Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Molecular genetic and biochemical characterization of a putative family of zinc metalloproteins in Caenorhabditis elegans†
Poulami
Chaudhuri
a,
Hasan Tanvir
Imam
b,
Yona
Essig
a,
Jovaras
Krasauskas
a,
Samuel M.
Webb
c,
Claudia A.
Blindauer
 b and
Stephen R.
Stürzenbaum
b and
Stephen R.
Stürzenbaum
 *a
*a
aKing's College London, School of Population Health & Environmental Sciences, Faculty of Life Sciences & Medicine, 150 Stamford Street, London, SE1 9NH, UK. E-mail: stephen.sturzenbaum@kcl.ac.uk
bUniversity of Warwick, Department of Chemistry, Gibbet Hill, Coventry, CV4 7AL, UK
cStanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, 2575 Sand Hill Road, Menlo Park, CA 94025, USA
First published on 16th November 2018
Abstract
Four highly similar genes (W08E12.2, W08E12.3, W08E12.4 and W08E12.5) which are consecutively aligned on chromosome IV of the C. elegans genome are predicted to code for small (120–141aa) yet cysteine rich (18–19 cysteines) proteins. Cloning and sequencing of the genomic regions of the isoforms confirmed the presence and order of all genes. The generation of transgenic worms strains with an integrated single copy or extrachromosomal multi-copy PW08E12.3;W08E12.4::GFP uncovered that W08E12.3 and W08E12.4 are constitutively expressed in the pharynx and significantly induced in worms exposed to 100 μM Zn. Knockdown by RNAi did not have a marked consequence on reproductive performance nor was a Zn-dependent effect on nematode growth observed. However, RNAi of these genes led to an accumulation of Zn in the intestinal cells. W08E12.3 was recombinantly expressed in E. coli and the purified protein was shown to be able to bind up to 6.5 Zn molecules at neutral pH. Zn-binding was acid-labile and the apo protein was observed at pH < 4.3. This characterization suggests W08E12.2, W08E12.3, W08E12.4 and W08E12.5 belong to a family of putative Metalloproteins which, akin to metallothioneins, may play an important role in Zn-sensing, homeostasis and/or detoxification.
Significance to metallomicsC. elegans metallothionein and phytochelatin synthase mutants are surprisingly resilient when challenged with a toxic load of heavy metals. This suggests that other, yet to be identified, metalloproteins may also be involved in essential metal homeostasis and non-essential metal detoxification. This paper provides the first characterization of a putative family of metalloproteins, focusing primarily on molecular genetic and biochemical studies to define their involvement in Zn metabolism/detoxification. |
1. Introduction
C. elegans is an ideal model to study toxicity and toxicological mechanisms of heavy metals.1 Most studies have focused on toxic endpoints including reproduction, life span, lethality and protein expression. More specifically, the effect of metals on the nervous system has been studied by investigating the behaviour, reporter expression and neuronal morphology.2 Moreover, silencing methodologies such as knockdown by RNAi or the generation of chromosomal deletions provide opportunities for a targeted manipulation of specific genes at the molecular level. This has helped to unveil information about how certain mutants are more sensitive to metal toxicity.3–5 The transparent body of C. elegans allows the visualization of fluorescently labelled molecules, genes and proteins in vivo,6 a feature that can aid in the investigation of metal detoxification pathways and is amenable for assessing the interaction between the genes and environmental factors. The role of the nematode as a biosensor for environmental risk assessment has been explored by exploiting the current knowledge of established biomarkers such as glutathione (GSH), metallothionein (MT), heat shock proteins (HSPs) and pumps and transporters involved in metal detoxification. C. elegans is thus an indispensable model to complement more classical (mammalian) toxicological research.2,7The homeostatic control of heavy metals is biologically complex as it requires an intricate balance between maintaining essential micronutrients (e.g. Zn and Cu) and the detoxification of harmful metals (e.g. Cd). Two prominent pathways involved in this challenge are the phytochelatins (PCs)/phytochelatin synthase (pcs-1)8–11 and the MTs (in C. elegans nomenclature MTs are referred to as MTLs).12–16 PCs are a family of metal-inducible thiol-rich peptides that are synthesised enzymatically and play a prominent role in the detoxification of heavy metals by acting as chelators.17 The PCs form complexes with toxic metals in the cytosol of the cell which are transported into the vacuole, protecting the organism from heavy metal toxicosis18 and the biosynthesis of PCs is auto-regulated by this metal chelation.19 Unlike PCs, expression of metallothionein is driven by transcriptional activation and the resultant proteins bind metals and act as antioxidants.20C. elegans possesses two metallothionein (MT) genes, mtl-1 and mtl-2, which have been shown to play an important role in the protection from metal toxicity. They are thought to have distinct functions. The mtl-1 is constitutively expressed in the lower pharyngeal bulb in the absence of metal exposure and thus may act as a metal sensor. The expression of mtl-1 and mtl-2 is elevated in the gut region upon metal exposure. Both isoforms have preferences for metal binding, where mtl-1 is biased towards Zn(II) and mtl-2 towards Cd(II).16,21 The single and double knockout of mtl-1 and/or mtl-2 as well as the mtl-1;mtl-2;pcs-1 triple knockout mutants are all characterized by an increased sensitivity (compared to wild type) when challenged with high doses of Zn or Cd, at least in terms of development and reproduction.10,22 However, given that the mutants are all viable and capable of producing offspring suggests that other, hitherto uncharacterized metalloproteins must play equally important roles in essential metal homeostasis and non-essential metal detoxification.
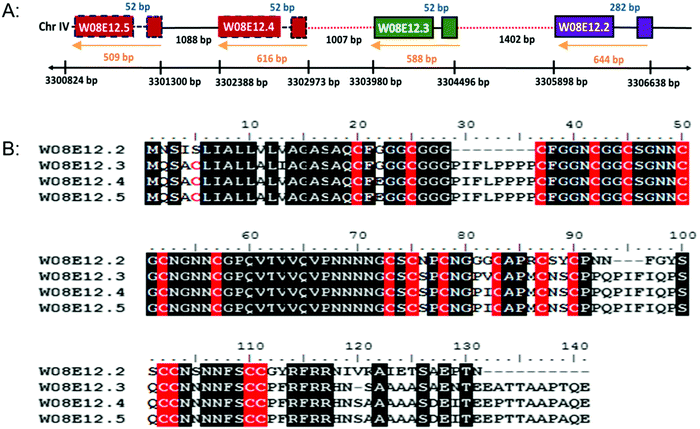
The C. elegans whole genome sequencing effort23 has allowed the building of WormBase (http://www.wormbase.org), an exquisitely detailed database. A manual screen of the database identified four highly conserved genes that are chronologically arranged on chromosome IV. Annotated as W08E12.2, W08E12.3, W08E12.4, and W08E12.5 (based on their position within the sequenced cosmid W08E12) they are predicted to encode cysteine-rich proteins. The sequences are highly similar to each other as well as to uncharacterized genes present in other Caenorhabditis species (http://www.wormbase.org). Given that the C. elegans genome is compact (with just over 100 million bp encoding for about 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 proteins), multi-copy isoforms/isomers are less frequent compared to higher organisms with more complex genomes. The aligned (sequential) occurrence of numerous members of a highly similar gene family within a compact genome is therefore relatively rare and highlights that their role within the nematode is likely to be significant. Moreover, the abundance and conservation of cysteine residues within W08E12.2, W08E12.3, W08E12.4 and W08E12.5 suggests that this family may be involved in the binding of heavy metals, a notion this paper sets out to explore in more detail.
