Open Access Article
Open Access ArticleFungal siderophore metabolism with a focus on Aspergillus fumigatus
Hubertus
Haas
*
Division of Molecular Biology/Biocenter, Innsbruck Medical University, Innrain 80-82, A-6020 Innsbruck, Austria. E-mail: hubertus.haas@i-med.ac.at
First published on 20th August 2014
Abstract
Covering: up to 2014
Siderophores are chelators synthesized by microbes to sequester iron. This article summarizes the knowledge on the fungal siderophore metabolism with a focus on Aspergillus fumigatus. In recent years, A. fumigatus became a role model for fungal biosynthesis, uptake and degradation of siderophores as well as regulation of siderophore-mediated iron handling and the elucidation of siderophore functions. Siderophore functions comprise uptake, intracellular transport and storage of iron. This proved to be crucial not only for adaptation to iron starvation conditions but also for germination, asexual and sexual propagation, antioxidative defense, mutual interaction, microbial competition as well as virulence in plant and animal hosts. Recent studies also indicate the high potential of siderophores and its biosynthetic pathway to improve diagnosis and therapy of fungal infections.
1. Introduction
Fungi play a major role in ecology and affect human life in many ways positively and negatively: they are major players in saprobic decomposition, important symbionts of plants (mycorrhiza), pathogens of plants and animals affecting health as well as food production and preservation (decay, toxin production), serve as food or in food production (e.g., mushrooms, alcohol, bread, mould cheese), and are important work horses in biotechnology as producers of widely used enzymes as well as primary and secondary metabolites (e.g. citric acid, statins, cyclosporine, β-lactam antibiotics). A crucial nutrient affecting all of these functions is iron and in most fungal species, iron handling involves fungal-specific ferric iron chelators, termed siderophores.2. Aspergillus fumigatus
Aspergillus fumigatus is a ubiquitous saprophytic fungus typically residing in the soil or decaying organic matter. As an efficient recycler in nature, this mould possesses a versatile metabolism that meets its nutritional requirements under a wide range of environmental conditions.1A. fumigatus produces huge numbers of minute (2–3 μm diameter) asexual spores that are efficiently released into the environment, particularly into the air. These conidia are constantly inhaled and rapidly eliminated by a functional immune system. However, immunosuppression such as in patients undergoing bone marrow transplantation, cancer chemotherapy or suffering from chronic granulomatous disease, increases the risk of invasive systemic infection, termed aspergillosis. Due to the increase in immunosuppressive regiments in medical interventions, A. fumigatus has become the most common air-borne fungal pathogen of humans.2 The latter boosted the interest in this fungus, leading to the still ongoing detailed characterization of its physiology and metabolism, particularly of course of its virulence determinants. As a result, A. fumigatus is now one of the best-studied filamentous fungal species and a fungal role model including a sequenced genome, which facilitates molecular analysis and allows “genome mining”.33. Iron homeostasis
Due to the ability to coordinate and activate oxygen and the ideal redox chemistry for electron transport, iron is an indispensable cofactor for a variety of cellular processes in all eukaryotes and most prokaryotes including respiration, amino acid metabolism, and biosynthesis of DNA and sterols. On the other hand, iron excess has the potential to catalyse the formation of cell-damaging reactive oxygen species via the Fenton and Haber–Weiss reactions.4 The iron-dependence of several enzymes involved in oxidative stress detoxification such as heme-dependent catalases and peroxidases emphasizes the complexity of cellular iron handling.Despite its high abundance in the Earth's crust, the bioavailability of iron is low owing to its oxidation by atmospheric oxygen into sparely soluble ferric hydroxides. For pathogens in plant as well as animal hosts, iron is not freely available but tightly sequestered, e.g. in vertebrates most of the iron is associated with proteins such as transferrin, lactoferrin and ferritin and with heme that is bound by hemoglobin. The preclusion of invading microbes from iron represents a key pathway in host defence as a component of the so called “nutritional immunity”.5,6 Consequently, pathogenic as well as non-pathogenic organisms had to evolve sophisticated strategies to ensure iron supply, while at the same time avoiding iron toxicity. This includes diverse mechanisms for uptake and storage of iron as well as fine-tuned regulation of acquisition, storage and consumption of iron.
4. Fungal iron acquisition and storage
In fungi, four different mechanisms for acquisition of iron have been identified: (i) ferric iron uptake mediated by ferric iron-specific chelators, termed siderophores, (ii) reductive iron assimilation (RIA), (iii) low-affinity ferrous iron uptake and (iv) heme uptake and degradation. A. fumigatus, similar to most fungal species, employs siderophore-mediated iron uptake, RIA and low affinity iron uptake.7A. fumigatus appears unable to utilize heme as an iron source.8 Interestingly, the fungal species studied in most detail lack siderophore biosynthesis (SB) including the opportunistic pathogenic basidiomycete Cryptococcus neoformans and the Saccharomycotina including the fungal role model Saccharomyces cerevisiae and the opportunistic pathogenic genus Candida spp.7 Nevertheless, these siderophore non-producers are able to take up and utilize iron chelated by siderophores from other organisms, termed xenosiderophores.9 Interestingly, all these siderophore non-producers display predominantly yeast morphology. However, there are also yeast species that produce siderophores such as Schizosaccharomyces pombe or Aureobasidium pullulans.10,11 The siderophore non-producers utilize RIA, heme-bound iron and low affinity iron uptake.12 There are also examples for species lacking RIA such as Aspergillus nidulans and Coprinus spp.7,13To overcome periods of iron-limitation as well as to avoid iron toxicity, all cells require means to store iron. In fungi, two strategies for iron storage are known: (i) vacuolar iron deposition and (ii) siderophore-mediated iron storage.7,12 In contrast to bacteria, plants and animals, most fungi appear to lack ferritin-mediated iron storage and detoxification. A. fumigatus employs both intracellular siderophores and vacuolar iron deposition for iron storage.14,15
5. Fungal siderophores
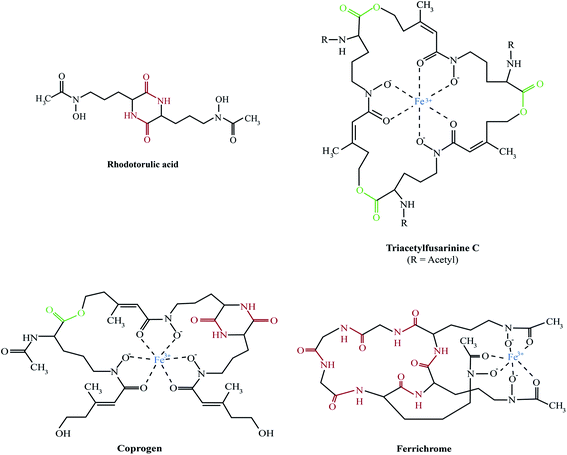
