Open Access Article
Open Access ArticleSample-to-answer COVID-19 nucleic acid testing using a low-cost centrifugal microfluidic platform with bead-based signal enhancement and smartphone read-out†
Ruben R. G.
Soares
 a,
Ahmad S.
Akhtar
a,
Ahmad S.
Akhtar
 a,
Inês F.
Pinto
a,
Inês F.
Pinto
 a,
Noa
Lapins
a,
Donal
Barrett
b,
Gustaf
Sandh
c,
Xiushan
Yin
bde,
Vicent
Pelechano
a,
Noa
Lapins
a,
Donal
Barrett
b,
Gustaf
Sandh
c,
Xiushan
Yin
bde,
Vicent
Pelechano
 b and
Aman
Russom
b and
Aman
Russom
 *af
*af
aKTH Royal Institute of Technology, Division of Nanobiotechnology, Department of Protein Science, Science for Life Laboratory, Solna, Sweden. E-mail: aman@kth.se
bScience for Life Laboratory, Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet, Solna, Sweden
cDepartment of Clinical Microbiology, Karolinska University Hospital, Stockholm, Sweden
dApplied Biology Laboratory, Shenyang University of Chemical Technology, Shenyang, China
eBiotech and Biomedicine Science Co. Ltd, Shenyang, China
fAIMES – Center for the Advancement of Integrated Medical and Engineering Sciences at Karolinska Institutet and, KTH Royal Institute of Technology, Stockholm, Sweden
First published on 7th June 2021
Abstract
With its origin estimated around December 2019 in Wuhan, China, the ongoing SARS-CoV-2 pandemic is a major global health challenge. The demand for scalable, rapid and sensitive viral diagnostics is thus particularly pressing at present to help contain the rapid spread of infection and prevent overwhelming the capacity of health systems. While high-income countries have managed to rapidly expand diagnostic capacities, such is not the case in resource-limited settings of low- to medium-income countries. Aiming at developing cost-effective viral load detection systems for point-of-care COVID-19 diagnostics in resource-limited and resource-rich settings alike, we report the development of an integrated modular centrifugal microfluidic platform to perform loop-mediated isothermal amplification (LAMP) of viral RNA directly from heat-inactivated nasopharyngeal swab samples. The discs were pre-packed with dried n-benzyl-n-methylethanolamine modified agarose beads used to selectively remove primer dimers, inactivate the reaction post-amplification and allowing enhanced fluorescence detection via a smartphone camera. Sample-to-answer analysis within 1 hour from sample collection and a detection limit of approximately 100 RNA copies in 10 μL reaction volume were achieved. The platform was validated with a panel of 162 nasopharyngeal swab samples collected from patients with COVID-19 symptoms, providing a sensitivity of 96.6% (82.2–99.9%, 95% CI) for samples with Ct values below 26 and a specificity of 100% (90–100%, 95% CI), thus being fit-for-purpose to diagnose patients with a high risk of viral transmission. These results show significant promise towards bringing routine point-of-care COVID-19 diagnostics to resource-limited settings.
1. Introduction
Simple, rapid, and sensitive analytical methods for viral detection at the point-of-care have been increasingly in demand in recent decades. Such devices are especially relevant in resource-limited settings (RLS) in Africa, Asia and South America, where the prevalence of ubiquitous viral pathogens such as human immunodeficiency virus (HIV) and other tropical viruses including Zika, Ebola, Crimean-Congo hemorrhagic fever (CCHF), chikungunya and dengue claim tens of thousands of human lives per year mostly due to the lack of effective diagnostics and subsequent disease containment and therapy.1–3 However, few occasions in modern history have been more pressing for developments in this field than the present SARS-CoV-2 coronavirus pandemic4 which from December 2019 to May 2021 has infected more than 160 million people in over 210 countries and territories, resulting in more than 3.3 million deaths.The first cases of SARS-CoV-2 infection were reported in December 2019 in Wuhan with a probable zoonotic origin considering its genome sequence is closely related to bat coronaviruses.5 Although other human coronaviruses associated with epidemic outbreaks, i.e. SARS-CoV and MERS-CoV, had a higher mortality rate, the much higher infectivity and long asymptomatic incubation period associated with SARS-CoV-2 infections, resulting in the pathologic condition designated as coronavirus disease-19 (COVID-19), allowed the rapid spreading of the virus around the globe.6 In some cases, a detectable viral load in nasopharyngeal swabs was observed prior to symptom onset7 and as long as 20 days after the first symptoms.6 Furthermore, considering the significant differences in symptoms and their severity, it is estimated that as many as 90% of all infections were undocumented early in the pandemic before travel restrictions were imposed.8
Aiming at controlling the spread of the virus, millions of viral load tests have been performed in centralized labs supported by sample collection in the field to screen symptomatic and asymptomatic infections. The sampling methods of choice are nasal or throat swabs stored in viral collection medium prior to RT-PCR,9 with primers/probes targeting the ORF1ab, RNA dependent RNA polymerase (RdRp), envelope (E), spike protein (S) or nucleocapsid (N) genes of the SARS-CoV-2 single stranded RNA genome.10–12 Viral load values during the course of the infection typically range between ∼103 to ∼1011 copies per mL6,7,9,13 and >106 copies per mL are common early upon the onset of symptoms,7 with a peak in the order of 109 copies per throat swab on day 4 after onset of symptoms.14 However, in the context of viral viability and risk of transmission, it has been observed that the probability of isolating infectious SARS-CoV-2 is less than 5% when the viral load in the respiratory tract is below ∼4.3 × 106 RNA copies per mL.15 Thus, it has been recently advocated that more frequent and cost effective testing with lower sensitivity, instead of lengthy and sparse PCR testing in centralized labs, can provide a more efficient strategy for containment by (1) rapidly detecting the viral load peak early upon infection and (2) avoiding unnecessary isolation during the long viral load decrease period detectable only with high analytical sensitivity and with minimal to zero risk of cross-human infection.16 In this context, it is expected that nucleic acid amplification tests can still provide higher sensitivity and potentially specificity than antigen rapid tests,17 thus being the ideal solution if costs and equipment complexity can be kept low.
Within the landscape of the ongoing pandemic, it has become clear that testing is paramount to reduce the spread of infection and ease the impact on health systems combined with a smart and data-driven management of social distancing policies. However, while developed countries can economically and logistically support an exponential increase in lab-based testing and social distancing measures, such a course of action is hardly feasible in RLS.18 Thus, to support routine SARS-CoV-2 diagnostics on a global scale but particularly in RLS, a cost-effective, rapid, sensitive, portable and simple to use device to detect RNA directly from biological specimens would have a significant impact.
