Open Access Article
Open Access ArticleTips and turns of bacteriophytochrome photoactivation
Heikki
Takala†
 *ab,
Petra
Edlund†
c,
Janne A.
Ihalainen
*ab,
Petra
Edlund†
c,
Janne A.
Ihalainen
 a and
Sebastian
Westenhoff
*c
a and
Sebastian
Westenhoff
*c
aDepartment of Biological and Environmental Science, Nanoscience Center, University of Jyvaskyla, Box 35, 40014 Jyvaskyla, Finland. E-mail: heikki.p.takala@jyu.fi
bDepartment of Anatomy, Faculty of Medicine, University of Helsinki, Box 63, 00014 Helsinki, Finland
cDepartment of Chemistry and Molecular Biology, University of Gothenburg, Box 462, 40530 Gothenburg, Sweden. E-mail: westenho@cmb.gu.se
First published on 27th October 2020
Abstract
Phytochromes are ubiquitous photosensor proteins, which control the growth, reproduction and movement in plants, fungi and bacteria. Phytochromes switch between two photophysical states depending on the light conditions. In analogy to molecular machines, light absorption induces a series of structural changes that are transduced from the bilin chromophore, through the protein, and to the output domains. Recent progress towards understanding this structural mechanism of signal transduction has been manifold. We describe this progress with a focus on bacteriophytochromes. We describe the mechanism along three structural tiers, which are the chromophore-binding pocket, the photosensory module, and the output domains. We discuss possible interconnections between the tiers and conclude by presenting future directions and open questions. We hope that this review may serve as a compendium to guide future structural and spectroscopic studies designed to understand structural signaling in phytochromes.
Discovery and function of phytochromes
Brief history of the discovery of plant and fungal phytochromes
Multiple responses of plants depend on environmental light conditions. First observed in the 1930s,1 and further rationalized in the 1950s,2 the photoperiodic control of flowering time and seed germination in plants were found to be sensitive to illumination by red and far-red light. The responsible photoconvertible protein was identified in 1959 by absorption spectroscopy and termed phytochrome.3 In 1983, an oat phytochrome was purified4 and two years later, the primary amino acid sequence was revealed.5 Phytochromes have been shown to be important for many properties of plants, such as shade avoidance, plant cycle, stem elongation, and flowering time.6A characteristic feature of phytochromes is that they can photoswitch between two photochemical states. Depending on the light conditions, they adopt a state that absorbs red light, termed “Pr” and one that absorbs far-red light, termed “Pfr” (Fig. 1c). The two states have different biochemical activity, which leads to different cellular responses.
The hypothesis that plant chloroplasts have developed from a photosynthetic bacterium led to the speculations that prokaryotic genes represent the evolutionary origin of plant phytochromes.7–9 Indeed, the first prokaryotic phytochrome to be discovered was Cph1 in the cyanobacterium Synechocystis sp. PCC6803 in 1997, when two groups showed that it could be expressed in Escherichia coli and autoassembled with a bilin cofactor to show phytochrome-typical photoswitching.10,11 In 1999, the rather more divergent group of bacteriophytochromes (BphP) was discovered in the non-photosynthetic bacterium Deinoccocus radiodurans.12 Although the gene rcaE in the cyanobacterium Fremyella displosisphon was proposed to encode a phytochrome in 1996 on the basis of genetic evidence,13 it was shown only recently on the basis of a revised sequence that it indeed encodes a bona fide photoreceptor.14 In 2005, a phytochrome was described in the fungus Aspergillus nidulans.15 Phytochrome-related proteins exist in eukaryotic algae, where their absorption maxima can span the entire visible region.16
Plant phytochromes are important because they control the growth, reproduction, and development of virtually all vegetation on Earth. Uncovering in detail how phytochromes achieve this, is fundamentally important for understanding the basis of life on Earth. Moreover, the knowledge could enable modification of plants to control their growth and development.17 Bacteriophytochromes are particularly interesting, because they have a relatively simple modular architecture and photoswitch with red light. This makes them interesting targets for optogenetic applications, imaging in tissues, light-dependent gene expression, and possible medical applications.18–23 These applications can be achieved by coupling the light-sensing part of phytochrome to the proteins of interest with a desired output signal.21 Out of all of these motivations, it is important to understand how phytochromes function at the atomic level.
Plant phytochromes are involved in a complex signaling network within the cell. They function as serine/threonine kinases,24 probably facilitated by a family of nuclear protein kinases (photoregulatory protein kinases 1–4, also known as MUT9-like kinases)25 and also have several additional biological outputs in vivo.26–28 Curiously, the physiological function of cyanobacterial and bacteriophytochromes is less well understood compared to plant phytochromes. For example, the Rhodopseudomonas palustris phytochrome regulates the photosynthetic apparatus,29 and in the cyanobacterium Leptolyngbya sp. strain JSC-1, a phytochrome controls remodeling of the photosynthetic apparatus to adapt to far-red light conditions.30 In non-photosynthetic prokaryotes, the physiological function of BphPs remains even more unclear, with some notable exceptions.31,32 Interestingly, prokaryotic phytochromes have a variety of different output domains, which include histidine kinases (HK and HWE), diguanylyl cyclases (GGDEF), phosphodiesterases (EAL), metal-dependent Ser/Thr protein phosphatases (PPM), as well as domains without any enzymatic activity (e.g., PAS and HOS).33
The discovery of prokaryotic phytochromes marked a turning point in the structural biology of phytochromes, because it was now possible to express and purify the proteins at larger quantities.34 Thereby, crystal structures could be determined, and more extensive spectroscopic and biochemical analyses could be undertaken.35,36 Information obtained on bacteriophytochromes can often be transferred to plant phytochromes. This is especially true for the photosensing part of the phytochrome, which is homologous across the kingdoms of life in which phytochromes exist. Indeed, the first crystal structure of a phytochrome fragment from the plant Arabidopsis thaliana shows similarities to its prokaryotic counterparts in terms of the domain structure and arrangement.37 Differences are also observed, for example in the thioether linkage of the chromophore38 and the photoactivation mechanisms of the D-ring, which in Pfr is the likely oriented in a β-facial position in plant phytochromes, compared to α-facial in bacterial phytochromes.39
The extensive interest in phytochromes has led to decades of research and several reviews on phytochromes.9,32,33,40–44 Specific reviews on plant phytochromes28,45–48 and fungal phytochromes49 are also available. Here we summarize the current understanding of how phytochromes remodel their three-dimensional structure when they photoconvert between Pr and Pfr. We concentrate on bacteriophytochromes because the most structural dynamic data has been reported for these species. We start by describing the photocycle and chromophore and we continue to discuss the structural features that change upon photoactivation. It is currently not fully understood how the biochemical activity of phytochromes is controlled, but we discuss a few potential explanations. We end by providing an outlook on the future of the structural biology of phytochromes.
Chromophore and photocycle
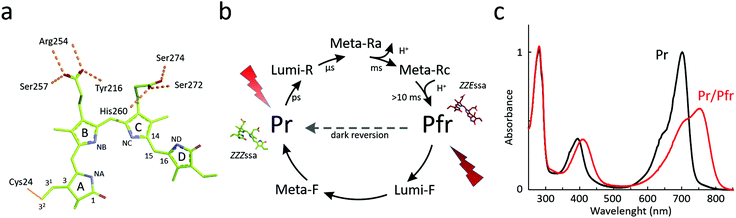
Chromophore
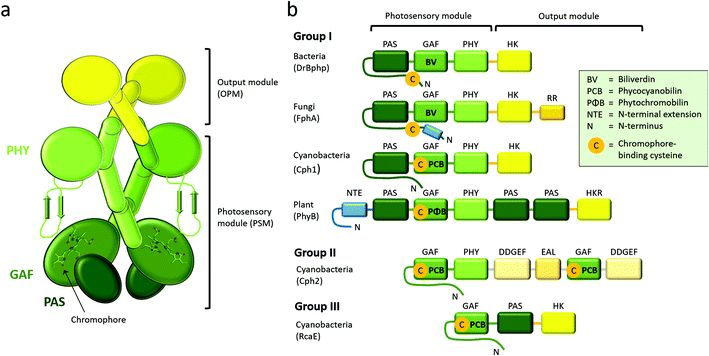
Phytochromes carry a bilin chromophore that absorbs light at the red/far-red wavelength region. It is an open tetrapyrrole, which originates from heme catabolism in all organisms (Fig. 1a). The phytochromes from bacteria, cyanobacteria and plants bind biliverdin (BV), phycocyanobilin (PCB) or phytochromobilin (PΦB), respectively.44 The cofactor is covalently attached to a cysteine residue that resides in the PAS domain in bacteria and fungi, and in the GAF domain in cyanobacteria and plants (Fig. 2), but is embedded inside the GAF domain in all occasions. In bacteria, the gene for expression of the enzyme hemeoxygenase, which converts the heme precursor into a biliverdin, is located in the same operon as BphPs.34,50,51 In recombinant expression, hemeoxygenase can be co-expressed with phytochrome to yield the holoprotein.34 An extensive review on the synthesis of bilin is available.52The photocycle
The photocycle of phytochromes has been studied extensively with spectroscopy. A hallmark feature of the superfamily is that the proteins can adopt two spectroscopically different metastable states (Fig. 1b). For canonical phytochromes, these are called the red light-absorbing Pr state and the far-red light-absorbing Pfr state. The absorption maximum of bacteriophytochrome is usually at around 700 nm for the Pr state and 750 nm for the Pfr state (Fig. 1c). This however may vary between the phytochrome species, reaching the blue-most absorption for a Pr state at 610 nm and Pfr state at 670 nm,53 albeit in this extreme case the bilin chromophore is phycocyanobilin instead of biliverdin. Phytochromes can be actively switched between the Pr and Pfr states with red/far-red light, or by thermal reversion in the dark. This dark reversion occurs in minutes, hours, or even days depending on the species and construct. The dark reversion time depends on cellular conditions, such as pH, ionic strength, reducing agents concentrations of metal ions,54 and on temperature.55,56Generally, the resting state of phytochromes is the Pr state. However, a number of bacteriophytochromes show reversed thermal dark reversion into the Pfr state. These are called bathy phytochromes, discovered first in phytochromes from Bradyrhizobium and Agrobacterium fabrum.29,32,50 Bathy phytochromes are primarly found in nitrogen-fixing plants, which is logical since far-red light penetrates the soil more effectively and can be transduced in the roots of plants.57 Some bacterial species express both canonical and bathy phytochromes. One widely studied example is from the soil bacterium A. fabrum, with a canonical phytochrome Agp1 and a bathy phytochrome Agp2. These proteins have been proposed to complement each other.58 The structural and spectroscopic properties of the Pr and Pfr states are retained in bathy phytochromes and it is merely the relative free energy of the two states which is interchanged.59
Intermediate states
When switching between the two metastable states, phytochromes pass through a number of intermediate states. A different number of intermediates have been reported for plant, cyanobacterial, and bacteriophytochromes, with plant phytochromes having the most complex photocycle.60 For bacteriophytochromes, a minimal and practical consensus is to consider two intermediate states, Lumi and Meta, as shown in Fig. 1b.36Here we primarily describe the photocycle of bacteriophytochromes. Photoexcitation of the Pr state (or Pfr state) prepares an excited state, which relaxes into a Lumi-R (or Lumi-F) intermediate. It is currently not entirely clear, how this relaxation proceeds. The predominant view is that the excited state is relatively long-lived (tens of picoseconds) in Pr and short-lived (around one picosecond) in Pfr.61–65 The biliverdin D-ring undergoes Z-to-E isomerization during the Pr-to-Pfr transition, and E-to-Z isomerization during the Pfr-to-Pr transition.66,67 Polarization-resolved mid-infrared spectroscopy with structural refinement has confirmed that the isomerization occurs by a rotation over the C15–C16 methine-bridge between ring C and D.68 The quantum yield of reaching the Lumi states is relatively low at less than 15%. Even though the transitions in Fig. 1b are indicated as linear forward reactions, back-reactions from intermediate states to the resting state have been recorded as well.69,70
In bacteriophytochromes, the Lumi-R state typically lives for tens of microseconds until it converts into a Meta state, which in turn translates into the Pfr state on millisecond time scale. The Meta-R intermediate(s) has been shown to involve deprotonation and re-protonation events in several species.71–74 This splits the Meta-R state into Meta-Ra and Meta-Rc (Fig. 1b).75 Importantly, the chromophore is fully protonated in both Pr and Pfr states.76 For the back reaction from Pfr to Pr, evidence by NMR suggests that the chromophore and protein stay protonated.77 From the same study, it was also concluded that the back-isomerization occurs in two steps, which is in agreement with the crystallography of cryo-trapped intermediates of PaBphP78 and with the fact that there are changes in the hydrogen-bonding network around the biliverdin D-ring in Meta-F. Possibly because there are no particular proton transfer reactions, the photoinduced reaction times from Pfr to Pr are somewhat faster. The Lumi-F state decays bi-phasic on a microsecond time-scale and the Meta-F state converts to Pr state within a millisecond time-scale.79
Most information about the intermediate states has been obtained from spectroscopic studies, and direct structural information is unfortunately scarce. A notable exception is a recent structure of a cryo-trapped Meta-F intermediate, which reveals a twist of the biliverdin D-ring by virtually 180°, significant movements of the C- and B-ring propionates and side chain adjustments around the chromophore.80 We note that the studied phytochrome fragment contains 24 mutations and an aberrant photocycle, raising a question whether the intermediate structure reflects the wild-type situation. In the study of cryo-trapped PaBphP intermediates,78 the acquisition temperature resembles time, with lower temperatures trapping earlier intermediates. However, it was not possible to correlate the cryo-trapped intermediates to photocycle intermediates. These studies are a welcome start to understanding the structural mechanism of photoconversion. In the next section, we discuss what is known about this process.