The majority of fungal siderophores belong to the hydroxamate-class. Exceptions are the carboxylate-type siderophore rhizzoferrin produced by various Mucorales and the catecholate pistillarin produced by the marine species Penicillium bilaii.16,17 Fungal hydroxamate siderophores can be grouped into four structural families: fusarinines, coprogens, ferrichromes and rhodotorulic acid. Representatives of each family are shown in Fig. 1. The chemistry of siderophores has been comprehensively reviewed.18,19 Universal to all is the hydroxamate group that forms a strong iron(III)-binding bidentate. The hydroxamate group is built by acylation of the non-proteinogenic amino acid N5-hydroxy-L-ornithine, which is derived by hydroxylation of L-ornithine, with acetyl or more complex groups such as anhydromevalonyl. To increase affinity for Fe(III), most fungal siderophores include three of these moieties linked by ester or peptide bonds to form hexadentate structures. Cyclisation of the siderophore, as found in ferrichromes and some fusarinines, improves the chemical stability.Rhodotorulic acid, produced by basidomycetous yeasts such as Rhodotorula spp., is a dipeptide consisting of two N5-acetyl-N5-hydroxy-L-ornithine units linked head-to-head to form a diketopiperazine ring. Fusarinines consist of three N5-cis-anhydromevalonyl-N5-hydroxy-L-ornithine units linked head-to-tail by ester bonds in linear or cyclic form. Coprogens are linear di- or tri-hydroxamates composed of trans-fusarinine units (i.e. the acyl groups are trans-anhydromevalonyl moieties). Coprogen consist of three N5-trans-anhydromevalonyl-N5-hydroxy-L-ornithine units, with two units linked head-to-head to form a diketopiperazine ring (dimerum acid) and a third unit esterified to the C-terminal OH group of dimerum acid. N2-Acetylation and N2-methylation allow for the diversity of the coprogen family. Ferrichromes are cyclic hexapeptides consisting of three N5-acyl-N5-hydroxy-L-ornithines and three additional amino acids, with two residues consisting of alanine, serine or glycine and the third residue always being a glycine.20 A number of different acyl groups have been found in this family (acetyl, malonyl, trans-β-ethylglutaconyl, trans-anhydromevalonyl, and cis-anhydromevalonyl). Recently, a ferrichrome-type siderophore with a novel structure comprised of four glycines, a glutamine and three N5-trans-anhydromevalonyl-N5-hydroxy-L-ornithine moieties, produced by the endophyte Epichloë festucae, termed epichloënin A, was identified.21 The function of hexadehydro-astechrome, a tryptophan-derived and iron-binding hydroxamate recently isolated from A. fumigatus, is still unknown.22
6. The siderophore biosynthetic pathway
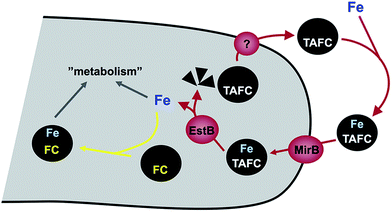
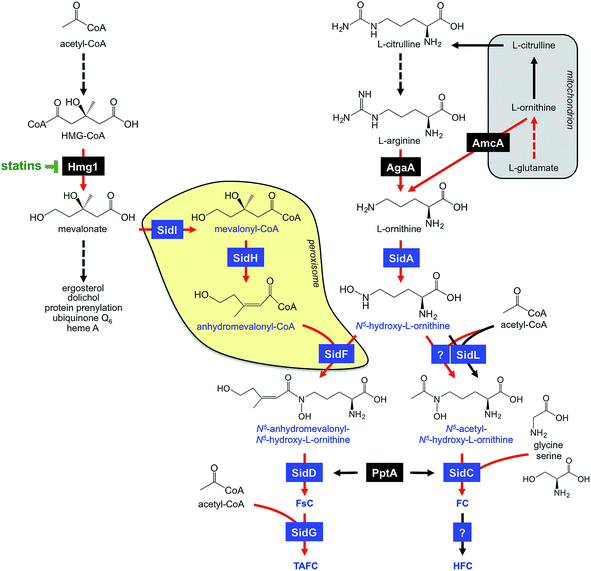
A. fumigatus produces four siderophores: two extracellular fusarinine-type siderophores, fusarinine C (FsC) and its derivative triacetylfusarinine C (TAFC) for iron uptake as well as two intracellular ferrichrome-type siderophores, ferricrocin (FC) for distribution and storage of iron in hyphae and hydroxyferricrocin (HFC) for storage of iron in asexually-produced spores, termed conidia.23 A scheme of TAFC-mediated iron uptake and FC-mediated iron storage is displayed in Fig. 2.FsC consists of three N5-anhydromevalonyl-N5-hydroxy-L-ornithine residues cyclically linked by ester bonds. TAFC is the N2-acetylated FsC. FC is a cyclic hexapeptide with the structure Gly-Ser-Gly-(N5-acetyl-N5-hydroxy-L-ornithine)3 and HFC is the hydroxylated FC with a single unidentified hydroxylation site.7 Based on biochemical studies, the fungal siderophore biosynthetic pathway was postulated by Plattner and Diekmann.24 In A. fumigatus, the siderophore biosynthetic pathway has been characterized at the molecular level by reverse genetics, i.e. generation and characterization of mutant strains with defects in putative siderophore biosynthetic genes identified by their upregulation during iron starvation.23 The siderophore biosynthetic pathway of A. fumigatus is depicted in Fig. 3. The first committed step in the biosynthesis of all four siderophores is the N5-hydroxylation of L-ornithine catalysed by the L-ornithine-N5-monooxygenase SidA.8 SidA orthologues have been shown to be responsible for SB in various other fungal species including the ascomycetes A. nidulans, Aspergillus oryzae, Fusarium graminearum, Magnaporthe grisea, Histoplasma capsulatum, A. pullulans and the basidiomycete Ustilago maydis.11,13,25–29 These fungal ornithine-N5-monooxygenases show significant similarity at the protein level to bacterial siderophore-biosynthetic enzymes such as the ornithine-N5-monooxygenases PvdA from Pseudomonas aeruginosa and the lysine-N6-monooxygenases IucD from Escherichia coli.30,31 PvdA and IucD are essential for biosynthesis of pyoverdine and aerobactin, respectively. All these enzymes require oxygen as substrate and NADPH and FAD as co-factors.29
Subsequently, the pathways for biosynthesis of extra- and intracellular siderophores, i.e. fusarinines and ferrichromes, split due to attachment of different acyl residues. For extracellular fusarinine-type siderophores, the transacylase SidF transfers anhydromevalonyl to N5-hydroxy-L-ornithine.23 The required anhydromevalonyl-CoA moiety is derived from mevalonate by CoA-ligation and dehydration catalysed by the enzymes SidI and SidH, respectively, thereby linking isoprenoide and siderophore biosynthetic pathways.32 SidI belongs to the acyl-CoA synthetase protein family with most members being involved in lipid metabolism. In Neurospora crassa, the SidI ortholog Acs5 was shown to be involved in coprogen biosynthesis.33 SidH belongs to the enoyl-CoA hydratase protein family, again, with most members being involved in lipid metabolism.34 The N5-acetylation of N5-hydroxy-L-ornithine for the intracellular ferrichrome-type siderophores requires two transacetylases: the constitutively expressed enzyme SidL and an unidentified enzyme, the activity of which is upregulated by iron starvation.35 Both SidF and SidL belong to the “GCN5-related N-acetyltransferases (GNAT)”. A. fumigatus possesses several GNAT proteins, but SidF and SidL appear to be the only two carrying the pfam10331/AlcB domain that is also conserved in bacterial siderophore biosynthetic enzymes such as the N5-hydroxylysine:acetyl coenzyme A N5-transacetylase IucB, which is involved in biosynthesis of the hydroxamate aerobactin.36 The only other functionally characterized fungal SidF orthologue is Fer5 from U. maydis, which catalyses N5-acylation of N5-hydroxy-L-ornithine with methylglutaconyl-CoA for biosynthesis of ferrichrome A.37 Similar to fusarinine biosynthesis in A. fumigatus, ferrichrome A biosynthesis is also linked to isoprenoide biosynthesis as methylglutaconyl-CoA is derived from HMG-CoA via dehydration by the enoyl-CoA-hydratase Fer4.37
In A. fumigatus, fusarinines and ferrichromes are then assembled by two different non-ribosomal peptide synthetases (NRPSs), an enzyme family that is best known from secondary metabolism. The NRPS SidD links three N5-cis-anhydromevalonyl-N5-hydroxy-L-ornithine residues by ester bonds, yielding FsC.23 The NRPS SidC links three N5-acetyl-N5-hydroxy-L-ornithine, two L-glycine and one L-serine residue by peptide bonds, yielding the hexapeptide ferricrocin. In other fungal species, SidC homologs were confirmed to be essential for synthesis of ferricrocin or other ferrichrome-type siderophores such as ferrichrome, ferrichrome A and ferrirhodin in S. pombe, U. maydis and Fusarium sacchari, respectively.13,37–40 Consistently, phylogenetic analysis predicts monogenetic origin of ferrichrome-type NRPS.41 Similarly, SidD orthologs in other species were found to be responsible not only for synthesis of fusarinine- but also coprogen-type siderophores, e.g. Alternaria brassicicola, C. heterostrophus and Colletotrichum graminicola.42,43 This indicates a common evolutionary origin of fusarinine- and coprogen-synthetic NRPSs. SidE, another NRPS predicted to be involved in SB in A. fumigatus,44 was recently demonstrated to be responsible for synthesis of fumarylalanine but not siderophores.45
NRPSs, similar to polyketide synthetases and the α-aminoadipate reductase, which is involved in lysine-biosynthesis, depend on activation by the Sfp-type 4′-phosphopantetheinyl transferase, termed PptA in A. fumigatus.46 Consistently, this enzyme was confirmed to be essential for SB in various fungal species, including A. nidulans, Cochliobolus sativus, Fusarium fujikuroi and C. graminicola.47–50
TAFC is derived by triple N2-acetylation of FsC catalysed by SidG.23 SidG belongs to the GNAT protein family and its activity is particularly required for biosynthesis of TAFC but not FsC, coprogens and ferrichromes. In agreement, SidG orthologs are found only in few Aspergillus, Fusarium and Nectria species but not in species lacking TAFC production.