Concerning the development of portable analytical devices, the use of RT-PCR is suboptimal considering the intrinsic technical complexity of performing several precise heating–cooling cycles up to >94 °C.19 Thus, in the context of pathogen detection, several groups have been actively exploring the use of loop-mediated isothermal amplification (LAMP) as a potential alternative, requiring only a constant and relatively lower temperature of ∼65 °C.19 Concerning the miniaturization of LAMP towards integrated analytical platforms, its combination with centrifugal microfluidics20–25 and/or smartphone-based signal readout26–33 has been showing significant promise to achieve a true sample-to- answer operation. A few remarkable examples of LAMP-based miniaturized modules using either of these approaches are: (1) capture of LAMP amplification products with anti-DIG antibodies followed by generation of a TMB precipitate measured using a standard light source and a smartphone camera;26 (2) digital microfluidic platform with temperature monitoring/control provided by a thermal imaging camera and SYBR Green I derived fluorescence transduction by naked eye or smartphone camera;28 (3) microfluidic cartridge combining immune-capture, lysis and LAMP to detect viable bacteria using a reader platform comprising two light sources for fluorometric and/or turbidimetric analysis resorting to a smartphone camera;31 (4) a hermetic container providing power-free chemical-based heating for LAMP amplification followed by detection using a smartphone flashlight and camera for fluorometric detection;33 (5) centrifugal platform combining silica-based DNA extraction and integrated LFA strips to multiplex the detection of multiple LAMP products using anti-DIG antibodies and colorimetric detection;33 (6) centrifugal platform with automated bead-beating lysis followed by direct RT-LAMP by continuous measurement of fluorescence with UVC illumination and a standard camera;22 and (7) centrifugal platform incorporating non-contact heating of the disc and colorimetric detection of LAMP products using a white LED for illumination and filtered photodiodes for signal acquisition.24
Here, LAMP, centrifugal microfluidics, smartphone-based detection and recent developments in RT-LAMP applied to the detection of SARS-CoV-2 RNA34–40 are combined to develop a novel cost-effective and fully integrated platform for COVID-19 diagnostics directly from heat-inactivated nasopharyngeal samples. The direct detection from heat-inactivated samples was achieved using (1) a one-pot combination of reverse transcriptase and polymerase enzymes for robust isothermal amplification and (2) an agarose bead-based signal enhancement strategy for improved fluorometric detection, thus avoiding the impact of collection media on weakly-buffered pH responsive colorimetric amplification mixtures.40 This signal enhancement strategy relies on a previously unreported molecular size-selectivity of N-benzyl-N-methylethanolamine (NBNM) agarose beads, trade named Capto adhere, allowing the separation of short single stranded primer oligos and large double stranded LAMP amplification products.
2. Methods
2.1. Fabrication of PDMS microchannels and discs
The PDMS microchannels used for fluorescence microscopy characterization were fabricated as described in detail elsewhere.41 Briefly, microchannels comprising a 700 × 100 μm2 cross section converging into a 200 × 20 μm2 cross-section aimed at trapping agarose beads with an average diameter of 90 μm, were fabricated using standard SU-8 mold replication techniques. 18-Gauge inlet and outlet access holes were punched using a blunt syringe and the channels were sealed against Corning glass slides after an oxygen plasma treatment (30 s, Femto Science CUTE, 100 W, 80 Pa O2).The discs were designed using Autodesk® Fusion 360 (education license) and cut into 1 mm thick PMMA sheets using computer-numerical-control milling machine (Roland Modela MDX-40A). The disc layers were bonded together using clear medical grade pressure-sensitive adhesive (PSA) (ARcare® 92712). The microchannels on the PSA were cut using a cutter plotter (Graphtec CE6000-40). After aligning the PMMA layers with the PSA layer, the disc was placed in a manual press machine overnight to ensure uniform bonding. Detailed dimensions of each microchannel are shown in Fig. S3.†
2.2. Assembly of the heating module
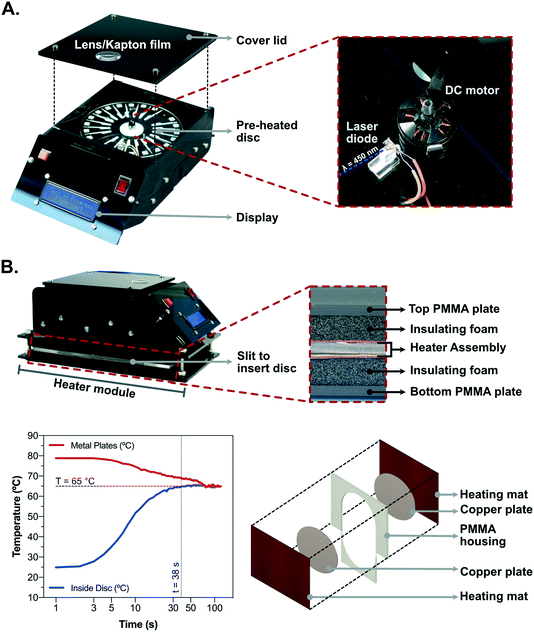
The heating module consisted of two silicone heater mats (30 W, 150 × 200 mm, 12 V DC; RS PRO Article # 731-366) which were used to heat up 1 mm thick copper plates (120 mm diameter). The heater mats are self-adhesive and the copper plates were attached to the mats. A 4 mm thick PMMA sheet served as a chamber for the heating module, having a slot, made with a CNC milling machine, to insert the disc. Heater mats with the attached copper plates in the center were attached on either side of the PMMA sheet, thus forming a heating chamber for the disc with an open slit on the side for inserting disc. The thickness of the copper plates and PMMA sheets was chosen to ensure direct contact with the two copper plates upon insertion of the disc into the chamber. Finally, the heating chamber was closed to ensure rapid and uniform heating using a PMMA part cut to fit into the slit opening. The heating module was lined with insulating foam on either side and placed between two 4 mm thick PMMA sheets (150 × 250 mm) which were then closed using M6 bolts on the 4 corners. A thermocouple (Type K; RS PRO Article # 621-2170) was embedded between the copper plate and the heater mat to measure the temperature. Temperature regulation was achieved by using N-channel MOSFET (IRF540PBF, 28A, 100 V; RS PRO Article # 708-5143) in combination with Arduino UNO Rev 3 MCU development board controlling the power supply to the heater mats.2.3. Assembly of the centrifugal and signal acquisition modules
The centrifugal module (140 × 230 × 80 mm) was fabricated using 4 mm thick PMMA sheets which were cut using CNC milling machine and assembled using M3 bolts and nuts. It consisted of two chambers; the bottom chamber served as the housing for the electronic components and the top chamber is where the rotation of the disc and the signal acquisition was done. The control of the whole platform is achieved using an Arduino UNO Rev 3 MCU development board. In the center of the top chamber there was a motor (Emax MT-2204; 2300 kv brushless DC motor) used to rotate the disc. A small piece of magnetic tape was attached to the body of the motor and a bipolar hall effect sensor (Honeywell SS411A; RS Pro Article # 181-1463) was used to measure the rotation speed in real time while sending feedback to the Arduino board. The top chamber also included a blue laser diode (Osram Opto PL 450 nm, 80 mW; RS PRo Article # 758-7810) which was used for signal acquisition. A 16 × 2 pixel display module attached at the front provides information to the user regarding ongoing assay steps. Further technical details and exploded view is shown in Fig. S4.†2.4. Packing, drying and characterization of N-benzyl-N-methylethanolamine (NBNM) agarose beads
The NBNM agarose beads were packed in the discs by first diluting the bead resin stock (Capto Adhere, Cytiva) in DI-water at 10% (v/v). 20 μL of the diluted bead stock were then added to each channel on the disc and subsequently centrifuged at 6000 rpm for 10 s. The excess solution was manually removed from the outlet and the disc was subsequently placed in a vacuum chamber at 15 Pa for 8 min to dehydrate the beads. After dehydration, the disc was centrifuged a second time at 6000 rpm for 10 s to ensure a homogeneous packing. The bp-cutoff of the beads packed inside the discs was characterized by microfluidic capillary electrophoresis in a Bioanalyzer 2100 system with a DNA 1000 kit (Agilent Technologies, USA).The packing in the PDMS microchannels was performed by first preparing a suspension of beads in a 20% PEG 8000 (w/w) solution by adding 1 μL bead stock to 19 μL PEG solution. The suspension was flowed into the microchannels at 10 μL min−1 using a NE-1200 syringe pump (New Era Pump Systems, USA). The channels were then rinsed with DI-water flowed at 10 μL min−1 for 5 min to ensure removal of any residual PEG and salts. To dehydrate the beads, the PDMS device was placed in a vacuum chamber at 15 Pa for 5 min and subsequently stored at room temperature until further usage. The beads inside the PDMS devices were characterized by fluorescence microscopy in a Nikon Ti-Eclipse inverted microscope equipped with a Lumencor SOLA light engine and a FITC filter cube. The acquired fluorescence microscopy images were analyzed using ImageJ software (NIH, USA).
