Open Access Article
Open Access ArticleImpact of CeO2 nanoparticles on the microbiota of the S. flos-cuculi L. (Caryophyllaceae) rhizosphere†
M.
Civilini
a,
A.
Colautti
b,
A.
Brunello
c,
N.
Saccomanno
 c,
L.
Marchiol
a,
A.
Foscari
a and
L.
Iacumin
c,
L.
Marchiol
a,
A.
Foscari
a and
L.
Iacumin
 *b
*b
aDepartment of Agriculture, Food, Environmental and Animal Sciences, University of Udine, Via delle Scienze 206, 33100 Udine, Italy
bDepartment of Agriculture, Food, Environmental and Animal Sciences, University of Udine, Via Sondrio 2/A, 33100 Udine, Italy. E-mail: lucilla.iacumin@uniud.it; Tel: +39 338 1378082
cDepartment of Mathematics, Computer Science, and Physics, University of Udine, Via delle Scienze 206, 33100 Udine, Italy
First published on 9th April 2024
Abstract
The aim of this study was to correlate the influence of cerium oxide nanoparticles (nCeO2) resulting from pollution sources, on the root bacterial composition and the associated substrate (root zone soil) of Silene flos-cuculi (L.) using a metabarcoding technique. Currently, limited information is available regarding the direct effects of nCeO2 on plants and the rhizosphere microbiota, where changes in turn could positively or negatively influence plant performance. To assess the distribution of the main bacterial phyla in the culture substrates, analyses were conducted considering both intracellular DNA (iDNA) contained within intact and live bacterial cells, and extracellular DNA (eDNA) from lysed cells. The impact of various nCeO2 dosages on phyla, families, and genera was then investigated with a detailed examination of all detected members at the family and genus levels to differentiate the nCeO2 treatment effects. The results revealed that 25 out of 641 identified bacterial genera, primarily anaerobic and strictly anaerobic, exhibited reduced presence in nCeO2 treated samples compared to the controls. This decrease was particularly evident in species belonging to the phylum Firmicutes. Metabolic function analysis performed using FAPROTAX indicated a decline in fermentative, nitrogen fixation, and iron/nitrate respiration metabolisms in nCeO2 treated samples, especially at higher concentrations. Conversely, there was an increase in chemo-heterotrophy and aerobic chemoheterotrophic-related functions in these samples.
Environmental significanceNanoparticles are even more present in the environment, for many reasons. However, little is known about their impact on the different environmental microbiota and also in relation to humans, animals, and plants. For this reason, the present work aims to evaluate the effect of nCeO2 particles on the Silene flos-cuculi (L.) root bacterial composition and its associated substrate (root zone soil). |
1 Introduction
Thanks to the advent of nanotechnology, new nanomaterials useful in numerous applications have been engineered.1 However, the diffusion of these new materials in the environment must be studied to evaluate their possible impact on ecosystems.2 In this regard, cerium (Ce) nanoparticles are one of the most important, already widespread into the environment because of their use as an anti-corrosion component in aluminium alloys to replace hexavalent chromium as well as an additive in painting systems.3,4 Ce has been also studied for decades as a catalyst and material for electrolytes and electrodes in fuel-cell systems, and nanostructured cerium oxide (NS-CeO2) materials have been developed to meet the high energy and environmental demands because of their superior redox and catalytic properties compared to bulk materials.5,6 Ce nanoparticles (CeNPs) in water suspensions also found application as polishing supports, anticorrosive systems, oxidants for electrode sensors, and redox agents for medical treatment. In the last case, although Ce itself has no biological role in mammalian physiology, Ce3+ soluble salts (nitrate, acetate, and chloride) have traditionally been used for biomedical purposes due to their antiemetic, bacteriostatic and bactericidal properties.7 CeNPs have been also reported to beneficially modulate excess reactive oxygen species (ROS) in cells; however, their toxicological effects associated with their structure and stability should be considered.8The effects of CeNPs in complex environments are influenced by several variables, including the manufacturing method, nanocrystal shape and size,9,10 and redox potential.8,11,12 Biological studies involving CeNPs revealed significant variability and conflicting results.13,14In vivo studies have shown that CeNPs are bio-persistent for long periods after inhalation15,16 and oral administration,17 and can be internalized by macrophages and epithelial cells.18 Furthermore, given the irreversibility of their accumulation as well as the potential long-term effects and interactions of Ce oxide nanoparticles (nCeO2) with abiotic and biotic entities, it is critical to conduct research on the consequences of the large-scale use of these materials.13
For this reason, given their already present diffusion and their high persistence in soil, one of the most important aspects to evaluate are the effects of these nanoparticles on soil structure and plants, especially those intended for human and animal consumption, also in relation to the microbial population that lives symbiotically on the roots.19,20 Synchrotron spectroscopic techniques have revealed that plants can improve CeNP reduction in soil and hydroponic systems, and Ce absorption and translocation differed among rice, wheat, and barley grown in soils enriched with CeNPs. However, while Ce does not accumulate in the grains of wheat, it accumulates in the tissues and grains of rice and barley.21
There are conflicting reports regarding the growth and health of plants exposed to this nanomaterial. Some studies report an improvement in factors, such as growth and increases in biomass, others testify to the induction of physiological and morphological anomalies.22–28
Some evidence suggests a possible toxic effect of this element on the microbial flora of the soil29 and Ce microbial accumulation in microorganisms isolated from rare earth-rich environments.30,31 For this reason, given the fundamental role of bacteria in the health and growth of plants,32,33 it is important to expand knowledge on the possible effects of an accumulation of this compound in relation to the effects induced on the soil microbiota. In fact, it is possible to hypothesize that some of the effects previously observed on plants could be correlated to modifications of the symbiotic microflora, which, given the high importance and interconnection, would be reflected in the physiology of the plant.
In this regard, starting from a previous study, which reported that nCeO2 is absorbed by plant roots and translocated toward aerial plant fractions, inducing differences in the development of the treated plants,34 this work focused on the modification induced by different concentrations of nCeO2 soil amendments on substrates on the rhizospheric microbiota of S. flos-cuculi L. (Caryophyllaceae). For this purpose, a next-generation sequencing technology (NGS) approach was applied.
2 Materials and methods
2.1 Experimental design and sampling
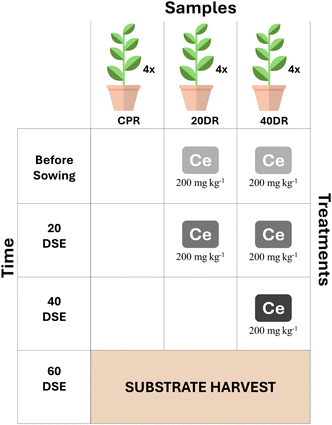
The characteristics of nCeO2, plant materials and the experimental design were set up as described in a previous study34 and are summarized in Fig. 1. Briefly, Silene flos-cuculi (L.) was grown in an organic substrate (“Compo Sana”, pH = 6.8–7.2), with four replicates for each trial, in growing pots filled with 500 g of substrate. Control samples (CPR) were sown in the substrate without any addition of nCeO2 and were irrigated with pure tap water. In contrast, the potting substrate of samples 20DR and 40DR was irrigated with water suspensions of nCeO2 with a dimension of 25 nm to achieve an nCeO2 substrate concentration of 200 mg kg−1. Then, growing pots were seeded with S. flos-cuculi (L.) and placed in full sunlight at 18–27 °C (night/day) with a relative humidity of around 60%, irrigated every three days, and seedlings were thinned to two for each pot after two weeks. In trial 20DR, a further nCeO2 application of 200 mg kg−1 was performed 20 days after seedling emergence (DSE), whereas for trial 40DR, two applications of 200 mg kg−1 nCeO2 were performed after 20 and 40 days from DSE. At 60 days from DSE, all plants were harvested. The nCeO2 doses were chosen on the basis of previous results,34 which demonstrated that 200 and 400 mg kg−1 were found to be the most effective in affecting plant growth parameters, with a shortage in biomass of roots and stems, and in nCeO2 translocation towards aerial plant fractions. The purpose of this protocol was to simulate conditions closer to reality, where plants are probabilistically exposed to CeNPs over a more extended period, at relatively lower concentrations, but through repeated pulses.35Before being separated from the stems and leaves, the roots were gently shaken in an empty sterile glass beaker to collect the substrate in four replicates. A sterilized cleaned trowel was used for each sample to collect the substrate for microbial community analyses. All samples were transported to the laboratory on ice, and the substrates for molecular characterization were immediately stored at −80 °C until analysis.