HFC is derived from FC by single hydroxylation catalysed by an unidentified gene product.23 The hydroxylation site in HFC remains to be identified. So far, HFC has been detected only in A. fumigatus and appears to be missing in other FC-producing fungal species such as A. nidulans and N. crassa.51,52
7. Subcellular localization of SB
In agreement with the presence of putative peroxisomal targeting motifs, the three enzymes linking isoprenoide and TAFC biosynthetic pathways, SidI, SidH and SidF, were recently found to be sequestered in peroxisomes.34 This study also demonstrated that artificial delocalization of one or two but not of all three enzymes into the cytosol impairs TAFC biosynthesis suggesting that not all TAFC biosynthesis intermediates can efficiently cross the peroxisomal membrane. The peroxisomal targeting motifs are conserved in fungal orthologues of SidF, SidH and SidI.34 In agreement with the evolutionary conservation of the partial peroxisomal compartmentalization of fungal SB, the SidI orthologue of N. crassa and the SidF ortholog of Penicillium chrysogenum, which are both involved in coprogen synthesis, were identified to be peroxisomal matrix proteins.34,53 Exceptions are the SidH and SidF orthologs Fer4 and Fer5 from U. maydis, which lack peroxisomal targeting motifs.378. Siderophore uptake and degradation
Cellular uptake of siderophore–iron complexes is mediated by “siderophore–iron transporters (SITs)”, which belong to a subfamily of the major facilitator protein superfamily. SITs act most likely as proton symporters energized by the plasma membrane potential.54,55 SIT-mediated iron uptake is universally conserved in the fungal kingdom, even in species not producing siderophores such as S. cerevisiae, Candida spp. and C. neoformans.7,55–57 Functional characterization of SITs started in S. cerevisiae, which possesses four SITs with different substrate specificity: Sit1p/Arn3p for ferrioxamine B (a hydroxamate-type siderophore produced by various Streptomycetes species), Arn1p for ferrichromes, Taf1p/Arn2p for TAFC, Enb1p/Arn4p for enterobactin (a catecholate-type siderophore produced by various Enterobacteriaceae). In contrast, C. albicans contains only a single broad range substrate specific SIT, CaArn1p/CaSit1p.58 Similar to the siderophore non-producer S. cerevisiae, most fungal species including siderophore producers possess multiple SIT, e.g. A. fumigatus and A. nidulans encode seven and ten putative SITs, respectively.7 Heterologous expression in S. cerevisiae indicated that the A. nidulans SITs MirB and MirA transport TAFC and enterobactin, respectively.54,55 Similarly, the A. fumigatus MirB orthologue was confirmed to transport TAFC and FgSit1 from F. graminearum was suggested to transport ferrichrome.59,60Subsequent to uptake, the ester bonds of TAFC- and FsC-iron complexes are hydrolysed in the cytosol. Two of the enzymes involved have been identified, the esterases EstB and SidJ, which belong to two different protein families.61,62 Interestingly, EstB is homologous to bacterial siderophore degrading enzymes such as E. coli Fes, which is involved in release of iron from the catecholate-type siderophore enterobactin.63 EstB is specific for TAFC and SidJ is specific for FsC.61,62 However, both enzymes are not essential for but rather optimize siderophore hydrolysis and consequently release of iron; e.g. inactivation of EstB (ΔestB mutant) decreases the growth rate during iron starvation, decreases the transfer of iron from TAFC to the metabolism and to the intracellular siderophore FC and consequently delays iron sensing. Lack of FC, vacuolar storage, and in particular lack of both increases the cellular content of iron chelated by siderophore breakdown products,15 indicating that the transfer of iron from extracellular siderophores to the metabolism, FC or the vacuole precedes recycling of siderophore breakdown products.