2.5. Real-time loop mediated isothermal amplification (RT-LAMP) of SARS-CoV-2 RNA
The RT-LAMP was performed in a Mic qPCR cycler (Biomolecular systems, Australia) with a total of 10 μL per reaction mixture. The reaction mixture containing 1 μL template RNA or heat-inactivated transport medium was prepared with final concentrations of 1× isothermal amplification buffer II (New England Biolabs, USA), 6 mM dNTPs (RP65, Blirt, Poland), 6 mM MgSO4 (New England Biolabs, USA), 1× primer mixture (six primers described in detail below), 1× SYBR Green I (S7585, Thermo Fisher Scientific), 320 mU Bst 3.0 polymerase (New England Biolabs, USA) and 4 U SuperScript IV (SSIV) reverse transcriptase (Thermo Fisher Scientific). The primer mixture at 10× concentration was prepared with 2 μM F and B, 16 μM FIP and BIP and 4 μM LF and LB. Two primer sets were tested, namely the iLACO set reported by Yu et al.34 and the As1e set reported by Rabe et al.42 The iLACO primer sequences were the following: F-5′-CCA CTA GAG GAG CTA CTG TA-3′; B-5′-TGA CAA GCT ACA ACA CGT-3′; FIP-5′-AGG TGA GGG TTT TCT ACA TCA CTA TAT TGG AAC AAG CAA ATT CTA TGG-3′; BIP-5′-ATG GGT TGG GAT TAT CCT AAA TGT GTG CGA GCA AGA ACA AGT G-3′; LF-5′-CAG TTT TTA ACA TGT TGT GCC AAC C-3′; LB-5′-TAG AGC CAT GCC TAA CAT GCT-3′. The As1e primer sequences were the following: F-5′-CGG TGG ACA AAT TGT CAC-3′; B-5′-CTT CTC TGG ATT TAA CAC ACT T-3′; FIP-5′-TCA GCA CAC AAA GCC AAA AAT TTA TTT TTC TGT GCA AAG GAA ATT AAG GAG-3′; BIP-5′-TAT TGG TGG AGC TAA ACT TAA AGC CTT TTC TGT ACA ATC CCT TTG AGT G-3′; LF-5′-TTA CAA GCT TAA AGA ATG TCT GAA CAC T-3′; LB-5′-TTG AAT TTA GGT GAA ACA TTT GTC ACG-3′. All primers were synthesized by Thermo Fisher Scientific. The RNA fragment (226 bp) matching the sequence of the ORF1ab of SARS-CoV-2 (MT883505.1, 15118–15343) used to characterize and optimize the LAMP reaction with the iLACO primer set was produced by in vitro transcription and stored at −80 °C in DI-water at 5 ng μL−1. Synthetic full-length SARS-CoV-2 RNA (GenBank MN908947.3) at a stock concentration of 106 copies per μL was purchased from Twist Bioscience (CA, USA).2.6. Collection and processing of nasopharyngeal swab samples
The nasopharyngeal samples were obtained from Karolinska University Hospital, Huddinge (Stockholm) collected between May 20th and June 1st 2020 from hospitalized patients suspected of COVID-19. All samples were collected using Sigma-Transwab (MWE, UK), Sigma-Virocult (MWE, UK) or COPAN-eSwab (Copan Diagnostics Inc., CA, USA) kits as described in detail elsewhere.43 We used pseudo-anonymized surplus material previously collected for clinical diagnostics of SARS-CoV-2. This is in accordance with the Swedish Act concerning the ethical review of research involving humans, which allows development and improvement of diagnostic assays using patient samples which were collected to perform the testing in question. Additional ethical approval for RT-LAMP diagnosis was obtained by the appropriate Swedish Authority (Dnr 2020-01945, Etikproevningsnaemnden). All samples were pre-analysed on the GeneXpert Xpress SARS-CoV-2 system (Cepheid, USA) and Ct values for E and N2 genes were obtained. All Ct values referred in the text are the average of E and N2 genes. For the LAMP experiments, 50 μl of each sample was heat-inactivated at 95 °C for 15 minutes and stored at −80 °C until further processing.2.7. Detection of SARS-CoV-2 RNA using the integrated centrifugal platform
The sample containing the RNA template (in vitro synthesized RNA fragment or inactivated SARS-CoV-2) was first combined with the LAMP master mix at a ratio of 10% (v/v) and 10 μL were subsequently added to each of the 20 channels in the disc. Both inlet and outlet access holes of all channels were sealed with a sheet of PSA (ARSeal™ 90880) covering the entire disc surface. The PSA sheet prevents evaporation of the solutions during LAMP and cross-contamination via aerosols. The sealed disc was subsequently inserted into the heater module and incubated at 65 °C for 25–30 min. The disc was then immediately (within 30 s) transferred to the centrifugal platform and subjected to 4 acceleration/deceleration cycles from 0 to 6000 rpm taking 1 min each. After centrifugation, the laser diode was turned on and each channel was sequentially imaged using a mid-range smartphone camera (OnePlus 6T, China) with constant ISO. The acquired RGB images were processed using ImageJ software (NIH, USA) measuring the grayscale profile of the green channel along the interface between the solution and the packed beads (100 pixels long and average of 30 pixels perpendicular to the line). The grayscale intensity on the beads was normalized relative to the solution for each channel and was smoothed with a 10 pixel moving average before further processing.3. Results and discussion
3.1. Characterization of RT-LAMP sensitivity and development of a bead-based strategy for fluorescence signal enhancement
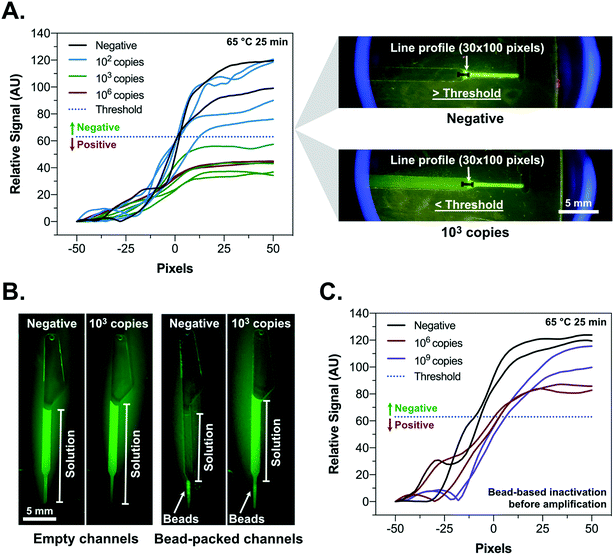
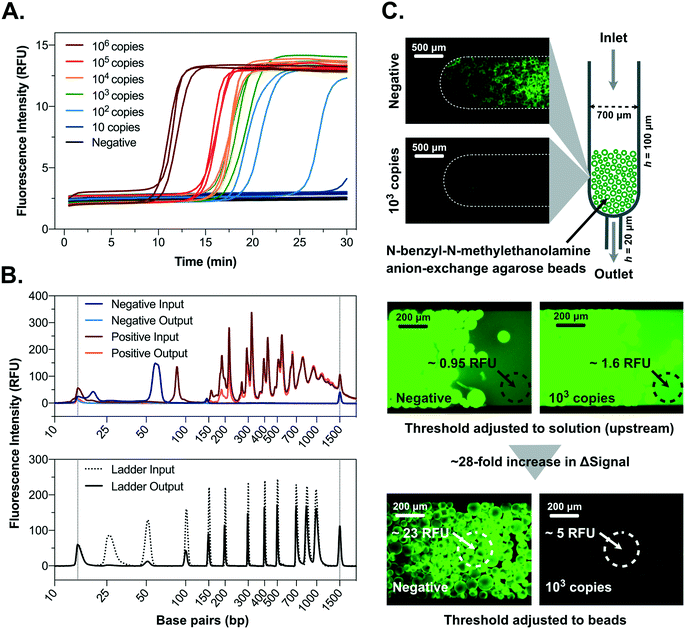