Three structural tiers of signal transduction
Phytochromes are generally homodimeric complexes. Each subunit contains an N-terminal photosensory module (PSM), which senses light and transfers the signal, to the C-terminal output module (OPM) (Fig. 2a). Plant and fungal phytochromes have an additional N-terminal extension (NTE). The NTE has shown to stabilize their Pfr state and to participate in phosphorylation events in plants.81–85 Plant phytochromes contain also two C-terminal PAS domains and a histidine kinase-like domain. Similar to other photosensor proteins, phytochromes are built up from a limited number of conserved domains.45 Based on the composition of the PSM, the phytochrome superfamily can be divided into three classes (Fig. 2b).46 Group I is the biggest group which describes phytochromes with three PSM domains: PAS (Per/Arnt/Sim), GAF (cGMP phosphodiesterase/adenylyl cyclase/Fhl1), and PHY (phytochrome-specific). Group I comprises of phytochromes from plants, fungi, bacteria and cyanobacteria. Group II contains phytochrome-like proteins without the PAS domain. These PAS-less phytochromes, like cyanobacterial Cph2, have a dimeric GAF-PHY structure. Group III proteins comprise of a single GAF domain and represents the cyanobacteriochromes, CBCRs.86 PAS domains are present in many sensing proteins, in protein–protein interaction scaffolds and in transcription factors, which points towards a similar evolutionary origin.87 Here, we focus on the group I phytochromes, which has the widest distribution across the kingdoms of life.A few years after their discovery,10,13 several crystal and NMR structures of prokaryotic phytochromes were solved. The first crystal structure of a phytochrome was uncovered for the PAS-GAF fragment of the phytochrome from Deinococcus radiodurans.88 The first structures of the complete phytochrome PSMs were from Cph1 from cyanobacterium Synechocystis sp. PCC680389 and PaBphP from Pseudomonas aeruginosa,90 after which several other species have followed. More recently, PAS-GAF and PSM structures of plant phytochromes have been disclosed,37,38 as well as bacteriophytochromes with an output module.91–94 The presently solved crystal structures of PAS-GAF, PSM and full-length phytochromes from group I are summarized in Tables 1–3.
| Name | Organism | PDB code | Release year | Resolution (Å) | Pr/Pfr | Mutations | Comment | Ref. |
|---|---|---|---|---|---|---|---|---|
| DrBphP | D. radiodurans | 1ZTU | 2005 | 2.5 | Pr | P240T | 88 | |
| DrBphP | D. radiodurans | 2O9B | 2007 | 2.15 | Pr | Y307S | 101 | |
| DrBphP | D. radiodurans | 2O9C | 2007 | 1.45 | Pr | Y307S | 101 | |
| RpBphP3 | R. palustris | 2OOL | 2007 | 2.2 | Pr | 97 | ||
| DrBphP | D. radiodurans | 3S7N | 2012 | 2.45 | Pr | D207H, Y263F | Fluorescent | 102 |
| DrBphP | D. radiodurans | 3S7O | 2012 | 1.24 | Pr | D207H (IFP1.0) | Fluorescent | 102 |
| DrBphP | D. radiodurans | 3S7P | 2012 | 1.72 | Pr | D207H (IFP1.0) | Fluorescent | 102 |
| DrBphP | D. radiodurans | 3S7Q | 2012 | 1.75 | Pr | F145S, D207H, L311E, L314E | Fluorescent, monomeric | 102 |
| DrBphP | D. radiodurans | 4CQH | 2014 | 1.14 | Pr | IFP2.0 | Fluorescent | 103 |
| RpBphP2 | R. palustris | 4E04 | 2012 | 1.79 | Pr | 16 mutations | Packing mutations | 104 |
| DrBphP | D. radiodurans | 4O8G | 2014 | 1.65 | Pr | IFP1.4 | Fluorescent | 105 |
| DrBphP | D. radiodurans | 4IJG | 2012 | 1.7 | Pr | F145S, L311E, L314E | Monomeric | 105 |
| DrBphP | D. radiodurans | 4Q0H | 2014 | 1.16 | Pr | Y307S | 96 | |
| DrBphP | D. radiodurans | 4Q0I | 2014 | 1.74 | Pr | Y307S, D207A | 96 | |
| SaBphP1 | S. aurantiaca | 4RQ9 | 2016 | 2.5 | Pr | T289H | See structure ‘6BAK’ | — |
| RpBphP1 | R. palustris | 4XTQ | 2015 | 1.64 | Pr | BphP1-FP, C20S | Fluorescent | 19 |
| DrBphP | D. radiodurans | 4Y3I | 2015 | 1.69 | Pr | Y307S | Low X-ray | 106 |
| DrBphP | D. radiodurans | 4Y5F | 2015 | 1.7 | Pr | Y307S | High X-ray | 106 |
| DrBphP | D. radiodurans | 4Z1W | 2015 | 1.3 | Pr | D207L, Y263F | Fluorescent | 107 |
| DrBphP | D. radiodurans | 4ZRR | 2015 | 1.5 | Pr | D207L Y263F | Fluorescent | 107 |
| DrBphP | D. radiodurans | 5AJG | 2016 | 1.11 | Pr | IFP1.4 | Fluorescent | 108 |
| DrBphP | D. radiodurans | 5K5B | 2016 | 1.35 | Pr | 109 | ||
| DrBphP | D. radiodurans | 5L8M | 2016 | 2.1 | Pr | SFX | 109 | |
| DrBphP | D. radiodurans | 5LBR | 2016 | 2.2 | Pr | SFX | 109 | |
| DrBphP | D. radiodurans | 5MG0 | 2017 | 1.65 | Pr | Y307S | SFX | 110 |
| DrBphP | D. radiodurans | 5NFX | 2018 | 1.34 | Pr | Y263F | 111 | |
| RpBphP1 | R. palustris | 5VIK | 2017 | 1.35 | Pr | miRFP703 | Fluorescent, bathy | 112 |
| RpBphP1 | R. palustris | 5VIQ | 2017 | 1.34 | Pr | miRFP709 | Fluorescent, bathy | 112 |
| RpBphP1 | R. palustris | 5VIV | 2017 | 1.33 | Pr | miRFP670, monomeric | Fluorescent, bathy, two BV linkages | 112 |
| SaBphP1 | S. aurantiaca | 6BAF | 2017 | 2.73 | Pr | 31 | ||
| SaBphP1 | S. aurantiaca | 6BAK | 2017 | 2.5 | Pr | T289H | 31 | |
| DrBphP | D. radiodurans | 6FTD | 2018 | 1.4 | Pr | H290T | 113 | |
| IsPadC | Idiomarina sp. A28L | 6SAX | 2019 | 2.4 | Pr | Monomeric | 114 | |
| IsPadC | Idiomarina sp. A28L | 6SAW | 2019 | 3.0 | Pfr-like | Dimeric | 114 | |
| DrBphP | D. radiodurans | 6T3L | 2020 | 2.07 | Pr | SFX | 95 | |
| DrBphP | D. radiodurans | 6T3U | 2020 | 2.21 | I0 | SFX, 1 ps after photoexcitation | 95 | |
| phyB | Sorghum bicolor | 6TBY | 2020 | 1.80 | Pr | Plant, with PCB | 38 | |
| phyB | Sorghum bicolor | 6TC5 | 2020 | 2.10 | Pr | Plant, with PΦB | 38 | |
| phyA | Glycine max | 6TC7 | 2020 | 2.13 | Pr | Plant, with PCB | 38 |
| Name | Organism | PDB code | Release year | Resolution (Å) | Pr/Pfr | Mutations | Comment | Ref. |
|---|---|---|---|---|---|---|---|---|
| Cph1 | Synechocystis sp. PCC6803 | 2VEA | 2008 | 2.21 | Pr | Antiparallel | 89 | |
| PaBphP | P. aeruginosa | 3C2W | 2008 | 2.9 | Pfr | Bathy | 90 | |
| PaBphP | P. aeruginosa | 3G6O | 2009 | 2.85 | Mixed | Q188L | Bathy | 132 |
| PaBphP | P. aeruginosa | 3IBR | 2009 | 2.97 | Mixed | Q188L | Bathy | 132 |
| PaBphP | P. aeruginosa | 3NHQ | 2011 | 2.55 | Pfr | Bathy | 78 | |
| PaBphP | P. aeruginosa | 3NOP | 2011 | 2.8 | L1 | Bathy | 78 | |
| PaBphP | P. aeruginosa | 3NOT | 2011 | 2.7 | L2 | Bathy | 78 | |
| PaBphP | P. aeruginosa | 3NOU | 2011 | 3 | L3 | Bathy | 78 | |
| Cph1 | Synechocystis sp. PCC6803 | 3ZQ5 | 2011 | 1.95 | Pr | Y263F | Antiparallel | 125 |
| Cph2 | Synechocystis sp. PCC6803 | 4BWI | 2013 | 2.6 | Pr | PAS-less | 133 | |
| DrBphP | D. radiodurans | 4O01 | 2014 | 3.24 | Pr/Pfr | Illuminated | 100 | |
| DrBphP | D. radiodurans | 4O0P | 2014 | 3.8 | Pr | Dark | 100 | |
| phyB | A. thaliana | 4OUR | 2014 | 3.4 | Pr | Plant | 37 | |
| DrBphP | D. radiodurans | 4Q0J | 2014 | 2.75 | Pr | 96 | ||
| RpBphP2 | R. palustris | 4R6L/4S21 | 2015 | 3.39 | Pr | 98 | ||
| RpBphP3 | R. palustris | 4R70 | 2015 | 2.85 | Pr | Antiparallel | 98 | |
| DrBphP | D. radiodurans | 5C5K | 2016 | 3.31 | Pfr | F469W | 119 | |
| Agp1 | A. fabrum | 5HSQ | 2016 | 1.85 | Pr | E86A, E87A, E336A, K337A | Antiparallel | 99 |
| Agp1 | A. fabrum | 5I5L | 2016 | 2.7 | Pr | 99 | ||
| IsPadC | Idiomarina sp. A28L | 5LLX | 2017 | 2.8 | Pr | With GTP | 92 | |
| IsPadC | Idiomarina sp. A28L | 5LLY | 2017 | 2.4 | Pr | 92 | ||
| DrBphP | D. radiodurans | 5MG1 | 2017 | 3.3 | Pr | Y307S | SFX | 110 |
| DrBphP | D. radiodurans | 5NM3 | 2018 | 3.3 | Pr/Pfr | Y263F | BV in Pr/Pfr, protein Pfr | 111 |
| DrBphP | D. radiodurans | 5NWN | 2018 | 3.6 | Pr/Pfr | Y263F | BV in Pr, protein Pfr | 111 |
| RpBphP1 | R. palustris | 5OY5 | 2019 | 2.6 | Pfr | Monomeric | — | |
| SaBphP1 | S. aurantiaca | 6BAO | 2017 | 2.5 | Pr | 31 | ||
| SaBphP1 | S. aurantiaca | 6BAP | 2017 | 2.5 | Pr | T289H | 31 | |
| SaBphP1 | S. aurantiaca | 6BAY | 2017 | 2.5 | Pr | T289H | SFX | 31 |
| Agp2 | A. fabrum | 6G1Y | 2018 | 2.5 | Pfr | Bathy | 80 | |
| Agp2 | A. fabrum | 6G1Z | 2018 | 2.03 | Pfr | PAiRFP2, 24 mutations | Fluorescent bathy, antiparallel | 80 |
| Agp2 | A. fabrum | 6G20 | 2018 | 2.16 | Meta-F | PAiRFP2, 24 mutations | Fluorescent bathy, antiparallel | 80 |
| SaBphP2 | S. aurantiaca | 6PTQ | 2019 | 2.1 | Pr | SFX | 134 | |
| SaBphP2 | S. aurantiaca | 6PTX | 2019 | 1.65 | Pr | 134 | ||
| SaBphP2 | S. aurantiaca | 6PU2 | 2019 | 2.2 | Pr | H275T | 134 | |
| Agp1 | A. fabrum | 6R26 | 2020 | 3.11 | Pr-like | Synthetic chromophore | — | |
| Agp1 | A. fabrum | 6R27 | 2020 | 3.40 | Pr-like | Synthetic chromophore | — | |
| phyB | Glycine max | 6TL4 | 2020 | 2.90 | Pr | Plant | 38 |
| Name | Organism | PDB code | Release year | Resolution (Å) | Pr/Pfr | OPM | Comment | Ref. |
|---|---|---|---|---|---|---|---|---|
| RpBphP1 | R. palustris | 4GW9 | 2012 | 2.9 | Pfr | PAS/PAC | Bathy-like, antiparallel | 91 |
| XccBphP | X. campestris | 5AKP | 2016 | 3.25 | Pr/mixed | PAS9 | Bathy-like | 93 |
| IsPadC | Idiomarina sp. A28L | 5LLW | 2017 | 3 | Pr | GGDEF | 92 | |
| XccBphP | X. campestris | 5UYR | 2018 | 3.45 | Pr | PAS9 | Dark-adapted mutant (D199A) | — |
| IsPadC | Idiomarina sp. A28L | 6ET7 | 2018 | 2.85 | Pr + Pfr heterodimer | GGDEF | Heterodimer, constantly active mutation (S505V, A526V) | 94 |
| PaaCΔC | D. radiodurans/Synechocystis sp. PCC6803 | 6FHT | 2018 | 2.35 | Pr | GC/AC | Artificial chimera | 153 |
| XccBphP | X. campestris | 6NDO | 2019 | 3.58 | Pr | PAS9 | Dark-adapted mutant (L193N) | — |
| XccBphP | X. campestris | 6NDP | 2019 | 3.89 | Pr | PAS9 | Dark-adapted mutant (L193Q) | — |
Whereas many structures of the Pr and Pfr states of the PSM of bacteriophytochromes are available, the structures of the intermediate states remain to be disclosed. This means that the structural activation of phytochromes is starting to be understood, but that the mechanism of the changes is unclear.
Tier one: the chromophore-binding pocket
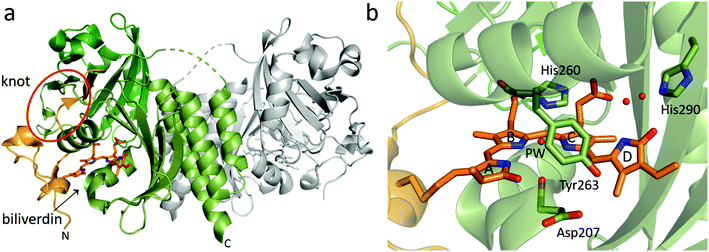 | ||
| Fig. 3 Overview structure and chromophore binding of the BphP PAS-GAF fragment. (a) The structural features of a PAS-GAF dimer from Deinococcus radiodurans phytochrome (DrBphP, PDB id 6T3L95). Subunit 1 with its PAS and GAF domain is colored in different shades of green whereas subunit 2 is grey. N-Terminal end of the PAS domain as well as biliverdin chromophore are colored in orange. The biliverdin position and knot region are indicated. (b) Zoom in on the biliverdin and selected surrounding amino acid residues. Residue numbers are from DrBphP, and the ring names (A–D) are indicated. The so-called pyrrole water (PW) and two well-defined waters between His290, biliverdin D-ring, and C-ring propionate group are also shown as red spheres (PDB id: 4Q0H96). | ||
Curiously, the peptide chain in PAS-GAF forms a figure-of-eight knot (Fig. 3a), in which the N-terminal end of the chain is threaded through a loop consisting of amino acids that reside approximately several hundred residues C-terminal to them.88 The knot has been confirmed in all PAS-GAF phytochrome structures.31,37,89–93,96–99 So far, the knot has not been assigned a clear function for signal transduction, as structures in the Pr and Pfr states show hardly any rearrangements in this region.89,90,100
Interactions between the chromophore, conserved amino acid side chains and water molecules in the chromophore-binding pocket are important for photochemistry. The crystal structures of PAS-GAF fragments in the Pr state (Table 1) have provided structural basis for understanding these interactions (Fig. 3b). PAS-GAF crystal structures,88 which now have reached a resolution close to 1 Å,96,109 show that the D-ring of the biliverdin has substantially more space than the other pyrrole rings and thereby greater freedom to move within the chromophore-binding pocket.