9. Genomic organization of genes involved in siderophore metabolism
In A. fumigatus, most of the identified siderophore metabolic genes are organized in three gene clusters, i.e. they are neighbouring each other in the genome, which most likely facilitates co-regulation of their expression.14 Exceptions are the genes encoding SidA, SidL, PptA and five of the seven putative siderophore transporters. In this respect interesting to note, expression of the genes encoding SidL and PptA is, in contrast to the other siderophore metabolic genes, not regulated by iron availability. Moreover, PptA is not exclusively involved in SB. Siderophore metabolic genes are usually also genomically clustered in other fungal species, whereby the number of clusters and gene combinations vary significantly,14,34e.g., SidA ortholog-encoding genes are clustered with siderophore NRPS in various other fungi including U. maydis and N. crassa.10. Regulation of the siderophore system
Cellular adaptation to iron starvation requires upregulation of high-affinity iron acquisition and downregulation of both iron storage and iron consumption, while adaptation to iron excess requires the inverse response. In A. fumigatus, changes in iron availability cause an extensive transcriptional remodeling affecting about 13% of the protein-encoding genes.14 The two central regulators mediating iron regulation at the transcriptional level in this fungus are the transcription factors SreA and HapX.14,64 During iron sufficiency, the GATA-factor SreA, which binds to the consensus sequence ATCWGATAA, represses high-affinity iron uptake, including the siderophore system, to avoid iron toxicity. During iron starvation, the bZIP-transcription factor HapX represses iron-consuming pathways such as respiration, heme biosynthesis and iron–sulfur cluster-dependent processes to spare iron. Furthermore, HapX activates synthesis of the ribotoxin AspF1 and the siderophore system. HapX functions via physical interaction with the DNA-binding CCAAT-binding complex (CBC).65 The CBC is a heterotrimeric DNA-binding complex, which is conserved in all eukaryotes. In A. nidulans, inactivation of either one of its subunits phenocopies HapX-deficiency with respect to the defects in adaptation to iron starvation.65 However, the CBC has HapX-independent functions and is speculated to affect expression of about 30% of all genes. SreA and HapX are interconnected in a negative feed-back loop: SreA represses expression of hapX during iron sufficiency, while HapX represses sreA during iron starvation. Additionally, both SreA and HapX are regulated posttranslationally with iron blocking HapX function and activating SreA function.65,66 Inactivation of both HapX and SreA is synthetically lethal underlining the critical role of iron homeostasis in cellular survival.14,64,65Most fungal species possess orthologs to SreA and HapX and the role of SreA in regulation of SB has been confirmed in N. crassa, H. capsulatum, C. heterostrophus, S. pombe, A. pullulans and U. maydis.67–72 In S. pombe, posttranslational iron sensing by the HapX and SreA orthologs involves glutathione and the monothiol glutaredoxin Grx4, indicating sensing of the cellular iron status via sensing the cellular iron–sulfur cluster availability.70 In agreement with a similar mode of iron-sensing in A. fumigatus, extreme starvation for sulfur, which is essential for biosynthesis of both iron–sulfur clusters and glutathione, was found to cause derepression of the siderophore system leading to detrimental excessive iron uptake.73 Remarkably, S. cerevisiae lacks orthologs of SreA and HapX. It employs entirely different regulators, Aft1/2 and Cth1/2, which are conserved only in closely related Saccharomycotina species.12 Nevertheless, S. cerevisiae Aft1/2 also senses the cellular iron status via iron sulfur cluster biogenesis involving glutathione and glutaredoxin.74 These data indicate a universal conservation of the iron-sensing machinery executed by different types of transcription factors in different organisms.
Due to the central metabolic role of iron, it is not surprising that a variety of regulatory circuits affect cellular iron handling. For example, adaptation to hypoxia of the obligate aerobic A. fumigatus includes upregulation of glycolysis, the TCA-cycle, respiration and ergosterol biosynthesis and as a consequence increased cellular iron and heme contents.75,76 Hypoxic adaptation in A. fumigatus is coordinated by the transcription factor SrbA, a member of the “sterol regulatory element binding protein (SREBP)” family, which is conserved in most eukaryotes.76 These transcription factors are proteolytically activated by cellular sterol-depletion to maintain sterol homeostasis.77 In A. fumigatus, SrbA was found to be crucial not only for ergosterol-feedback regulation, resistance against triazole drugs and hypoxic growth, but also for activation of siderophore-mediated iron uptake in response to hypoxia and iron starvation.78 The defects in hypoxic growth and azole resistance caused by SrbA-deficiency were partially cured by high-iron supplementation, which underlines the involvement of SrbA in iron homeostasis. SrbA is transcriptionally upregulated during both hypoxia and iron starvation, most likely in response to sterol-depletion. These data implicate that A. fumigatus might sense iron not only via HapX and SreA but also via SrbA. S. cerevisiae lacks an SrbA ortholog and the regulators mediating hypoxic adaptation in this yeast are not conserved in most other fungal species.76
Further regulatory proteins suggested to affect siderophore metabolism are the pH regulatory transcription factor PacC, the gluconeogenesis-activating transcription factor AcuM, the unfolded protein-responsive IreA-HacA pathway, the cross-pathway control mediating transcription factor CpcA, and the mitogen-activated protein kinase (MAPK) MpkA, which is involved in maintaining cell wall integrity, protection against ROS, and secondary metabolism.79–83
11. Precursor supply for SB
During iron starvation, siderophore production reaches up to 10% of the biomass. This condition dramatically remodels the free amino acid pool of A. fumigatus with eight amino acids increasing and three amino acids decreasing more than 1.5-fold.64 The 7-fold increase of the siderophore precursor L-ornithine during iron starvation compared to sufficiency indicates that the massive L-ornithine demand for SB is matched by active up-regulation of its biosynthesis. In agreement, blocking siderophore-mediated ornithine consumption by inactivation of SidA (ΔsidA mutant) increases the cellular L-ornithine pool another 2.9-fold.64The biosynthesis of L-ornithine in the mitochondrial matrix as well as its export via the transporter AmcA to the cytosol are transcriptionally upregulated under iron deprivation.64L-Ornithine is a precursor of L-arginine, and the cytosolic localized conversion of L-arginine to ornithine by the arginase AgaA is likewise upregulated in response to iron starvation (Fig. 3). Nevertheless, the metabolic commitment for siderophore production must be balanced against other essential cellular functions, e.g.L-arginine is vital for protein biosynthesis and ornithine is a precursor for polyamine biosynthesis. Recently, L-arginine was found to allosterically activate SidA enzyme activity,84i.e. is stimulated only if the cytosolic L-arginine pool is sufficient for all cellular needs. In agreement, studies with arginine-auxotrophic A. fumigatus mutants, which allowed manipulation of the cellular L-arginine and L-ornithine levels, revealed prioritization of these amino acids for cellular needs other than SB, e.g. arginine for protein biosynthesis and ornithine for polyamine biosynthesis.82 The rational for this regulation might be that A. fumigatus possesses RIA as an alternative high-affinity iron acquisition system that is independent of L-ornithine. Furthermore, these studies indicated that SB is mainly fueled by mitochondrial rather than cytosolic ornithine production, at least with L-glutamine as the nitrogen source.
Consistent with mevalonate being a precursor for TAFC biosynthesis (Fig. 2), overexpression of the hydroxymethylglutaryl-CoA (HMG-CoA) reductase Hmg1 increases TAFC production, while inhibition of Hmg1 with lovastatin reduces TAFC biosynthesis in A. fumigatus.32 Mevalonate is an intermediate of the isoprenoide biosynthetic pathway with ergosterol as one important product. Ergosterol is the major target of antifungal treatment, i.e. amphotericine B and azoles. In contrast to siderophore production, iron starvation decreases the cellular ergosterol level due to the iron-requirement of ergosterol biosynthesis.32 Concordant with bilateral demand for mevalonate, genetic inactivation of mevalonate consumption for TAFC biosynthesis altered the sterol composition and increased lovastatin resistance during iron starvation.32 These data also revealed a novel consequence of statin treatment in fungi that produce siderophores, the synthesis of which depend on HMG-CoA, such as fusarinines and coprogens.