2.2 Extraction and purification of intra- and extracellular DNA
For each trial, DNA was directly extracted from equal aliquots (5 g) of substrate obtained from the four replicates performed for each trial using the following protocol:36 extracellular DNA (eDNA) was extracted by gentle substrate washing with 5 mL of 0.12 M Na2HPO4 (Sigma-Aldrich, Italy) at pH 8 in 50 mL Falcon tubes horizontally shaken for 30 min (80 rpm). Tubes were then centrifuged (4 °C, 30 min, 7500 × g), and the 5 mL supernatant was collected. The same procedure was repeated twice, and the resulting supernatants were pooled to obtain a final volume of 15 mL of unpurified eDNA. Then, the eDNA solution was purified using a commercial extraction kit (DNeasy® PowerMax® Soil kit, Qiagen, USA) following the manufacturer's instructions but avoiding incubation in cell lysis buffer. Intracellular DNA (iDNA) was extracted from the residual substrate pellets resulting from the alkaline washing. The pellet was collected in a 50 mL Falcon tube and processed using the extraction kit according to the manufacturer's instructions, including incubation in cell lysis buffer. At the end of purification, all DNA samples were suspended in 5 mL of 10 mM Tris solution (Sigma-Aldrich, Italy).To remove undesired RNA residues, eDNA and iDNA solutions were incubated with RNAse A (25 μL, 1 ng μL−1, Sigma-Aldrich, Italy) and shaken at 37 °C (1 h, 300 rpm); then sodium acetate (3 M, 10% v/v), NaCl (4 M, 30% v/v), and isopropyl alcohol (66% v/v) were added prior to overnight incubation at −20 °C and centrifugation (4 °C, 30 min, 7500×g). After removing the supernatant, the residual pellet was washed twice with 2 mL ethanol (80%). Finally, the eDNA and iDNA pellets were dried in a stove (10–15 min at 37 °C) and resuspended in 1 mL of DNAse free sterile water. Purified DNA samples were quantified using a Qubit 3.0 (Life Technology, Carlsbad, California, USA), and quality was assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The fragment length distribution was assessed using 0.8% agarose gel electrophoresis.
2.3 Microbiota identification by 16S rRNA gene – amplification, sequencing, and data analysis
The 16S rRNA gene sequences were amplified from the extracted DNA, according to a previous reported protocol.37 Briefly, for the amplification of the V3–V4 region, the primer pair Probio_Uni (forward primer 5′-CCTACGGGRSGCAGCAG-3′) and Probio_Rev (reverse primer 5′-ATTACCGCGGCTGCT-3′) was used, verifying the integrity of amplicons by electrophoresis with an Experion workstation (BioRad, Italy). The amplicons were purified using electrophoretic separation on a 1.5% agarose gel of a Wizard SV Gen PCR Clean-Up System (Promega, Italy), with a subsequent purification step to remove primer dimers made with Agencourt AMPure XP DNA purification beads (Beckman Coulter Genomics GmbH, Germany). The 16S rRNA gene sequencing was performed using the MiSeq Platform (Illumina) at the GenProbio s.r.l. DNA sequencing facility (http://www.genprobio.com). The obtained raw reads were processed using the QIIME2 software suite.38 The complete Probio_Uni/Probio_Rev amplicons were reconstructed from the assembled paired-end reads. Sequences with a length between 140 and 400 bp and a mean sequence quality score >20 were retained, whereas sequences with homopolymers >7 bp and mismatched primers were omitted. 16S rRNA amplicon sequence variants (ASVs) were identified with DADA2, which defines variants based on unique sequences,39 and singletons were removed. All reads were classified to the lowest possible taxonomic rank using QIIME2 (ref. 38 and 40) using SILVA as the database.41 Statistical analyses were conducted using R (v 4.2.1). Observed ASVs and Good's coverage were calculated using ggrare (ranacapa package 0.1.0) and represented as rarefaction curves for species richness. After rarefaction of the samples implemented through the phyloseq package,42 the mean species diversity for each treatment (Alpha-diversity) was assessed using the Shannon index. Principal coordinate analysis (PCoA) was used to analyze the patterns of prokaryotic communities using Bray–Curtis and Jaccard dissimilarity matrixes with the vegan R package (v 2.5–6).32 Likewise, to evaluate the similarities among samples from a qualitative point of view, principal component analysis (PCA) was applied to the bacterial group distributions at the genus granularity level, using the scikit-learn Python package (v. 0.23.2). To better understand the behavior of the microbiological groups in the different samples, deviations in their distributions compared to the control samples at the phylum, family, and genus granularity levels were evaluated. Finally, the ranking trends of various bacterial groups in the different samples were studied. To such an extent, other than presenting a series of qualitative results concerning the changes in the bacterial rankings in the different samples, the Spearman correlation was calculated pairwise between the samples to evaluate the degree of concordance (between −1 and 1) of the ranks. The ranking, which sorts the bacterial groups from the most to the least present in a sample, provides an overview of the bacterial group distribution. Given two rankings pertaining to two different samples, their similarity through the Spearman correlation was compared. In short, the Spearman correlation measures the strength and direction of the monotonic relationship between the two rankings. Instead of comparing raw data values, it assesses how well the relationship between the rankings of one sample predict the rankings of the other sample. When both samples have perfectly similar rankings, the Spearman correlation is 1 (thus the two bacterial group distributions are similar); when the rankings are completely opposite, it is −1; and when the rankings are unrelated, it is close to 0. Furthermore, the metabolic functions of the microbiota in different samples were predicted using the FAPROTAX database.433 Results
3.1 Sequencing results
The total DNA that could be extracted from soil could be distinguished in the iDNA, found within cell membranes, and eDNA, representing DNA located outside cell membranes.36,44 In this study, iDNA and eDNA were differentiated as considering the whole DNA could have led to bias as considering not only the present microbial community but also paleome,45 overestimating the actual diversity.46,47The obtained sequences were published in National Centre for Biotechnology Information (NCBI) under the BioProject PRJNA888088 and are available at the BioSample accession numbers shown in Table 1.
| Sample | Biosample | SRA |
|---|---|---|
| CPR_iDNA | SAMN31207732 | SRR21875482 |
| CPR_eDNA | SAMN31207733 | SRR21875481 |
| 20DR_iDNA | SAMN31207734 | SRR21875480 |
| 20DR_eDNA | SAMN31207735 | SRR21875479 |
| 40DR_iDNA | SAMN31207736 | SRR21875478 |
| 40DR_eDNA | SAMN31207737 | SRR21875477 |
3.2 Quality control report of the high-throughput sequencing
The validity and reproducibility of the sequencing data were verified using rarefaction curves. As shown in Fig. S1,† each curve reached plateau, indicating that the number of reads obtained was sufficient to effectively represent the entire bacterial population.3.3 Microbial composition by high-throughput sequencing
Looking at the composition of the microbiota at a phylum level, whose percentages are reported in Table S1,†Proteobacteria was the phylum present with the highest percentage, with an average relative abundance of 47.11%, with a minimum percentage of 41.83% in the CPRi sample and a maximum percentage of 51.92% in the 20DRi sample, thus constituting alone almost half of the phyla present (Fig. 2). The others more abundant phyla (threshold above 1%) resulted in Gemmatimonadetes (9.82%), Bacteroidetes (9.60%), Chloroflexi (7.50%), Acidobacteria (6.46%), Actinobacteria (5.59%), Verrucomicrobia (5.02%), Firmicutes (4.05%) and Patescibacteria (1.35%).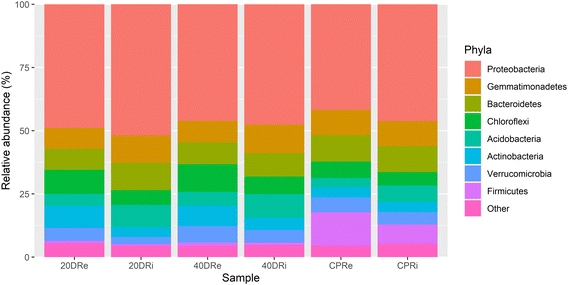 | ||
| Fig. 2 Phylum level report of the high-throughput sequencing. 16S rDNA sequences of bacterial rhizosphere communities (stacked bar plot). | ||
3.4 Prokaryotic community diversity
Analysing the mean species diversity induced by treatments in the soil (α-diversity), the Shannon–Wiener H index highlighted biodiversity differences among the samples. From the values reported in Table 2, albeit all H values were close to the average value of 5.053, CPR samples showed higher average biodiversity values than the nCeO2 treated samples (CP = 5119, 20DR = 5023, 40DR = 5035), however, this difference was not statistically significant (p > 0.05). Maximum H values (5228) were observed for CPR_iDNA, while lower values for Ce-treated samples, with a minimum value of 4.941 for sample 20DR_iDNA. eDNA samples showed a higher average biodiversity than those obtained from iDNA (iDNA= 5057, eDNA = 5061); however, also in this case the difference was not significant (p > 0.05).| Sample | Shannon index | Treatment | DNA origin |
|---|---|---|---|
| CPR_iDNA | 5.228 | CP | iDNA |
| CPR_eDNA | 5.009 | CP | eDNA |
| 20DR_iDNA | 4.941 | 20DR | iDNA |
| 20DR_eDNA | 5.105 | 20DR | eDNA |
| 40DR_iDNA | 5.001 | 40DR | iDNA |
| 40DR_eDNA | 5.069 | 40DR | eDNA |
Through PCoA, it was also possible to observe how the different iDNA and eDNA samples were arranged according to the treatment. In Fig. 3A, the Bray–Curtis dissimilarity, which considers both the number of species and the frequency with which they occur, showed that nCeO2 treated samples clearly separated from CPR control samples, and grouped according to the origin of the DNA (iDNA or eDNA), but independently from the nCeO2 dose (20DR and 40DR). The same results could be also observed from the Jaccard dissimilarity PCoA (Fig. 3B), which, unlike the Bray–Curtis index, does not consider the frequency with which different species occur. However, the percentages of variance explained by these PCoAs were minimal, indicating that although separated, the samples had very low diversity values.