A number of conserved residues surround the chromophore. Mutational investigations show that most of them are important for proper photo-switching, but that only a few of them completely disrupt photo-conversion.73,90,102,120,121 We note that the majority of the results here have evaluated the mutations against photoconversion as measured by optical spectroscopy. This may be appropriate as long as the investigated residue is located close to the chromophore. It would be useful to repeat some of the studies in the future by probing the direct structural changes or biochemical activity of the phytochromes.59,92 The incorporation of the chromophore appears to be relatively robust against single mutations. Next, we name a few prominent amino acids that interact with the chromophore (see Fig. 3b):
Two residues, an aspartate and a histidine, are shown to be crucial for proper phytochrome photochemistry.73 The aspartate (Asp207 in DrBphP) is part of the highly conserved DIP motif and resides at close proximity of pyrrole rings A and D. When this aspartate was removed in plant phytochrome B, phytochrome failed to photoconvert and instead became fluorescent.122 The same mutation in DrBphP has been used to generate fluorescent phytochrome variants.102 The histidine (His260 in DrBphP) lies face-to-face on the B- and C-rings and forms a hydrogen network with the pyrrole nitrogens of the biliverdin. The histidine is also important for buffering the deprotonation and protonation events of the chromophore in different states of the photocycle.123,124
The aromatic character of a conserved tyrosine (Tyr263 in DrBphP) has been demonstrated to be important for Pfr formation in Cph1 and its absence affects the position of the PHY domain and its interactions.125 In DrBphP, the absence of the hydroxyl group of Tyr263 destabilizes the β-sheet conformation of the PHY-tongue in the Pr state and makes the protein more prone to adopt an α-helical structure regardless of the chromophore configuration.111 One of the two phytochromes in Stigmatella aurantiaca, SaBphP1, has a threonine instead of the highly conserved histidine (His290 in DrBphP) that is in H-binding distance to the D-ring carbonyl. In terms of absorption spectra, this protein shows an incomplete photoconversion but an unusually high quantum yield of reaching the first intermediate state. In this case, normal photoconversion can be obtained when the threonine is mutated to a histidine.59,69,113 It is noteworthy that regardless of whether a histidine or threonine occupies this position, the structural changes of the PSM (PHY separation, see Fig. 4) remain similar to other bacteriophytochromes in the Pr-to-Pfr transition.59
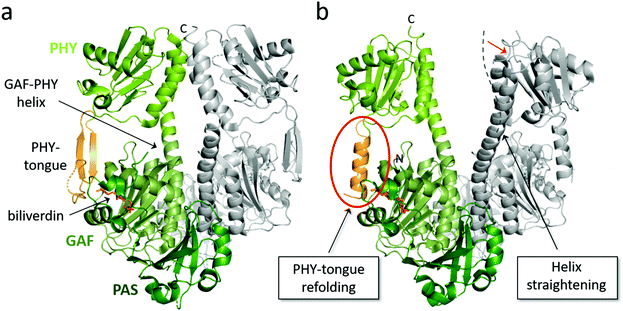 | ||
Fig. 4 Overview of structural features of the PSM of BphPs. The structural features of a PSM dimer from D. radiodurans (a) in Pr state (PDB id: 4O0P100) and (b) in mixed Pr/Pfr state after red light illumination (PDB id: 4O01![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100). Subunit 1 is colored with its PAS, GAF and PHY domain in different shades of green whereas subunit 2 is grey. The biliverdin position, PHY tongue and GAF-PHY helix in subunit 1 are indicated by arrows. Two structural changes that accompany light activation are indicated in boxes. Dashed line in panel b denotes the orientation of the GAF-PHY helix in the Pr state and the red arrow the movements of the helix during in Pr-to-Pfr transition. 100). Subunit 1 is colored with its PAS, GAF and PHY domain in different shades of green whereas subunit 2 is grey. The biliverdin position, PHY tongue and GAF-PHY helix in subunit 1 are indicated by arrows. Two structural changes that accompany light activation are indicated in boxes. Dashed line in panel b denotes the orientation of the GAF-PHY helix in the Pr state and the red arrow the movements of the helix during in Pr-to-Pfr transition. | ||
The crystal structures of phytochromes reveal various consistent waters in close contact to the biliverdin chromophore. The waters are thought to be involved in the structural mechanism upon photoactivation as they act as fast structural mediators between biliverdin and surrounding amino acids. Although their potential role in the photoactivation mechanism has been recognized, the detailed role of these waters remains unclear. The most prominent water is the so-called pyrrole water88 (Fig. 3b). The pyrrole water is located in the center of the bilin and in hydrogen bond distance to the nitrogens of pyrrole rings A, B and C. It also resides at the hydrogen bond distance to the backbone carbonyl group of the aspartate in the DIP motif (Asp207 in DrBphP). There are two highly ordered waters forming a hydrogen-bonding network between the D-ring carbonyl, the C-ring propionate and the conserved histidine (His290 in DrBphP). If the His290 position is occupied by a threonine, this water molecule network is extended.113 Furthermore, a water positioned between the pyrrole water and the tyrosine (Tyr263 in DrBphP) has been identified to exchange rapidly at room temperature.109 In addition, a dynamic water molecule has been observed by molecular dynamics simulations of a simulated Lumi-R state close to the D-ring carbonyl.126
Crystal structures of the same phytochrome in Pr and Pfr state are now available, in which the position of the D-ring is clearly resolved.78,96,100,119 This has revealed new chromophore interactions showing that the bilin slides in the chromophore pocket and the propionate side chains of ring B and C forms new interactions with the adjacent amino acid in the Pfr state compared to the Pr state. A crystallographic study of the bathy phytochrome PaBphP resolved the structure of three intermediates between Pfr and Pr by cryo-trapping.78 It was found that in the first intermediate, the D-ring twists around the C15-methine bridge, the twist is partially released, the C-ring adjusts in the second intermediate. In addition, the C- and B-ring propionates were found to detach from the surrounding protein pocket in the second and third intermediate, respectively. Some surrounding residues were found to move in accordance with these structural changes, but overall the protein structural changes were small. This may be an artifact of the confinement of the protein by the crystal packing.
Effects of the chromophore-binding pocket configuration can also be observed in the early photocycle. The evolution from the Pr to Lumi-R state appears much slower than what would have been the case for free isomerization in solution.68 This is consistent with that the chromophore is tightly held in the binding pocket and that structural changes in the binding pocket have to occur before the isomerization reaction can proceed. For example, a study with mid-infrared spectroscopy of RpBphP2 and RpBphP3 showed that additional hydrogen bonds with the D-ring in Pr state prolong the excited-state lifetime.127 This trend was also observed for SaBphP1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 69 but not for a DrBphP variant.113 Contrary to these results, the formation of the Lumi-F state from Pfr in a bathy phytochrome was found to take place already within femtoseconds,128 potentially because the protein structure does not have to adjust as much as in the Pr to Lumi-R transition.
69 but not for a DrBphP variant.113 Contrary to these results, the formation of the Lumi-F state from Pfr in a bathy phytochrome was found to take place already within femtoseconds,128 potentially because the protein structure does not have to adjust as much as in the Pr to Lumi-R transition.
Tier two: the photosensory module
The PHY domain connects back to the chromophore-binding pocket via the so-called PHY tongue (Fig. 4), whose length varies between different species. The interactions of the PHY tongue and the chromophore occur through bridge residues, most notably the conserved Asp residue of the DIP motif (Asp207 in DrBphP). The addition of the PHY domain to PAS-GAF fragment makes the chromophore-binding pocket slightly tighter and shows a small displacement of the D-ring of the chromophore.96 In some structures, the angle of the helical bundle at the dimer interface of the PAS-GAF domains becomes tilted by the addition of the PHY domain.31 The presence of the PHY domain appears important for formation of the Pfr state in bacteriophytochromes.51 For these proteins, the photoactivated state is less stable when lacking the PHY domain, which results a roughly 100 times faster thermal back reversion of the PAS-GAF domain compared to the PSM and full-length phytochrome.96,130 Moreover, the PHY tongue changes its secondary structure when transforming from Pr (β-sheet) to Pfr (α-helix).100,131 We will discuss this in detail in the next section.
Further aspects of the tongue refolding were proposed with the release of the Pr structure from the PAS-less Cph2 from Synechocystis sp. and called the “tryptophan switch”.133 In the proposal, two conserved tryptophans flanking the PRxSF motif in the tongue were suggested to switch places upon tongue refolding and the 180° rotation of the PHY tongue around its own axis. An extensive comparison and clustering of the tongue fold can be found in a review by Burgie and Vierstra.44
The crystal structures of DrBphP PSM both in its dark (Pr) and illuminated (mixed Pfr/Pr) form100 directly confirmed that the PHY tongue refolds upon photoconversion from β-sheet to α-helical conformation. The refolding occurs every time the phytochrome photoswitches and is an integral part of the phytochrome photoconversion mechanism. Two years after the initial discovery, a structure of a DrBphP F469W mutant was presented, which had a high Pfr content (87%).119 Due to this, the electron density of the chromophore and surrounding residues stabilizing the Pfr state were clearer. This structure confirmed the PHY tongue refolding mechanism, and revealed some more details on the interactions between biliverdin and the surrounding amino acids in the Pfr state.
The tongue region contains conserved motifs, with PRxSF being the most prominent one. It makes close contacts with the GAF domain close to the chromophore. In the Pr state, the arginine in this motif (Arg466 in DrBphP) forms a salt bridge with the aspartate in the DIP motif of the GAF domain (Asp207 in DrBphP), which stabilizes the β-sheet arrangement of the PHY tongue.133 Upon photoactivation, the salt bridge between the aspartate and the arginine breaks. This leads to the release of the tongue and the formation of new interaction between of the DIP aspartate and the PRxSF serine (Ser468 in DrBphP) in the Pfr state.
The importance of this conserved PRxSF motif is supported by mutational studies. In DrBphP, many mutations in the motif cause incomplete photoconversion and alter dark reversion rates, especially in the case of serine (Ser468 in DrBphP).96 Proline forms packing interactions with the biliverdin A-ring in the Pr state and stabilizes the Pfr state in Agp1.99RpBphP3 has a threonine instead of proline in this site, which leads to a formation of a near-red-absorbing Pnr state instead of a Pfr state.98 Finally, a phenylalanine in the motif (Phe469 in DrBphP) stabilizes the Pfr state by impeding the thermal dark reversion.73
The PSM crystal structures show high plasticity of the PHY domain orientation, which indicates that they are highly sensitive to both internal and external triggers such as mutations, crystal contacts and freezing. For example, the PSM crystal structure of PaBphP in the Pfr state shows a straight GAF-PHY helix but not a distinct separation of the PHY domains.90 In SaBphP1 PSM, a Thr-to-His mutation at the conserved histidine position close to the D-ring (His290 in DrBphP) led to a displacement of the PHY domain in the crystal structure, although the tongue fold remained unaltered from the wild-type PSM.31 As a third example, the PSM of DrBphP in its Pr state can be forced to adopt an overall Pfr-like conformation by the crystallization conditions.111 Crystallization conditions may also affect dimer packing, as the PSM crystal structures with the highest resolution have unnatural anti-parallel dimerization contacts.99,125 The anti-parallel dimerization, which is observed in some crystal structures,89,99 potentially restricts the natural flexibility of GAF-PHY helix, leading to increased stability and higher resolution. As such, the crystal structures should be evaluated with care, as they do not necessarily represent the distribution of conformations that likely exist in solution. This conformational discrepancy between crystals and solution has been exemplified by DrBphP PSM.100
It has been speculated that the degree of bending of the GAF-PHY helix correlates with the photoactive state of the phytochrome, leading to a conclusion that this helix would be straight in the Pfr state and relatively bent in the Pr state.99 Although the GAF-PHY helix is generally straighter in the Pfr structures, no prominent difference in PHY orientation to the Pr state can be observed. The orientations vary greatly in different PSM structures, but this does not directly reflect the photoactive state. For example, the positioning of the PHY domain can be arranged along a trajectory that corresponds to a rotation about the dimer interface with the two Pfr structures (DrBphP and PaBphP) adopting extreme position on both ends. In all other Pr structures, the PHY domains are positioned in between these two extremes.92
X-ray solution scattering experiments of a monomeric PSM domain of DrBphP further identifies that the separation of the PHY domains is actually a combination of a straightening of the GAF-PHY helix and a rotational motion that pulls the PHY domain closer to the GAF domain.135 This kind of movement has been recently confirmed in the PSM of the group II phytochrome Cph2 from Synechocystis sp. PCC 6803.136 The angled arrangement of the dimer might be crucial for signaling in the full-length BphPs and play an important role in the flexible regulation various OPMs.
In the PHY domain, the GAF-PHY helix connects with a PHY-OPM linker helix.137 The pronounced plasticity of the PHY domain in the PSM structures is therefore likely to be an artefact resulting from the missing OPM and interactions of the linker regions. For example, the light-induced PHY separation revealed by Takala et al. has not been observed in full-length Agp1.100,138 Although the existing full-length structures agree quite well with PSM structure when overlaid to corresponding PSM structures,92,93 they generally show tighter packing of the PHY domains.
Light activation leads to refolding of the PHY tongue, and we believe that this will also occur in full-length phytochromes. Larger scale conformational changes of the PHY domains may also occur in them, but the trajectories will be different when the OPM is present. Thus, PSM fragments provide valuable information regarding these changes, but to map the domain rearrangements of phytochromes it is advisable to focus on structural investigation of full-length BphPs.