Considerable advances in LAMP-based detection of SARS-CoV-2 RNA have been achieved since the beginning of the pandemic, with several primer sets and potential detection strategies being reported. In this context, particular attention has been given to pH-based colorimetric signal transduction considering the simple visual interpretation of the results.40 However, mildly buffered systems are highly prone to interference from different biological samples and collection media, typically requiring a previous silica-based extraction and elution with DI-water for improved robustness.40 Fluorescence detection using DNA intercalator dyes is an equally simple and more interference-forgiving approach but requires the tackling of two key limitations, namely (1) the relatively more complex signal transduction, particularly in miniaturized systems concerning low fluorescence signal intensities and filtering requirements44 and (2) the high numbers (typically 6 different sequences) and design complexity of LAMP primers results very often in a variable degree of intra- (hairpins) and cross-primer hybridization (primer dimers), which result in significant non-specific fluorescence signal.45 This non-specific signal is particularly relevant at room temperature due to the lower hybridization stringency, hindering a simple, non-real-time, post-LAMP measurement.46 Several approaches have been developed to overcome this limitation involving sequence specific detection of LAMP products in a homogeneous manner, such as detection of amplification by release of quenching (DARQ), quenching of unincorporated amplification signal reporters (QUASR) and one-step strand displacement (OSD).47 However, all of these methods imply additional specific oligos and their modification with organic fluorophores and quenchers, significantly increasing efforts in assay design and intrinsic costs per test. As a specific example, considering a typical fluorophore–quencher hairpin probe used as a molecular beacon at a concentration of 200 nM per 10 μL reaction volume, its cost for 100 tests would be ∼1.2 USD, whereas SYBR Green I dye at 1× concentration costs less than 0.02 USD, a 60-fold reduction.Here, to strike an innovative compromise between fit-for-purpose performance and cost-effectiveness, the developed device incorporates an agarose bead-based strategy combined with centrifugal microfluidics to fundamentally tackle these limitations and significantly enhance the negative-to-positive signal ratio at room-temperature. Using a set of primers that we have previously developed34 (referred below as iLACO primer set) and resorting to a combination of SSIV and Bst 3.0 for LAMP and SYBR Green I for fluorescence generation, the obtained RT-LAMP results are shown in Fig. 1A. Performing a dilution series of an RNA fragment with a 226 bp sequence matching the ORF1ab of SARS-CoV-2 in water, 100 copies per 10 μL reaction could be detected within 30 min of amplification time, in agreement with the typical LoD requirements of FDA-approved COVID-19 diagnostic technologies.4 Strikingly, while the increase in fluorescence signal magnitude from negative to positive measured at 65 °C was about 5-fold (Fig. 1A), the difference was reduced to ∼1.5-fold at room temperature (∼25 °C) according to the microscopy measurements in Fig. 1C, due to an increase in signal in the non-amplified samples. This fluorescence signal background, arising from hybridization of primer dimers/hairpins at lower temperatures after the amplification, was addressed using the novel bead-based sample processing strategy described in Fig. 1B and C. This strategy dramatically improved the signal-to-noise ratio at room temperature, thus facilitating end-point signal acquisition when portability and cost reduction are a priority. Multimodal ligands as is the case of N-benzyl-N-methylethanolamine (NBNM) combining anion exchange and other interactions, i.e. hydrophobic and hydrogen bonding, are typically used in chromatography to capture host cell DNA as an impurity48 or plasmid DNA as target product49 directly from complex matrices. Here used for sample processing, this ligand, when modified on agarose beads with an average porosity of ∼100 nm, was found to have a remarkable selectivity for short oligonucleotides with lengths of 25–100 bp and a cutoff region between 200–400 bp (Fig. 1B). According to the results in Fig. 1B, 1.3 μL of beads could deplete virtually all the primers in 10 μL of LAMP mixture (negative input vs. negative output), while the titers of LAMP products amplified from 106 copies of template RNA remained constant above 400 bp (positive input vs. positive output). These observations were confirmed using fluorescence microscopy in the presence of SYBR Green I as dsDNA selective intercalator. According to the results in Fig. 1C, the primers in a LAMP mixture without RNA template (negative) result in a very high fluorescence measured at bead level, which is ∼4.6-fold higher compared to a positive LAMP mixture (103 copies of RNA template). This observation is in agreement with the electropherograms in Fig. 1B which highlight the complete depletion of all primers (<50 bp) in the presence of template (negative input vs. positive input). The fluorescence microscopy measurements also highlight an increase of ∼28-fold in signal magnitude between negative and positive, comparing the measurement performed directly on solution before bead processing (upstream of the beads) relative to the measurement performed directly on the beads. Such an increase in signal magnitude shows promise to implement a robust signal acquisition strategy with minimal instrumentation complexity. The versatility of the bead-based enrichment as a general post nucleic acid amplification signal amplification strategy was further validated for up-concentrating SARS-CoV-2 PCR amplicons (100 bp long) detected with a molecular beacon (Fig. S1†). In agreement with the characterization in Fig. 1B, the short PCR amplicons were effectively captured within the pores of the NBNM beads, contrarily to the longer LAMP products, thus generating a linear and directly proportional correlation between bead fluorescence and RNA copies with Ct ranging from 25 to 36 (R2 = 0.99) (Fig. S1†). Schematics illustrating the bead capture behavior in the presence of LAMP or PCR positive and negative samples are shown as ESI† (Fig. S2).