12. The role of siderophores in physiology and interaction with other organisms
In A. fumigatus, genetic elimination of extracellular siderophores (ΔsidI, ΔsidH, ΔsidF, and ΔsidD mutants) decreases growth, conidiation and oxidative stress resistance during iron limitation but not during iron sufficiency, which enables compensation by other iron acquisition systems, mainly RIA.23 In agreement, simultaneous inactivation of RIA and SB blocks growth without high ferrous iron supplementation, which supports growth via low-affinity ferrous iron uptake.8 Eliminating intracellular siderophores (ΔsidC and ΔsidL, mutants) reduces conidiation and blocks sexual development, as shown in A. nidulans, Cochliobolus heterostrophus and Gibberella zeae,51,85 most likely due to the crucial role of FC in intracellular iron transport from substrate-contacting hyphae into aerial hyphae.86 The lack of HFC (ΔsidC and ΔsidL, mutants) decreases the conidial iron content by about 50%, which impairs iron-dependent enzymes such as aconitase and catalase A, and thereby decreases conidial size and conidial resistance to oxidative stress.23,35,86 Moreover, the lack of HFC delays germination during iron starvation.23 Intracellular iron excess, e.g. due to derepression of iron uptake caused by SreA-deficiency, induces massive hyphal accumulation of ferri-FC and transcriptional induction of the transporter CccA, which mediates vacuolar iron deposition in both A. fumigatus and A. nidulans.14,51,87 Nevertheless, vacuolar iron deposition rather than FC-mediated iron storage appears to represent the major iron detoxification system.15 Inactivation of the entire siderophore system (ΔsidA mutant) combines the defects caused by inactivation of either extra- or intracellular SB and renders A. fumigatus extremely sensitive to iron starvation.8,23Both extra- and intracellular siderophores are crucial for virulence as elimination of the entire SB (ΔsidA mutant) results in absolute avirulence of A. fumigatus in a murine model of invasive pulmonary aspergillosis,8,88 while deficiency in either extracellular (ΔsidI, ΔsidH, ΔsidF or ΔsidD mutants) or intracellular siderophores (ΔsidC mutants) causes partial attenuation of virulence.23,32 The fact that mutants lacking extracellular siderophores are not avirulent indicates partial compensation by RIA in vivo. Consistently, both RIA and the siderophore system are transcriptionally upregulated during murine infection.89 Conidial HFC appears to play a particularly crucial role during initiation of infection because restoration of the conidial HFC content by supplementation with FC during conidiation partially cures the virulence defect of ΔsidA conidia.23 SidG-deficiency, which eliminates TAFC production with concomitant increase of FsC production, affects neither growth nor virulence, indicating that the structural differences between these two siderophores are irrelevant in the settings analysed.23 The fact that A. fumigatus siderophores are able to remove iron from host sources, such as transferrin underlines their crucial role in virulence.90,91
During infection, the siderophore system is important not only for extra-, but most likely also for intracellular growth as defects in the siderophore system decrease intracellular growth and survival of A. fumigatus after phagocytosis by murine alveolar macrophages.92 Consistently, impairment of SB alters the immune response of macrophages after phagocytosis of A. fumigatus,93 and the gene encoding SidA was among the highest upregulated genes after internalization of A. fumigatus by epithelial cells.94 Moreover, the siderophore system was also found to be important for virulence of Histoplasma capsulatum, a dimorphic fungal pathogen replicating in the yeast form within macrophages.28 The evolutionary conserved role of siderophores in virulence of A. fumigatus has been confirmed in various infection models apart from pulmonary murine infection, i.e. a murine cutaneous model, Drosophila melanogaster and Galleria mellonella.95–97
SB is also a virulence determinant of a number of phytopathogenic ascomycetes with a necrotrophic life style such as C. heterostrophus, Cochliobolus miyabeanus, F. graminearum, A. brassicicola, Alternaria alternata and M. grisea on different plant hosts.25,42,98,99 In contrast, SB is dispensable and RIA is essential for phytopathogenicity of biotrophic U. maydis.29,100 Recent studies with the hemibiotrophic fungus C. graminicola shed light on the apparent discrepancy of the role of siderophores in virulence of necro- and biotrophic pathogens.43,101 In this maize pathogen, SB was found to be induced and important for virulence during the necrotrophic phase but to be specifically downregulated during the biotrophic phase, compensated by RIA, in order to repress the plant immune response induced by fungal siderophores.
Consistent with the crucial role of adaptation to iron starvation, including SB as virulence determinant, involved regulators such as HapX and SrbA as well as NRPS-activating Sfp-type 4′-phosphopantetheinyl transferase is essential for virulence of different siderophore producers.49,102
In Epichloë festucae, extracellular siderophore biosynthesis is important to maintain mutualistic symbiotic grass–endophyte associations as its inactivation transformed this mutualistic fungus into an antagonistic one.103
Apart from their roles in virulence and symbiosis, siderophores might also play a role in competitive and cooperative interactions among microorganisms because chelation of environmental iron by siderophore-types that are not recognized by competitors might be used to starve them for iron. This is counteracted by the evolvement of transporters recognizing these xenosiderophores. The latter turns a growth-suppressing into a growth-promoting molecule. This scenario is supported by the fact that most siderophore-producing bacteria and fungi possess xenosiderophore-specific SITs.7 The central role of iron and siderophores in microbial interaction is underlined by two recent findings.
Patients with cystic fibrosis contain a mixed microbial community including the bacterium P. aeruginosa and A. fumigatus. In vitro mass spectrometry imaging revealed that P. aeruginosa stimulates production by A. fumigatus via secretion of 1-hydroxyphenazine TAFC.104 The mechanism and rational of this microbial interaction is currently unknown but it underlines the central role of siderophores. Along these lines, HapX-mediated iron regulation was demonstrated to be crucial for iron competition of F. oxysporum with siderophores of a biocontrol strain of Pseudomonas putida.105
13. Applied aspects: potential use in treatment and diagnosis of fungal infections
Fungal infections such as aspergillosis are difficult to diagnose and treat, which is reflected by the high mortality rates that continue to be associated with this disease.106 The crucial role of the siderophore system for fungal virulence and the differences to the mammalian iron acquisition mechanisms might help to improve therapy and diagnosis of fungal infections. Specifically, the fungal siderophore biosynthetic pathway represents a promising target for selective therapeutic intervention. In this respect, the structural elucidation of the A. fumigatus SidA enzyme might facilitate design of selective inhibitors.107 Recently, treatment of fungal keratitis in a murine model by dual topical therapy with the iron chelator deferiprone and statins, which target biosynthesis of both isoprenoides and extracellular siderophores, showed promising restriction of fungal growth.108Furthermore, SITs represent one of few protein families that are unique to the fungal kingdom and are not present in prokaryotes or other eukaryotes.109 Consequently, this protein family is an attractive drug target by potentially mediating fungal-specific drug delivery by a Trojan horse approach,110 in which antifungal agents are covalently attached to siderophores for selective import into fungi.
Moreover SITs represent an attractive target to improve diagnosis of fungal infections. 68Gallium chelated by TAFC or the bacterial siderophore ferrioxamine E was found to enable in vivo imaging of aspergillosis in animal models by Positron Emission Tomography (PET) due to selective accumulation in fungal cells.111–114
The understanding of the fungal siderophore system has advanced significantly in recent years. Nevertheless, its potential application in treatment and diagnosis of fungal infections still requires deeper insights.
Moreover, the recombinant A. fumigatus enzymes SidI and SidH were employed for biosynthesis of anhydromevalonolacton, an intermediate in the production of bioplastic in E. coli, which indicates the biotechnological potential of enzymes involved in the fungal siderophore pathway.115
14. Abbreviations
| CBC | CCAAT binding complex |
| CoA | Coenzyme A |
| FC | Ferricrocin |
| FsC | Fusarinine C |
| GNAT | GCN5-related N-acetyltransferases |
| HFC | Hydroxyferricrocin |
| RIA | Reductive iron assimilation |
| SB | Siderophore biosynthesis |
| SIT | Siderophore–iron transporter |
| TAFC | Triacetylfusarinine C |
Acknowledgements
Work in the author’s laboratory on fungal iron and siderophore metabolism is currently supported by Austrian Science Fund (FWF I-1346-B21). I thank Thomas Müller and Beatrix E. Lechner for assistance in Figure production. I apologize to the authors whose work could not be cited because of space limitations.References
- K. J. Kwon-Chung and J. A. Sugui, PLoS Pathog., 2013, 9, e1003743 Search PubMed.
- F. Tekaia and J. P. Latge, Curr. Opin. Microbiol, 2005, 8, 385–392 CrossRef CAS PubMed.