Tier three: the output module
Many bacteriophytochromes are histidine kinases32,140 and we will describe them briefly. For an extensive review on photoreceptor histidine kinases, see a recent review.141 Phytochromes with a histidine kinase (HK) OPM are part of a two-component signaling system.32,34,141,142 Two-component system constitutes of a histidine kinase sensor and a response regulator protein. The HK domains consist of two subdomains: a phosphotransferase (DHp) and a catalytic ATP-binding (CA) domain.51 The DHp domain consists of two helices that together with its sister subunit forms a four-helix bundle, which is the site of three catalytic reactions: histidine phosphorylation, phosphotransfer to an interacting response regulator, and a phosphatase reaction. The catalytic histidine is conserved and is located around the center of the first helix of the DHp bundle.139
The response regulator is phosphorylated by the histidine kinase, and in many cases binds then to DNA to mediate cellular response.143–145 BphP and its response regulator are typically encoded in the same operon and positioned closely in the genome. There are also BphPs, like Agp2 from Agrobacterium fabrum, which have their response regulator attached as an additional C-terminal domain of the phytochrome.146 Activity studies of different phytochromes have been performed in the Pr and Pfr states. In Cph1, the binding of the chromophore stimulates the activity of the phytochrome. Red light inhibits autophosphorylation and the phosphotransfer to the response regulator, meaning that the Pr state is catalytically more active than the Pfr state.11,147–149 From the bacteriophytochromes, Agp1, RpBphP2 and RpBphP3 are also shown to be catalytically more active in the Pr state.50,58,150
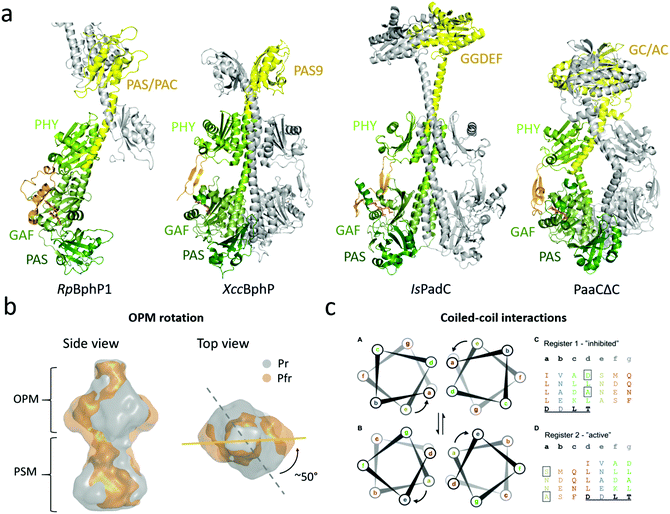 | ||
| Fig. 5 Structures and mechanisms of full-length phytochromes. (a) Four BphP structures that contain output modules. The PSMs are colored in different shades of green and the OPM (with PHY-OPM linker) is in yellow. RpBphP1 shows an antiparallel arrangement whereas XccBphP, IsPadC, and PaaCΔC are aligned as parallel dimers with a left hand rotation of the long helices at the dimer interface. (b) Reconstruction of SAXS envelope of full length DrBphP, obtained by time-resolved X-ray scattering.60 The Pr structure is shown in gray and the Pfr structure in gold. Red light illumination leads to a 50° rotation of the OPM relative to the PSM. (c) The heptad registers of the coiled coil linker region between the PHY-GAF helices of IsPadC. The upper “register 1” found in the crystal structure of the Pr from whereas lower “register 2” accommodates a rotation of the heptad position e to a. Left subpanels show the heptad units of the registers rainbow colored to the heptad repeats of “register 1”. A conserved DxLT motif is underlined. The boxed amino acids destabilize the coiled-coil contacts. Panels B and C are reproduced with permission from Björling et al. 2016 and Gourinchas et al. 2017.60,92 | ||
The first near-full-length structure was the ‘bathy-like’ RpBphP1 from Rhodopseudomonas palustris.91 Under red-light conditions, RpBphP1 monomerizes and forms heterodimers with its gene repressor RpPpsR2. In the structure, the chromophore was in a Pfr state. The OPM of RpBphP1 consists of a PAS/PAC domain and a 10 kDa HOS domain, which was truncated for crystallization. The protein crystallized in an antiparallel dimer arrangement with the OPM forming the dimerization interactions and the PSM stretching out from each subunit in opposite directions (Fig. 5a). This arrangement has not since been confirmed for phytochromes and can be considered as an exception. It may arise from the requirement of RpBphP1 to form heterodimers.91
The second full-length phytochrome structure to be solved was XccBphP from the plant pathogen Xanthomonas campestris.93XccBphP is a ‘bathy-like’ protein involved in regulating the virulence of the pathogen and has a PAS9 domain as an OPM.154 The protein crystallized with the PHY tongue in a β-sheet configuration (Fig. 5a). This indicates a Pr conformation of the protein, but surprisingly the diffraction data, Raman spectroscopy and theoretical results indicated that the chromophore adopted a mixture of the Pr and Pfr state.93
The third full-length structure of a BphP was of the Pr state of the phytochrome IsPadC from Idomarina sp. A28L, which has an attached diguanylate cyclase (GGDEF) domain.92 The GGDEF domain executes diguanylate cyclase activity in cells, forming one cyclic-di-GMP and two diphosphates from two molecules of GTP.155 Similar to the XccBphP structure, IsPadC crystallized in head-to-head arrangement. The OPM is connected to the PHY domain via a coiled-coil linker constituting of two intertwined α-helices that also form interactions with the GAF-PHY helix of the PSM (Fig. 5a). This linker is considerably longer in IsPadC compared to XccBphP. Mutational analysis showed that the key for activation of the GGDEF domain lies in the coiled-coiled linker elements (Fig. 5c, see below). This notion has recently been supported by the structure of a mutant in the linker region, which renders the phytochrome constantly active.94
The fourth full-length structure shown in Fig. 5 shows an artificial phytochrome PaaCΔC, composed out of the PSM of DrBphP and a guanylate/adenylate cyclase (GC/AC) from Synechocystis sp.153 This dimer also adopts a head-to-head arrangement, with a short coiled-coil linker section between the PSM and OPM. The PSM resembled the overall arrangement of the DrBphP PSM structures with a characteristic kink in the GAF-PHY helix.96,100
The ‘opening’ model. Burgie et al. have proposed a structural signaling mechanism for the DrBphP, which is based on negative stain and cryo-electron microscopy of the phytochrome in the resting (Pr) and illuminated (Pr/Pfr) state.96,129 The data showed that photoactivation pulls the PHY domains outwards, creating ‘shoulders’ on the protein. The OPMs tease apart or completely release from each other in the Pfr state. The authors suggested that the splaying apart of the OPMs would alter the OPM activity or open up binding sites for phytochrome interaction partners.44 This model rests on relatively low resolution electron microscopy data and is not in line with how other histidine kinases are thought to work.139 The opening movement has also not been observed in an EPR study on Agp1, where no major changes in between subunits were detected upon red illumination.138
The ‘rotation’ model. In contrast to the ‘opening’ mechanism, X-ray solution scattering studies of DrBphP have indicated that the OPMs stay dimerized and that a twist of the histidine kinase OPM occurs with respect to the PSM when the phytochrome is photoactivated.60 The time-resolved study showed that this global rearrangement occurred after a few milliseconds in correlation with the Meta-R state and that the structural changes prior to this event are small and localized to the PAS-GAF domain. The reconstruction of the SAXS protein envelopes of both the Pr and Pfr state describes a 50° right-handed twist of the entire dimer when the protein is viewed from the OPM (Fig. 5b).
This rotational mechanism is also supported by studies with other histidine kinases. Most histidine kinases form parallel dimers, and this arrangement is vital for autophosphorylation.145,156,157 An X-ray scattering study with an YF1 chimera, which consists of a LOV domain combined with a HK domain, shows that the signal from the chromophore is translated into a left-handed rotation of the HK on the millisecond time scale.158 Another example stems from the structures of a nitrate sensing sensor histidine kinase, NarQ from E. coli. By crystallizing NarQ with and without its substrate, a piston-like displacement of the transmembrane helices was identified. This motion then would be converted into a rotational motion in its OPM, but splaying apart of the OPM subunits was not observed.159
The rotational mechanism requires that the phytochrome has two dimerization interfaces. In DrBphP, these interfaces reside between the GAF domains and between the HK domains. The interfaces have approximately equal energetic contribution to overall dimerization,152 which is proposed to put a strain in the phytochrome dimer. Indeed, the lack of strain alters the function of the photoreceptors. The removal of the HK domains in DrBphP slows down the thermal dark reversion and the absence of the GAF dimerization interactions completely impairs it.152 Furthermore, dimerization of phytochromes is crucial for proper phytochrome functionality in plant phytochrome B.160,161
The ‘register’ model. In agreement with the rotational mechanism, the activation of phytochrome IsPadC, which has GGDEF as an OPM, has been identified to include rotational motion upon light activation.92 The coiled-coil linker has been shown to be relevant for regulation of sensors with cyclase activity162,163 and for IsPadC the linker region showed a distinct increase in conformational dynamics upon red light illumination, highlighting the importance of this structural element for signal transduction.92 The investigation shows that this PHY-OPM helix can vary in lengths by heptads of amino acids, which correspond to approximately two full α-helical turns, meaning that the relative orientation of the OPM compared to the PSM is conserved to retain activity. Truncation studies within the heptads also showed that the amino acid composition of the helical spine has evolved to stabilize the dimer interactions of the PSM and the OPM.
Two different helical registers of amino acids have been associated with the resting and activated states (Fig. 5c). The registers differ by a rotation of the heptad, where the Pr state adopts the ‘register 1’ and the light-activated state adopts the ‘register 2’. The light activation leads to a rotation of the heptad positions ‘e’ to ‘a’. To test the effects of coiled-coil rearrangements on DCG activity, IsPadC was mutated to stabilize either register conformation. If the register 1 was stabilized, IsPadC could no longer be activated by red light, whereas the stabilization of register 2 led to constantly active IsPadC. This indicates that the regulation of the cyclase activity is fine-tuned by the equilibrium of the relative populations of signaling helix registers, which in turn affects the residues important for GGDEF function. In darkness, the PHY tongue restricts the configuration of the helical spine and the transition between the two registers. When the tongue refolds, the locking mechanism is released, leading to the population of register 2. This was further strengthened by the fact that deletion of the PHY tongue resulted in increased cyclase activity.92
The ‘register’ model is in line how histidine kinases are thought to work. Histidine kinases sensors, the linker region between the sensor module and the HK domains is a signaling helix bundle. The length of the linker region can vary between species, but the angular orientation of the OPM in relation to the PSM is mainly conserved.98 A shift of register is thought to be important for activation of the HK domains, but (partial) monomerization is not.139,141,158 The ‘register’ model is attractive, because it may work even if only one of the subunits is photoexcited.
Model summary. We suggest that photoregulation of OPM activity in BphPs is described by a combination of the ‘rotation’ and ‘register’ models. The OPMs rotate in relation to the PSM and this is connected to a shift of register of the signaling helical bundle (Fig. 5b). The driving forces is the photoinduced remodeling of the PSM. The ‘opening’ model of OPM activation is less likely to apply. Although it is a compelling extension to the ‘toggle’ model and PSM opening observed in truncated PSM proteins, it has not been supported by other studies with full-length bacteriophytochromes.
The events start at the bilin chromophore, where light absorption leads to an isomerization of its D-ring. The signal is then relayed to the protein, with the details largely unknown. The PHY tongue refolds and the PHY domains move, but not necessarily as observed for truncated PSM fragments alone. The two dimerization interfaces in the full-length phytochrome steers the PHY movement into a rotation of the OPM in relation to the PSM. The rotation changes the interaction register in the helical bundle,155 and finally leads to a change in the OPM activity.
Photosignalling occurs on three hierarchical levels
In the preceding sections, we have described the structural photoconversion of phytochromes from resting to an activated state. It is instructive to group the structural changes into three tiers, which we have defined as the chromophore and its binding pocket, the photosensory module, and the output module. Now the question arise on how these changes are connected.The simplest concept would be that of a linear cascade of structural events. Each change would lead to another change on the next tier, transducing the signal through the proteins from the chromophore to the output domain. This view is inspired by the photocycle of photosynthetic proteins, where concerted sequences of conformational changes and charge/proton dislocations are observed.
Here we argue that the regulation of the BphP activity is not well described by a concerted cascade of structural events. We have recently presented a concept in which the regulation of photoactivity is an interplay between structural elements on different hierarchal tiers.111 For each of the three tiers an equilibrium between two (or more) states exist, which can be shifted to one direction either by external factors or by changes in other tiers. The net activity of BphPs would therefore be fine-tuned by a complex interplay between changes in each tier, but also other factors, which influence the equilibria. We have shown that a single mutation or the crystallization conditions can decouple the chromophore (tier 1) from the overall structure of the PSM (tier 2).111 A mutation of a conserved tyrosine (Tyr263 in DrBphP) resulted in that the PSM could adopt a Pfr-like open conformation and α-helical PHY tongue fold although the chromophore remained in the Pr-state arrangement. This means that the mutation shifts the equilibrium within one level while also diminishes its coupling with other level. In a similar way, signaling between the PHY tongue and the PHY-OPM linker region could be decoupled in IsPadC, by either removal of the tongue or by stabilizing either conformation of the coiled-coil linker.92 Finally, a crystal structure of the full-length phytochrome from the plant pathogen Xanthomonas campestris pv. campestris (XccBphP) showed a β-sheet (Pr-like) tongue conformation, but a mixture of ZZZssa and a ZZEssa chromophore configurations.93 This also supports the notion that the states which the tiers adopt are at least partially uncoupled.
The above results can be rationalized by the concept of (weakly) coupled equilibria between structural changes on different tiers in the BphP,111 but not by a concerted, linear cascade of structural events as for example observed for photosynthetic proteins. The conformational dynamics of each structural level define the state of the protein, hence fine-tuning the optimal output activity of the BphPs in the bacteria. The signaling pathways are conserved, but are also redundant and partially dispensable for the net protein function. This is consistent with that different degrees of conformational heterogeneity exist in Pr and Pfr,164,165 which has been termed as a “soft-to-hard” transition.166 The concept of coupled equilibria may be important for the biological function of the sensor proteins and provide phytochromes the ability to integrate other signals than light into their photoresponse.55
Outlook
For better understanding of the signaling mechanisms of BphPs, it is desired to obtain full-length structures for phytochromes, both in their Pr and Pfr state. This would provide insight in the attachment of the OPM and further elucidate how the signaling is transferred from PSM to OPM. Moreover, the structures of PSM fragments are fairly well characterized in Pr and Pfr, but the molecular mechanism of the phototransition is not clear. Especially, knowledge is missing on how the signal is relayed from the chromophore to the protein matrix. We therefore propose that more structures of phytochrome intermediates are solved and reported. This would provide insight into the mechanism of photoactivation.As can be deduced from the previous chapters, X-ray crystallography is a powerful method for structural determination of phytochrome, but it also has restrictions. Conventional crystal structures may be subject to radiation damage or artifacts, which arise from freezing of the molecules. In the case of phytochromes, it has been shown that the biliverdin becomes deprotonated at higher X-ray radiation doses at synchrotrons and that the high dose eventually leads to rupture of the cysteine linkage to the chromophore.106 In addition, crystallography usually provides structural snapshots that are frozen in space and time. Proteins are locked in one conformation due to crystal contacts, which restricts potential structural changes.