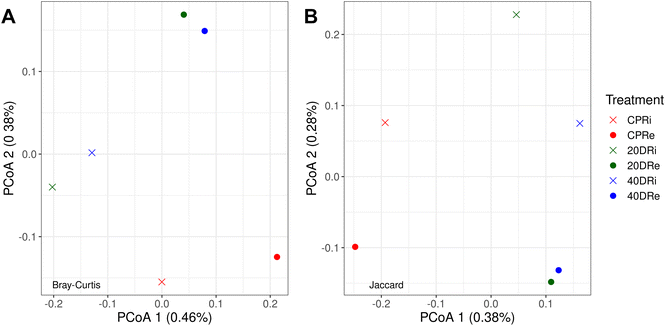 | ||
| Fig. 3 Bray–Curtis (A) and Jaccard (B) principal coordinates analysis (PCoA) arrangement of samples according to the treatments (CPR, 20DR, and 40DR) and DNA origin (iDNA or eDNA). | ||
Following a similar approach, PCA was performed on the bacterial distribution of the samples at the genus granularity level, retaining the first two components. As can be seen in Fig. 4, also in this case, the treated samples grouped according to the origin of the DNA, while the two control samples clearly separated from the treated ones as well as from each other. Overall, the explained variance of the two components was 87.39% (52.91% for the first component and 34.48% for the second component).
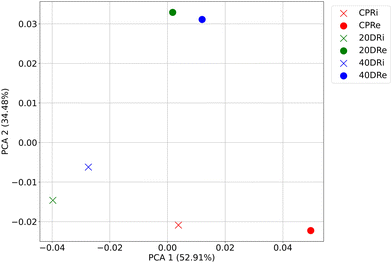 | ||
| Fig. 4 PCA (first two components) of bacterial distribution at the genus granularity level according to the treatments (CPR, 20DR, and 40DR) and DNA origin (iDNA or eDNA). | ||
3.5 Differences between detected ASVs
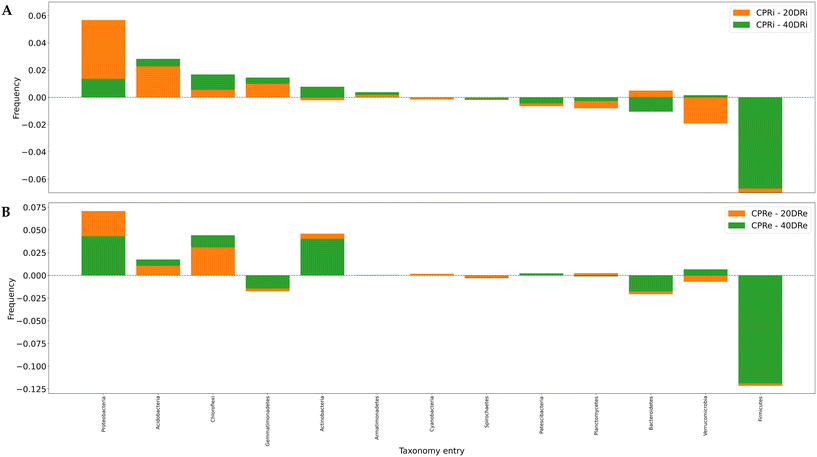
The differences induced on bacterial populations by the application of nCeO2 were then evaluated by comparing the increase or decrease in the relative percentages at phylum, family and genus level of the treated samples to the control.Of the 31 identified phyla, 13 were found to be present at >0.5% (iDNA Fig. 5A, eDNA Fig. 5B). One of the most significant differences observed was a consistent decrease of Firmicutes in substrates treated with nCeO2 at both dosages and both in iDNA and eDNA in comparison to the corresponding control samples. In fact, from an average relative percentage of 10.35% in CPR samples, mean percentages of 0.74% and 1.07% were observed in 20DR and 40DR samples. Another decrease in the percentage of the population was observed for Bacteroidetes, with an average percentage of 10.33% in CPR against an average percentage of 9.23% in 20DR and 40DR samples. Conversely, in nCeO2-treated samples, Proteobacteria increased from an average concentration of 44.04% in CPR samples to an average concentration of 48.64% in 20DR and 40DR, Acidobacteria moved from 5.14% to 7.12%, as well as Chloroflexi and Actinobacteria rose from 5.88% to 8.31% and 4.06% to 6.35%, respectively. As for the differences between iDNA and eDNA, a greater proportion of eDNA was found in Chloroflexi (6.03% vs. 8.96%), Actinobacteria (4.24% vs. 5.80%), Firmicutes (3.00% vs. 5.11%), Verrucomicrobia (4.24% vs. 5.80%), and Patescibacteria (1.12% vs. 1.58%). In summary, considering these 13 main phyla, in comparison to control samples, in the case of the 20DRi sample, 7 phyla decreased while 6 increased, while for 40DRi 6 decreased and 7 increased. For eDNA samples, in both 20DRe and 40DRe, compared to CPRe, 5 phyla decreased and 8 increased. Comparing the values of each phylum for the iDNA and eDNA of the controls, 6 couples behaved differently in samples 20DRi and 20Dre, whereas 7 couples performed equally. In samples 40DRi and 40DRe, 3 couples behaved differently and 10 had the same trend.
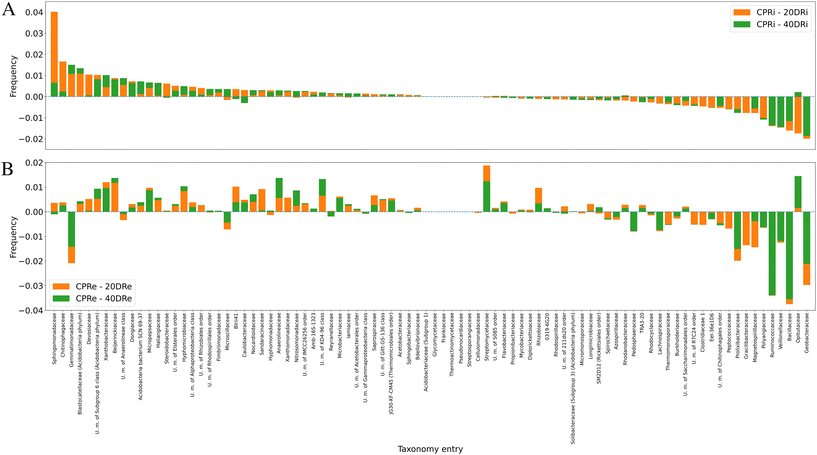
Moving to the family level (iDNA Fig. 6A, eDNA Fig. 6B), 88 out of 336 identified families were present at values >0.5% and considered for this analysis. These results highlighted that two nCeO2 dosages have similar effects to the triple dosage. Also, it was observed that comparing 20DRi to CPRi, 41 families decreased, 6 remained unchanged, and 41 increased, while comparing 40DRi, 36 families decreased, 7 remained unchanged, and 45 increased. Similarly, for eDNA samples, comparing 20DRe to CPRe, 31 families decreased, 6 remained unchanged, and 51 increased, whereas for 40DRe, 33 families decreased, 6 remained unchanged, and 49 increased. Furthermore, comparing 20DRi and 20DRe with respect to the corresponding controls, 23 couples behaved differently, and 65 couples showed the same behaviour, whereas in the case of the 40DRi and 40DRe samples, 21 couples behaved differently, and 67 couples behaved in the same way.
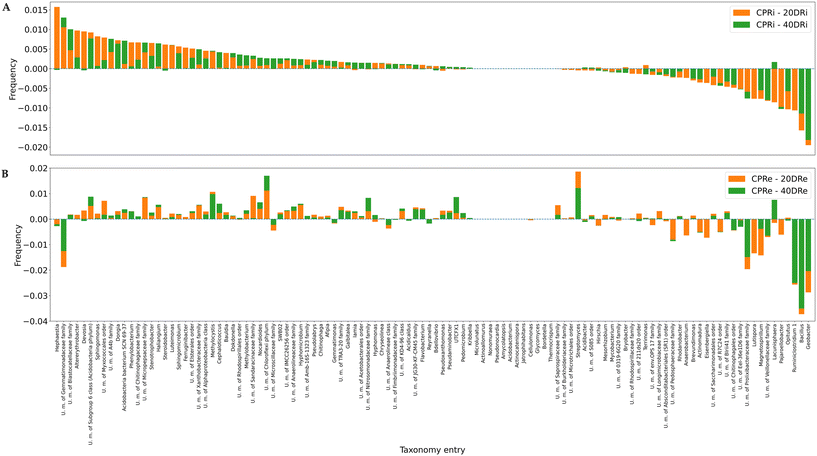
Fig. 7A and B depict the deviations compared with the control samples at the genus level for iDNA and eDNA, respectively. Out of 641 detected genera, 112, whose presence was >0.5%, were considered for the analysis. The comparison of the treatments with the controls revealed that in the case of 20DRi versus CPRi, 42 genera decreased, 13 did not change, and 57 increased, whereas in the case of 40DRi, 39 genera decreased, 12 remained unchanged, and 61 increased. Similarly, considering the eDNA, in 20DRe versus CPRe, 31 genera decreased, 12 remained unchanged, and 69 increased, whereas in 40DRe, 34 genera increased, 11 were stable, and 67 increased. Furthermore, comparing 20DRi versus 20DRe with reference to the corresponding controls, 31 couples behaved in the opposite way, and 81 couples reported the same trend. For 40DRi versus 40DRe, 23 couples behaved in the opposite way, while 89 couples had the same behaviour.