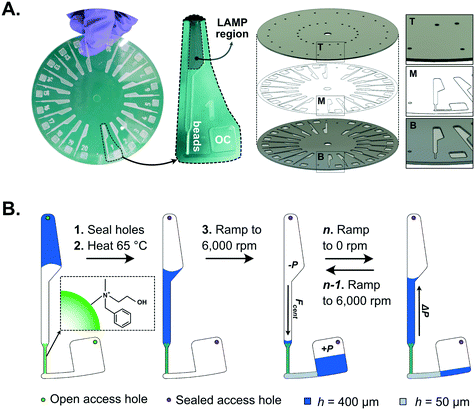
3.2. Development of an integrated platform for point-of-care COVID-19 diagnostics using a smartphone camera for fluorometric signal transduction
Aiming at bringing cost-effective, rapid and sensitive COVID-19 diagnostics to RLS, the main goal of this work was to develop a robust but minimally complex and portable platform for SARS-CoV-2 RNA detection directly from heat inactivated biological samples. Taking advantage of the signal enhancement strategy reported in section 3.1. The concept was based on designing a PMMA disc with 20 independent channels (Fig. 2A) each comprising (1) a region to perform LAMP of 10 μL sample (LAMP section), (2) a section containing packed NBNM beads at the interface between 400 and 50 μm deep channels and (3) an outlet chamber (OC) connected to the previous two components via the narrow 50 μm deep section. To process the sample added to the disc, the sequence of steps schematized in Fig. 2B was followed. The heat-inactivated swab sample containing template RNA was mixed with the LAMP reagents and a total of 10 μL of the mixture was added to the channel, the access holes were sealed and the whole disc incubated at 65 °C. After amplification, the disc was subjected to four cycles of ramping the rotation linearly from 0 to 6000 rpm and vice-versa (1 cycle meaning 0–6000–0 rpm). During the ramp-up, the liquid is forced through the beads due to the centrifugal force and held back by an increase in pressure inside the sealed OC. During ramp-down, the decrease in centrifugal force allows the OC to decompress pushing the liquid backwards, while the denser agarose beads are still held in place by the centrifugal force. This back-and-forth motion with 4 cycles allows a complete capture of the primers in solution according to the previous results in Fig. 1B.The portable platform developed to combine the centrifugal and heating modules required to perform the LAMP followed by the bead-based signal enhancement is shown in Fig. 3. The top centrifugal module shown in Fig. 3A uses a DC motor controlled by a microcontroller board and a hall-effect sensor to complete the rotation protocol. After the rotation protocol, the measurement is performed through a PMMA lens in the cover lid covered on the backside with a 100 μm polyimide film which is aligned with the smartphone camera with a custom-made adapter. The polyimide film serves as an emission absorption filter to block the excitation light from the 450 nm 80 mW laser embedded on the platform.50 The laser diode is aligned at an angle of ∼17.5° relative to the disc (Fig. S5†) to take advantage of total internal reflection of the PMMA–air interface between the disc and the lens to minimize residual leakage of blue light into the camera sensor.
The heating module assembled below the centrifugal platform is shown in Fig. 3B. This module comprises a PMMA housing with a removable front piece to insert the disc. The disc enclosed by the housing is sandwiched in contact with two copper plates which are actively heated by two resistive silicone mats. The temperature control is achieved with a feedback loop measuring the temperature between the copper plates and the heating mats on both sides of the stack. The PMMA housing and the insulating foam on each side of the heating mats serves to minimize convective and conductive heat dissipation. To minimize non-specific amplification of hybridized primer dimer and primer-template pairs at lower temperatures, rapid heating is achieved by initially setting the temperature of the copper plates at 79 °C according to the plot in Fig. 3B. The plates are passively cooled down by the disc and active heating is only actuated once the temperature of the plates decreases below 65 °C.
3.3. Characterization of the integrated platform and data acquisition
The developed platform was subsequently characterized by testing a dilution series of ORF1ab RNA fragment spiked in DI-water (Fig. 4). A dilution series using the complete platform was tested and the results in Fig. 4A show a limit of detection between 102–103 copies per reaction (0% positive at 102 copies and 100% positive for 103 copies performing 4 independent measurements for each concentration). The analysis of the signal in each channel was performed by measuring the grayscale intensity profile of the green channel (average of 30 pixels) 100 pixels along the interface of the beads and the solution. Centering the 30 × 100 pixels line profile at the bead–solution interface (negative pixel values −50 to 0 in solution and positive values 0 to 50 on the beads), the response was described as a 10 pixel moving average relative to the intensity of the solution. A negative/positive decision threshold was defined according to 3.29 times the standard deviation of the relative signal obtained at the end of the line profile (pixel 50) for 5 independent negative controls (0.01% probability of false positive below the threshold). The obtained sensitivity for the on-disc LAMP was lower than that obtained in the RT-LAMP tests (Fig. 1A), where 100% of the samples with 102/copies per reaction showed a positive signal. The relatively lower sensitivity is hypothesized to arise from non-specific adsorption of enzymes and/or template to the PMMA channel, having a significantly higher surface area in contact with the 10 μL of solution compared to a standard reaction tube. Efforts to improve channel passivation strategies or application of alternative materials are envisioned to maximize performance. Fig. 4B highlights the enhanced signal (negative vs. 103 copies of target RNA fragment) provided by the beads. While for the previous fluorescence microscopy analysis discussed in section 3.1. the focus was placed on the beads and LAMP solution before processing, the photographs shown here highlight also the complete removal of non-specific fluorescence signal in solution for the negative control. These observations support the combination of both types of measurements, i.e., directly on the beads and on the solution, to allow the maximization of the signal-to-noise ratio. The bead-based primer depletion was also validated as a simultaneous means of inactivating the reaction post-LAMP, thus avoiding higher temperatures and longer assay times for enzyme inactivation (Fig. 4C). It was observed that when the primer depletion cycles were performed before amplification (65 °C for 25 min), no LAMP products were obtained with initial RNA template titers as high as 109 copies per reaction. However, for this feature to be used without risk of false-positives, the processing of the disc post-heating has to be performed immediately at t = 30 min. Overall these results confirm the triple functionality of the agarose beads for (1) signal intensity enhancement, (2) sample preparation to remove non-specific background and (3) simple reaction inactivation at room temperature.3.4. Detection of SARS-CoV-2 in heat-inactivated nasopharyngeal swab samples