One interesting method for future studies is serial femtosecond crystallography (SFX). SFX is relatively new method, in which a stream of protein microcrystals are exposed to X-ray pulses. The experiment is performed at room temperature and was first developed by using ultrashort and ultrabright X-ray pulses at a free electron X-ray laser,167 and now also at synchrotrons.168–170 At the free electron laser sources, the X-ray pulses are brilliant and so short that the diffraction can be recorded before the protein molecules are destroyed.167,171 The data acquisition strategy minimizes radiation damage and freezing artifacts in the crystallographic data.
Several structures of BphPs in their resting states have been solved by SFX.31,109,110,134 Overall, the SFX structures agree well with conventional crystallography. This indicates that the bias due to radiation damage and cryo-distortion is small. The first room temperature structure of a bacteriophytochrome solved by SFX showed that the cysteine linkage of the chromophore is subject to radiation damage in conventional crystallography.109 Fuller et al. detected a moderate shift of the position of the PHY domain in relation to the PAS-GAF bidomain in the room temperature structure compared to cryogenic structures of DrBphP.96,100,110 Similarly, one of the subunits in the asymmetric unit of SaBphP1 crystals showed substantially improved electron density at room temperature compared to cryo conditions.31
An interesting direction is the use of time-resolved SFX, which resolves the structural changes as a function of time after a laser pulse triggers the photoreaction.172 Time resolution of down to femtoseconds is possible, but care has to be taken that the structural changes are not inhibited by the crystal packing. One indication is that the protein can be photoconverted in the crystal. This has been achieved with a GAF domain from cyanobacteriochrome PixJ in Thermosynechococcus elongatus.173 However, it is harder to determine if regions, which are more distant from the chromophore, exhibit the biologically relevant structural changes, because their impact on the optical spectra may be negligible. We have very recently revealed the structural changes in the excited state of the PAS-GAF fragment of DrBphP which occur within picoseconds after photoexcitation.95 These changes in this first time-resolved structural snapshot involve partial rotation of the D-ring and an unexpected de-localization of the pyrrole water. The structural changes are assigned to events that precede Lumi-R state formation. This provides a starting point for understanding the structural photoactivation of phytochromes with much greater precision.
Phytochromes present structural and functional heterogeneity at several levels, which are mostly inaccessible by crystallography. These levels include heterogeneity in the Pr-state chromophore, photocycle, chromophore surroundings, the remainder of the protein, and dimer packing. Structural heterogeneity of the chromophore has been identified for proteins in crystalline form and in solution. An NMR investigation has identified two Pr isoforms of Cph1 in solution with distinguished hydrogen bonding networks and charge distribution in the chromophore-binding pocket.76 In Pr-state crystal structures, the chromophore may adopt alternate conformations with different crystal packing.109 Raman spectroscopy indicates different chromophore structures between protein in solution and in crystals174 and spectroscopic evolutions furthermore support heterogeneous chromophore conformations with up to three different conformations for some BphPs.175 The Pfr state has also been shown to be heterogeneous in some cases, with a temperature-dependent equilibrium that is suggested to be important for the dark reversion.176 Solution-based methods have been used to provide this information and should also be applied in the future to make further progress.
The structural biology of phytochromes is currently dominated by prokaryotic phytochromes. In the future, it is necessary that more plant phytochrome structures are solved, both in resting and activated states. Progress into this direction will require the use of several methods. NMR, X-ray crystallography, optical spectroscopy, single-particle cryo electron microscopy, and theoretical modelling should all be used and combined to find out how phytochromes are photoactivated at the atomic level.
Conflicts of interest
There are no conflicts to declare.References
- L. H. Flint and E. D. McAlister, Wave lengths of radiation in the visible spectrum inhibiting the germination of light-sensitive lettuce seed, Smithson. Misc. Collect., 1935, 94, 1–11 Search PubMed.
- H. A. Borthwick, S. B. Hendricks, M. W. Parker, E. H. Toole and V. K. Toole, A Reversible Photoreaction Controlling Seed Germination, Proc. Natl. Acad. Sci. U. S. A., 1952, 38, 662–666, DOI:10.1073/pnas.38.8.662.
- W. L. Butler, K. H. Norris, H. W. Siegelman and S. B. Hendricks, Detection, Assay, and Preliminary Purification of the Pigment Controlling Photoresponsive Development of Plants, Proc. Natl. Acad. Sci. U. S. A., 1959, 45, 1703–1708, DOI:10.1073/pnas.45.12.1703.
- R. D. Vierstra and P. H. Quail, Purification and initial characterization of 124 kDalton phytochrome from Avena, Biochemistry, 1983, 22, 2498–2505, DOI:10.1021/bi00279a029.
- H. P. Hershey, R. F. Barker, K. B. Idler, J. L. Lissemore and P. H. Quail, Analysis of cloned cDNA and genomic sequences for phytochrome: complete amino acid sequences for two gene products expressed in etiolated Avena, Nucleic Acids Res., 1985, 13, 8543–8559, DOI:10.1093/nar/13.23.8543.
- J. J. Casal, Photoreceptor signaling networks in plant responses to shade, Annu. Rev. Plant Biol., 2013, 64, 403–427, DOI:10.1146/annurev-arplant-050312-120221.
- H. Schneider-Poetsch, Ü. Kolukisaoglu, D. H. Clapham, J. Hughes and T. Lamparter, Non-angiosperm phytochromes and the evolution of vascular plants, Physiol. Plant., 1998, 102, 612–622, DOI:10.1034/j.1399-3054.1998.1020417.x.
- Z. Jiang, L. R. Swem, B. G. Rushing, S. Devanathan, G. Tollin and C. E. Bauer, Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes, Science, 1999, 285, 406–409, DOI:10.1126/science.285.5426.406.
- N. C. Rockwell and J. C. Lagarias, Phytochrome evolution in 3D: deletion, duplication, and diversification, New Phytol., 2020, 225, 2283–2300, DOI:10.1111/nph.16240.
- J. Hughes, T. Lamparter, F. Mittmann, E. Hartmann, W. Gartner, A. Wilde and T. Borner, A prokaryotic phytochrome, Nature, 1997, 386, 663, DOI:10.1038/386663a0.
- K. C. Yeh, S. H. Wu, J. T. Murphy and J. C. Lagarias, A cyanobacterial phytochrome two-component light sensory system, Science, 1997, 277, 1505–1508, DOI:10.1126/science.277.5331.1505.
- S. J. Davis, A. V. Vener and R. D. Vierstra, Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria, Science, 1999, 286, 2517–2520, DOI:10.1126/science.286.5449.2517.
- D. M. Kehoe and A. R. Grossman, Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors, Science, 1996, 273, 1409–1412, DOI:10.1126/science.273.5280.1409.
- Y. Hirose, T. Shimada, R. Narikawa, M. Katayama and M. Ikeuchi, Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 9528–9533, DOI:10.1073/pnas.0801826105.
- A. Blumenstein, K. Vienken, R. Tasler, J. Purschwitz, D. Veith, N. Frankenberg-Dinkel and R. Fischer, The Aspergillus nidulans phytochrome FphA represses sexual development in red light, Curr. Biol., 2005, 15, 1833–1838, DOI:10.1016/j.cub.2005.08.061.
- N. C. Rockwell, D. Duanmu, S. S. Martin, C. Bachy, D. C. Price, D. Bhattacharya, A. Z. Worden and J. C. Lagarias, Eukaryotic algal phytochromes span the visible spectrum, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3871–3876, DOI:10.1073/pnas.1401871111.
- P. R. Robson, A. C. McCormac, A. S. Irvine and H. Smith, Genetic engineering of harvest index in tobacco through overexpression of a phytochrome gene, Nat. Biotechnol., 1996, 14, 995–998, DOI:10.1038/nbt0896-995.
- D. M. Shcherbakova, M. Baloban and V. V. Verkhusha, Near-infrared fluorescent proteins engineered from bacterial phytochromes, Curr. Opin. Chem. Biol., 2015, 27, 52–63, DOI:10.1016/j.cbpa.2015.06.005.
- D. M. Shcherbakova, M. Baloban, S. Pletnev, V. N. Malashkevich, H. Xiao, Z. Dauter and V. V. Verkhusha, Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins, Chem. Biol., 2015, 22, 1540–1551, DOI:10.1016/j.chembiol.2015.10.007.
- D. M. Shcherbakova, M. Baloban, A. V. Emelyanov, M. Brenowitz, P. Guo and V. V. Verkhusha, Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging, Nat. Commun., 2016, 7, 12405, DOI:10.1038/ncomms12405.
- A. Moglich and K. Moffat, Engineered photoreceptors as novel optogenetic tools, Photochem. Photobiol. Sci., 2010, 9, 1286–1300, 10.1039/c0pp00167h.
- A. A. Kaberniuk, A. A. Shemetov and V. V. Verkhusha, A bacterial phytochrome-based optogenetic system controllable with near-infrared light, Nat. Methods, 2016, 13, 591–597, DOI:10.1038/nmeth.3864.
- K. G. Chernov, T. A. Redchuk, E. S. Omelina and V. V. Verkhusha, Near-Infrared Fluorescent Proteins, Biosensors, and Optogenetic Tools Engineered from Phytochromes, Chem. Rev., 2017, 117, 6423–6446, DOI:10.1021/acs.chemrev.6b00700.
- K. C. Yeh and J. C. Lagarias, Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 13976–13981, DOI:10.1073/pnas.95.23.13976.
- W. Ni, S. L. Xu, E. Gonzalez-Grandio, R. J. Chalkley, A. F. R. Huhmer, A. L. Burlingame, Z. Y. Wang and P. H. Quail, PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3, Nat. Commun., 2017, 8, 15236, DOI:10.1038/ncomms15236.
- G. Bae and G. Choi, Decoding of light signals by plant phytochromes and their interacting proteins, Annu. Rev. Plant Biol., 2008, 59, 281–311, DOI:10.1146/annurev.arplant.59.032607.092859.
- K. A. Franklin and P. H. Quail, Phytochrome functions in Arabidopsis development, J. Exp. Bot., 2010, 61, 11–24, DOI:10.1093/jxb/erp304.
- J. Hughes, Phytochrome cytoplasmic signaling, Annu. Rev. Plant Biol., 2013, 64, 377–402, DOI:10.1146/annurev-arplant-050312-120045.
- E. Giraud, J. Fardoux, N. Fourrier, L. Hannibal, B. Genty, P. Bouyer, B. Dreyfus and A. Vermeglio, Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria, Nature, 2002, 417, 202–205, DOI:10.1038/417202a.
- F. Gan, S. Zhang, N. C. Rockwell, S. S. Martin, J. C. Lagarias and D. A. Bryant, Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light, Science, 2014, 345, 1312–1317, DOI:10.1126/science.1256963.
- N. C. Woitowich, A. S. Halavaty, P. Waltz, C. Kupitz, J. Valera, G. Tracy, K. D. Gallagher, E. Claesson, T. Nakane, S. Pandey, G. Nelson, R. Tanaka, E. Nango, E. Mizohata, S. Owada, K. Tono, Y. Joti, A. C. Nugent, H. Patel, A. Mapara, J. Hopkins, P. Duong, D. Bizhga, S. E. Kovaleva, R. St Peter, C. N. Hernandez, W. B. Ozarowski, S. Roy-Chowdhuri, J. H. Yang, P. Edlund, H. Takala, J. Ihalainen, J. Brayshaw, T. Norwood, I. Poudyal, P. Fromme, J. C. H. Spence, K. Moffat, S. Westenhoff, M. Schmidt and E. A. Stojkovic, Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome, IUCrJ, 2018, 5, 619–634, DOI:10.1107/S2052252518010631.
- M. E. Auldridge and K. T. Forest, Bacterial phytochromes: more than meets the light, Crit. Rev. Biochem. Mol. Biol., 2011, 46, 67–88, DOI:10.3109/10409238.2010.546389.
- G. Gourinchas, S. Etzl and A. Winkler, Bacteriophytochromes - from informative model systems of phytochrome function to powerful tools in cell biology, Curr. Opin. Struct. Biol., 2019, 57, 72–83, DOI:10.1016/j.sbi.2019.02.005.
- S. H. Bhoo, S. J. Davis, J. Walker, B. Karniol and R. D. Vierstra, Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore, Nature, 2001, 414, 776–779, DOI:10.1038/414776a..
- R. D. Vierstra and S. J. Davis, Bacteriophytochromes: new tools for understanding phytochrome signal transduction, Semin. Cell Dev. Biol., 2000, 11, 511–521, DOI:10.1006/scdb.2000.0206.
- R. D. Vierstra and J. Zhang, Phytochrome signaling: solving the Gordian knot with microbial relatives, Trends Plant Sci., 2011, 16, 417–426, DOI:10.1016/j.tplants.2011.05.011.
- E. S. Burgie, A. N. Bussell, J. M. Walker, K. Dubiel and R. D. Vierstra, Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 10179–10184, DOI:10.1073/pnas.1403096111.
- S. Nagano, K. Guan, S. M. Shenkutie, C. Feiler, M. Weiss, A. Kraskov, D. Buhrke, P. Hildebrandt and J. Hughes, Structural insights into photoactivation and signalling in plant phytochromes, Nat. Plants, 2020, 6, 581–588, DOI:10.1038/s41477-020-0638-y.
- N. C. Rockwell, L. Shang, S. S. Martin and J. C. Lagarias, Distinct classes of red/far-red photochemistry within the phytochrome superfamily, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 6123–6127, DOI:10.1073/pnas.0902370106 DOI:10.1073/pnas.0902370106.
- N. C. Rockwell, Y. S. Su and J. C. Lagarias, Phytochrome structure and signaling mechanisms, Annu. Rev. Plant Biol., 2006, 57, 837–858, DOI:10.1146/annurev.arplant.56.032604.144208.
- B. L. Montgomery, Sensing the light: photoreceptive systems and signal transduction in cyanobacteria, Mol. Microbiol., 2007, 64, 16–27, DOI:10.1111/j.1365-2958.2007.05622.x.
- M. A. van der Horst, J. Key and K. J. Hellingwerf, Photosensing in chemotrophic, non-phototrophic bacteria: let there be light sensing too, Trends Microbiol., 2007, 15, 554–562, DOI:10.1016/j.tim.2007.09.009.