000 proteins), multi-copy isoforms/isomers are less frequent compared to higher organisms with more complex genomes. The aligned (sequential) occurrence of numerous members of a highly similar gene family within a compact genome is therefore relatively rare and highlights that their role within the nematode is likely to be significant. Moreover, the abundance and conservation of cysteine residues within W08E12.2, W08E12.3, W08E12.4 and W08E12.5 suggests that this family may be involved in the binding of heavy metals, a notion this paper sets out to explore in more detail.
2. Materials and methods
2.1 Nematode strains and culturing
C. elegans were grown at 20 °C on nematode growth media (NGM) plates with E. coli OP50 as food source. The nematodes were age-synchronized by treating gravid adults with alkaline hypochlorite. The isolated eggs were rotated overnight in M9 buffer (KH2PO4 (22 mM), Na2HPO4 (42 mM), NaCl (85.5 mM)) in distilled water, autoclaved then adding MgSO4 (1 mM) to allow nematodes to hatch and arrest at L1 stage. The age-synchronized (L1) nematodes were placed on NGM plates ready for subsequent experiments. Wild type nematodes were sourced from the Caenorhabditis Genetics Centre (University of Minnesota) and the transgenic strain cop-136(PW08E12.3/4::GFP) was created by Mos1-mediated single copy insertion (KnudraTransgenics, USA). In addition, an extrachromosomal, multi-copy version (zsEx6(PW08E12.3/4::GFP)) was obtained by injecting the pPD95.75 vector containing 1115 bp of the W08E12.3/4 promoter fused in-frame to the coding sequence of the GFP gene. A plasmid containing rol-6 was used as co-injection marker to allow the identification of transgenic organisms.2.2 Metal exposure and sample collection
Age-synchronized nematodes were exposed to Zn by adding the heavy metal in equimolar concentrations to the NGM agar and the bacterial food source (E. coli OP50). The cop-136 (PW08E12.3/4::GFP) strain was exposed to concentrations ranging from 0–800 μM Zn for 48 h for fluorescence quantitation from bulk samples. For X-ray fluorescence imaging, control (wild type) and W08E12.3/4 RNAi worms were grown in the presence of 150 μM Zn. Developmental growth and brood size experiments were performed for control (wild type) and W08E12.3/4 RNAi worms challenged with 200 μM Zn. The concentration ranges were selected based on publications that characterized other C. elegans metalloproteins.11,162.3 Quantification of gene expression using GFP alleles
GFP-tagged transgenic worms cop-136(PW08E12.3/4::GFP) and zsEx6(PW08E12.3/4::GFP) were exposed to 100, 400, 600, 800 μM Zn to evaluate changes in GFP signal intensity and also to determine the localization of expression. For single worm imaging, the transgenic worms were maintained on Zn-containing plates/OP50 from L1 to L4 stage, then imaged. L4 worms (4–8) were picked onto a glass slide carrying a drop of M9 solution to which sodium azide (2%) was added to immobilize the animals. The worms were imaged by means of an inverted fluorescence Nikon microscope, using a blue laser scanning fluorescence (λex = 450–490 nm) and standard phase contrast microscopy. The images were captured using a Nikon camera (Nikon UK Ltd, Kingston upon Thames, UK) at 10× magnification and analysed by dVision software or Image J software.For the nematode based bulk assay, transgenic PW08E12.3/4::GFP-tagged worms were exposed to Zn (as described above) from stage L1 to L4 stage and collected. At least three M9 washes were carried out to remove the OP50 bacteria. Octyl phenyl-polyethylene glycol (IGEPAL) was added to the tube (1 μL) to prevent the worms sticking to the pipette tips. Following a titre, 5000 worms were aliquoted into a black 96 well microplate with three repeats for each condition. The microplate was scanned at 355 nm excitation and 510 nm emission wavelength using an automated plate reader (Fluoroskan Ascent FL, ThermoFisher). Following the plate reading, the worms were imaged using a blue laser scanning fluorescence (λex = 450–490 nm) and standard phase contrast microscopy in order to test the reproducibility between the bulk assay and single worm imaging.
2.4 X-ray fluorescence imaging (XFI)
The metal accumulation pattern in individual worms was determined by X-ray fluorescence imaging (XFI) conducted at the Stanford Synchrotron Radiation Laboratory (SSRL). Briefly, the nematodes were immobilised by adding sodium azide on a plastic slide. Zn was measured at 10 keV incident energy selected with a Si(111) double crystal monochromator. The X-ray beam was focussed at a spot size of 5 × 5 μm using a Rh-coated Kirkpatrick–Baez (KB) mirror pair (Xradia Inc). The worms were rastered continuously across the beam and Zn was monitored at each pixel by recording the intensity of the fluorescence lines using a silicon drift Vortex detector (Hitachi) equipped with Xspress3 electronics (Quantum Detectors) with a dwell time/pixel of approximately 75 ms. Calibration was performed with an X-ray fluorescence thin film standard (MicroMatter). The data were quantitatively analysed with the Micro Analysis Toolkit (Webb 2011, Palo Alto, CA, USA), which allowed the visualisation and the determination of the zinc concentration in individual nematodes.2.5 Life history traits (growth and brood size)
Two toxicity assays (growth over time and brood size) were performed on wild type and W08E(12.3–12.5) knockdown worms in the presence or absence of Zn (200 μM). For both assays, L1 worms were plated onto control and metal supplemented NGM plates seeded with HT115 bacteria (containing either the empty RNAi vector pPD129.36 or pPD129.36 expressing W08E(12.3–12.5)).For the brood size assay, after the worms reached the L4 stage, single worms were picked at L4 stage into individual wells of a 12-well plate containing NGM with or without Zn. Each worm was transferred each day at the same time onto a new well until the egg-laying period was completed. The hatched progeny from each worm was scored once the progeny reached the L2/L3 stage. The worms were maintained at 20 °C throughout the assay.