- W. C. Nierman, A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell and D. W. Denning, Nature, 2005, 438, 1151–1156 CrossRef CAS PubMed.
- B. Halliwell and J. M. Gutteridge, Biochem. J., 1984, 219, 1–14 CAS.
- T. Ganz, Curr. Opin. Immunol., 2009, 21, 63–67 Search PubMed.
- E. D. Weinberg, Biochim. Biophys. Acta, 2009, 1790, 600–605 CrossRef CAS PubMed.
- H. Haas, M. Eisendle and B. G. Turgeon, Ann. Rev. Phytopathol., 2008, 46, 149–187 CrossRef CAS PubMed.
- M. Schrettl, E. Bignell, C. Kragl, C. Joechl, T. Rogers, H. N. Arst, Jr, K. Haynes and H. Haas, J. Exp. Med., 2004, 200, 1213–1219 CrossRef CAS PubMed.
- C. C. Philpott, Biochim. Biophys. Acta, 2006, 1763, 636–645 CrossRef CAS PubMed.
- M. Schrettl, G. Winkelmann and H. Haas, Biometals, 2004, 17, 647–654 CrossRef CAS.
- Z. Chi, X. X. Wang, Z. C. Ma, M. A. Buzdar and Z. M. Chi, Biometals, 2012, 25, 219–230 CrossRef CAS PubMed.
- C. D. Kaplan and J. Kaplan, Chem. Rev., 2009, 109, 4536–4552 CrossRef CAS PubMed.
- M. Eisendle, H. Oberegger, I. Zadra and H. Haas, Mol. Microbiol., 2003, 49, 359–375 CrossRef CAS.
- M. Schrettl, H. S. Kim, M. Eisendle, C. Kragl, W. C. Nierman, T. Heinekamp, E. R. Werner, I. Jacobsen, P. Illmer, H. Yi, A. A. Brakhage and H. Haas, Mol. Microbiol., 2008, 70, 27–43 CrossRef CAS PubMed.
- F. Gsaller, M. Eisendle, B. E. Lechner, M. Schrettl, H. Lindner, D. Muller, S. Geley and H. Haas, Metallomics, 2012, 4, 1262–1270 RSC.
- A. Thieken and G. Winkelmann, FEMS Microbiol. Lett., 1992, 73, 37–41 CrossRef CAS PubMed.
- R. J. Capon, M. Stewart, R. Ratnayake, E. Lacey and J. H. Gill, J. Nat. Prod., 2007, 70, 1746–1752 CrossRef CAS PubMed.
- J. C. Renshaw, G. D. Robson, A. P. J. Trinci, M. G. Wiebe, F. R. Livens, D. Collison and R. J. Taylor, Mycol. Res., 2002, 106, 1123–1142 CrossRef CAS.
- R. C. Hider and X. Kong, Nat. Prod. Rep., 2010, 27, 637–657 RSC.
- M. B. Hossain, M. A. Jalal and D. van der Helm, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1997, 53(Pt 6), 716–718 Search PubMed.
- A. Koulman, T. V. Lee, K. Fraser, L. Johnson, V. Arcus, J. S. Lott, S. Rasmussen and G. Lane, Phytochemistry, 2012, 75, 128–139 CrossRef CAS PubMed.
- W. B. Yin, J. A. Baccile, J. W. Bok, Y. Chen, N. P. Keller and F. C. Schroeder, J. Am. Chem. Soc., 2013, 135, 2064–2067 CrossRef CAS PubMed.
- M. Schrettl, E. Bignell, C. Kragl, Y. Sabiha, O. Loss, M. Eisendle, A. Wallner, H. N. Arst, K. Haynes and H. Haas, PLoS Pathog., 2007, 3, e128 Search PubMed.
- H. J. Plattner and H. Diekmann, in Metal ions in fungi, ed. G. Winkelmann and D. R. Winge, Marcel Decker, Inc., New York, N.Y., 1994, pp. 99–117 Search PubMed.
- D. Greenshields, L. Guosheng, J. Feng, G. Selvaraj and Y. Wei, Mol. Plant Pathol., 2007, 8, 411–421 CrossRef CAS PubMed.
- O. Yamada, S. Na Nan, T. Akao, M. Tominaga, H. Watanabe, T. Satoh, H. Enei and O. Akita, J. Biosci. Bioeng., 2003, 95, 82–88 CrossRef CAS.
- C. Hof, K. Eisfeld, L. Antelo, A. J. Foster and H. Anke, Fungal Genet. Biol., 2009, 46, 321–332 CrossRef CAS PubMed.
- L. H. Hwang, J. A. Mayfield, J. Rine and A. Sil, PLoS Pathog., 2008, 4, e1000044 Search PubMed.
- B. Mei, A. D. Budde and S. A. Leong, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 903–907 CrossRef CAS.
- P. Visca, A. Ciervo and N. Orsi, J. Bacteriol., 1994, 176, 1128–1140 CAS.
- M. Herrero, V. de Lorenzo and J. B. Neilands, J. Bacteriol., 1988, 170, 56–64 CAS.
- S. Yasmin, L. Alcazar-Fuoli, M. Grundlinger, T. Puempel, T. Cairns, M. Blatzer, J. F. Lopez, J. O. Grimalt, E. Bignell and H. Haas, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E497–E504 CAS.
- C. M. Roche, H. W. Blanch, D. S. Clark and N. L. Glass, Eukaryot. Cell, 2013, 12, 1244–1257 CrossRef CAS PubMed.
- M. Grundlinger, S. Yasmin, B. E. Lechner, S. Geley, M. Schrettl, M. Hynes and H. Haas, Mol. Microbiol., 2013, 88, 862–875 CrossRef PubMed.
- M. Blatzer, M. Schrettl, B. Sarg, H. H. Lindner, K. Pfaller and H. Haas, Appl. Environ. Microbiol., 2011, 77, 4959–4966 CrossRef CAS PubMed.
- V. de Lorenzo, A. Bindereif, B. H. Paw and J. B. Neilands, J. Bacteriol., 1986, 165, 570–578 CAS.
- B. Winterberg, S. Uhlmann, U. Linne, F. Lessing, M. A. Marahiel, H. Eichhorn, R. Kahmann and J. Schirawski, Mol. Microbiol., 2010, 75, 1260–1271 CrossRef CAS PubMed.
- W. M. Yuan, G. D. Gentil, A. D. Budde and S. A. Leong, J. Bacteriol., 2001, 183, 4040–4051 CrossRef CAS PubMed.
- T. Schwecke, K. Gottling, P. Durek, I. Duenas, N. F. Kaufer, S. Zock-Emmenthal, E. Staub, T. Neuhof, R. Dieckmann and H. von Dohren, ChemBioChem, 2006, 7, 612–622 CrossRef CAS PubMed.
- A. Munawar, J. W. Marshall, R. J. Cox, A. M. Bailey and C. M. Lazarus, ChemBioChem, 2013, 14, 388–394 CrossRef CAS PubMed.
- K. E. Bushley, D. R. Ripoll and B. G. Turgeon, BMC Evol. Biol., 2008, 8, 328 CrossRef PubMed.
- S. Oide, W. Moeder, H. Haas, S. Krasnoff, D. Gibson, K. Yoshioka and B. G. Turgeon, Plant Cell, 2006, 18, 2836–2853 CrossRef CAS PubMed.
- E. Albarouki, L. Schafferer, F. Ye, N. von Wiren, H. Haas and H. B. Deising, Mol. Microbiol., 2014, 92, 338–355 CAS.