- P. Scheerer, N. Michael, J. H. Park, S. Nagano, H. W. Choe, K. Inomata, B. Borucki, N. Krauss and T. Lamparter, Light-induced conformational changes of the chromophore and the protein in phytochromes: bacterial phytochromes as model systems, ChemPhysChem, 2010, 11, 1090–1105, DOI:10.1002/cphc.200900913.
- E. S. Burgie and R. D. Vierstra, Phytochromes: an atomic perspective on photoactivation and signaling, Plant Cell, 2014, 26, 4568–4583, DOI:10.1105/tpc.114.131623.
- A. Moglich, X. Yang, R. A. Ayers and K. Moffat, Structure and function of plant photoreceptors, Annu. Rev. Plant Biol., 2010, 61, 21–47, DOI:10.1146/annurev-arplant-042809-112259.
- N. C. Rockwell and J. C. Lagarias, A brief history of phytochromes, ChemPhysChem, 2010, 11, 1172–1180, DOI:10.1002/cphc.200900894.
- H. Wang and H. Wang, Phytochrome signaling: time to tighten up the loose ends, Mol. Plant, 2015, 8, 540–551, DOI:10.1016/j.molp.2014.11.021.
- M. Legris, Y. C. Ince and C. Fankhauser, Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants, Nat. Commun., 2019, 10, 5219, DOI:10.1038/s41467-019-13045-0.
- J. Rodriguez-Romero, M. Hedtke, C. Kastner, S. Muller and R. Fischer, Fungi, hidden in soil or up in the air: light makes a difference, Annu. Rev. Microbiol., 2010, 64, 585–610, DOI:10.1146/annurev.micro.112408.134000.
- B. Karniol and R. D. Vierstra, The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 2807–2812, DOI:10.1073/pnas.0437914100.
- B. Karniol, J. R. Wagner, J. M. Walker and R. D. Vierstra, Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors, Biochem. J., 2005, 392, 103–116, DOI:10.1042/BJ20050826.
- T. Dammeyer and N. Frankenberg-Dinkel, Function and distribution of bilin biosynthesis enzymes in photosynthetic organisms, Photochem. Photobiol. Sci., 2008, 7, 1121–1130, 10.1039/b807209b.
- M. Jaubert, J. Lavergne, J. Fardoux, L. Hannibal, L. Vuillet, J. M. Adriano, P. Bouyer, D. Pignol, E. Giraud and A. Vermeglio, A singular bacteriophytochrome acquired by lateral gene transfer, J. Biol. Chem., 2007, 282, 7320–7328, DOI:10.1074/jbc.M611173200.
- B. Frankland, Biosynthesis and dark transformations of phytochrome, 1972, vol. 1972, pp. 195–225 Search PubMed.
- M. Legris, C. Klose, E. S. Burgie, C. C. Rojas, M. Neme, A. Hiltbrunner, P. A. Wigge, E. Schafer, R. D. Vierstra and J. J. Casal, Phytochrome B integrates light and temperature signals in Arabidopsis, Science, 2016, 354, 897–900, DOI:10.1126/science.aaf5656.
- M. Legris, C. Nieto, R. Sellaro, S. Prat and J. J. Casal, Perception and signalling of light and temperature cues in plants, Plant J., 2017, 90, 683–697, DOI:10.1111/tpj.13467.
- G. Rottwinkel, I. Oberpichler and T. Lamparter, Bathy phytochromes in rhizobial soil bacteria, J. Bacteriol., 2010, 192, 5124–5133, DOI:10.1128/JB.00672-10.
- T. Lamparter, N. Michael, F. Mittmann and B. Esteban, Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11628–11633, DOI:10.1073/pnas.152263999.
- A. Björling, O. Berntsson, H. Takala, K. D. Gallagher, H. Patel, E. Gustavsson, R. St Peter, P. Duong, A. Nugent, F. Zhang, P. Berntsen, R. Appio, I. Rajkovic, H. Lehtivuori, M. R. Panman, M. Hoernke, S. Niebling, R. Harimoorthy, T. Lamparter, E. A. Stojkovic, J. A. Ihalainen and S. Westenhoff, Ubiquitous Structural Signaling in Bacterial Phytochromes, J. Phys. Chem. Lett., 2015, 6, 3379–3383, DOI:10.1021/acs.jpclett.5b01629.
- A. Björling, O. Berntsson, H. Lehtivuori, H. Takala, A. J. Hughes, M. Panman, M. Hoernke, S. Niebling, L. Henry, R. Henning, I. Kosheleva, V. Chukharev, N. V. Tkachenko, A. Menzel, G. Newby, D. Khakhulin, M. Wulff, J. A. Ihalainen and S. Westenhoff, Structural photoactivation of a full-length bacterial phytochrome, Sci. Adv., 2016, 2, e1600920, DOI:10.1126/sciadv.1600920.
- J. Matysik, P. Hildebrandt, W. Schlamann, S. E. Braslavsky and K. Schaffner, Fourier-transform resonance Raman spectroscopy of intermediates of the phytochrome photocycle, Biochemistry, 1995, 34, 10497–10507, DOI:10.1021/bi00033a023.
- F. Andel III, K. C. Hasson, F. Gai, P. A. Anfinrud and R. A. Mathies, Femtosecond time-resolved spectroscopy of the primary photochemistry of phytochrome, Biospectroscopy, 1997, 3, 421–433 CrossRef , DOI: 10.1002/(SICI)1520-6343(1997)3:63.0.CO;2-3.
- A. Remberg, I. Lindner, T. Lamparter, J. Hughes, C. Kneip, P. Hildebrandt, S. E. Braslavsky, W. Gartner and K. Schaffner, Raman spectroscopic and light-induced kinetic characterization of a recombinant phytochrome of the cyanobacterium Synechocystis, Biochemistry, 1997, 36, 13389–13395, DOI:10.1021/bi971563z.
- J. J. van Thor, K. L. Ronayne and M. Towrie, Formation of the early photoproduct lumi-R of cyanobacterial phytochrome cph1 observed by ultrafast mid-infrared spectroscopy, J. Am. Chem. Soc., 2007, 129, 126–132, DOI:10.1021/ja0660709.
- J. Dasgupta, R. R. Frontiera, K. C. Taylor, J. C. Lagarias and R. A. Mathies, Ultrafast excited-state isomerization in phytochrome revealed by femtosecond stimulated Raman spectroscopy, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 1784–1789, DOI:10.1073/pnas.0812056106.
- M. A. Mroginski, D. H. Murgida and P. Hildebrandt, The chromophore structural changes during the photocycle of phytochrome: a combined resonance Raman and quantum chemical approach, Acc. Chem. Res., 2007, 40, 258–266, DOI:10.1021/ar6000523.
- U. Robben, I. Lindner and W. Gartner, New open-chain tetrapyrroles as chromophores in the plant photoreceptor phytochrome, J. Am. Chem. Soc., 2008, 130, 11303–11311, DOI:10.1021/ja076728y.
- Y. Yang, M. Linke, T. von Haimberger, J. Hahn, R. Matute, L. Gonzalez, P. Schmieder and K. Heyne, Real-time tracking of phytochrome's orientational changes during Pr photoisomerization, J. Am. Chem. Soc., 2012, 134, 1408–1411, DOI:10.1021/ja209413d.
- T. Mathes, J. Ravensbergen, M. Kloz, T. Gleichmann, K. D. Gallagher, N. C. Woitowich, R. St Peter, S. E. Kovaleva, E. A. Stojkovic and J. T. Kennis, Femto- to Microsecond Photodynamics of an Unusual Bacteriophytochrome, J. Phys. Chem. Lett., 2015, 6, 239–243, DOI:10.1021/jz502408n.
- D. Buhrke, U. Kuhlmann, N. Michael and P. Hildebrandt, The Photoconversion of Phytochrome Includes an Unproductive Shunt Reaction Pathway, ChemPhysChem, 2018, 19, 566–570, DOI:10.1002/cphc.201701311.
- J. J. van Thor, B. Borucki, W. Crielaard, H. Otto, T. Lamparter, J. Hughes, K. J. Hellingwerf and M. P. Heyn, Light-induced proton release and proton uptake reactions in the cyanobacterial phytochrome Cph1, Biochemistry, 2001, 40, 11460–11471, DOI:10.1021/bi002651d.
- B. Borucki, Proton transfer in the photoreceptors phytochrome and photoactive yellow protein, Photochem. Photobiol. Sci., 2006, 5, 553–566, 10.1039/b603846h.
- J. R. Wagner, J. Zhang, D. von Stetten, M. Gunther, D. H. Murgida, M. A. Mroginski, J. M. Walker, K. T. Forest, P. Hildebrandt and R. D. Vierstra, Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes, J. Biol. Chem., 2008, 283, 12212–12226, DOI:10.1074/jbc.M709355200.
- F. Velazquez-Escobar, P. Piwowarski, J. Salewski, N. Michael, M. Fernandez Lopez, A. Rupp, B. M. Qureshi, P. Scheerer, F. Bartl, N. Frankenberg-Dinkel, F. Siebert, M. A. Mroginski and P. Hildebrandt, A protonation-coupled feedback mechanism controls the signalling process in bathy phytochromes, Nat. Chem., 2015, 7, 423–430, DOI:10.1038/nchem.2225.
- B. Borucki, D. von Stetten, S. Seibeck, T. Lamparter, N. Michael, M. A. Mroginski, H. Otto, D. H. Murgida, M. P. Heyn and P. Hildebrandt, Light-induced proton release of phytochrome is coupled to the transient deprotonation of the tetrapyrrole chromophore, J. Biol. Chem., 2005, 280, 34358–34364, DOI:10.1074/jbc.M505493200.
- C. Song, G. Psakis, C. Lang, J. Mailliet, W. Gartner, J. Hughes and J. Matysik, Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3842–3847, DOI:10.1073/pnas.1013377108.
- T. Rohmer, C. Lang, C. Bongards, K. B. Gupta, J. Neugebauer, J. Hughes, W. Gartner and J. Matysik, Phytochrome as molecular machine: revealing chromophore action during the Pfr –> Pr photoconversion by magic-angle spinning NMR spectroscopy, J. Am. Chem. Soc., 2010, 132, 4431–4437, DOI:10.1021/ja9108616.
- X. Yang, Z. Ren, J. Kuk and K. Moffat, Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome, Nature, 2011, 479, 428–432, DOI:10.1038/nature10506.
- E. Consiglieri, A. Gutt, W. Gartner, L. Schubert, C. Viappiani, S. Abbruzzetti and A. Losi, Dynamics and efficiency of photoswitching in biliverdin-binding phytochromes, Photochem. Photobiol. Sci., 2019, 18, 2484–2496, 10.1039/c9pp00264b.
- A. Schmidt, L. Sauthof, M. Szczepek, M. F. Lopez, F. V. Escobar, B. M. Qureshi, N. Michael, D. Buhrke, T. Stevens, D. Kwiatkowski, D. von Stetten, M. A. Mroginski, N. Krauss, T. Lamparter, P. Hildebrandt and P. Scheerer, Structural snapshot of a bacterial phytochrome in its functional intermediate state, Nat. Commun., 2018, 9, 4912–4917, DOI:10.1038/s41467-018-07392-7.
- S. Brandt, D. von Stetten, M. Gunther, P. Hildebrandt and N. Frankenberg-Dinkel, The fungal phytochrome FphA from Aspergillus nidulans, J. Biol. Chem., 2008, 283, 34605–34614, DOI:10.1074/jbc.M805506200.
- M. Medzihradszky, J. Bindics, E. Adam, A. Viczian, E. Klement, S. Lorrain, P. Gyula, Z. Merai, C. Fankhauser, K. F. Medzihradszky, T. Kunkel, E. Schafer and F. Nagy, Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis, Plant Cell, 2013, 25, 535–544, DOI:10.1105/tpc.112.106898.
- K. Nito, C. C. Wong, J. R. Yates and J. Chory, Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors, Cell Rep., 2013, 3, 1970–1979, DOI:10.1016/j.celrep.2013.05.006.
- S. von Horsten, S. Strass, N. Hellwig, V. Gruth, R. Klasen, A. Mielcarek, U. Linne, N. Morgner and L. O. Essen, Mapping light-driven conformational changes within the photosensory module of plant phytochrome B, Sci. Rep., 2016, 6, 34366, DOI:10.1038/srep34366.
- F. Velazquez-Escobar, D. Buhrke, M. Fernandez Lopez, S. M. Shenkutie, S. von Horsten, L. O. Essen, J. Hughes and P. Hildebrandt, Structural communication between the chromophore-binding pocket and the N-terminal extension in plant phytochrome phyB, FEBS Lett., 2017, 591, 1258–1265, DOI:10.1002/1873-3468.12642.
- M. Ikeuchi and T. Ishizuka, Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria, Photochem. Photobiol. Sci., 2008, 7, 1159–1167, 10.1039/b802660m.
- B. L. Taylor and I. B. Zhulin, PAS domains: internal sensors of oxygen, redox potential, and light, Microbiol. Mol. Biol. Rev., 1999, 63, 479–506 CrossRef CAS.
- J. R. Wagner, J. S. Brunzelle, K. T. Forest and R. D. Vierstra, A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome, Nature, 2005, 438, 325–331, DOI:10.1038/nature04118.
- L. O. Essen, J. Mailliet and J. Hughes, The structure of a complete phytochrome sensory module in the Pr ground state, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14709–14714, DOI:10.1073/pnas.0806477105.
- X. Yang, J. Kuk and K. Moffat, Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14715–14720, DOI:10.1073/pnas.0806718105.
- D. Bellini and M. Z. Papiz, Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor, Structure, 2012, 20, 1436–1446, DOI:10.1016/j.str.2012.06.002.
- G. Gourinchas, S. Etzl, C. Gobl, U. Vide, T. Madl and A. Winkler, Long-range allosteric signaling in red light-regulated diguanylyl cyclases, Sci. Adv., 2017, 3, e1602498, DOI:10.1126/sciadv.1602498.
- L. H. Otero, S. Klinke, J. Rinaldi, F. Velazquez-Escobar, M. A. Mroginski, M. Fernandez Lopez, F. Malamud, A. A. Vojnov, P. Hildebrandt, F. A. Goldbaum and H. R. Bonomi, Structure of the Full-Length Bacteriophytochrome from the Plant Pathogen Xanthomonas campestris Provides Clues to its Long-Range Signaling Mechanism, J. Mol. Biol., 2016, 428, 3702–3720, DOI:10.1016/j.jmb.2016.04.012.
- G. Gourinchas, U. Heintz and A. Winkler, Asymmetric activation mechanism of a homodimeric red light-regulated photoreceptor, eLife, 2018, 7, e34815, DOI:10.7554/eLife.34815.