To record the change in the growth of the nematodes upon metal exposure, the flat volumetric body area was recorded from day 2 of plating until day 6 (i.e. from stage L2 to adult). Changes in growth were captured by taking digital pictures of each nematode every day at the same time under the same magnification of the microscope throughout the development period. The images were analysed using the Image-Pro Express software.
2.6 Statistics
The data derived from fluorescent quantification, XFI, life history analyses were scrutinized by one-way ANOVA testing followed by a Dunnett's multiple comparison test (Prism, Graphpad).2.7 Recombinant expression of the W08E12.3 protein
W08E12.3 was cloned, in frame, into the NcoI and SalI restriction sites of the pET29a vector system. Due to the cloning strategy, six additional nucleotides (encoding for a methionine and a glutamate) were inserted between the S-tag and the start codon. The construct sequence was confirmed by standard Sanger sequencing. The calculated theoretical mass of the recombinantly expressed protein was 17280.51 Da.The protein was recombinantly expressed in BL21 (DE3pLysS) cells in a similar manner to that previously used for C. elegans metallothioneins.16 A single colony was picked and grown in LB broth in the presence of Kanamycin (50 ng mL−1) for 8 h at 37 °C. This starter culture was used to inoculate (with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) a larger volume of LB broth with Kanamycin. The cultures were grown to OD600 0.6 and then induced with Isopropylthiogalactoside (IPTG, 1 mM). After 30 min of induction, ZnCl2 (100 μM) was added to the culture and cells were grown for 5 h at 37 °C with shaking. After induction, the cells were harvested by centrifugation at 1000g for 20 min. The pelleted cells were frozen at −80 °C until further use.
10) a larger volume of LB broth with Kanamycin. The cultures were grown to OD600 0.6 and then induced with Isopropylthiogalactoside (IPTG, 1 mM). After 30 min of induction, ZnCl2 (100 μM) was added to the culture and cells were grown for 5 h at 37 °C with shaking. After induction, the cells were harvested by centrifugation at 1000g for 20 min. The pelleted cells were frozen at −80 °C until further use.
2.8 Protein purification
The frozen cell pellets were resuspended in 15 mL resuspension buffer (200 mM Tris–HCl, pH 7.5; 150 mM NaCl, 1 mM DTT, 0.02% sodium azide, 0.1% TritonX) including an EDTA-free protease inhibitor cocktail (Roche). The cell suspension was processed in a cell disruptor (Constant Systems, 20.3 PSI) and the resulting mixture was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min in a floor-standing centrifuge. The supernatant was discarded and the pellet was resuspended (10 mL resuspension buffer), processed a second time in the cell disruptor, and centrifuged again. The pellet, containing inclusion bodies, was collected. The protein was extracted from inclusion bodies following the protocol by Oneda and Inouye, 1999.24 The inclusion bodies were dissolved in 5 mL of solubilisation buffer (0.1 M Tris HCl buffer, 6 M guanidine HCl, 0.1 M DTT, pH 7.5) and the resulting mixture was left on an orbital shaker at 4 °C overnight. The solubilised protein was added dropwise (with constant stirring) to a 100-fold volume of refolding buffer (50 mM Tris HCl, 0.1 mM Zn acetate, 1 mM DTT, 0.2 M NaCl, 10 mM CaCl2, 0.02% sodium azide, pH 7.5, 0.25 M L-arginine) at 4 °C. The resulting mixture contained some precipitate, which was removed by centrifugation (3000g for 15–20 min).
000g for 30 min in a floor-standing centrifuge. The supernatant was discarded and the pellet was resuspended (10 mL resuspension buffer), processed a second time in the cell disruptor, and centrifuged again. The pellet, containing inclusion bodies, was collected. The protein was extracted from inclusion bodies following the protocol by Oneda and Inouye, 1999.24 The inclusion bodies were dissolved in 5 mL of solubilisation buffer (0.1 M Tris HCl buffer, 6 M guanidine HCl, 0.1 M DTT, pH 7.5) and the resulting mixture was left on an orbital shaker at 4 °C overnight. The solubilised protein was added dropwise (with constant stirring) to a 100-fold volume of refolding buffer (50 mM Tris HCl, 0.1 mM Zn acetate, 1 mM DTT, 0.2 M NaCl, 10 mM CaCl2, 0.02% sodium azide, pH 7.5, 0.25 M L-arginine) at 4 °C. The resulting mixture contained some precipitate, which was removed by centrifugation (3000g for 15–20 min).
The solubilised protein from the inclusion bodies was concentrated and buffer exchanged into 20 mM NH4HCO3 by ultracentrifugation (MWCO 3 kDa, Amicon Ultra, Millipore), loaded onto a gel filtration column (FPLC 16/60 HiLoad 75 Superdex prep grade, Amersham Biosciences) connected to an FPLC system (Pharmacia Akta Purifier) and eluted using 20 mM NH4HCO3. The elution of protein fractions was monitored at 220 nm (peptide bonds and Zn–S ligand-to-metal charge transfer) and 280 nm (aromatic residues). The eluted fractions containing the pure protein were pooled and re-concentrated by ultracentrifugation (MWCO 3 kDa, Amicon Ultra, Millipore). During purification, the S-tagged protein was detected by SDS-PAGE and Western blotting employing an S-tag monoclonal primary antibody and polyclonal goat-anti-mouse immunoglobulins/HRP secondary antibody.