The developed platform was tested for the detection of SARS-CoV-2 viral RNA directly from nasopharyngeal swab samples collected in viral transport media resorting to a simple heat inactivation step (95 °C, 15 min). A set of 31 clinical samples (Table S1†) spanning Ct values from 11 to 27, including 6 negative controls, was initially tested using the conditions optimized above (Fig. S6†). It was observed that the iLACO primers are highly prone to false-positive results at 30 min of amplification time required to maximize sensitivity. To improve the assay performance, the alternative primer set developed by Rabe and Cepko42 (As1e), minimizing primer dimer amplification according to previous reports,43 was tested and directly compared with the iLACO primer set for the amplification of full-length SARS-CoV-2 synthetic RNA (GenBank MN908947.3) spiked in water. The results of RT-LAMP shown in Fig. 5A highlight two critical improvements in assay performance. Firstly, non-specific amplification of primer dimers starts occurring only after 40 min of amplification time, a positive shift of more than 10 min relative to the iLACO primers. Secondly, lower RNA titers are amplified more rapidly with a loss in quantitative resolution, suggesting that a higher sensitivity can be achieved for the clinical samples keeping the amplification time constant at 30 min. However, despite the lower degree of non-specific amplification, the As1e primers result in an equally high background fluorescence signal resulting in positive to negative signal ratio of only 1.6-fold when measured on the disc at room temperature without the bead-based enhancement (Fig. S7†). Remarkably, NBNM agarose beads increase the positive to negative signal ratio more than 10 times up to approximately 17-fold (Fig. S7†).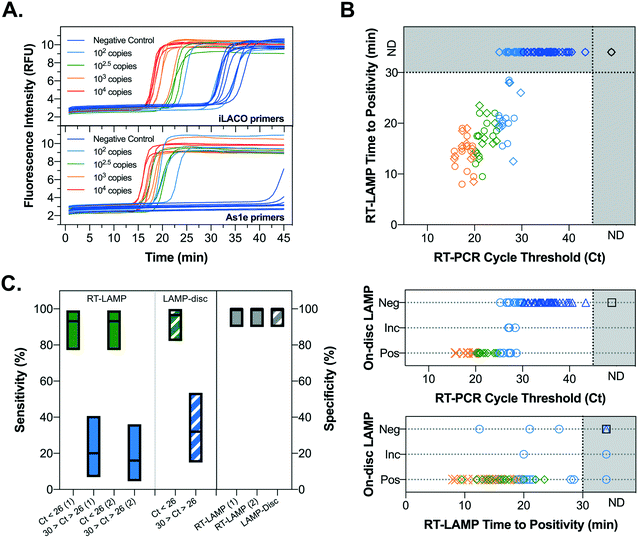 | ||
| Fig. 5 Optimization of primer set, estimation of sensitivity/specificity and benchmarking of the integrated platform with an expanded panel of heat-inactivated nasopharyngeal swab samples. A – Comparison iLACO and As1e primers with RT-LAMP (benchtop thermocycler) in the presence of increasing copies of full-length synthetic SARS-CoV-2 RNA (GenBank MN908947.3) spiked in water. B – RT-LAMP and on-disc LAMP measurements of 131 heat inactivated nasopharyngeal swab samples (96 samples with Ct values ranging from 15.7 to 43.45 and 35 RT-PCR negative samples) using the As1e primer set. Ct values were measured in a Cepheid GeneXpert system and are the average of E and N genes. Each symbol in the top plot corresponds to a technical replicate of each sample. Color codes (all plots) and symbol shapes (bottom plots) correspond to different Ct value ranges according to the legend of Fig. 5. The abbreviations “Neg”, “Inc” and “Pos” refer to negative, inconclusive or positive result using the integrated platform. C – Sensitivity and specificity for the RT-LAMP (benchtop thermocycler) and LAMP-disc (on-disc LAMP using the integrated platform) measurements. The plotted mean values and Clopper–Pearson 95% confidence intervals were calculated using MedCalc software (Ostend, Belgium) diagnostic test evaluation calculator. RT-LAMP (1) and (2) refer to the sensitivity determined for two technical replicates. To determine the specificity, only the PCR negative samples were considered (n = 35). Filled and dashed boxes refer to RT-LAMP and LAMP-disc measurements, respectively. | ||
The As1e primer set was subsequently used to test a wider panel of 131 nasopharyngeal swab samples (Table S1†) using RT-LAMP and the integrated platform in parallel. The results from the integrated platform were classified as (1) positive, (2) negative or (3) inconclusive according to the criteria detailed in Fig. S8,† highlighting a significant improvement in positive–negative result discrimination. The compiled results correlating the output of each LAMP assay with the Ct values of each sample and the time to positivity of RT-LAMP are shown in Fig. 5B (representative smartphone images shown as ESI,† Fig. S9). In both assays, all samples with Ct values below 25 (n = 23) resulted in a positive response, while all samples with Ct values above 30 (n = 77) resulted in a negative response. Comparing the output of the RT-LAMP and the integrated platform, deviations in positive, inconclusive or negative outcomes were observed only for samples with Ct values between 25 and 30 around the detection limit of the assay. The sensitivity for high (Ct < 26) and low (30 > Ct > 26) viral titers and overall specificity was then calculated for each replicate of the RT-LAMP and for the integrated platform according to Fig. 5C. The performance of the integrated platform was comparable to the RT-LAMP performed on a benchtop thermocycler for the same samples, resulting in a sensitivity of 96.6% (82.2–99.9%, 95% CI) and a specificity of 100% (90–100%, 95% CI) for samples with Ct values < 26. In both cases the sensitivity dropped significantly for Ct values > 26. While LAMP has been reported to provide high sensitivity at higher Ct values in the range of 25–30 for COVID-19 diagnostics, such performance can only be achieved with complex RNA extraction or sample clean-up procedures, which were not used in this case.40,51 Furthermore, considering that Ct values for RT-PCR vary significantly between laboratories (as much as 5 Ct for the 30–35 range),52 it was not possible to accurately quantify the absolute sensitivity of the developed platform for unprocessed clinical samples. Nevertheless, a Ct value of 26 using Cepheid GeneXpert corresponds broadly to a range of 1 × 104–1 × 105 copies per mL in transport medium according to previous reports using spiked samples.52
4. Conclusions