- E. Claesson, W. Y. Wahlgren, H. Takala, S. Pandey, L. Castillon, V. Kuznetsova, L. Henry, M. Panman, M. Carrillo, J. Kubel, R. Nanekar, L. Isaksson, A. Nimmrich, A. Cellini, D. Morozov, M. Maj, M. Kurttila, R. Bosman, E. Nango, R. Tanaka, T. Tanaka, L. Fangjia, S. Iwata, S. Owada, K. Moffat, G. Groenhof, E. A. Stojkovic, J. A. Ihalainen, M. Schmidt and S. Westenhoff, The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser, eLife, 2020, 9, e53514, DOI:10.7554/eLife.53514.
- E. S. Burgie, T. Wang, A. N. Bussell, J. M. Walker, H. Li and R. D. Vierstra, Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion, J. Biol. Chem., 2014, 289, 24573–24587, DOI:10.1074/jbc.M114.571661.
- X. Yang, E. A. Stojkovic, J. Kuk and K. Moffat, Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 12571–12576, DOI:10.1073/pnas.0701737104.
- X. Yang, E. A. Stojkovic, W. B. Ozarowski, J. Kuk, E. Davydova and K. Moffat, Light Signaling Mechanism of Two Tandem Bacteriophytochromes, Structure, 2015, 23, 1179–1189, DOI:10.1016/j.str.2015.04.022.
- S. Nagano, P. Scheerer, K. Zubow, N. Michael, K. Inomata, T. Lamparter and N. Krauss, The Crystal Structures of the N-terminal Photosensory Core Module of Agrobacterium Phytochrome Agp1 as Parallel and Anti-parallel Dimers, J. Biol. Chem., 2016, 291, 20674–20691, DOI:10.1074/jbc.M116.739136.
- H. Takala, A. Björling, O. Berntsson, H. Lehtivuori, S. Niebling, M. Hoernke, I. Kosheleva, R. Henning, A. Menzel, J. A. Ihalainen and S. Westenhoff, Signal amplification and transduction in phytochrome photosensors, Nature, 2014, 509, 245–248, DOI:10.1038/nature13310.
- J. R. Wagner, J. Zhang, J. S. Brunzelle, R. D. Vierstra and K. T. Forest, High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution, J. Biol. Chem., 2007, 282, 12298–12309, DOI:10.1074/jbc.M611824200.
- M. E. Auldridge, K. A. Satyshur, D. M. Anstrom and K. T. Forest, Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein, J. Biol. Chem., 2012, 287, 7000–7009, DOI:10.1074/jbc.M111.295121.
- D. Yu, W. C. Gustafson, C. Han, C. Lafaye, M. Noirclerc-Savoye, W. P. Ge, D. A. Thayer, H. Huang, T. B. Kornberg, A. Royant, L. Y. Jan, Y. N. Jan, W. A. Weiss and X. Shu, An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging, Nat. Commun., 2014, 5, 3626, DOI:10.1038/ncomms4626.
- D. Bellini and M. Z. Papiz, Dimerization properties of the RpBphP2 chromophore-binding domain crystallized by homologue-directed mutagenesis, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 1058–1066, DOI:10.1107/S0907444912020537.
- S. Bhattacharya, M. E. Auldridge, H. Lehtivuori, J. A. Ihalainen and K. T. Forest, Origins of fluorescence in evolved bacteriophytochromes, J. Biol. Chem., 2014, 289, 32144–32152, DOI:10.1074/jbc.M114.589739.
- F. Li, E. S. Burgie, T. Yu, A. Heroux, G. C. Schatz, R. D. Vierstra and A. M. Orville, X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome, J. Am. Chem. Soc., 2015, 137, 2792–2795, DOI:10.1021/ja510923m.
- H. Lehtivuori, S. Bhattacharya, N. M. Angenent-Mari, K. A. Satyshur and K. T. Forest, Removal of Chromophore-Proximal Polar Atoms Decreases Water Content and Increases Fluorescence in a Near Infrared Phytofluor, Front. Mol. Biosci., 2015, 2, 65, DOI:10.3389/fmolb.2015.00065.
- M. Feliks, C. Lafaye, X. Shu, A. Royant and M. Field, Structural Determinants of Improved Fluorescence in a Family of Bacteriophytochrome-Based Infrared Fluorescent Proteins: Insights from Continuum Electrostatic Calculations and Molecular Dynamics Simulations, Biochemistry, 2016, 55, 4263–4274, DOI:10.1021/acs.biochem.6b00295.
- P. Edlund, H. Takala, E. Claesson, L. Henry, R. Dods, H. Lehtivuori, M. Panman, K. Pande, T. White, T. Nakane, O. Berntsson, E. Gustavsson, P. Bath, V. Modi, S. Roy-Chowdhury, J. Zook, P. Berntsen, S. Pandey, I. Poudyal, J. Tenboer, C. Kupitz, A. Barty, P. Fromme, J. D. Koralek, T. Tanaka, J. Spence, M. Liang, M. S. Hunter, S. Boutet, E. Nango, K. Moffat, G. Groenhof, J. Ihalainen, E. A. Stojkovic, M. Schmidt and S. Westenhoff, The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography, Sci. Rep., 2016, 6, 35279, DOI:10.1038/srep35279.
- F. D. Fuller, S. Gul, R. Chatterjee, E. S. Burgie, I. D. Young, H. Lebrette, V. Srinivas, A. S. Brewster, T. Michels-Clark, J. A. Clinger, B. Andi, M. Ibrahim, E. Pastor, C. de Lichtenberg, R. Hussein, C. J. Pollock, M. Zhang, C. A. Stan, T. Kroll, T. Fransson, C. Weninger, M. Kubin, P. Aller, L. Lassalle, P. Brauer, M. D. Miller, M. Amin, S. Koroidov, C. G. Roessler, M. Allaire, R. G. Sierra, P. T. Docker, J. M. Glownia, S. Nelson, J. E. Koglin, D. Zhu, M. Chollet, S. Song, H. Lemke, M. Liang, D. Sokaras, R. Alonso-Mori, A. Zouni, J. Messinger, U. Bergmann, A. K. Boal, J. M. Bollinger, C. Krebs, M. Hogbom, G. N. Phillips, R. D. Vierstra, N. K. Sauter, A. M. Orville, J. Kern, V. K. Yachandra and J. Yano, Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers, Nat. Methods, 2017, 14, 443–449, DOI:10.1038/nmeth.4195.
- H. Takala, H. K. Lehtivuori, O. Berntsson, A. Hughes, R. Nanekar, S. Niebling, M. Panman, L. Henry, A. Menzel, S. Westenhoff and J. A. Ihalainen, On the (un)coupling of the chromophore, tongue interactions, and overall conformation in a bacterial phytochrome, J. Biol. Chem., 2018, 293, 8161–8172, DOI:10.1074/jbc.RA118.001794.
- M. Baloban, D. M. Shcherbakova, S. Pletnev, V. Z. Pletnev, J. C. Lagarias and V. V. Verkhusha, Designing brighter near-infrared fluorescent proteins: insights from structural and biochemical studies, Chem. Sci., 2017, 8, 4546–4557, 10.1039/c7sc00855d.
- N. Lenngren, P. Edlund, H. Takala, B. Stucki-Buchli, J. Rumfeldt, I. Peshev, H. Hakkanen, S. Westenhoff and J. A. Ihalainen, Coordination of the biliverdin D-ring in bacteriophytochromes, Phys. Chem. Chem. Phys., 2018, 20, 18216–18225, 10.1039/c8cp01696h.
- D. Buhrke, G. Gourinchas, M. Muller, N. Michael, P. Hildebrandt and A. Winkler, Distinct chromophore-protein environments enable asymmetric activation of a bacteriophytochrome-activated diguanylate cyclase, J. Biol. Chem., 2020, 295, 539–551, DOI:10.1074/jbc.RA119.011915.
- F. Andel, J. C. Lagarias and R. A. Mathies, Resonance raman analysis of chromophore structure in the lumi-R photoproduct of phytochrome, Biochemistry, 1996, 35, 15997–16008, DOI:10.1021/bi962175k.
- C. Kneip, P. Hildebrandt, W. Schlamann, S. E. Braslavsky, F. Mark and K. Schaffner, Protonation state and structural changes of the tetrapyrrole chromophore during the Pr –> Pfr phototransformation of phytochrome: a resonance Raman spectroscopic study, Biochemistry, 1999, 38, 15185–15192, DOI:10.1021/bi990688w.
- F. Andel, J. T. Murphy, J. A. Haas, M. T. McDowell, I. van der Hoef, J. Lugtenburg, J. C. Lagarias and R. A. Mathies, Probing the photoreaction mechanism of phytochrome through analysis of resonance Raman vibrational spectra of recombinant analogues, Biochemistry, 2000, 39, 2667–2676, DOI:10.1021/bi991688z.
- W. Rudiger, F. Thummler, E. Cmiel and S. Schneider, Chromophore structure of the physiologically active form (P(fr)) of phytochrome, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 6244–6248, DOI:10.1073/pnas.80.20.6244.
- E. S. Burgie, J. Zhang and R. D. Vierstra, Crystal Structure of Deinococcus Phytochrome in the Photoactivated State Reveals a Cascade of Structural Rearrangements during Photoconversion, Structure, 2016, 24, 448–457, DOI:10.1016/j.str.2016.01.001.
- A. J. Fischer and J. C. Lagarias, Harnessing phytochrome’s glowing potential, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17334–17339, DOI:10.1073/pnas.0407645101.
- J. Hahn, H. M. Strauss, F. T. Landgraf, H. F. Gimenez, G. Lochnit, P. Schmieder and J. Hughes, Probing protein-chromophore interactions in Cph1 phytochrome by mutagenesis, FEBS J., 2006, 273, 1415–1429, DOI:10.1111/j.1742-4658.2006.05164.x.
- J. Zhang, R. J. Stankey and R. D. Vierstra, Structure-guided engineering of plant phytochrome B with altered photochemistry and light signaling, Plant Physiol., 2013, 161, 1445–1457, DOI:10.1104/pp.112.208892.
- F. Velazquez-Escobar, C. Lang, A. Takiden, C. Schneider, J. Balke, J. Hughes, U. Alexiev, P. Hildebrandt and M. A. Mroginski, Protonation-Dependent Structural Heterogeneity in the Chromophore Binding Site of Cyanobacterial Phytochrome Cph1, J. Phys. Chem. B, 2017, 121, 47–57, DOI:10.1021/acs.jpcb.6b09600.
- J. A. Rumfeldt, H. Takala, A. Liukkonen and J. A. Ihalainen, UV-Vis Spectroscopy Reveals a Correlation Between Y263 and BV Protonation States in Bacteriophytochromes, Photochem. Photobiol., 2019, 95, 969–979, DOI:10.1111/php.13095.
- J. Mailliet, G. Psakis, K. Feilke, V. Sineshchekov, L. O. Essen and J. Hughes, Spectroscopy and a high-resolution crystal structure of Tyr263 mutants of cyanobacterial phytochrome Cph1, J. Mol. Biol., 2011, 413, 115–127, DOI:10.1016/j.jmb.2011.08.023.
- J. A. Ihalainen, E. Gustavsson, L. Schroeder, S. Donnini, H. Lehtivuori, L. Isaksson, C. Thoing, V. Modi, O. Berntsson, B. Stucki-Buchli, A. Liukkonen, H. Hakkanen, E. Kalenius, S. Westenhoff and T. Kottke, Chromophore-Protein Interplay during the Phytochrome Photocycle Revealed by Step-Scan FTIR Spectroscopy, J. Am. Chem. Soc., 2018, 140, 12396–12404, DOI:10.1021/jacs.8b04659.
- K. C. Toh, E. A. Stojkovic, A. B. Rupenyan, I. H. van Stokkum, M. Salumbides, M. L. Groot, K. Moffat and J. T. Kennis, Primary reactions of bacteriophytochrome observed with ultrafast mid-infrared spectroscopy, J. Phys. Chem. A, 2011, 115, 3778–3786, DOI:10.1021/jp106891x.
- Y. Yang, K. Heyne, R. A. Mathies and J. Dasgupta, Non-Bonded Interactions Drive the Sub-Picosecond Bilin Photoisomerization in the P(fr) State of Phytochrome Cph1, ChemPhysChem, 2016, 17, 369–374, DOI:10.1002/cphc.201501073.
- H. Li, J. Zhang, R. D. Vierstra and H. Li, Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 10872–10877, DOI:10.1073/pnas.1001908107.
- H. Takala, H. Lehtivuori, H. Hammarén, V. P. Hytönen and J. A. Ihalainen, Connection between absorption properties and conformational changes in Deinococcus radiodurans phytochrome, Biochemistry, 2014, 53, 7076–7085, DOI:10.1021/bi501180s.
- E. A. Stojkovic, K. C. Toh, M. T. Alexandre, M. Baclayon, K. Moffat and J. T. Kennis, FTIR Spectroscopy Revealing Light-Dependent Refolding of the Conserved Tongue Region of Bacteriophytochrome, J. Phys. Chem. Lett., 2014, 5, 2512–2515, DOI:10.1021/jz501189t.
- X. Yang, J. Kuk and K. Moffat, Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 15639–15644, DOI:10.1073/pnas.0902178106.
- K. Anders, G. Daminelli-Widany, M. A. Mroginski, D. von Stetten and L. O. Essen, Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling, J. Biol. Chem., 2013, 288, 35714–35725, DOI:10.1074/jbc.M113.510461.
- J. C. Sanchez, M. Carrillo, S. Pandey, M. Noda, L. Aldama, D. Feliz, E. Claesson, W. Y. Wahlgren, G. Tracy, P. Duong, A. C. Nugent, A. Field, V. Srajer, C. Kupitz, S. Iwata, E. Nango, R. Tanaka, T. Tanaka, L. Fangjia, K. Tono, S. Owada, S. Westenhoff, M. Schmidt and E. A. Stojkovic, High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures, Struct. Dyn., 2019, 6, 054701, DOI:10.1063/1.5120527.
- H. Takala, S. Niebling, O. Berntsson, A. Björling, H. Lehtivuori, H. Hakkanen, M. Panman, E. Gustavsson, M. Hoernke, G. Newby, F. Zontone, M. Wulff, A. Menzel, J. A. Ihalainen and S. Westenhoff, Light-induced structural changes in a monomeric bacteriophytochrome, Struct. Dyn., 2016, 3, 054701, DOI:10.1063/1.4961911.
- T. E. Assafa, K. Anders, U. Linne, L. O. Essen and E. Bordignon, Light-Driven Domain Mechanics of a Minimal Phytochrome Photosensory Module Studied by EPR, Structure, 2018, 26, 1534–1545 CrossRef CAS , pii: S0969-2126(18)30291-0.