3.6 Ranking analysis: Spearman correlation
The ranking of various bacterial groups in the different samples was considered. To such an extent, the Spearman correlation was calculated pairwise between the samples at different granularity levels to evaluate the degree of concordance (between −1 and 1) of the ranks (Table 3).| Granularity | Samples | Corr | p-Value | Granularity | Samples | Corr | p-Value |
|---|---|---|---|---|---|---|---|
| Phylum | CPRi/20DRi | 0.86 | 1.50 × 10−4 | Family | CPRe/20DRe | 0.6 | 6.30 × 10−10 |
| Phylum | CPRi/40DRi | 0.85 | 2.20 | Family | CPRe/40DRe | 0.7 | 3.90 × 10−14 |
| Phylum | 20DRi/40DRi | 0.99 | 1.70 × 10−10 | Family | 20DRe/40DRe | 0.93 | 2.40 × 10−38 |
| Phylum | CPRe/20DRe | 0.73 | 5.00 × 10−3 | Genus | CPRi/20DRi | 0.72 | 7.50 × 10−19 |
| Phylum | CPRe/40DRe | 0.82 | 6.20 × 10−4 | Genus | CPRi/40DRi | 0.75 | 4.00 × 10−21 |
| Phylum | 20DRe/40DRe | 0.97 | 2.60 × 10−8 | Genus | 20DRi/40DRi | 0.93 | 1.00 × 10−49 |
| Family | CPRi/20DRi | 0.75 | 2.50 × 10−17 | Genus | CPRe/20DRe | 0.6 | 4.20 × 10−12 |
| Family | CPRi/40DRi | 0.77 | 1.40 × 10−18 | Genus | CPRe/40DRe | 0.7 | 1.50 × 10−17 |
| Family | 20DRi/40DRi | 0.95 | 4.10 × 10−45 | Genus | 20DRe/40DRe | 0.88 | 2.70 × 10−38 |
Generally, among the samples analysed using the same technique and treated with different quantities of nCeO2, the concordance between rankings was sufficiently high (higher than 0.92, except for one case with 0.88). In contrast, the agreement was significantly lower among the samples treated with nCeO2 and the related control samples (always with the same analysis). This confirmed the qualitative results previously provided by the PCoA and PCA. Moreover, from Table 3, it can be observed that for the same pairs of samples, passing from the phylum to families and genera levels, the 20DRi/40DRi correlation was more homogeneous and similar, with values between 0.93 and 0.99. At the same time, the CPRe/20DRe showed the greatest diversity with values between 0.60 and 0.73.
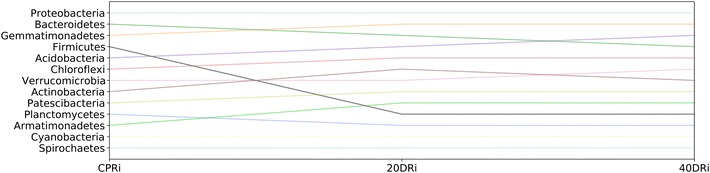
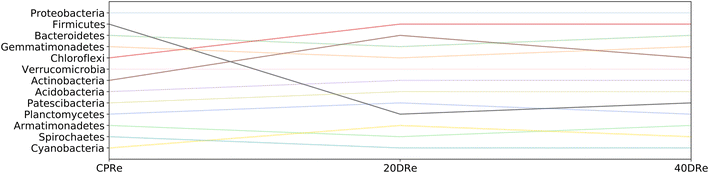
Fig. 8 and 9 highlight how the rankings of the different bacteria changed among the samples, reported the position in the rank for each bacterial group at the phylum granularity level, considering iDNA and eDNA. In both cases, in the treated samples a marked reduction in the presence of Firmicutes with respect to the untreated samples was observed. In contrast, there was an increase in both Chloroflexi and Actinobacteria (more pronounced in the iDNA samples than in the eDNA cases). Acidobacteria and Patescibacteria showed similar increases in both cases. Interestingly, Armatimonadetes decreased when eDNA was considered and increased when iDNA was considered.
3.7 Metabolisms
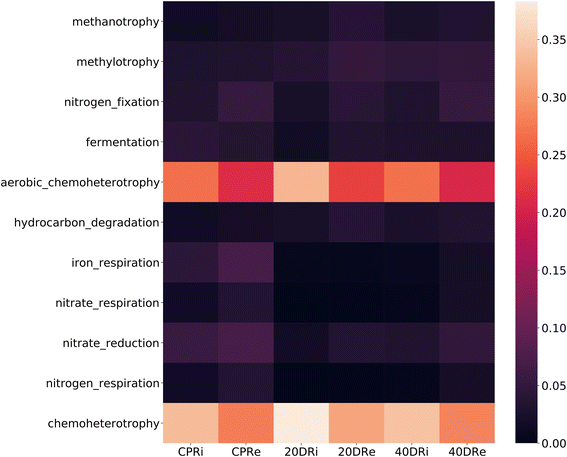
The analysis of the metabolic functions, reconstructed using the FAPROTAX database, revealed differences induced by nCeO2 to the reconstructed metabolic functions. As observed from the heatmap shown in Fig. 10, metabolic functions related to fermentation, nitrogen fixation, and iron, nitrate, and nitrogen respiration decreased in the samples treated with cerium (20DR and 40DR) compared to those in the control samples (CPR), in association with an increase in functions related to chemo-heterotrophy.4 Discussion
The aim of this study was to deepen the knowledge of the influence of CeNPs on the soil microbiota, and more specifically on the rhizosphere microbiota during the vegetative growth. For this purpose, iDNA and eDNA obtained from the rhizospheric soil of Silene flos-cuculi amended with different nCeO2 concentrations in a controlled environment were considered. To date, studies have focused mainly on the relationships among soil, plant and CeNPs, but not on the relationship between CeNPs and microbiota.48,49 Soil microbial communities are the poorly ununderstood driver of many processes that directly or indirectly govern the fluxes of carbon, nitrogen, and other elements through ecosystems.47,50–52 In particular, rhizospheric microbiota and the symbiotic processes triggered by plant growth-promoting rhizobacteria (PGPR) play a fundamental role in improving productivity by reducing fertilization and pesticide utilization.53In this regard, nCeO2 can impact this mutualistic relationship in different ways, both directly and indirectly. Due to their physicochemical characteristics, CeNPs are translocated in plants through the root system to leaves, determining a specific plant modulable adaptation response that also influences the microbiota.29 Furthermore, the retention or release of oxygen from the nCeO2 structure8,9,54 can modify the biogeochemical conditions, inhibiting gas flow across the sediment/water interface profile55–57 and therefore modifying the oxygen gradient, thus directly reducing the aerobic and facultative bacterial microflora.58Several studies have also proved the direct antimicrobial effect of CeNPs, and their possible application for medical use59,60 against both Gram-positive and Gram-negative bacteria.61,62 At low pH values, nanomaterials (positively charged) adhere to microorganisms (negatively charged), acquiring an electron, which converts Ce4+ to Ce3+, deranging the cell wall structure. Gram negatives and fungi are more susceptible to this effect, while Gram positives, thanks to a thick peptidoglycan layer, can modulate this effect.9,16,62,63
Therefore, biological spheres are subject to complex interactions related to the dosage and physical properties of nanoparticles present in the environment.8,10,64 Lizzi et al. (2020)27 demonstrated that CeNPs with a nominal dimension of 25 nm and 99.95% purity showed an average dimension of 126.7 ± 1.0 nm but CeNPs translocated to roots and leaves were respectively 33 ± 2 nm and 31 ± 1.5 nm. This underlined the greater catalytic action of the smaller particles,65 and demonstrated that the reduction of CeNPs to Ce3+ is necessary to facilitate the absorption of cerium at the root level.21,66
Given the complexity of these biotic and environmental factors, to evaluate the differences induced in this study, a NGS approach was chosen for its capable of evaluating the composition of the entire population with a high resolution degree and also allowing detection of minimal differences. In this regard, the evaluation of alpha diversity analysis results underlined a lower Shannon–Viener H index value in nCeO2 treated samples. The same evidence of an effect on microbial populations emerged from the results of PCoA. In fact, both in Bray–Curtis and Jaccard dissimilarity matrices, the treated samples clustered separately from the control samples. However, these differences, although present, were minimal, indicating a limited effect of the treatments on the differences in the biodiversity level of the samples. It should also be underlined that the differences induced by the presence of nCeO2 were poorly correlated with its concentration, as suggested by the similarity of the results obtained between samples 20DR and 40DR. From these results we can therefore observe a non-significant effect on the overall biodiversity level induced by the amendment with nCeO2. Although there was an effect on the proportion of some species present, no impoverishment of the soil was therefore observed at a biological level. In contrast, it was possible to observe a clear separation between the iDNA and eDNA samples, testifying to the importance of distinguishing these two components of the total DNA to obtain an effective analysis, capable of distinguishing the still vital microbial population from the paleome. Subsequently, focusing the analysis on the present phyla, a drastic decrease in Firmicutes was observed. This difference has certainly induced significant effects on a biological level, in fact Firmicutes are a phylum of bacteria that exerts important metabolic functions in the soil, contributing to terrestrial ecosystems in different ways. Several of these bacteria are involved in the decomposition of organic matter and nitrogen fixation67,68 producing beneficial chemicals and competing with plant pathogens. Therefore their disruption and imbalance in soil could result in the emergence of several plant diseases.69
These results were in contrast with other studies, in which this difference was not reported70,71 or even reported an increase of this phylum.72 This evidence, however, was obtained under completely different conditions from those applied in this study. For instance, some experiments were conducted on activated sludge, in a liquid environment maintained under aerobic conditions, and the increase in Firmicutes was observed due to an increment of Bacilli. In contrast, in this study the reduction of Firmicutes was attributed to a decrease in Clostridia, reflecting disparate environmental and microbial dynamics.