2.9 Electrospray ionisation mass spectrometry (ESI-MS)
The S-tagged W08E12.3 protein was desalted in 10 mM NH4HCO3 (pH 7.9) using a PD-10 Sephadex G-25 column (GE Healthcare) and subsequent ESI-MS was performed at acidic pH to obtain the metal-free (apo) form of the protein. The sample was made up of 40 μM protein in 10 mM NH4HCO3 and 10% methanol, with the addition of 2% formic acid. Mass spectra were recorded on a Bruker Daltonics MicroTOF fitted with an electrospray ionization source operating in positive mode. The samples were injected into the spectrometer at a flow rate of 240 μL hour−1. Instrument conditions were as follows: source temperature 195 °C, nebulizer 0.6 Bar, dry gas 4.5 L minute−1, capillary exit 100 V, skimmer1 50 V, skimer2 25.2 V, hexapole1 24.2 V, hexapole2 22.4 V, hexapole RF 450 V, transfer time 81 μs and detector TOF 2300 V. The data were recorded for 0.4–2 min over the range of 500–5000 m/z. The experimental data were smoothed and deconvoluted by means of the Bruker DataAnalysis software v. 4.0 (Bruker Daltonics).2.10 Inductively coupled plasma optical emission spectroscopy (ICP-OES)
Inductively Coupled Plasma-Optical Emission Spectroscopy (ICP-OES) (Perkin-Elmer Optima 5300 DV, Model S10) was applied to determine the S and Zn content of the proteins by measuring S at 180.669 nm and 181.975 nm, and Zn at 206.200 nm and 213.857 nm. Plasma operating conditions were: argon (Ar) flow rate 13.0 L minute−1, auxiliary gas flow rate 0.2 L minute−1, nebuliser flow rate 0.8 L minute−1 and RF power at 1300 W. About 30 mg (1/50th of the total pellet) of the inclusion body fraction was dissolved in 600 μL of 1 M nitric acid and then diluted 10-fold for ICP-OES sample preparation. Samples from gel filtration fractions were prepared for ICP-OES in 0.1 M nitric acid at the ratio of 1 part of sample![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 parts of ultrapure nitric acid (prepared from 72% distilled HNO3). The ICP-OES instrument was calibrated using S and Zn standards (1–5 ppm) prepared from 1000 ppm stocks (TraceCERT, Sigma-Aldrich). All samples were analysed using three replicate readings, with a washing time of 60 seconds for each sample. Data were analysed using the WinLab 32 software (Perkin-Elmer). The concentration of the protein was determined via S content from these ICP-OES measurements, based on 22 sulfur atoms from 19 cysteine and 3 methionine residues.25
3 parts of ultrapure nitric acid (prepared from 72% distilled HNO3). The ICP-OES instrument was calibrated using S and Zn standards (1–5 ppm) prepared from 1000 ppm stocks (TraceCERT, Sigma-Aldrich). All samples were analysed using three replicate readings, with a washing time of 60 seconds for each sample. Data were analysed using the WinLab 32 software (Perkin-Elmer). The concentration of the protein was determined via S content from these ICP-OES measurements, based on 22 sulfur atoms from 19 cysteine and 3 methionine residues.25
2.11 pH titration
A pH titration was conducted to estimate the strength of metal binding to the S-tagged W08E12.3 protein. The protein was buffer-exchanged into 10 mM NH4HCO3 buffer (pH 7.9). pH titration of the Zn-loaded protein was monitored by UV-visible spectroscopy (Cary 50, Varian) in the range of 200–300 nm at 298 K. Respective protein samples (1.5 mL, 5–10 μM, 10 mM NH4HCO3 buffer, pH 7.9) were titrated with HCl (1–5 μL, 0.01–5 M). Each absorbance measurement was taken after 10 min of equilibration time. pH readings were taken before and after the UV spectrometric measurement.3. Results and discussion
W08E12.2, W08E12.3, W08E12.4 and W08E12.5 are consecutively aligned on chromosome IV of C. elegans (http://www.wormbase.org). The genomic region spanning W08E12.2 to W08E12.5 was cloned and sequenced, which confirmed the information (position and sequences) provided by the WormBase database (data not shown). All genes possess similar intron and exon structures and are evenly spaced out (Fig. 1A). A sequence alignment depicts the high level of sequence similarity within the respective coding regions, notably with W08E12.4 and W08E12.5 being 100% identical. Likewise, the promoter regions of W08E12.3 and W08E12.4 are identical. The predicted (in silico translated) protein sequences revealed that all four proteins are cysteine-rich (18–19 cysteines with a total length of 120–141aa) (Fig. 1B). A bioinformatic screen was conducted in PERL using the UVA FASTA SERVER to define how often a chain of four (or more) highly similar genes are sequentially positioned within the nematode genome. Three criteria were set, namely the identified transcripts need to be highly similar (e ≤ 0.0001), each chain should contain at least four genes and the gap between two consecutive transcripts should be ≤2000 bp. This identified 116 chains comprising clusters of 4–12 transcripts, 53 of which were composed of four transcripts (Table S1, ESI†). This similarity suggested that a recent duplication event may have taken place. In contrast, W08E12.2 was found to be more different in certain parts of the coding region, possibly due to a more distant divergence. Gene duplication resulting in a tandem repeat is estimated at 60.5 and 58.8 × 10−11 per gene per generation in D. melanogaster and C. elegans.26 First, this demonstrates that gene duplication rates are comparable between flies and worms and second, emphasizes that a second duplication (resulting in a string of four genes) must be rare and perhaps underlines the functional importance of this family as the expression of tandem repeats often exceeds the sum of duplications.27The gene family shares a significant sequence homology to uncharacterised genes present in other Caenorhabditis species, including C. briggsae, C. remanei but also with keratin associated proteins (65–75%) of Homo sapiens and Mus musculus. The hair keratin intermediate filaments are embedded in a matrix comprising keratin associated proteins, which form disulphide bonds with abundant cysteine residues of hair keratins.28 Cytokeratins have proven to resist Cd induced apoptosis by representing an adaptive survival mechanism.29 In the case of As poisoning, a temporary accumulation of the metal takes place in the soft tissue organs but the major long term accumulation takes place in keratin rich tissues such as nails, hair and skin.30 Cu, Hg, Ag, Cd, Pb, Cr and Al are taken up by wool keratin.31 Keratin protein fibres from hair, wool, horns and feather have been used to absorb heavy metals in industry for water purification and air cleaning.32 The homology of W08E12.2 to W08E12.5 with keratin associated protein confirms the possible association of the gene family with heavy metals. The JASPAR CORE database was chosen to screen for putative metal binding sites within 1000 bp of the promoter region which revealed a list of Zn-coordinating transcription factor binding sites, many of which are in common with the metallothioneins (MTs) including ELT-3, CHE-1, EOR-1 and BLMP-1. The C. elegans metallothioneins (MTL-1 and MTL-2) are small (75 and 63aa, respectively) cysteine-rich (19/18aa) proteins involved in essential metal homeostasis and toxic metal detoxification.23 Unlike most other animal genomes, the nematode does not encode for a classical MTF-1 transcription factor, and expression is believed to be modulated by ELT-2 and GATA.13 W08E12.2 to W08E12.5 are longer than MTLs (120–141aa), however with a comparable cysteine content (18–19 cysteines), therefore providing circumstantial evidence of a similar function (Table S2, ESI†).
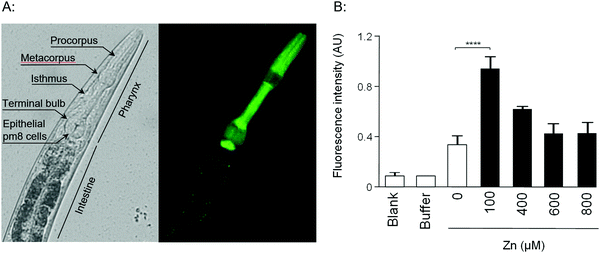
Given that the promoter regions of W08E12.3 and W08E12.4 are identical, they were used to design a PW08E12.3/4::GFP construct which was introduced into position ttTi5605 of chromosome II by Mos1-mediated single copy insertion yielding cop-136(PW08E12.3/4::GFP). In addition, an extrachromosomal, multi-copy version was created (zsEx6(PW08E12.3/4::GFP)) by injecting the pPD95.75 vector. Both constructs contained 1115 bp of the W08E12.3/4 promoter fused in-frame to the coding sequence of GFP. The transgenic strain cop-136(PW08E12.3/4::GFP) was visualized at L4 stage using an inverted fluorescence microscope, which revealed that the expression of W08E12.3/W08E12.4 is, at large, confined to the pharynx, and in particular pronounced in the dorsal epithelial pm8 cells present above the pharyngeal-intestinal valve (Fig. 2A), which incidentally aligns with the location of the constitutive expression (i.e. in the absence of metal supplementation) of mtl-113,14 and supports the notion that mtl-1 and W08E12.3/4 may possibly function in a similar or complementary way. The location, albeit with a significantly stronger signal multi-copy expression, was confirmed with the extrachromosomal variant zsEx6(PW08E12.3/4::GFP) (Fig. S1, ESI†). The cop-136(PW08E12.3/4::GFP) transgenic strain was exposed to a concentration range (100–800 μM) of Zn from L1 to L4 stage (i.e. for 48 h) and changes in fluorescence were measured via a bulk assay using a fluorescence plate reader. Whilst the fluorescent signal of unexposed worms was statistically indistinguishable to samples chronically exposed to high concentrations of Zn (400 μM to 800 μM), a statistically significant increase in fluorescence intensity was observed in transgenic worms exposed to 100 μM Zn. (Fig. 2B), possibly due to the zinc-induced toxicity as reported by others.33–35
The location of expression remained within the pharyngeal area and thus is, at least in this context, fundamentally different to MTLs which are induced within the intestinal cells of heavy metal exposed worms.13,14 At this stage we are not able to stipulate whether the observed transcriptional induction of W08E12.3/4 is due to a direct (e.g. via a zinc bound transcription factor) or indirect effect, a line of research warranting further investigations.
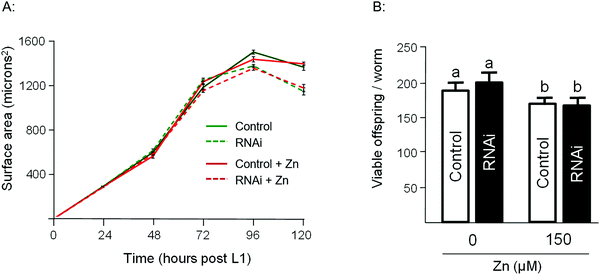
To assess the Zn-link further, the expression of W08E12.3 to W08E12.5 was knocked down by RNAi. Due to the sequence similarity it was possible to design a construct that was able to knock-down, in concert, W08E12.3 to W08E12.5 and a second construct that was specific to the knock-down of W08E12.3. In both cases the knock-down efficiency exceeded 50%, however due to the sequence divergence neither of the two RNAi constructs was able to knock-down W08E12.2 (data not shown). Wild type N2 (control worms fed with HT115 bacteria containing the empty RNAi vector) and the RNAi counterparts were exposed to a dose of Zn (150 μM) for 48 h. X-ray Fluorescence Imaging (XFI) was applied to quantify the concentration of Zn in individual worms at 5 × 5 μm resolution.36 Upon knock-down by RNAi (using the construct targeting W08E12.3 to W08E12.5 and the more specific W08E12.3), a statistically significant increase in Zn was observed in the gut tissue of the worms (Fig. 3A and B). The knockdown efficiency is known to decrease when multiple transcripts are targeted by RNAi37 thus possibly explaining the differences in metal accumulation between the two RNAi experiments. This should not distract from the fact that independent RNAi with different constructs yielded similar trends in metal accumulation, although at this point it is not possible to determine if all proteins contribute equally to the phenotype. The location of accumulation at this concentration was notably not the pharyngeal region but rather the alimentary canal, which matches the expression pattern of both metallothioneins in worms exposed to heavy metals. The metal accumulation pattern is therefore comparable to other proteins which are involved in the homeostasis/detoxification of metals, despite of the fact that the expression of W08E12.3/4 remains within the pharyngeal region.14 RNAi knockdown experiments were also applied to assess whether the silencing of the genes exerts an effect on growth and reproductive output, two sensitive phenotypic endpoints previously linked to Zn exposure in worms.38 The growth (flat volumetric surface area) of wild type and RNAi knock-down worms was comparable for the first 72 h (post L1 stage). Thereafter, growth retardation was observed in worms subjected to the knock-down of W08E12.3 to W08E12.5 in comparison to wild type worms fed the (empty) RNAi vector. The observed inhibition of growth was however independent of Zn exposure (Fig. 4A). In contrast, Zn exposure resulted in a significant reduction in reproductive performance which was not further affected by the knock-down of W08E12.3–W08E12.5 (Fig. 4B). No significant changes in lethality were observed in worms subjected to the knockdown by RNAi (data not shown).