- V. Anantharaman, S. Balaji and L. Aravind, The signaling helix: a common functional theme in diverse signaling proteins, Biol. Direct, 2006, 1, 25, DOI:10.1186/1745-6150-1-25.
- S. Kacprzak, I. Njimona, A. Renz, J. Feng, E. Reijerse, W. Lubitz, N. Krauss, P. Scheerer, S. Nagano, T. Lamparter and S. Weber, Intersubunit distances in full-length, dimeric, bacterial phytochrome Agp1, as measured by pulsed electron-electron double resonance (PELDOR) between different spin label positions, remain unchanged upon photoconversion, J. Biol. Chem., 2017, 292, 7598–7606, DOI:10.1074/jbc.M116.761882.
- M. P. Bhate, K. S. Molnar, M. Goulian and W. F. DeGrado, Signal transduction in histidine kinases: insights from new structures, Structure, 2015, 23, 981–994, DOI:10.1016/j.str.2015.04.002.
- S. Nagano, From photon to signal in phytochromes: similarities and differences between prokaryotic and plant phytochromes, J. Plant Res., 2016, 129, 123–135, DOI:10.1007/s10265-016-0789-0.
- A. Moglich, Signal transduction in photoreceptor histidine kinases, Protein Sci., 2019, 28, 1923–1946, DOI:10.1002/pro.3705.
- L. E. Ulrich and I. B. Zhulin, The MiST2 database: a comprehensive genomics resource on microbial signal transduction, Nucleic Acids Res., 2010, 38, 401, DOI:10.1093/nar/gkp940.
- J. S. Parkinson, Signal transduction schemes of bacteria, Cell, 1993, 73, 857–871, DOI:10.1016/0092-8674(93)90267-t.
- A. H. West and A. M. Stock, Histidine kinases and response regulator proteins in two-component signaling systems, Trends Biochem. Sci., 2001, 26, 369–376, DOI:10.1016/s0968-0004(01)01852-7.
- P. Casino, V. Rubio and A. Marina, Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction, Cell, 2009, 139, 325–336, DOI:10.1016/j.cell.2009.08.032.
- B. Zienicke, I. Molina, R. Glenz, P. Singer, D. Ehmer, F. V. Escobar, P. Hildebrandt, R. Diller and T. Lamparter, Unusual spectral properties of bacteriophytochrome Agp2 result from a deprotonation of the chromophore in the red-absorbing form Pr, J. Biol. Chem., 2013, 288, 31738–31751, DOI:10.1074/jbc.M113.479535.
- T. Hubschmann, H. J. Jorissen, T. Borner, W. Gartner and N. Tandeau de Marsac, Phosphorylation of proteins in the light-dependent signalling pathway of a filamentous cyanobacterium, Eur. J. Biochem., 2001, 268, 3383–3389, DOI:10.1046/j.1432-1327.2001.02229.x.
- T. Lamparter, B. Esteban and J. Hughes, Phytochrome Cph1 from the cyanobacterium Synechocystis PCC6803. Purification, assembly, and quaternary structure, Eur. J. Biochem., 2001, 268, 4720–4730, DOI:10.1046/j.1432-1327.2001.02395.x.
- B. Esteban, M. Carrascal, J. Abian and T. Lamparter, Light-induced conformational changes of cyanobacterial phytochrome Cph1 probed by limited proteolysis and autophosphorylation, Biochemistry, 2005, 44, 450–461, DOI:10.1021/bi0484365.
- E. Giraud, S. Zappa, L. Vuillet, J. M. Adriano, L. Hannibal, J. Fardoux, C. Berthomieu, P. Bouyer, D. Pignol and A. Vermeglio, A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris, J. Biol. Chem., 2005, 280, 32389–32397, DOI:10.1074/jbc.M506890200.
- K. Evans, J. G. Grossmann, A. P. Fordham-Skelton and M. Z. Papiz, Small-angle X-ray scattering reveals the solution structure of a bacteriophytochrome in the catalytically active Pr state, J. Mol. Biol., 2006, 364, 655–666, DOI:10.1016/j.jmb.2006.09.045.
- H. Takala, A. Björling, M. Linna, S. Westenhoff and J. A. Ihalainen, Light-induced Changes in the Dimerization Interface of Bacteriophytochromes, J. Biol. Chem., 2015, 290, 16383–16392, DOI:10.1074/jbc.M115.650127.
- S. Etzl, R. Lindner, M. D. Nelson and A. Winkler, Structure-guided design and functional characterization of an artificial red light-regulated guanylate/adenylate cyclase for optogenetic applications, J. Biol. Chem., 2018, 293, 9078–9089, DOI:10.1074/jbc.RA118.003069.
- H. R. Bonomi, L. Toum, G. Sycz, R. Sieira, A. M. Toscani, G. E. Gudesblat, F. C. Leskow, F. A. Goldbaum, A. A. Vojnov and F. Malamud, Xanthomonas campestris attenuates virulence by sensing light through a bacteriophytochrome photoreceptor, EMBO Rep., 2016, 17, 1565–1577, DOI:10.15252/embr.201541691.
- N. Ausmees, R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman and M. Lindberg, Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity, FEMS Microbiol. Lett., 2001, 204, 163–167, DOI:10.1111/j.1574-6968.2001.tb10880.x.
- A. Marina, C. D. Waldburger and W. A. Hendrickson, Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein, EMBO J., 2005, 24, 4247–4259 CrossRef CAS , pii: 7600886.
- S. Yamada, H. Sugimoto, M. Kobayashi, A. Ohno, H. Nakamura and Y. Shiro, Structure of PAS-linked histidine kinase and the response regulator complex, Structure, 2009, 17, 1333–1344, DOI:10.1016/j.str.2009.07.016.
- O. Berntsson, R. P. Diensthuber, M. R. Panman, A. Björling, E. Gustavsson, M. Hoernke, A. J. Hughes, L. Henry, S. Niebling, H. Takala, J. A. Ihalainen, G. Newby, S. Kerruth, J. Heberle, M. Liebi, A. Menzel, R. Henning, I. Kosheleva, A. Moglich and S. Westenhoff, Sequential conformational transitions and alpha-helical supercoiling regulate a sensor histidine kinase, Nat. Commun., 2017, 8, 284–285, DOI:10.1038/s41467-017-00300-5.
- I. Gushchin, I. Melnikov, V. Polovinkin, A. Ishchenko, A. Yuzhakova, P. Buslaev, G. Bourenkov, S. Grudinin, E. Round, T. Balandin, V. Borshchevskiy, D. Willbold, G. Leonard, G. Buldt, A. Popov and V. Gordeliy, Mechanism of transmembrane signaling by sensor histidine kinases, Science, 2017, 356(6342), eaah6345, DOI:10.1126/science.aah6345.
- T. Matsushita, N. Mochizuki and A. Nagatani, Dimers of the N-terminal domain of phytochrome B are functional in the nucleus, Nature, 2003, 424, 571–574, DOI:10.1038/nature01837.
- Y. Oka, T. Matsushita, N. Mochizuki, T. Suzuki, S. Tokutomi and A. Nagatani, Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis, Plant Cell, 2004, 16, 2104–2116, DOI:10.1105/tpc.104.022350.
- N. De, M. V. Navarro, R. V. Raghavan and H. Sondermann, Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR, J. Mol. Biol., 2009, 393, 619–633, DOI:10.1016/j.jmb.2009.08.030.
- T. Schirmer and U. Jenal, Structural and mechanistic determinants of c-di-GMP signalling, Nat. Rev. Microbiol., 2009, 7, 724–735, DOI:10.1038/nrmicro2203.
- S. Lim, Q. Yu, S. M. Gottlieb, C. W. Chang, N. C. Rockwell, S. S. Martin, D. Madsen, J. C. Lagarias, D. S. Larsen and J. B. Ames, Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4387–4392, DOI:10.1073/pnas.1720682115.
- E. Gustavsson, L. Isaksson, C. Persson, M. Mayzel, U. Brath, L. Vrhovac, J. A. Ihalainen, B. G. Karlsson, V. Orekhov and S. Westenhoff, Modulation of Structural Heterogeneity Controls Phytochrome Photoswitching, Biophys. J., 2020, 118, 415–421, DOI:10.1016/j.bpj.2019.11.025.
- C. Song, T. Rohmer, M. Tiersch, J. Zaanen, J. Hughes and J. Matysik, Solid-state NMR spectroscopy to probe photoactivation in canonical phytochromes, Photochem. Photobiol., 2013, 89, 259–273, DOI:10.1111/php.12029; 10.1111/php.12029.
- H. N. Chapman, P. Fromme, A. Barty, T. A. White, R. A. Kirian, A. Aquila, M. S. Hunter, J. Schulz, D. P. DePonte, U. Weierstall, R. B. Doak, F. R. Maia, A. V. Martin, I. Schlichting, L. Lomb, N. Coppola, R. L. Shoeman, S. W. Epp, R. Hartmann, D. Rolles, A. Rudenko, L. Foucar, N. Kimmel, G. Weidenspointner, P. Holl, M. Liang, M. Barthelmess, C. Caleman, S. Boutet, M. J. Bogan, J. Krzywinski, C. Bostedt, S. Bajt, L. Gumprecht, B. Rudek, B. Erk, C. Schmidt, A. Homke, C. Reich, D. Pietschner, L. Struder, G. Hauser, H. Gorke, J. Ullrich, S. Herrmann, G. Schaller, F. Schopper, H. Soltau, K. U. Kuhnel, M. Messerschmidt, J. D. Bozek, S. P. Hau-Riege, M. Frank, C. Y. Hampton, R. G. Sierra, D. Starodub, G. J. Williams, J. Hajdu, N. Timneanu, M. M. Seibert, J. Andreasson, A. Rocker, O. Jonsson, M. Svenda, S. Stern, K. Nass, R. Andritschke, C. D. Schroter, F. Krasniqi, M. Bott, K. E. Schmidt, X. Wang, I. Grotjohann, J. M. Holton, T. R. Barends, R. Neutze, S. Marchesini, R. Fromme, S. Schorb, D. Rupp, M. Adolph, T. Gorkhover, I. Andersson, H. Hirsemann, G. Potdevin, H. Graafsma, B. Nilsson and J. C. Spence, Femtosecond X-ray protein nanocrystallography, Nature, 2011, 470, 73–77, DOI:10.1038/nature09750.
- F. Stellato, D. Oberthur, M. Liang, R. Bean, C. Gati, O. Yefanov, A. Barty, A. Burkhardt, P. Fischer, L. Galli, R. A. Kirian, J. Meyer, S. Panneerselvam, C. H. Yoon, F. Chervinskii, E. Speller, T. A. White, C. Betzel, A. Meents and H. N. Chapman, Room-temperature macromolecular serial crystallography using synchrotron radiation, IUCrJ, 2014, 1, 204–212, DOI:10.1107/S2052252514010070.
- N. Coquelle, A. S. Brewster, U. Kapp, A. Shilova, B. Weinhausen, M. Burghammer and J. P. Colletier, Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2015, 71, 1184–1196, DOI:10.1107/S1399004715004514.
- P. Nogly, D. James, D. Wang, T. A. White, N. Zatsepin, A. Shilova, G. Nelson, H. Liu, L. Johansson, M. Heymann, K. Jaeger, M. Metz, C. Wickstrand, W. Wu, P. Bath, P. Berntsen, D. Oberthuer, V. Panneels, V. Cherezov, H. Chapman, G. Schertler, R. Neutze, J. Spence, I. Moraes, M. Burghammer, J. Standfuss and U. Weierstall, Lipidic cubic phase serial millisecond crystallography using synchrotron radiation, IUCrJ, 2015, 2, 168–176, DOI:10.1107/S2052252514026487.
- R. Neutze, R. Wouts, D. van der Spoel, E. Weckert and J. Hajdu, Potential for biomolecular imaging with femtosecond X-ray pulses, Nature, 2000, 406, 752–757, DOI:10.1038/35021099.
- J. Tenboer, S. Basu, N. Zatsepin, K. Pande, D. Milathianaki, M. Frank, M. Hunter, S. Boutet, G. J. Williams, J. E. Koglin, D. Oberthuer, M. Heymann, C. Kupitz, C. Conrad, J. Coe, S. Roy-Chowdhury, U. Weierstall, D. James, D. Wang, T. Grant, A. Barty, O. Yefanov, J. Scales, C. Gati, C. Seuring, V. Srajer, R. Henning, P. Schwander, R. Fromme, A. Ourmazd, K. Moffat, J. J. Van Thor, J. C. Spence, P. Fromme, H. N. Chapman and M. Schmidt, Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein, Science, 2014, 346, 1242–1246, DOI:10.1126/science.1259357.
- E. S. Burgie, J. A. Clinger, M. D. Miller, A. S. Brewster, P. Aller, A. Butryn, F. D. Fuller, S. Gul, I. D. Young, C. C. Pham, I. S. Kim, A. Bhowmick, L. J. O'Riordan, K. D. Sutherlin, J. V. Heinemann, A. Batyuk, R. Alonso-Mori, M. S. Hunter, J. E. Koglin, J. Yano, V. K. Yachandra, N. K. Sauter, A. E. Cohen, J. Kern, A. M. Orville, G. N. Phillips and R. D. Vierstra, Photoreversible interconversion of a phytochrome photosensory module in the crystalline state, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 300–307, DOI:10.1073/pnas.1912041116.
- D. von Stetten, M. Gunther, P. Scheerer, D. H. Murgida, M. A. Mroginski, N. Krauss, T. Lamparter, J. Zhang, D. M. Anstrom, R. D. Vierstra, K. T. Forest and P. Hildebrandt, Chromophore heterogeneity and photoconversion in phytochrome crystals and solution studied by resonance Raman spectroscopy, Angew. Chem., Int. Ed., 2008, 47, 4753–4755, DOI:10.1002/anie.200705716.
- P. W. Kim, N. C. Rockwell, S. S. Martin, J. C. Lagarias and D. S. Larsen, Dynamic inhomogeneity in the photodynamics of cyanobacterial phytochrome Cph1, Biochemistry, 2014, 53, 2818–2826, DOI:10.1021/bi500108s.
- F. Velazquez Escobar, D. von Stetten, M. Gunther-Lutkens, A. Keidel, N. Michael, T. Lamparter, L. O. Essen, J. Hughes, W. Gartner, Y. Yang, K. Heyne, M. A. Mroginski and P. Hildebrandt, Conformational heterogeneity of the Pfr chromophore in plant and cyanobacterial phytochromes, Front. Mol. Biosci., 2015, 2, 37, DOI:10.3389/fmolb.2015.00037.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry and Owner Societies 2020 |