Going into further detail to the genus level, of the 112 genera identified at a percentage >0.5% (out of 641 total), 25 of these were present in the controls but totally absent in the 20D and 40D treated samples (for both iDNA and eDNA) (Table S2†). Of these, 10 were Gram-negative, 12 were Gram-positive, while Lutispora, Eisenbergiella, and Anaerovorax, despite the resulting Gram-negative bacteria from Gram staining possessing a cell wall typical of Gram-positive bacteria. In light of these results, it was therefore not possible to associate the different antimicrobial effects of nCeO2 based on the different cell wall structure as reported in the literature.
On the other hand, considering the possible differences induced by the modification of oxygen exchange due to the presence of nCeO2, the oxygen demand of the 25 genera was analyzed: 4 were aerobic, 1 facultatively anaerobic and the remaining 19 genera corresponded to anaerobic and strictly anaerobic bacteria. Currently, few studies focus on the redox cycle between the Ce3+ and Ce4+ states on the surface of nCeO2 in the proximity of the rhizosphere, and its effects on oxygen uptake/release,73 and further research is required to understand the modifications of the aerobic/facultative/anaerobic microorganism ratio from the nCeO2 induced ROS.
Focusing on the effects of nCeO2 on PGPR, important bacteria for soil and plant health,53,74–82 the majority of which with their relative functions are summarized in Table S3,† it was possible to identify the presence of 13 PGPR genera (Allorhizobium, Azospirillum, Pseudomonas, Bacillus, Bradyrhizobium, Chryseobacterium, Enterobacter, Mesorhizobium, Paenibacillus, Pantoea, Pseudomonas, Rhizobium, and Stenotrophomonas). From the comparison of their presence in the different samples (Table S4†), it was possible to observe lower percentages in iDNA samples compared to eDNA. Additionaly, it was possible to observe an increase in 5 genera and a decrease in 8 genera in nCeO2 treated samples for iDNA.
Overall, these induced differences in the PGPR population could be directly responsible for the differences in plant growth, and given the discordant results between different studies, could indicate a specific effect between certain PGPRs and different plant species. Other studies report effects on plant growth, demonstrating both beneficial and detrimental outcomes depending on the doses employed. At high concentrations of nCeO2, an increase in Azotobacter, Clostridium, Rhodospirillaceae, and Ensifer was observed in the soybean-cultivated soil, while the populations of Sphingomonas and Rhizobium significantly decreased. Furthermore, the impact of nCeO2 was notable only in the soil cultivated with soybean and not to the unplanted soil, where bacterial communities remained unaffected.83 However, with the currently available knowledge, which often consists of experiments conducted on a laboratory scale, with results currently at a preliminary level, which are often contradictory, it is not yet possible to associate a specific effect on plant and soil health with the variation of each individual microbial group given the complexity of the interactions, and the functional redundancy in soil microbial community.84,85 The importance of further in-depth studies on nanomaterials to comprehend their actual effects on the environment, soil, and plants has also been emphasized by the Organization for Economic Co-operation and Development (OECD).86
To analyse the impact on all metabolic functions induced by diversity changes, the FAPROTAX database was used. Through this useful tool, it is in fact possible to investigate and assign in a more in-depth and effective way the ecological functions of soil bacteria.87 This analysis showed that nCeO2 treatment in 20DR and 40DR samples decreased fermentation, nitrogen fixation, iron, nitrate, and the nitrogen respiration function compared to the controls. The predominant function in all samples was chemo-heterotrophy, followed by aerobic chemo-heterotrophy, that was found to be comparable among CPRi and CPRe and 40DRi and 40DRe treated samples. Instead, in the 20DRi sample, both functionalities increased compared to those in 20DRe and the respective controls. This positive selection of aerobic heterotrophs was in accordance with the previously discussed reduction of anaerobic and strictly anaerobic genera in treated samples. Previous studies also reported that exposure to nCeO2 leads to the degradation of the denitrifying process. This observation aligns with the inhibition of enzyme activities associated with nitrite oxidoreductase and nitrate reductase. Concurrently, a reduction in microbial diversity, a decrease in the network complexity of the bacterial community, and enhanced competitive dynamics among bacterial populations were observed. Specifically, there was a notable decrease in Flexibacter and Acinetobacter.88 However, different results were observed for the sediment/water interface.58 In this scenario, with the presence of halogens in the rhizosphere (e.g., micromolar concentrations of bromides, chlorides, or reactive oxygen species), CeNPs may have exhibited a mimetic action similar to peroxidase enzymes. This action involves the production of hypo-halogen acids (HOCl and HOBr), which interfere with quorum sensing molecules, thereby influencing the metabolism of both Gram-positive and Gram-negative bacteria.89 Under these conditions, CeNPs present in soils with low chloride concentrations and reduced water content can therefore interact with the rhizosphere, inhibiting microbial quorum sensing molecules and altering the density of bacterial populations, the production of extracellular enzymes, and nitrogen and phosphorus cycles. The actions of mycorrhizae and PGPR can mitigate these effects by intervening in the ROS compounds produced in the root system, thereby reducing the mimetic and catalytic peroxidase-like activity of CeNPs.90–96 Considering climate change, these conditions characterized by reduced water and humidity content in soils are expected to become increasingly prevalent. In such scenarios, in addition to the abovementioned effects, different studies reported a direct beneficial effect of nCeO2. Under water stress conditions, nCeO2 affected the activity of Rubisco, thereby enhancing photosynthetic activity, mitigating oxidative stress induced by drought-related ROS, and consequently improving plant growth under these challenging conditions.97,98 However, in addition to potential applications in agronomy, the direct effects on humans resulting from the consumption of products grown in the presence of nCeO2 must be thoroughly assessed. From the initial studies conducted, these nanoparticles appear to influence the gut microbiota, potentially having negative implications and thereby compromising their use as agronomical adjuvants.99–101
5 Conclusions
This study stems from a previous study whose results showed that nCeO2 may have caused a slowdown in the vegetative growth rate of S. flos-cuculi (L.). The possible causes of this phenomenon were therefore sought in a modification induced by this compound in the rhizosphere microbiota.The most abundant bacterial phylum found in the rhizosphere of the S. flos-cuculi substrate was Proteobacteria followed by Gemmatimonadetes, Bacteroidetes, Chloroflexi, Acidobacteria, Actinobacteria, and Verrucomicrobia. In contrast, Firmicutes which constituted 7.58% and 13.12% in the CPRi and CPRe untreated samples were greatly reduced in soil treated with nCeO2 amendments, reaching thresholds lower than 1% in nCeO2 treated soils.
By extending the comparison also to the phyla present in a lower percentage, the inhibition of some anaerobic and strictly anaerobic genera was observed.
Considering the families affected by the same effects both in iDNA and eDNA samples, approximately 50% of them showed inhibition compared with the control, particularly for Prolixibacteraceae, Bacillaceae, Peptococcaceae, Ruminococcaceae, Veillonellaceae, Geobacteraceae, Thermomonosporaceae, Clostridiaceae, Azospirillaceae, Magnetospirillaceae, Gracilibacteraceae, Lachnospiraceae, Polyangiaceae and Burkholderiaceae.
From these results, and in consideration of the existing studies, nCeO2 particles have an influence on soil microbiota, which impacts plant health. However, the definition of a general trend of impact is not possible, due to so many variables that can affect the complex soil–root-microbiota system and the few studies present in the literature on the topic. For this reason, we strongly encourage investigations on the topic to improve the knowledge on the effects on different cultivations, which is extremely important in the case of crop rotations given the nCeO2 soil accumulation, but also in consideration of better understanding the plant translocation mechanisms and its build-up in plant edible parts that could affect human and animal health.
Data availability
The data are available, and the hyperlink to the published data is present in the manuscript.Author contributions
MC, LI, LM: conceptualization; MC, LI: resources; AB, MC, AC, LI, NS: methodology; MC, LI: supervision; AB, AC, AF, NS: formal analysis; AB, AC, NS: data curation; AB, AC, NS: visualization; AB, MC, AC, NS: writing – original draft; AC, LI: writing – draft revision; MC, AC, LI: writing – review and editing; MC, LI, LM: funding acquisition.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors are deeply grateful to all those who played a role in the success of this project. In particular, we would like to thank Daniel Lizzi who contributed to manage the plant growth experiment, donating the material for the experiments.References
- W. S. Bainbridge and M. C. Roco, Science and technology convergence: with emphasis for nanotechnology-inspired convergence, J. Nanopart. Res., 2016, 18(7), 1–19, DOI:10.1007/s11051-016-3520-0.
- M. Mortimer and P. A. Holden, Chapter 3 - Fate of engineered nanomaterials in natural environments and impacts on ecosystems, in Exposure to Engineered Nanomaterials in the Environment, Elsevier, 2019, pp. 61–103, DOI:10.1016/B978-0-12-814835-8.00003-0.
- P. Yu, S. A. Hayes, T. J. O'Keefe, M. J. O'Keefe and J. O. Stoffer, The Phase Stability of Cerium Species in Aqueous Systems, J. Electrochem. Soc., 2006, 153(1), C74, DOI:10.1149/1.2130572.
- P. J. Denissen, P. J. Denissen, V. Shkirskiy, P. Volovitch and S. J. Garcia, Corrosion Inhibition at Scribed Locations in Coated AA2024-T3 by Cerium- And DMTD-Loaded Natural Silica Microparticles under Continuous Immersion and Wet/Dry Cyclic Exposure, ACS Appl. Mater. Interfaces, 2020, 12(20), 23417–23431, DOI:10.1021/acsami.0c03368.
- C. Sun, H. Li and L. Chen, Nanostructured ceria-based materials: Synthesis, properties, and applications, Energy Environ. Sci., 2012, 5(9), 8475–8505, 10.1039/C2EE22310D.
- A. Vita, Catalytic Applications of CeO2-Based Materials, Catalysts, 2020, 5–8, DOI:10.3390/catal10050576.
- M. A. Jakupec, P. Unfried and B. K. Keppler, Pharmacological properties of cerium compounds, Rev. Physiol., Biochem. Pharmacol., 2005, 153, 101–111, DOI:10.1007/s10254-004-0024-6.
- K. Reed, A. Cormack, A. Kulkarni, M. Mayton, D. Sayle and F. Klaessig, et al., Exploring the properties and applications of nanoceria: Is there still plenty of room at the bottom?, Environ. Sci.: Nano, 2014, 1(5), 390–405, 10.1039/C4EN00079J.
- I. A. P. Farias, C. C. L. Dos Santos and F. C. Sampaio, Antimicrobial activity of cerium oxide nanoparticles on opportunistic microorganisms: A systematic review, BioMed Res. Int., 2018, 1923606, DOI:10.1155/2018/1923606.
- E. Grulke, K. Reed, M. Beck, X. Huang, A. Cormack and S. Seal, Nanoceria: Factors affecting its pro- and anti-oxidant properties, Environ. Sci.: Nano, 2014, 1(5), 429–444, 10.1039/C4EN00105B.
- L. Chen, P. Fleming, V. Morris, J. D. Holmes and M. A. Morris, Size-related lattice parameter changes and surface defects in ceria nanocrystals, J. Phys. Chem. C, 2010, 114(30), 12909–12919, DOI:10.1021/jp1031465.
- F. Esch, S. Fabris, L. Zhou, T. Montini, C. Africh and P. Fornasiero, et al., Chemistry: Electron localization determines defect formation on ceria substrates, Science, 2005, 309(5735), 752–755, DOI:10.1126/science.1111568.
- OECD, Assessment of Biodurability of Nanomaterials and their Surface ligands Series on the Safety of Manufactured Nanomaterials, Oecd 86, 2018, (86), available from: https://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=env/jm/mono(2018)11&doclanguage=en Search PubMed.
- R. A. Yokel, S. Hussain, S. Garantziotis, P. Demokritou, V. Castranova and F. R. Cassee, The yin: An adverse health perspective of nanoceria: Uptake, distribution, accumulation, and mechanisms of its toxicity, Environ. Sci.: Nano, 2014, 1(5), 406–428, 10.1039/C4EN00039K.
- L. Geraets, A. G. Oomen, J. D. Schroeter, V. A. Coleman and F. R. Cassee, Tissue distribution of inhaled micro- and nano-sized cerium oxide particles in rats: Results from a 28-day exposure study, Toxicol. Sci., 2012, 127(2), 463–473, DOI:10.1093/toxsci/kfs113.
- C. Guo, S. Robertson, R. J. M. Weber, A. Buckley, J. Warren and A. Hodgson, et al., Pulmonary toxicity of inhaled nano-sized cerium oxide aerosols in Sprague–Dawley rats, Nanotoxicology, 2019, 13(6), 733–750, DOI:10.1080/17435390.2018.1554751.
- E. J. Park, Y. K. Park and K. Park, Acute toxicity and tissue distribution of cerium oxide nanoparticles by a single oral administration in rats, Toxicol. Res., 2009, 25(2), 79–84, DOI:10.5487/TR.2009.25.2.079.
- T. Xia, M. Kovochich, M. Liong, L. Mädler, B. Gilbert and H. Shi, et al., Comparison of the mechanism of toxicity of zinc oxide and cerium oxide nanoparticles based on dissolution and oxidative stress properties, ACS Nano, 2008, 2(10), 2121–2134, DOI:10.1021/nn800511k.
- X. Pang, D. Li and A. Peng, Application of rare-earth elements in the agriculture of china and its environmental behavior in soil, J. Soils Sediments, 2001, 1(2), 124–129, DOI:10.1007/BF02987462.
- W. Zhou, G. Han, M. Liu, C. Song and X. Li, Geochemical distribution characteristics of rare earth elements in different soil profiles in Mun River Basin, Northeast Thailand, Sustainability, 2020, 12(2), 457, DOI:10.3390/su12020457.
- C. M. Rico, M. G. Johnson and M. A. Marcus, Cerium oxide nanoparticles transformation at the root-soil interface of barley (Hordeum vulgare L.), Environ. Sci.: Nano, 2018, 5(8), 1807–1812, 10.1039/C8EN00316E.
- P. Miralles, T. L. Church and A. T. Harris, Toxicity, uptake, and translocation of engineered nanomaterials in vascular plants, Environ. Sci. Technol., 2012, 46(17), 9224–9239, DOI:10.1021/es202995d.
- N. Zuverza-Mena, D. Martínez-Fernández, W. Du, J. A. Hernandez-Viezcas, N. Bonilla-Bird and M. L. López-Moreno, et al., Exposure of engineered nanomaterials to plants: Insights into the physiological and biochemical responses-A review, Plant Physiol. Biochem., 2017, 110, 236–264, DOI:10.1016/j.plaphy.2016.05.037.
- G. V. Lowry, A. Avellan and L. M. Gilbertson, Opportunities and challenges for nanotechnology in the agri-tech revolution, Nat. Nanotechnol., 2019, 14(6), 517–522, DOI:10.1038/s41565-019-0461-7.
- M. Kah, N. Tufenkji and J. C. White, Nano-enabled strategies to enhance crop nutrition and protection, Nat. Nanotechnol., 2019, 14(6), 532–540, DOI:10.1038/s41565-019-0439-5.
- L. Marchiol, M. Iafisco, G. Fellet and A. Adamiano, Nanotechnology support the next agricultural revolution: Perspectives to enhancement of nutrient use efficiency, Adv. Agron., 2020, 1(161), 27–116, DOI:10.1016/bs.agron.2019.12.001.
- D. Lizzi, A. Mattiello, B. Piani, G. Fellet, A. Adamiano and L. Marchiol, Germination and early development of three spontaneous plant species exposed to nanoceria (nCeO2) with different concentrations and particle sizes, Nanomaterials, 2020, 10(12), 1–16, DOI:10.3390/nano10122534.
- Q. Wang, S. D. Ebbs, Y. Chen and X. Ma, Trans-generational impact of cerium oxide nanoparticles on tomato plants, Metallomics, 2013, 5(6), 753–759, 10.1039/C3MT00033H.
- V. Prakash, J. Peralta-Videa, D. K. Tripathi, X. Ma and S. Sharma, Recent insights into the impact, fate and transport of cerium oxide nanoparticles in the plant-soil continuum, Ecotoxicol. Environ. Saf., 2021, 221, 112403, DOI:10.1016/j.ecoenv.2021.112403.
- G. Tyler, Rare earth elements in soil and plant systems - A review, Plant Soil, 2004, 267(1–2), 191–206, DOI:10.1016/j.ecoenv.2021.112403.
- E. S. C. Emmanuel, V. Vignesh, B. Anandkumar and S. Maruthamuthu, Bioaccumulation of cerium and neodymium by Bacillus cereus isolated from rare earth environments of Chavara and Manavalakurichi, India, Indian J. Microbiol., 2011, 51(4), 488–495, DOI:10.1007/s12088-011-0111-8.
- A. Colautti, M. Civilini, M. Contin, E. Celotti and L. Iacumin, Organic vs. conventional: impact of cultivation treatments on the soil microbiota in the vineyard, Front. Microbiol., 2023, 14, 1242267, DOI:10.3389/fmicb.2023.1242267.
- L. J. Gómez-Godínez, J. L. Aguirre-Noyola, E. Martínez-Romero, R. I. Arteaga-Garibay, J. Ireta-Moreno and J. M. Ruvalcaba-Gómez, A Look at Plant-Growth-Promoting Bacteria, Plants, 2023, 12(8), 1668, DOI:10.3390/plants12081668.
- D. Lizzi, A. Mattiello, B. Piani, E. Gava, G. Fellet and L. Marchiol, Single and repeated applications of cerium oxide nanoparticles differently affect the growth and biomass accumulation of silene flos-cuculi l (caryophyllaceae), Nanomaterials, 2021, 11(1), 1–15, DOI:10.3390/nano11010229.
- A. D. Servin, A. D. Servin and J. C. White, Nanotechnology in agriculture: Next steps for understanding engineered nanoparticle exposure and risk, NanoImpact, 2016, 1, 9–12, DOI:10.1016/j.impact.2015.12.002.
- J. Ascher, M. T. Ceccherini, O. L. Pantani, A. Agnelli, F. Borgogni and G. Guerri, et al., Sequential extraction and genetic fingerprinting of a forest soil metagenome, Appl. Soil Ecol., 2009, 42(2), 176–181, DOI:10.1016/j.apsoil.2009.03.005.
- C. Milani, A. Hevia, E. Foroni, S. Duranti, F. Turroni and G. A. Lugli, et al., Assessing the Fecal Microbiota: An Optimized Ion Torrent 16S rRNA Gene-Based Analysis Protocol, PLoS One, 2013, 8(7), e68739, DOI:10.1371/journal.pone.0068739.
- J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman and E. K. Costello, et al., QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 2010, 7(5), 335–336, DOI:10.1038/nmeth.f.303.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13(7), 581–583, DOI:10.1038/nmeth.3869.
- I. Amir, P. Bouvet, C. Legeay, U. Gophna and A. Weinberger, Eisenbergiella tayi gen. nov., sp. nov., isolated from human blood, Int. J. Syst. Evol. Microbiol., 2014, 64(3), 907–914, DOI:10.1099/ijs.0.057331-0.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer and P. Yarza, et al., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucleic Acids Res., 2013, 41(D1), 590–596, DOI:10.1093/nar/gks1219.
- P. J. McMurdie and S. Holmes, Phyloseq: A bioconductor package for handling and analysis of high-throughput phylogenetic sequence data, Pacific Symp. Biocomput., 2012, 235–246, DOI:10.1142/9789814366496_0023.
- S. Louca, L. W. Parfrey and M. Doebeli, Decoupling function and taxonomy in the global ocean microbiome, Science, 2016, 353(6305), 1272–1277, DOI:10.1126/science.aaf4507.
- M. Nagler, S. M. Podmirseg, M. Mayr, J. Ascher-Jenull and H. Insam, Quantities of Intra- and Extracellular DNA Reveal Information About Activity and Physiological State of Methanogenic Archaea, Front. Microbiol., 2020, 11, 1894, DOI:10.3389/fmicb.2020.01894.
- F. Inagaki, H. Okada, A. I. Tsapin and K. H. Nealson, MICROBIAL SURVIVAL: The Paleome: A Sedimentary Genetic Record of Past Microbial Communities, Astrobiology, 2005, 5(2), 141–153, DOI:10.1089/ast.2005.5.141.
- M. Alawi, B. Schneider and J. Kallmeyer, A procedure for separate recovery of extra- and intracellular DNA from a single marine sediment sample, J. Microbiol. Methods, 2014, 104, 36–42, DOI:10.1016/j.mimet.2014.06.009.
- M. Nagler, H. Insam, G. Pietramellara and J. Ascher-Jenull, Extracellular DNA in natural environments: features, relevance and applications, Appl. Microbiol. Biotechnol., 2018, 102(15), 6343–6356, DOI:10.1007/s00253-018-9120-4.
- J. Hong, L. Wang, Y. Sun, L. Zhao, G. Niu and W. Tan, et al., Foliar applied nanoscale and microscale CeO2 and CuO alter cucumber (Cucumis sativus) fruit quality, Sci. Total Environ., 2016, 563–564, 904–911, DOI:10.1016/j.scitotenv.2015.08.029.
- A. C. Barrios, C. M. Rico, J. Trujillo-Reyes, I. A. Medina-Velo, J. R. Peralta-Videa and J. L. Gardea-Torresdey, Effects of uncoated and citric acid coated cerium oxide nanoparticles, bulk cerium oxide, cerium acetate, and citric acid on tomato plants, Sci. Total Environ., 2016, 563–564, 956–964, DOI:10.1016/j.scitotenv.2015.11.143.
- E. B. Graham, J. E. Knelman, A. Schindlbacher, S. Siciliano, M. Breulmann, A. Yannarell, J. M. Beman, G. Abell, L. Philippot, J. Prosser, A. Foulquier, J. C. Yuste, H. C. Glanville, D. L. Jones, R. Angel, J. Salminen, R. J. Newton, H. Bürgmann, L. J. Ingram, U. Hamer, H. M. P. Siljanen, K. Peltoniemi, K. Potthast, L. Bañeras, M. Hartmann, S. Banerjee, R.-Q. Yu, G. Nogaro, A. Richter, M. Koranda, S. C. Castle, M. Goberna, B. Song, A. Chatterjee, O. C. Nunes, A. R. Lopes, Y. Cao, A. Kaisermann, S. Hallin, M. S. Strickland, J. Garcia-Pausas, J. Barba, H. Kang, K. Isobe, S. Papaspyrou, R. Pastorelli, A. Lagomarsino, E. S. Lindström, N. Basiliko and D. R. Nemergut, Microbes as engines of ecosystem function: When does community structure enhance predictions of ecosystem processes?, Front. Microbiol., 2016, 7, 214, DOI:10.3389/fmicb.2016.00214.
- B. Waring, A. Gee, G. Liang and S. Adkins, A quantitative analysis of microbial community structure-function relationships in plant litter decay, iScience, 2022, 25(7), 104523, DOI:10.1016/j.isci.2022.104523.
- A. Colautti, F. Golinelli, L. Iacumin, D. Tomasi, P. Cantone and G. Mian, Triacontanol (long-chain alcohol) positively enhances the microbial ecology of berry peel in Vitis vinifera cv. ‘Glera’ yet promotes the must total soluble sugars content, OENO One, 2023, 57(2), 477–488, DOI:10.20870/oeno-one.2023.57.2.7507.
- D. Bhardwaj, M. W. Ansari, R. K. Sahoo and N. Tuteja, Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity, Microb. Cell Fact., 2014, 13(1), 1–10, DOI:10.1186/1475-2859-13-66.
- S. Deshpande, S. Patil, S. V. Kuchibhatla and S. Seal, Size dependency variation in lattice parameter and valency states in nanocrystalline cerium oxide, Appl. Phys. Lett., 2005, 87(13), 1–3, DOI:10.1063/1.2061873.
- S. Assouline and K. Narkis, Effect of Long-Term Irrigation with Treated Wastewater on the Root Zone Environment, Vadose Zone J., 2013, 128(2), 4–10, DOI:10.2136/vzj2012.0216.
- J. Pineda Pineda, M. de J. Roblero Moreno, M. T. Colinas León and J. Sahagún Castellanos, El oxígeno en la zona radical y su efecto en las plantas, Rev. Mex. De Cienc. Agric., 2020, 11(4), 931–943, DOI:10.29312/remexca.v11i4.2128.
- F. J. Cook and J. H. Knight, Oxygen Transport to Plant Roots, Soil Sci. Soc. Am. J., 2003, 67(1), 20–31, DOI:10.2136/sssaj2003.2000.
- L. Miao, P. Wang, C. Wang, J. Hou, Y. Yao and J. Liu, et al., Effect of TiO2 and CeO2 nanoparticles on the metabolic activity of surficial sediment microbial communities based on oxygen microelectrodes and high-throughput sequencing, Water Res., 2018, 129, 287–296, DOI:10.1016/j.watres.2017.11.014.
- C. Kaittanis, S. Santra, A. Asati and J. M. Perez, A cerium oxide nanoparticle-based device for the detection of chronic inflammation via optical and magnetic resonance imaging, Nanoscale, 2012, 4(6), 2117–2123, 10.1039/C2NR11956K.
- M. P. Nikolova and M. S. Chavali, Metal oxide nanoparticles as biomedical materials, Biomimetics, 2020, 5(2), 27, DOI:10.3390/biomimetics5020027.
- O. L. Pop, A. Mesaros, D. C. Vodnar, R. Suharoschi, F. Tăbăran and L. Mageruşan, et al., Cerium oxide nanoparticles and their efficient antibacterial application in vitro against gram-positive and gram-negative pathogens, Nanomaterials, 2020, 10(8), 1–15, DOI:10.3390/nano10081614.
- E. Alpaslan, B. M. Geilich, H. Yazici and T. J. Webster, pH-Controlled Cerium Oxide Nanoparticle Inhibition of Both Gram-Positive and Gram-Negative Bacteria Growth, Sci. Rep., 2017, 7, 1–12, DOI:10.1038/srep45859.
- E. Barker, J. Shepherd and I. O. Asencio, The Use of Cerium Compounds as Antimicrobials for Biomedical Applications, Molecules, 2022, 27(9), 2678, DOI:10.3390/molecules27092678.
- G. Pulido-Reyes, I. Rodea-Palomares, S. Das, T. S. Sakthivel, F. Leganes and R. Rosal, et al., Untangling the biological effects of cerium oxide nanoparticles: The role of surface valence states, Sci. Rep., 2015, 5, 1–14, DOI:10.1038/srep15613.
- M. R. Khan, V. Adam, T. F. Rizvi, B. Zhang, F. Ahamad and I. Jośko, et al., Nanoparticle–Plant Interactions: Two-Way Traffic, Small, 2019, 15(37), 1–20, DOI:10.1002/smll.201901794.
- C. M. Rico, M. G. Johnson, M. A. Marcus and C. P. Andersen, Intergenerational responses of wheat (Triticum aestivum L.) to cerium oxide nanoparticles exposure, Environ. Sci.: Nano, 2017, 4(3), 700–711, 10.1039/C7EN00057J.
- M. Ren, Z. Zhang, X. Wang, Z. Zhou, D. Chen, H. Zeng, S. Zhao, L. Chen, Y. Hu, C. Zhang, Y. Liang, Q. She, Y. Zhang and N. Peng, et al., Diversity and contributions to nitrogen cycling and carbon fixation of soil salinity shaped microbial communities in Tarim Basin, Front. Microbiol., 2018, 9, 431, DOI:10.3389/fmicb.2018.00431.
- X. Lou, J. Zhao, X. Lou, X. Xia, Y. Feng and H. Li, The Biodegradation of Soil Organic Matter in Soil-Dwelling Humivorous Fauna, Front. Bioeng. Biotechnol., 2022, 9, 1–8, DOI:10.3389/fbioe.2021.808075.
- S. M. Lee, H. G. Kong, G. C. Song and C. M. Ryu, Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease, ISME J., 2021, 15(1), 330–347, DOI:10.1038/s41396-020-00785-x.
- J. Zhang, H.-S. Meng, Y.-M. Shang, J.-R. Lead, Z.-Z. Guo and J.-P. Hong, Response of Soil Bacterial Diversity, Predicted Functions and Co-Occurrence Patterns to Nanoceria and Ionic Cerium Exposure, Microorganisms, 2022, 10(10), 1982, DOI:10.3390/microorganisms10101982.
- Y. Feng, C. Wang, F. Chen, X. Cao, J. Wang and L. Yue, et al., Cerium oxide nanomaterials improve cucumber flowering, fruit yield and quality: the rhizosphere effect, Environ. Sci.: Nano, 2023, 10(8), 2010–2021, 10.1039/D3EN00213F.
- I. Kamika and M. Tekere, Impacts of cerium oxide nanoparticles on bacterial community in activated sludge, AMB Express, 2017, 7(1), 63, DOI:10.1186/s13568-017-0365-6.
- A. M. Negrescu, M. S. Killian, S. N. V. Raghu, P. Schmuki, A. Mazare and A. Cimpean, Metal Oxide Nanoparticles: Review of Synthesis, Characterization and Biological Effects, J. Funct. Biomater., 2022, 13(4), 274, DOI:10.3390/jfb13040274.
- P. Vejan, R. Abdullah, T. Khadiran, S. Ismail and B. A. Nasrulhaq, Role of plant growth promoting rhizobacteria in agricultural sustainability-A review, Molecules, 2016, 21(5), 1–17, DOI:10.3390/molecules21050573.
- R. de Souza, A. Ambrosini and L. M. P. Passaglia, Plant growth-promoting bacteria as inoculants in agricultural soils, Genet. Mol. Biol., 2015, 38(4), 401–419, DOI:10.1590/S1415-475738420150053.
- S. Chavan and V. Nadanathangam, Effects of nanoparticles on plant growth-promoting bacteria in Indian agricultural soil, Agronomy, 2019, 9(3), 1–18, DOI:10.3390/agronomy9030140.
- H. Mekonnen and M. Kibret, The roles of plant growth promoting rhizobacteria in sustainable vegetable production in Ethiopia, Chem. Biol. Technol. Agric., 2021, 8(1), 1–11, DOI:10.1186/s40538-021-00213-y.
- E. Eren, The Effect of Plant Growth Promoting Rhizobacteria (PGPRs) on Yield and Some Quality Parameters during Shelf Life in White Button Mushroom (Agaricus bisporus L.), J. Fungi, 2022, 8(10), 1016, DOI:10.3390/jof8101016.
- N. C. Adele, B. T. Ngwenya, K. V. Heal and J. F. W. Mosselmans, Role of plant growth promoting bacteria in driving speciation gradients across soil-rhizosphere-plant interfaces in zinc-contaminated soils, Environ. Pollut., 2021, 279, 116909, DOI:10.1016/j.envpol.2021.116909.
- C. E. M. Grossi, E. Fantino, F. Serral, M. S. Zawoznik, D. A. Fernandez Do Porto and R. M. Ulloa, Methylobacterium sp. 2A Is a Plant Growth-Promoting Rhizobacteria That Has the Potential to Improve Potato Crop Yield Under Adverse Conditions. Front, Plant Sci., 2020, 11, 1–15, DOI:10.3389/fpls.2020.00071.
- P. Singh, V. Kumar and S. Agrawal, Evaluation of phytase producing bacteria for their plant growth promoting activities. Int, J. Microbiol., 2014, 2014, 426483, DOI:10.1155/2014/426483.
- B. R. Glick, Bacteria with ACC deaminase can promote plant growth and help to feed the world, Microbiol. Res., 2014, 169(1), 30–39, DOI:10.1016/j.micres.2013.09.009.
- T. T. Ajiboye, T. O. Ajiboye and O. O. Babalola, Impacts of Binary Oxide Nanoparticles on the Soybean Plant and Its Rhizosphere, Associated Phytohormones, and Enzymes, Molecules, 2023, 28(3), 1326, DOI:10.3390/molecules28031326.
- A. Dias Samarajeewa, J. R. Velicogna, D. M. Schwertfeger, M. J. Meier, R. M. Subasinghe and J. I. Princz, et al., Cerium oxide nanoparticles (nCeO2) exert minimal adverse effects on microbial communities in soils with and without biosolids amendment, Environ. Sci. Pollut. Res., 2023, 30(28), 72336–72353, DOI:10.1007/s11356-023-27313-6.
- H. Chen, K. Ma, C. Lu, Q. Fu, Y. Qiu and J. Zhao, et al., Functional Redundancy in Soil Microbial Community Based on Metagenomics Across the Globe, Front. Microbiol., 2022, 13, 1–13, DOI:10.3389/fmicb.2022.878978.
- OECD, Nanomaterials in Waste Streams: Current Knowledge on Risks and Impacts, OECD, 2016, available from: https://www.oecd-ilibrary.org/environment/nanomaterials-in-waste-streams_9789264249752-en Search PubMed.
- C. Sansupa, S. F. M. Wahdan, S. Hossen, T. Disayathanoowat, T. Wubet and W. Purahong, Can we use functional annotation of prokaryotic taxa (Faprotax) to assign the ecological functions of soil bacteria?, Appl. Sci., 2021, 11(2), 1–17, DOI:10.3390/app11020688.
- X. Wang, M. Zhu, N. Li, S. Du, J. Yang and Y. Li, Effects of CeO2 nanoparticles on bacterial community and molecular ecological network in activated sludge system, Environ. Pollut., 2018, 238, 516–523, DOI:10.1016/j.envpol.2018.03.034.
- E. Pütz, A. Gazanis, N. G. Keltsch, O. Jegel, F. Pfitzner and R. Heermann, et al., Communication Breakdown: Into the Molecular Mechanism of Biofilm Inhibition by CeO2 Nanocrystal Enzyme Mimics and How It Can Be Exploited, ACS Nano, 2022, 16(10), 16091–16108, DOI:10.1021/acsnano.2c04377.
- K. M. Deangelis, S. E. Lindow and M. K. Firestone, Bacterial quorum sensing and nitrogen cycling in rhizosphere soil, FEMS Microbiol. Ecol., 2008, 66(2), 197–207, DOI:10.1111/j.1574-6941.2008.00550.x.
- B. R. Martins, R. Siani, K. Treder, D. Michałowska, V. Radl and K. Pritsch, et al., Cultivar-specific dynamics: unravelling rhizosphere microbiome responses to water deficit stress in potato cultivars, BMC Microbiol., 2023, 23(1), 1–16, DOI:10.1186/s12866-023-03120-4.
- J. Majdura, U. Jankiewicz, A. Gałązka and S. Orzechowski, The Role of Quorum Sensing Molecules in Bacterial–Plant Interactions, Metabolites, 2023, 13(1), 114, DOI:10.3390/metabo13010114.
- Y. M. Huang, Y. N. Zou and Q. S. Wu, Alleviation of drought stress by mycorrhizas is related to increased root H 2 O 2 efflux in trifoliate orange, Sci. Rep., 2017, 7, 1–9, DOI:10.1038/srep42335.
- P. Voothuluru, P. Mäkelä, J. Zhu, M. Yamaguchi, I. J. Cho, M. J. Oliver, J. Simmonds and R. E. Sharp, Apoplastic Hydrogen Peroxide in the Growth Zone of the Maize Primary Root. Increased Levels Differentially Modulate Root Elongation Under Well-Watered and Water-Stressed Conditions. Front, Plant Sci., 2020, 11, 392, DOI:10.3389/fpls.2020.00392.
- A. T. Poret-Peterson, N. Sayed, N. Glyzewski, H. Forbes, E. T. González-Orta and D. A. Kluepfel, Temporal Responses of Microbial Communities to Anaerobic Soil Disinfestation, Microb. Ecol., 2020, 80(1), 191–201, DOI:10.1007/s00248-019-01477-6.
- W. J. Blok, J. G. Lamers, A. J. Termorshuizen and G. J. Bollen, Control of soilborne plant pathogens by incorporating fresh organic amendments followed by tarping, Phytopathology, 2000, 90(3), 253–259, DOI:10.1094/PHYTO.2000.90.3.253.
- M. Djanaguiraman, R. Nair, J. P. Giraldo and P. V. V. Prasad, Cerium Oxide Nanoparticles Decrease Drought-Induced Oxidative Damage in Sorghum Leading to Higher Photosynthesis and Grain Yield, ACS Omega, 2018, 3(10), 14406–14416, DOI:10.1021/acsomega.8b01894.
- Z. Cao, L. Rossi, C. Stowers, W. Zhang, L. Lombardini and X. Ma, The impact of cerium oxide nanoparticles on the physiology of soybean (Glycine max (L.) Merr.) under different soil moisture conditions, Environ. Sci. Pollut. Res., 2018, 25(1), 930–939, DOI:10.1007/s11356-017-0501-5.
- X. Ma, X. Wang, L. Xu, H. Shi, H. Yang and K. K. Landrock, et al., Fate and distribution of orally-ingested CeO2-nanoparticles based on a mouse model: Implication for human health, Soil Environ. Health, 2023, 1(2), 100017, DOI:10.1016/j.seh.2023.100017.
- A. Ladaycia, C. Passirani and E. Lepeltier, Microbiota and nanoparticles: Description and interactions, Eur. J. Pharm. Biopharm., 2021, 169, 220–240, DOI:10.1016/j.ejpb.2021.10.015.
- E. E. Fröhlich and E. Fröhlich, Cytotoxicity of nanoparticles contained in food on intestinal cells and the gut microbiota, Int. J. Mol. Sci., 2016, 17(4), 509, DOI:10.3390/ijms17040509.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3en00479a |
| This journal is © The Royal Society of Chemistry 2024 |