- K. Reiber, E. P. Reeves, C. M. Neville, R. Winkler, P. Gebhardt, K. Kavanagh and S. Doyle, FEMS Microbiol. Lett., 2005, 248, 83–91 CAS.
- W. Steinchen, G. Lackner, S. Yasmin, M. Schrettl, H. M. Dahse, H. Haas and D. Hoffmeister, Appl. Environ. Microbiol., 2013, 79, 6670–6676 CAS.
- G. Allen, M. Bromley, S. J. Kaye, D. Keszenman-Pereyra, T. D. Zucchi, J. Price, M. Birch, J. D. Oliver and G. Turner, Fungal Genet. Biol., 2011, 48, 456–464 Search PubMed.
- H. Oberegger, M. Eisendle, M. Schrettl, S. Graessle and H. Haas, Curr. Genet., 2003, 44, 211–215 Search PubMed.
- Y. Leng and S. Zhong, Mol. Plant Pathol., 2012, 13, 375–387 Search PubMed.
- R. Horbach, A. Graf, F. Weihmann, L. Antelo, S. Mathea, J. C. Liermann, T. Opatz, E. Thines, J. Aguirre and H. B. Deising, Plant Cell, 2009, 21, 3379–3396 Search PubMed.
- P. Wiemann, S. Albermann, E. M. Niehaus, L. Studt, K. W. von Bargen, N. L. Brock, H. U. Humpf, J. S. Dickschat and B. Tudzynski, PLoS One, 2012, 7, e37519 Search PubMed.
- M. Eisendle, M. Schrettl, C. Kragl, D. Muller, P. Illmer and H. Haas, Eukaryot. Cell, 2006, 5, 1596–1603 Search PubMed.
- B. F. Matzanke, E. Bill, A. X. Trautwein and G. Winkelmann, J. Bacteriol., 1987, 169, 5873–5876 Search PubMed.
- J. A. Kiel, M. A. van den Berg, F. Fusetti, B. Poolman, R. A. Bovenberg, M. Veenhuis and I. J. van der Klei, Funct. Integr. Genomics, 2009, 9, 167–184 Search PubMed.
- H. Haas, M. Schoeser, E. Lesuisse, J. F. Ernst, W. Parson, B. Abt, G. Winkelmann and H. Oberegger, Biochem. J., 2003, 371, 505–513 Search PubMed.
- C. C. Philpott and O. Protchenko, Eukaryot. Cell, 2008, 7, 20–27 Search PubMed.
- W. H. Jung and J. W. Kronstad, Cell. Microbiol., 2008, 10, 277–284 CrossRef CAS PubMed.
- T. Nevitt and D. J. Thiele, PLoS Pathog., 2011, 7, e1001322 CAS.
- O. Ardon, H. Bussey, C. Philpott, D. M. Ward, S. Davis-Kaplan, S. Verroneau, B. Jiang and J. Kaplan, J. Biol. Chem., 2001, 276, 43049–43055 CrossRef CAS PubMed.
- I. Raymond-Bouchard, C. S. Carroll, J. R. Nesbitt, K. A. Henry, L. J. Pinto, M. Moinzadeh, J. K. Scott and M. M. Moore, Eukaryot. Cell, 2012, 11, 1333–1344 CrossRef CAS PubMed.
- Y. S. Park, T. H. Kim, H. I. Chang, H. C. Sung and C. W. Yun, Biochem. Biophys. Res. Commun., 2006, 345, 1634–1642 CrossRef CAS PubMed.
- C. Kragl, M. Schrettl, B. Abt, B. Sarg, H. H. Lindner and H. Haas, Eukaryot. Cell, 2007, 6, 1278–1285 CrossRef CAS PubMed.
- M. Grundlinger, F. Gsaller, M. Schrettl, H. Lindner and H. Haas, Appl. Environ. Microbiol., 2013, 79, 7534–7536 CrossRef PubMed.
- T. J. Brickman and M. A. McIntosh, J. Biol. Chem., 1992, 267, 12350–12355 CAS.
- M. Schrettl, N. Beckmann, J. Varga, T. Heinekamp, I. D. Jacobsen, C. Jochl, T. A. Moussa, S. Wang, F. Gsaller, M. Blatzer, E. R. Werner, W. C. Niermann, A. A. Brakhage and H. Haas, PLoS Pathog., 2010, 6, e1001124 Search PubMed.
- P. Hortschansky, M. Eisendle, Q. Al-Abdallah, A. D. Schmidt, S. Bergmann, M. Thon, O. Kniemeyer, B. Abt, B. Seeber, E. R. Werner, M. Kato, A. A. Brakhage and H. Haas, EMBO J., 2007, 26, 3157–3168 CrossRef CAS PubMed.
- H. Haas, I. Zadra, G. Stoffler and K. Angermayr, J. Biol. Chem., 1999, 274, 4613–4619 CrossRef CAS PubMed.
- L. W. Zhou, H. Haas and G. A. Marzluf, Mol. Gen. Genet., 1998, 259, 532–540 CrossRef CAS.
- L. H. Hwang, E. Seth, S. A. Gilmore and A. Sil, Eukaryot. Cell, 2012, 11, 16–25 CrossRef CAS PubMed.
- N. Zhang, N. A. MohdZainudin, K. Scher, B. J. Condon, B. A. Horwitz and B. G. Turgeon, Mol. Plant-Microbe Interact., 2013, 26, 1473–1485 CrossRef CAS PubMed.
- S. Labbe, B. Pelletier and A. Mercier, Biometals, 2007, 20, 523–537 CrossRef CAS PubMed.
- Z. Chi, X. X. Wang, Q. Geng and Z. M. Chi, Biometals, 2013, 26, 955–967 CrossRef CAS PubMed.
- C. Voisard, J. Wang, J. L. McEvoy, P. Xu and S. A. Leong, Mol. Cell. Biol., 1993, 13, 7091–7100 CAS.
- J. Amich, L. Schafferer, H. Haas and S. Krappmann, PLoS Pathog., 2013, 9, e1003573 CAS.
- H. Li and C. E. Outten, Biochemistry, 2012, 51, 4377–4389 CrossRef CAS PubMed.
- M. Vodisch, K. Scherlach, R. Winkler, C. Hertweck, H. P. Braun, M. Roth, H. Haas, E. R. Werner, A. A. Brakhage and O. Kniemeyer, J. Proteome Res., 2011, 10, 2508–2524 CrossRef CAS PubMed.
- S. D. Willger, S. Puttikamonkul, K. H. Kim, J. B. Burritt, N. Grahl, L. J. Metzler, R. Barbuch, M. Bard, C. B. Lawrence and R. A. Cramer, Jr, PLoS Pathog., 2008, 4, e1000200 Search PubMed.
- C. M. Bien and P. J. Espenshade, Eukaryot. Cell, 2010, 9, 352–359 CrossRef CAS PubMed.
- M. Blatzer, B. M. Barker, S. D. Willger, N. Beckmann, S. J. Blosser, E. J. Cornish, A. Mazurie, N. Grahl, H. Haas and R. A. Cramer, PLoS Genet., 2011, 7, e1002374 CAS.
- M. Eisendle, H. Oberegger, R. Buttinger, P. Illmer and H. Haas, Eukaryot. Cell, 2004, 3, 561–563 CrossRef CAS.
- H. Liu, F. N. Gravelat, L. Y. Chiang, D. Chen, G. Vanier, D. E. Ejzykowicz, A. S. Ibrahim, W. C. Nierman, D. C. Sheppard and S. G. Filler, Mol. Microbiol., 2010, 78, 1038–1054 CrossRef CAS PubMed.