We introduce a novel portable bead-based centrifugal microfluidic platform and demonstrate LAMP-based viral RNA detection directly from heat-inactivated nasopharyngeal swab specimens. The platform achieves two major breakthroughs in the scope of point-of-care viral diagnostics. Firstly, a versatile agarose bead-based strategy was developed to significantly improve signal transduction after LAMP by removing the intrinsic background of primer dimer interactions when using intercalating fluorescent dyes. This strategy significantly improved fluorescence signal resolution between positive and negative samples compared with previous reports,53 allowing efficient detection without expensive band-pass filter setups and simple optics. Secondly, the developed centrifugal and heating modules serve as a complete package for sample-to-answer analysis directly from heat-inactivated nasopharyngeal samples in less than 1 hour of total processing time, combined with simple smartphone-based signal acquisition. In the context of the current SARS-CoV-2 pandemic, the full centrifugal platform is adaptable to any smartphone, costs less than 250 USD (Table S2†) and is compatible with cost-effective and scalable LAMP reagents under investigation.43 Furthermore, the reagents (including NBNM beads, buffers, intercalator and polymerase) for 20 parallel tests on a disc cost less than 2 USD excluding the relatively expensive SSIV reverse transcriptase which can be easily replaced by other cost-effective alternatives.43 The achieved sensitivity −96.6% (82.2–99.9%, 95% CI)- and specificity −100% (90–100%, 95% CI)- for Ct values below 26 (equivalent to approximately 1 × 104–1 × 105 copies per mL (ref. 52) according to previous estimates using the same PCR instrument) and inexpensive analysis are a suitable combination for frequent screening to allow detection of the spike in viral load early upon infection and minimize the risk of transmission.16 While the platform does not replace high-sensitivity PCR-based methods for confirmation and clinical follow-up of COVID-19 patients, its low cost allows tests to be performed more frequently and on the field. These features can potentially pave the way to bring routine and scalable diagnostics to RLS complemented by emerging portable platforms to determine disease severity,54 as well as expanding the current diagnostic capacities in high-income countries by bringing viral RNA detection directly to the field. Both these avenues are paramount to improve the containment of viral spread and simultaneously minimize the need for extreme lockdown policies.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work has been sponsored in part by the European Union's Horizon 2020 research and innovation programme “New Diagnostics for Infectious Diseases” (ND4ID) under the Marie Skłodowska-Curie Grant agreement No. 675412. The authors acknowledge the Innovative Medicines Initiative (IMI) 2 Joint Undertaking (iConsensus, grant agreement no. 777397) for partially funding this research. VP is supported by a SciLifeLab/KAW national COVID-19 research program project Grant (KAW 2020.0182), Yin is supported by Ganzhou COVID-19 Emergency Research Project, Key Special Project of “Technology Boosts Economy 2020” of Ministry of Science and Technology (SQ2020YFF0411358) for COVID-19 related work. The authors acknowledge Alisa Alekseenko and Yerma Pareja-Sanchez for technical assistance with the RT-LAMP.References
- L. D. Racsa, C. S. Kraft, G. G. Olinger and L. E. Hensley, Clin. Infect. Dis., 2016, 62, 214–219 Search PubMed.
- R. J. Shorten, C. S. Brown, M. Jacobs, S. Rattenbury, A. J. Simpson and S. Mepham, PLoS Neglected Trop. Dis., 2016, 10, e0004948 Search PubMed.
- K. Weidemaier, J. Carrino, A. Curry, J. H. Connor and A. Liebmann-Vinson, Future Virol., 2015, 10, 313–328 Search PubMed.
- M. J. MacKay, A. C. Hooker, E. Afshinnekoo, M. Salit, J. Kelly, J. V. Feldstein, N. Haft, D. Schenkel, S. Nambi, Y. Cai, F. Zhang, G. Church, J. Dai, C. L. Wang, S. Levy, J. Huber, H. P. Ji, A. Kriegel, A. L. Wyllie and C. E. Mason, Nat. Biotechnol., 2020, 38, 1021–1024, DOI:10.1038/s41587-020-0655-4.
- R. Lu, X. Zhao, J. Li, P. Niu, B. Yang, H. Wu, W. Wang, H. Song, B. Huang, N. Zhu, Y. Bi, X. Ma, F. Zhan, L. Wang, T. Hu, H. Zhou, Z. Hu, W. Zhou, L. Zhao, J. Chen, Y. Meng, J. Wang, Y. Lin, J. Yuan, Z. Xie, J. Ma, W. J. Liu, D. Wang, W. Xu, E. C. Holmes, G. F. Gao, G. Wu, W. Chen, W. Shi and W. Tan, Lancet, 2020, 395, 565–574 Search PubMed.
- K. K.-W. To, O. T.-Y. Tsang, W.-S. Leung, A. R. Tam, T.-C. Wu, D. C. Lung, C. C.-Y. Yip, J.-P. Cai, J. M.-C. Chan, T. S.-H. Chik, D. P.-L. Lau, C. Y.-C. Choi, L.-L. Chen, W.-M. Chan, K.-H. Chan, J. D. Ip, A. C.-K. Ng, R. W.-S. Poon, C.-T. Luo, V. C.-C. Cheng, J. F.-W. Chan, I. F.-N. Hung, Z. Chen, H. Chen and K.-Y. Yuen, Lancet Infect. Dis., 2020, 20, 565–574 Search PubMed.
- Y. Pan, D. Zhang, P. Yang, L. L. M. Poon and Q. Wang, Lancet Infect. Dis., 2020, 20, 411–412 Search PubMed.
- R. Li, S. Pei, B. Chen, Y. Song, T. Zhang, W. Yang and J. Shaman, Science, 2020, 368, 489–493 Search PubMed.
- K. A. Walsh, K. Jordan, B. Clyne, D. Rohde, L. Drummond, P. Byrne, S. Ahern, P. G. Carty, K. K. O'Brien, E. O'Murchu, M. O'Neill, S. M. Smith, M. Ryan and P. Harrington, J. Infect., 2020, 81, 357–371 Search PubMed.
- J. J. Waggoner, V. Stittleburg, R. Pond, Y. Saklawi, M. K. Sahoo, A. Babiker, L. Hussaini, C. S. Kraft, B. A. Pinsky, E. J. Anderson and N. Rouphael, Emerging Infect. Dis., 2020, 26, 1633–1635 Search PubMed.
- X. Wang, H. Yao, X. Xu, P. Zhang, M. Zhang, J. Shao, Y. Xiao and H. Wang, Clin. Chem., 2020, 66, 977–979 Search PubMed.
- V. M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer, D. K. Chu, T. Bleicker, S. Brunink, J. Schneider, M. L. Schmidt, D. G. Mulders, B. L. Haagmans, B. van der Veer, S. van den Brink, L. Wijsman, G. Goderski, J. L. Romette, J. Ellis, M. Zambon, M. Peiris, H. Goossens, C. Reusken, M. P. Koopmans and C. Drosten, Eurosurveillance, 2020, 25(3), 23–30 Search PubMed.
- F. Yu, L. Yan, N. Wang, S. Yang, L. Wang, Y. Tang, G. Gao, S. Wang, C. Ma, R. Xie, F. Wang, C. Tan, L. Zhu, Y. Guo and F. Zhang, Clin. Infect. Dis., 2020, 71, 793–798 Search PubMed.
- R. Wolfel, V. M. Corman, W. Guggemos, M. Seilmaier, S. Zange, M. A. Muller, D. Niemeyer, T. C. Jones, P. Vollmar, C. Rothe, M. Hoelscher, T. Bleicker, S. Brunink, J. Schneider, R. Ehmann, K. Zwirglmaier, C. Drosten and C. Wendtner, Nature, 2020, 581, 465–469 Search PubMed.