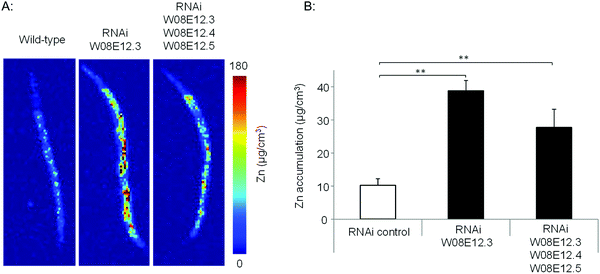 | ||
Fig. 3 (A) Wild type and worms subjected to the knockdown (by RNAi) of specifically W08E12.3, or W08E12.3 to W08E12.5 were exposed to Zn (150 μM) for 48 h and Zn accumulation was measured at 5-micron resolution by X-ray Fluorescence Imaging (XFI) at the Stanford Linear Accelerator Centre (SLAC). (B) Quantifications were conducted on 3 worms per condition and analyzed using the custom designed microprobe analysis software (SMAK).41 Note: knockdown of W08E12.3 in isolation or combined (W08E12.3 to W08E12.5) resulted in a statistically significant (**![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p < 0.01) increase in Zn levels, predominantly in the intestinal cells. p < 0.01) increase in Zn levels, predominantly in the intestinal cells. | ||
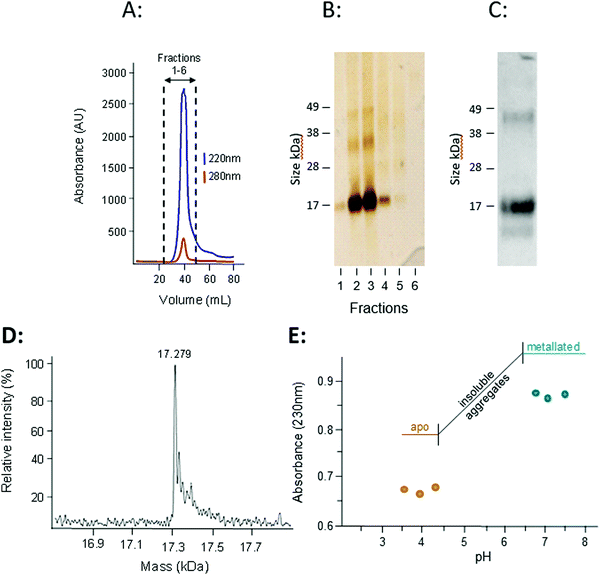
The presence of transcription factor binding sequences related zinc homeostasis in the promoter regions of the W08E12.2–W08E12.5 genes, together with the cysteine-rich nature of the translated proteins, raises the question whether the proteins possess zinc-binding ability. Thus, W08E12.3 was selected for further characterisation at the protein level. Protein was generated by means of recombinant expression in E. coli, with culture media being supplemented with 100 μM Zn(II). Analysis of the soluble and insoluble fractions of the cell lysates revealed that the W08E12.3 protein was predominantly present in the insoluble fraction, i.e. in inclusion bodies. Attempts to isolate protein expressed without any tags by classical chromatography were unsuccessful (data not shown), but the S-tagged version of W08E12.3 was found to be more stable and easier to purify. However, the subsequent cleavage of the S-tag with thrombin resulted in significant protein loss, a problem that was also encountered when S-tagged protein was captured via S-protein affinity chromatography (data not shown). Given that the length of the 15aa S-tag contributes only 10% of the recombinant protein and has no significant metal-binding capacity, it was assumed not to interfere with protein folding.39 The S-tag containing W08E12.3 protein extracted from the inclusion bodies was refolded by dilution under reducing conditions in the presence of Zn (100 μM) at pH 8 and purified by gel-filtration chromatography. Due to the cysteine content, all purification steps were conducted in the presence of reducing agents (DTT or TCEP) and Zn(II) to minimize oxidation. Protein-containing chromatography fractions were assessed by means of denaturing SDS-PAGE followed by western blotting using an S-tag antibody. This facilitated the identification of a prominent band that corresponded to the size of the monomeric W08E12.3 S-tagged protein, but also a small amount of dimer (Fig. 5A–C). Dimerization was not observed in subsequent MS experimentation, thus it was not possible to conclude whether the metallated protein was able to dimerize. The addition of Zn to the culture medium enhanced the protein yield, providing circumstantial evidence that Zn may interact with (and stabilize) the protein (Fig. S3, ESI†).
The subsequent analysis by mass spectrometry at pH 3.8 determined the mass of the metal-free apo-protein to be 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 279 Da (Fig. 5D), which is in reasonable agreement with the theoretical mass (17280.5 Da).
279 Da (Fig. 5D), which is in reasonable agreement with the theoretical mass (17280.5 Da).
Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES) was used to determine whether the protein expressed in E. coli was likely to bind Zn, and whether the purified protein had Zn-binding ability. The Zn and S contents of unprocessed inclusion bodies were determined after dissolution in nitric acid and assuming that all S originated from W08E12.3 protein the metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein ratio was calculated to be 5
protein ratio was calculated to be 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. A higher metal
1. A higher metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein ratio of 6.5
protein ratio of 6.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was observed in the purified S-tagged W08E12.3 protein (Table 1), which is similar to MTL-1 (7 Zn) and MTL-2 (6 Zn).15 This suggests that the protein is, in principle, capable of sequestering Zn, however is deposited in inclusion bodies of E. coli likely as a mis-folded protein.
1 was observed in the purified S-tagged W08E12.3 protein (Table 1), which is similar to MTL-1 (7 Zn) and MTL-2 (6 Zn).15 This suggests that the protein is, in principle, capable of sequestering Zn, however is deposited in inclusion bodies of E. coli likely as a mis-folded protein.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) metal ratio of the protein pool isolated from the E. coli inclusion bodies compared to the purified S-tagged W08E12.3 fractions
metal ratio of the protein pool isolated from the E. coli inclusion bodies compared to the purified S-tagged W08E12.3 fractions
| Sample name | Protein concentration (μM) | Zn concentration (μM) | Protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn ratio Zn ratio |
|---|---|---|---|
| S-tagged W08E12.3 within the inclusion bodies | 48 | 245 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
| Gel filtration column purified S-tagged W0812.3 | 35 | 244 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6.5 6.5 |
The zinc-binding strength of the metallated W08E12.3 protein was probed via a pH titration monitored by UV absorption spectroscopy, an approach widely used to characterise metallothioneins.40 The Zn–S Ligand-to-Metal-Charge-Transfer (LMCT) band was monitored at 230 nm (Fig. 5E). The Zn-bound protein was titrated with acid (HCl at different concentrations) across a pH range of 7.5 to 3.5; however the W08E12.3 protein became insoluble and aggregated between pH 6.4 and 4.3 (which coincides with its theoretical isoelectric point, pI = 5.55) and precluded the determination of a pH of half-displacement. However, the W08E12.3 protein re-dissolved at lower pH; the reduced absorption value at 230 nm confirmed that Zn loss was, in principle, pH dependent as expected. The likely complete demetallation by pH 4.3 suggest that if a pH of half-displacement could be determined, it would be at least one pH unit higher, i.e. significantly higher than those for most metallothioneins.21 Of course the above evidence does not rule out the possibility that other metals may be able to bind to these proteins and function in a zinc-independent manner. For example, using bioinformatics tools via the MyHits database Motif Scan revealed that W08E12.3 possesses a 2Fe–2S ferredoxin-type iron–sulfur binding region in the sequence. Thus, exposing the transgenic nematodes to different iron sources will unravel if this ferredoxin-type iron–sulfur binding region could be responsible for the metal ion-dependent transcriptional activation of W08E12.3. Preliminary data suggest that this is the case: supplementation of the media with 30–200 μM iron(II) induced the fluorescence in transgenic animals by 36–46% (24 h exposure) or 41–63% respectively (48 h exposure) (Fig. S2, ESI†).
4. Conclusions
The data obtained by the in silico metallomics screen, the in vivo gene expression analysis, targeted silencing by RNAi with follow-up studies investigating changes in metal accumulation and phenotypic endpoints as well as protein level studies have led to the identification and initial characterization of a closely related family of four metalloproteins, named W08E12.2, W08E12.3, W08E12.4 and W08E12.5 according to their position on a cosmid. The transcripts are metal-responsive and the resultant W08E12.3 protein is capable of binding Zn. The W08E12.2 to W08E12.5 family share some functional features of metallothioneins, namely several transcription factor binding sites within the promoters, the location of expression and metal responsive properties, changes in Zn metabolism following silencing by RNAi, and the capacity of binding and the proton-induced release of Zn from the protein. Taken together, this study has uncovered a new family of metalloproteins which may function in metal sensing, metal homeostasis and/or metal detoxification, not dissimilar to metallothioneins.Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
This work is an extension of a joint grant funded by the Biotechnology and Biological Sciences Research Council (BBSRC grants BB/E025099 (SRS) and BB/E025064 (CAB)). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The authors would also like to thank Phil Cunningham for his indispensable help with the bioinformatic screen. This paper is dedicated to Silvia Atrian who acted as external examiner of PC's PhD defense.References
- P. L. Williams and D. B. Dusenbery, Using the nematode Caenorhabditis elegans to predict mammalian acute lethality to metallic salts, Toxicol. Ind. Health, 1988, 4(4), 469–478 CrossRef CAS PubMed.
- M. C. Leung, P. L. Williams, A. Benedetto, C. Au, K. J. Helmcke, M. Aschner and J. N. Meyer, Caenorhabditis elegans: an emerging model in biomedical and environmental toxicology, Toxicol. Sci, 2008, 106(1), 5–28 CrossRef CAS PubMed.
- G. C. Jiang, S. Hughes, S. R. Stürzenbaum, L. Evje, T. Syversen and M. Aschner, Caenorhabditis elegans metallothioneins protect against toxicity induced by depleted uranium, Toxicol. Sci, 2009, 111(2), 345–354 CrossRef CAS PubMed.
- J. Lund, P. Tedesco, K. Duke, J. Wang, S. K. Kim and T. E. Johnson, Transcriptional profile of aging in C. elegans, Curr. Biol., 2002, 12(18), 1566–1573 CrossRef CAS PubMed.
- K. Reichert and R. Menzel, Expression profiling of five different xenobiotics using a Caenorhabditis elegans whole genome microarray, Chemosphere, 2005, 61(2), 229–237 CrossRef CAS PubMed.
- D. Dupuy, N. Bertin, C. A. Hidalgo, K. Venkatesan, D. Tu, D. Lee, J. Rosenberg, N. Svrzikapa, A. Blanc, A. Carnec, A. R. Carvunis, R. Pulak, J. Shingles, J. Reece-Hoyes, R. Hunt-Newbury, R. Viveiros, W. A. Mohler, M. Tasan, F. P. Roth, C. Le Peuch, I. A. Hope, R. Johnsen, D. G. Moerman, A. L. Barabási, D. Baillie and M. Vidal, Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans, Nat. Biotechnol., 2007, 25(6), 663–668 CrossRef CAS PubMed.
- E. J. Martinez-Finley and M. Aschner, Revelations from the nematode Caenorhabditis elegans on the complex interplay of metal toxicological mechanisms, J. Toxicol., 2011, 895236 Search PubMed.
- S. Clemens, J. I. Schroeder and T. Degenkolb, Caenorhabditis elegans expresses a functional phytochelatin synthase, Eur. J. Biochem., 2001, 268(13), 3640–3643 CrossRef CAS PubMed.
- O. K. Vatamaniuk, E. A. Bucher, J. T. Ward and P. A. Rea, A new pathway for heavy metal detoxification in animals. Phytochelatin synthase is required for cadmium tolerance in Caenorhabditis elegans, J. Biol. Chem., 2001, 276(24), 20817–20820 CrossRef CAS PubMed.
- S. L. Hughes, J. G. Bundy, E. J. Want, P. Kille and S. R. Stürzenbaum, The metabolomic responses of Caenorhabditis elegans to cadmium are largely independent of metallothionein status, but dominated by changes in cystathionine and phytochelatins, J. Proteome Res., 2009, 8(7), 3512–3519 CrossRef CAS PubMed.
- Y. J. Essig, S. M. Webb and S. R. Stürzenbaum, Deletion of phytochelatin synthase modulates the metal accumulation pattern of cadmium exposed C. elegans, Int. J. Mol. Sci., 2016, 17, 257 CrossRef PubMed.
- L. W. Slice, J. H. Freedman and C. S. Rubin, Purification, characterization, and cDNA cloning of a novel metallothionein-like, cadmium-binding protein from Caenorhabditis elegans, J. Biol. Chem., 1990, 265(1), 256–263 CAS.
- L. H. Moilanen, T. Fukushige and J. H. Freedman, Regulation of metallothionein gene transcription. Identification of upstream regulatory elements and transcription factors responsible for cell-specific expression of the metallothionein genes from Caenorhabditis elegans, J. Biol. Chem., 1999, 274(42), 29655–29665 CrossRef CAS PubMed.
- S. C. Swain, K. Keusekotten, R. Baumeister and S. R. Stürzenbaum, C. elegans metallothioneins: new insights into the phenotypic effects of cadmium toxicosis, J. Mol. Biol., 2004, 341, 951–959 CrossRef CAS PubMed.
- R. Bofill, R. Orihuela, M. Romagosa, J. Domènech, S. Atrian and M. Capdevila, Caenorhabditis elegans metallothionein isoform specificity--metal binding abilities and the role of histidine in CeMT1 and CeMT2, FEBS J., 2009, 276(23), 7040–7056 CrossRef CAS PubMed.
- S. Zeitoun-Ghandour, J. M. Charnock, M. E. Hodson, O. I. Leszczyszyn, C. A. Blindauer and S. R. Stürzenbaum, The two Caenorhabditis elegans metallothioneins (CeMT-1 and CeMT-2) discriminate between essential zinc and toxic cadmium, FEBS J., 2010, 277(11), 2531–2542 CrossRef CAS PubMed.
- C. S. Cobbett, Phytochelatins and their roles in heavy metal detoxification, Plant Physiol., 2000, 123(3), 825–832 CrossRef CAS PubMed.
- D. E. Salt and W. E. Rauser, MgATP-dependent transport of phytochelatins across the tonoplast of oat roots, Plant Physiol., 1995, 107(4), 1293–1301 CrossRef CAS PubMed.
- S. B. Ha, A. P. Smith, R. Howden, W. M. Dietrich, S. Bugg, M. J. O'Connell, P. B. Goldsbrough and C. S. Cobbett, Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe, Plant Cell, 1999, 11(6), 1153–1164 CrossRef CAS.
- E. J. Martinez-Finley and M. Aschner, Revelations from the nematode Caenorhabditis elegans on the complex interplay of metal toxicological mechanisms, J. Toxicol., 2011, 895236 Search PubMed.
- O. I. Leszczyszyn, S. Zeitoun-Ghandour, S. R. Stürzenbaum and C. A. Blindauer, Tools for metal ion sorting: in vitro evidence for partitioning of zinc and cadmium in C. elegans metallothionein isoforms, Chem. Commun., 2011, 47(1), 448–450 RSC.
- S. Hughes and S. R. Stürzenbaum, Single and double metallothionein knockout in the nematode C. elegans reveals cadmium dependent and independent toxic effects on life history traits, Environ. Pollut., 2007, 145(2), 395–400 CrossRef CAS PubMed.
- C. elegans Sequencing Consortium, Genome sequence of the nematode C. elegans: a platform for investigating biology, Science, 1998, 282(5396), 2012–2018 Search PubMed.
- H. Oneda and K. Inouye, Refolding and recovery of recombinant human matrix metalloproteinase 7 (matrilysin) from inclusion bodies expressed by Escherichia coli, J. Biochem., 1999, 126(5), 905–911 CrossRef CAS PubMed.
- M. Capdevila, J. Domenech, A. Pagani, L. Tio, L. Villarreal and S. Atrian, Zn– and Cd–Metallothionein Recombinant Species from the Most Diverse Phyla May Contain Sulfide (S2−) Ligands, Angew. Chem., Int. Ed., 2005, 44(29), 4618–4622 CrossRef CAS PubMed.
- V. Katju and U. Bergthorsson, Copy-number changes in evolution: rates, fitness effects and adaptive significance, Front. Genet., 2013, 4, 273 Search PubMed.
- D. W. Loehlin and S. B. Carroll, Expression of tandem gene duplicates is often greater than twofold, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(21), 5988–5992 CrossRef CAS PubMed.
- M. A. Rogers, L. Langbein, H. Winter, I. Beckmann, S. Praetzel and J. Schweizer, Hair keratin associated proteins: characterization of a second high sulfur KAP gene domain on human chromosome 21, J. Invest. Dermatol., 2004, 122(1), 147–158 CrossRef CAS PubMed.
- A. T. Lau and J. F. Chiu, The possible role of cytokeratin 8 in cadmium-induced adaptation and carcinogenesis, Cancer Res., 2007, 67(5), 2107–2113 CrossRef CAS PubMed.
- H. Hu, Human health and heavy metals exposure, Life Support: The Environment and Human Health Michael McCally, 2002, MIT press, pp. 65–81 Search PubMed.
- P. Kar and M. Misra, Use of keratin fiber for separation of heavy metals from water, J. Chem. Technol. Biotechnol., 2004, 79(11), 1313–1319 CrossRef CAS.
- A. Aluigi, C. Vineis, A. Varesano, G. Mazzuchetti, F. Ferrero and C. Tonin, Structure and properties of keratin/PEO blend nanofibres, Eur. Polym. J., 2008, 44(8), 2465–2475 CrossRef CAS.
- D. E. Davis, H. C. Roh, K. Deshmukh, J. J. Bruinsma, D. L. Schneider, J. Guthrie, J. D. Robertson and K. Kornfeld, The cation diffusion facilitator gene cdf-2 mediates zinc metabolism in Caenorhabditis elegans, Genetics, 2009, 182(4), 1015–1033 CrossRef CAS PubMed.
- A. Haegerbaeumer, S. Höss, K. Ristau, E. Claus, C. Möhlenkamp, P. Heininger and W. Traunspurger, A comparative approach using ecotoxicological methods from single-species bioassays to model ecosystems, Environ. Toxicol. Chem., 2016, 35(12), 2987–2997 CrossRef CAS PubMed.
- Y. Jiang, J. Chen, Y. Wu, Q. Wang and H. Li, Sublethal toxicity endpoints of heavy metals to the nematode Caenorhabditis elegans, PLoS One, 2016, 11(1), e0148014 CrossRef PubMed.
- M. J. Pushie, I. J. Pickering, M. Korbas, M. J. Hackett and G. N. George, Elemental and chemically specific X-ray fluorescence imaging of biological systems, Chem. Rev., 2014, 114(17), 8499–8541 CrossRef CAS PubMed.
- R. S. Kamath, M. Martinez-Campos, P. Zipperlen, A. G. Fraser and J. Ahringer, Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans, Genome Biol., 2001, 2(1), RESEARCH0002.1 Search PubMed.
- N. Polak, D. S. Read, K. Jurkschat, M. Matzke, F. J. Kelly, D. J. Spurgeon and S. R. Stürzenbaum, Metalloproteins and phytochelatin synthase may confer protection against zinc oxide nanoparticle induced toxicity in Caenorhabditis elegans, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2014, 160, 75–85 CAS.
- R. T. Raines, M. McCormick, T. R. Van Oosbree and R. C. Mierendorf, The S-tag fusion system for protein purification, Methods Enzymol., 2000, 326, 362–376 CAS.
- C. A. Blindauer, Metallothioneins, in Binding, Transport and Storage of Metal Ions in Biological Cells, ed. A. Wedd and W. Maret, The Royal Society of Chemistry, 2014, ch. 21, pp. 606–665 Search PubMed.
- S. M. Webb, The MicroAnalysis Toolkit: X-ray Fluorescence Image Processing Software, American Institute of Physics Conference Proceedings, 2011, 1365, 196–199.
Footnote |
| † Electronic supplementary information (ESI) available. According to the UK Research Councils' Common Principles on Data Policy, all data supporting this study are openly available. See DOI: 10.1039/c8mt00169c |
| This journal is © The Royal Society of Chemistry 2018 |