- X. Feng, K. Krishnan, D. L. Richie, V. Aimanianda, L. Hartl, N. Grahl, M. V. Powers-Fletcher, M. Zhang, K. K. Fuller, W. C. Nierman, L. J. Lu, J. P. Latge, L. Woollett, S. L. Newman, R. A. Cramer, Jr, J. C. Rhodes and D. S. Askew, PLoS Pathog., 2011, 7, e1002330 CAS.
- N. Beckmann, L. Schafferer, M. Schrettl, U. Binder, H. Talasz, H. Lindner and H. Haas, PLoS One, 2013, 8, e67426 CAS.
- R. Jain, V. Valiante, N. Remme, T. Docimo, T. Heinekamp, C. Hertweck, J. Gershenzon, H. Haas and A. A. Brakhage, Mol. Microbiol., 2011, 82, 39–53 CrossRef CAS PubMed.
- R. E. Frederick, J. A. Mayfield and J. L. DuBois, J. Am. Chem. Soc., 2011, 133, 12338–12341 CrossRef CAS PubMed.
- S. Oide, S. B. Krasnoff, D. M. Gibson and B. G. Turgeon, Eukaryot. Cell, 2007, 6, 1339–1353 CrossRef CAS PubMed.
- A. Wallner, M. Blatzer, M. Schrettl, B. Sarg, H. Lindner and H. Haas, Appl. Environ. Microbiol., 2009, 75, 4194–4196 CrossRef CAS PubMed.
- H. Oberegger, M. Schoeser, I. Zadra, B. Abt and H. Haas, Mol. Microbiol., 2001, 41, 1077–1089 Search PubMed.
- A. H. Hissen, A. N. Wan, M. L. Warwas, L. J. Pinto and M. M. Moore, Infect. Immun., 2005, 73, 5493–5503 CrossRef CAS PubMed.
- A. McDonagh, N. D. Fedorova, J. Crabtree, Y. Yu, S. Kim, D. Chen, O. Loss, T. Cairns, G. Goldman, D. Armstrong-James, K. Haynes, H. Haas, M. Schrettl, G. May, W. C. Nierman and E. Bignell, PLoS Pathog., 2008, 4, e1000154 Search PubMed.
- A. H. Hissen, J. M. Chow, L. J. Pinto and M. M. Moore, Infect. Immun., 2004, 72, 1402–1408 CrossRef CAS.
- A. H. Hissen and M. M. Moore, J. Biol. Inorg. Chem., 2005, 10, 211–220 Search PubMed.
- M. Schrettl, O. Ibrahim-Granet, S. Droin, M. Huerre, J. P. Latge and H. Haas, Microbes Infect., 2010, 12, 1035–1041 Search PubMed.
- M. Seifert, M. Nairz, A. Schroll, M. Schrettl, H. Haas and G. Weiss, Immunobiology, 2008, 213, 767–778 Search PubMed.
- J. L. Oosthuizen, P. Gomez, J. Ruan, T. L. Hackett, M. M. Moore, D. A. Knight and S. J. Tebbutt, PLoS One, 2011, 6, e20527 CAS.
- R. Ben-Ami, R. E. Lewis, K. Leventakos, J. P. Latge and D. P. Kontoyiannis, Antimicrob. Agents Chemother., 2010, 54, 1848–1854 CAS.
- G. Chamilos, E. M. Bignell, M. Schrettl, R. E. Lewis, K. Leventakos, G. S. May, H. Haas and D. P. Kontoyiannis, Med. Mycol., 2010, 48, 506–510 CAS.
- J. L. Slater, L. Gregson, D. W. Denning and P. A. Warn, Med. Mycol., 2011, 49 Suppl. 1, S107–113 CrossRef PubMed.
- L. H. Chen, C. H. Lin and K. R. Chung, Mol. Plant Pathol., 2013, 14, 497–505 Search PubMed.
- C. Hof, K. Eisfeld, K. Welzel, L. Antelo, A. J. Foster and H. Anke, Mol. Plant Pathol., 2007, 8, 163–172 CrossRef CAS PubMed.
- H. Eichhorn, F. Lessing, B. Winterberg, J. Schirawski, J. Kamper, P. Muller and R. Kahmann, Plant Cell, 2006, 18, 3332–3345 CrossRef CAS PubMed.
- E. Albarouki and H. B. Deising, Mol. Plant Microbe In., 2013, 26, 695–708 CAS.
- S. A. Leong and G. Winkelmann, Met. Ions Biol. Syst., 1998, 35, 147–186 CAS.
- L. J. Johnson, A. Koulman, M. Christensen, G. A. Lane, K. Fraser, N. Forester, R. D. Johnson, G. T. Bryan and S. Rasmussen, PLoS Pathog., 2013, 9, e1003332 CAS.
- W. J. Moree, V. V. Phelan, C. H. Wu, N. Bandeira, D. S. Cornett, B. M. Duggan and P. C. Dorrestein, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 13811–13816 CrossRef CAS PubMed.
- M. S. Lopez-Berges, J. Capilla, D. Turra, L. Schafferer, S. Matthijs, C. Jochl, P. Cornelis, J. Guarro, H. Haas and A. Di Pietro, Plant Cell, 2012, 24, 3805–3822 CrossRef CAS PubMed.
- W. J. Steinbach, PLoS Pathog., 2013, 9, e1003642 Search PubMed.
- S. Franceschini, M. Fedkenheuer, N. J. Vogelaar, H. H. Robinson, P. Sobrado and A. Mattevi, Biochemistry, 2012, 51, 7043–7045 CrossRef CAS PubMed.
- S. M. Leal, Jr, S. Roy, C. Vareechon, S. Carrion, H. Clark, M. S. Lopez-Berges, A. Di Pietro, M. Schrettl, N. Beckmann, B. Redl, H. Haas and E. Pearlman, PLoS Pathog., 2013, 9, e1003436 Search PubMed.
- T. Hsiang and D. L. Baillie, J. Mol. Evol., 2005, 60, 475–483 CrossRef CAS PubMed.
- M. J. Miller, H. Zhu, Y. Xu, C. Wu, A. J. Walz, A. Vergne, J. M. Roosenberg, G. Moraski, A. A. Minnick, J. McKee-Dolence, J. Hu, K. Fennell, E. Kurt Dolence, L. Dong, S. Franzblau, F. Malouin and U. Mollmann, Biometals, 2009, 22, 61–75 CrossRef CAS PubMed.
- M. Petrik, G. M. Franssen, H. Haas, P. Laverman, C. Hortnagl, M. Schrettl, A. Helbok, C. Lass-Florl and C. Decristoforo, Eur. J. Nucl. Med. Mol. Imaging, 2012, 39, 1175–1183 CrossRef CAS PubMed.
- M. Petrik, H. Haas, G. Dobrozemsky, C. Lass-Florl, A. Helbok, M. Blatzer, H. Dietrich and C. Decristoforo, J. Nucl. Med., 2010, 51, 639–645 CrossRef CAS PubMed.
- M. Petrik, H. Haas, P. Laverman, M. Schrettl, G. M. Franssen, M. Blatzer and C. Decristoforo, Mol. Imaging Biol., 2014, 16, 102–108 CrossRef PubMed.
- M. Petrik, H. Haas, M. Schrettl, A. Helbok, M. Blatzer and C. Decristoforo, Nucl. Med. Biol., 2012, 39, 361–369 CrossRef CAS PubMed.
- M. Xiong, D. K. Schneiderman, F. S. Bates, M. A. Hillmyer and K. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 8357–8362 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |