mout, M. Ray, G. Y. Tonga, Y. W. Lee, T. Tay, K. Sasaki and V. M. Rotello, ACS Nano, 2017, 11, 2452–2458 CrossRef CAS PubMed
.
- M. Ray, Y. W. Lee, J. Hardie, R. Mout, G. Y. Tonga, M. E. Farkas and V. M. Rotello, Bioconjugate Chem., 2018, 29, 445–450 CrossRef CAS PubMed
.
- S. K. Alsaiari, S. Patil, M. Alyami, K. O. Alamoudi, F. A. Aleisa, J. S. Merzaban, M. Li and N. M. Khashab, J. Am. Chem. Soc., 2018, 140, 143–146 CrossRef CAS PubMed
.
- A. L. Fu, R. Tang, J. Hardie, M. E. Farkas and V. M. Rotello, Bioconjugate Chem., 2014, 25, 1602–1608 CrossRef CAS PubMed
.
- R. Mout, M. Ray, Y. W. Lee, F. Scaletti and V. M. Rotello, Bioconjugate Chem., 2017, 28, 880–884 CrossRef CAS PubMed
.
- L. Y. Wang, F. F. Li, L. Dang, C. Liang, C. Wang, B. He, J. Liu, D. F. Li, X. H. Wu, X. G. Xu, A. P. Lu and G. Zhang, Int. J. Mol. Sci., 2016, 17, 626 CrossRef PubMed
.
- S. Altinoglu, M. Wang and Q. B. Xu, Nanomedicine, 2015, 10, 643–657 CrossRef CAS PubMed
.
- M. Wang, K. Alberti, S. Sun, C. L. Arellano and Q. B. Xu, Angew. Chem., Int. Ed., 2014, 53, 2893–2898 CrossRef CAS PubMed
.
- M. Wang, S. Sun, K. A. Alberti and Q. B. Xu, ACS Synth. Biol., 2012, 1, 403–407 CrossRef CAS PubMed
.
- M. Wang, S. Sun, C. I. Neufeld, B. Perez-Ramirez and Q. B. Xu, Angew. Chem., Int. Ed., 2014, 53, 13444–13448 CrossRef CAS PubMed
.
- X. Y. Wang, Y. M. Li, Q. S. Li, C. I. Neufeld, D. Pouli, S. Sun, L. Yang, P. Deng, M. Wang, I. Georgakoudi, S. Q. Tang and Q. B. Xu, J. Controlled Release, 2017, 263, 39–45 CrossRef CAS PubMed
.
- Y. Li, T. Yang, Y. Yu, N. Shi, L. Yang, Z. Glass, J. Bolinger, I. J. Finkel, W. Li and Q. Xu, Biomaterials, 2018 DOI:10.1016/j.biomaterials.2018.03.011
.
- A. Akinc, A. Zumbuehl, M. Goldberg, E. S. Leshchiner, V. Busini, N. Hossain, S. A. Bacallado, D. N. Nguyen, J. Fuller, R. Alvarez, A. Borodovsky, T. Borland, R. Constien, A. de Fougerolles, J. R. Dorkin, K. N. Jayaprakash, M. Jayaraman, M. John, V. Koteliansky, M. Manoharan, L. Nechev, J. Qin, T. Racie, D. Raitcheva, K. G. Rajeev, D. W. Y. Sah, J. Soutschek, I. Toudjarska, H. P. Vornlocher, T. S. Zimmermann, R. Langer and D. G. Anderson, Nat. Biotechnol., 2008, 26, 561–569 CrossRef CAS PubMed
.
- Y. Z. Dong, A. A. Eltoukhy, C. A. Alabi, O. F. Khan, O. Veiseh, J. R. Dorkin, S. Sirirungruang, H. Yin, B. C. Tang, J. M. Pelet, D. L. Chen, Z. Gu, Y. Xue, R. Langer and D. G. Anderson, Adv. Healthcare Mater., 2014, 3, 1392–1397 CrossRef CAS PubMed
.
- B. Li, X. Luo, B. B. Deng, J. B. Giancola, D. W. McComb, T. D. Schmittgen and Y. Z. Dong, Sci. Rep., 2016, 6, 22137 CrossRef CAS PubMed
.
- K. P. Mahon, K. T. Love, K. A. Whitehead, J. Qin, A. Akinc, E. Leshchiner, I. Leshchiner, R. Langer and D. G. Anderson, Bioconjugate Chem., 2010, 21, 1448–1454 CrossRef CAS PubMed
.
- K. A. Whitehead, J. R. Dorkin, A. J. Vegas, P. H. Chang, O. Veiseh, J. Matthews, O. S. Fenton, Y. L. Zhang, K. T. Olejnik, V. Yesilyurt, D. L. Chen, S. Barros, B. Klebanov, T. Novobrantseva, R. Langer and D. G. Anderson, Nat. Commun., 2014, 5, 4277 CrossRef CAS PubMed
.
- M. Wang, K. Alberti, A. Varone, D. Pouli, I. Georgakoudi and Q. B. Xu, Adv. Healthcare Mater., 2014, 3, 1398–1403 CrossRef CAS PubMed
.
- G. T. Zugates, D. G. Anderson, S. R. Little, I. E. B. Lawhorn and R. Langer, J. Am. Chem. Soc., 2006, 128, 12726–12734 CrossRef CAS PubMed
.
- A. Akbarzadeh, R. Rezaei-Sadabady, S. Davaran, S. W. Joo, N. Zarghami, Y. Hanifehpour, M. Samiei, M. Kouhi and K. Nejati-Koshki, Nanoscale Res. Lett., 2013, 8, 102 CrossRef PubMed
.
- Y. M. Li, G. H. Liu, X. R. Wang, J. M. Hu and S. Y. Liu, Angew. Chem., Int. Ed., 2016, 55, 1760–1764 CrossRef CAS PubMed
.
- K. Dan and S. Ghosh, Angew. Chem., Int. Ed., 2013, 52, 7300–7305 CrossRef CAS PubMed
.
- W. Cao, L. Wang and H. P. Xu, Nano Today, 2015, 10, 717–736 CrossRef CAS
.
- E. K. Lim, T. Kim, S. Paik, S. Haam, Y. M. Huh and K. Lee, Chem. Rev., 2015, 115, 327–394 CrossRef CAS PubMed
.
- S. Mitragotri, P. A. Burke and R. Langer, Nat. Rev. Drug Discovery, 2014, 13, 655–672 CrossRef CAS PubMed
.
- Q. H. Sun, Z. X. Zhou, N. S. Qiu and Y. Q. Shen, Adv. Mater., 2017, 29, 1606629 Search PubMed
.
- Y. Wei, Y. Wang, D. Xia, S. Guo, F. Wang, X. Zhang and Y. Gan, ACS Appl. Mater. Interfaces, 2017, 9, 25138–25151 CrossRef CAS PubMed
.
- S. P. Yoo, F. Pineda, J. C. Barrett, C. Poon, M. Tirrell and E. J. Chung, ACS Omega, 2016, 1, 996–1003 CrossRef CAS PubMed
.
- Y. Zhang, J. Wei, J. Xu, W. S. Leong, G. Liu, T. Ji, Z. Cheng, J. Wang, J. Lang, Y. Zhao, L. You, X. Zhao, T. Wei, G. J. Anderson, S. Qi, J. Kong, G. Nie and S. Li, ACS Appl. Mater. Interfaces, 2018, 10, 2347–2353 CrossRef CAS PubMed
.
- Y. M. Li, H. S. Yu, Y. F. Qian, J. M. Hu and S. Y. Liu, Adv. Mater., 2014, 26, 6734–6741 CrossRef CAS PubMed
.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed
.
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed
.
- Q. R. Ding, A. Strong, K. M. Patel, S. L. Ng, B. S. Gosis, S. N. Regan, C. A. Cowan, D. J. Rader and K. Musunuru, Circ. Res., 2014, 115, 488–492 CrossRef CAS PubMed
.
- X. W. Du, J. Zhou, J. F. Shi and B. Xu, Chem. Rev., 2015, 115, 13165–13307 CrossRef CAS PubMed
.
- X. W. Li, S. Y. Tzeng, X. Y. Liu, M. Tammia, Y. H. Cheng, A. Rolfe, D. Sun, N. Zhang, J. J. Green, X. J. Wen and H. Q. Mao, Biomaterials, 2016, 84, 157–166 CrossRef CAS PubMed
.
- S. Shen, J. X. Xia and J. Wang, Biomaterials, 2016, 74, 1–18 CrossRef CAS PubMed
.
- Z. Y. Song, Z. Y. Han, S. X. Lv, C. Y. Chen, L. Chen, L. C. Yin and J. J. Cheng, Chem. Soc. Rev., 2017, 46, 6570–6599 RSC
.
- Y. Wang, A. G. Cheetham, G. Angacian, H. Su, L. S. Xie and H. G. Cui, Adv. Drug Delivery Rev., 2017, 110, 112–126 CrossRef PubMed
.
- N. Bertrand, J. Wu, X. Y. Xu, N. Kamaly and O. C. Farokhzad, Adv. Drug Delivery Rev., 2014, 66, 2–25 CrossRef CAS PubMed
.
- J. Wu, N. Kamaly, J. J. Shi, L. L. Zhao, Z. Y. Xiao, G. Hollett, R. John, S. Ray, X. Y. Xu, X. Q. Zhang, P. W. Kantoff and O. C. Farokhzad, Angew. Chem., Int. Ed., 2014, 53, 8975–8979 CrossRef CAS PubMed
.
- H. Yin, C. Q. Song, S. Suresh, Q. Q. Wu, S. Walsh, L. H. Rhym, E. Mintzer, M. F. Bolukbasi, L. J. Zhu, K. Kauffman, H. W. Mou, A. Oberholzer, J. M. Ding, S. Y. Kwan, R. L. Bogorad, T. Zatsepin, V. Koteliansky, S. A. Wolfe, W. Xue, R. Langer and D. G. Anderson, Nat. Biotechnol., 2017, 35, 1179–1187 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic routes, 1H and 13C NMR spectra, DLS measurements, MTT assay, hemolysis test, TEM and fluorescence images. See DOI: 10.1039/c8bm00637g |
| This journal is © The Royal Society of Chemistry 2019 |