- J. J. A. V. Kampe, D. A. M. C. van de Vijver, P. L. A. Fraaij, B. L. Haagmans, M. M. Lamers, N. Okba, J. P. C. van den Akker, H. Endeman, D. A. M. P. J. Gommers, J. J. Cornelissen, R. A. S. Hoek, M. M. V. der Eerden, D. A. Hesselink, H. J. Metselaar, A. Verbon, J. E. M. D. Steenwinkel, G. I. Aron, E. C. M. V. Gorp, S. V. Boheemen, J. C. Voermans, C. A. B. Boucher, R. Molenkamp, M. P. G. Koopmans, C. Geurtsvankessel and A. A. V. D. Eijk, Nat. Commun., 2021, 12, 267, DOI:10.1101/2020.06.08.20125310.
- M. J. Mina, R. Parker and D. B. Larremore, N. Engl. J. Med., 2020, 383, e120, DOI:10.1056/NEJMp2025631.
- J. Dinnes, J. J. Deeks, A. Adriano, S. Berhane, C. Davenport, S. Dittrich, D. Emperador, Y. Takwoingi, J. Cunningham, S. Beese, J. Dretzke, L. Ferrante di Ruffano, I. M. Harris, M. J. Price, S. Taylor-Phillips, L. Hooft, M. M. Leeflang, R. Spijker, A. Van den Bruel and Cochrane COVID-19 Diagnostic Test Accuracy Group, Cochrane Database Syst. Rev., 2020, 8, CD013705 Search PubMed.
- A. Kalk and A. Schultz, Lancet Infect. Dis., 2020, 20, 1370, DOI:10.1016/s1473-3099(20)30563-6.
- H. Zhang, Y. Xu, Z. Fohlerova, H. Chang, C. Iliescu and P. Neuzil, Trends Analyt. Chem., 2019, 113, 44–53 Search PubMed.
- S. J. Oh, B. H. Park, J. H. Jung, G. Choi, D. C. Lee, D. H. Kim and T. S. Seo, Biosens. Bioelectron., 2016, 75, 293–300 Search PubMed.
- B. H. Park, S. J. Oh, J. H. Jung, G. Choi, J. H. Seo, D. H. Kim, E. Y. Lee and T. S. Seo, Biosens. Bioelectron., 2017, 91, 334–340 Search PubMed.
- H. Yan, Y. Zhu, Y. Zhang, L. Wang, J. Chen, Y. Lu, Y. Xu and W. Xing, Sci. Rep., 2017, 7, 1460 Search PubMed.
- A. A. Sayad, F. Ibrahim, S. M. Uddin, K. X. Pei, M. S. Mohktar, M. Madou and K. L. Thong, Sens. Actuators, B, 2016, 227, 600–609 Search PubMed.
- D. Liu, Y. Zhu, N. Li, Y. Lu, J. Cheng and Y. Xu, Sens. Actuators, B, 2020, 310, 127834 Search PubMed.
- F. Tian, C. Liu, J. Deng, Z. Han, L. Zhang, Q. Chen and J. Sun, Sci. China: Chem., 2020, 1–9, DOI:10.1007/s11426-020-9800-6.
- E. S. Yamanaka, L. A. Tortajada-Genaro, N. Pastor and A. Maquieira, Biosens. Bioelectron., 2018, 109, 177–183 Search PubMed.
- H. Wang, Z. Ma, J. Qin, Z. Shen, Q. Liu, X. Chen, H. Wang, Z. An, W. Liu and M. Li, Biosens. Bioelectron., 2019, 126, 373–380 Search PubMed.
- L. Wan, J. Gao, T. Chen, C. Dong, H. Li, Y. Z. Wen, Z. R. Lun, Y. Jia, P. I. Mak and R. P. Martins, Biomed. Microdevices, 2019, 21, 9 Search PubMed.
- K. R. Jackson, T. Layne, D. A. Dent, A. Tsuei, J. Li, D. M. Haverstick and J. P. Landers, Forensic Sci. Int.: Genet., 2020, 45, 102195 Search PubMed.
- J. Hui, Y. Gu, Y. Zhu, Y. Chen, S. J. Guo, S. C. Tao, Y. Zhang and P. Liu, Lab Chip, 2018, 18, 2854–2864 Search PubMed.
- S. Wang, N. Liu, L. Zheng, G. Cai and J. Lin, Lab Chip, 2020, 20, 2296–2305 Search PubMed.
- K. Kaarj, P. Akarapipad and J. Y. Yoon, Sci. Rep., 2018, 8, 12438 Search PubMed.
- S.-C. Liao, J. Peng, M. G. Mauk, S. Awasthi, J. Song, H. Friedman, H. H. Bau and C. Liu, Sens. Actuators, B, 2016, 229, 232–238 Search PubMed.
- L. Yu, S. Wu, X. Hao, X. Dong, L. Mao, V. Pelechano, W.-H. Chen and X. Yin, Clin. Chem., 2020, 66, 975–977 Search PubMed.
- C. Yan, J. Cui, L. Huang, B. Du, L. Chen, G. Xue, S. Li, W. Zhang, L. Zhao, Y. Sun, H. Yao, N. Li, H. Zhao, Y. Feng, S. Liu, Q. Zhang, D. Liu and J. Yuan, Clin. Microbiol. Infect., 2020, 26, 773–779 Search PubMed.
- Y. H. Baek, J. Um, K. J. C. Antigua, J. H. Park, Y. Kim, S. Oh, Y. I. Kim, W. S. Choi, S. G. Kim, J. H. Jeong, B. S. Chin, H. D. G. Nicolas, J. Y. Ahn, K. S. Shin, Y. K. Choi, J. S. Park and M. S. Song, Emerging Microbes Infect., 2020, 9, 998–1007 Search PubMed.
- G. S. Park, K. Ku, S. H. Baek, S. J. Kim, S. I. Kim, B. T. Kim and J. S. Maeng, J. Mol. Diagn., 2020, 22, 729–735 Search PubMed.
- W. E. Huang, B. Lim, C. C. Hsu, D. Xiong, W. Wu, Y. Yu, H. Jia, Y. Wang, Y. Zeng, M. Ji, H. Chang, X. Zhang, H. Wang and Z. Cui, Microb. Biotechnol., 2020, 13, 950–961 Search PubMed.
- L. E. Lamb, S. N. Bartolone, E. Ward and M. B. Chancellor, PLoS One, 2020, 15, e0234682 Search PubMed.
- V. L. D. Thi, K. Herbst, K. Boerner, M. Meurer, L. P. Kremer, D. Kirrmaier, A. Freistaedter, D. Papagiannidis, C. Galmozzi, M. L. Stanifer, S. Boulant, S. Klein, P. Chlanda, D. Khalid, I. B. Miranda, P. Schnitzler, H.-G. Kräusslich, M. Knop and S. Anders, Sci. Transl. Med., 2020, 12, eabc7075 Search PubMed.
- I. F. Pinto, C. R. F. Caneira, R. R. G. Soares, N. Madaboosi, M. R. Aires-Barros, J. P. Conde, A. M. Azevedo and V. Chu, Methods, 2017, 116, 112–124 Search PubMed.
- B. A. Rabe and C. Cepko, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 24450–24458 Search PubMed.
- A. Alekseenko, D. Barrett, Y. Pareja-Sanchez, R. J. Howard, E. Strandback, H. Ampah-Korsah, U. Rovsnik, S. Zuniga-Veliz, A. Klenov, J. Malloo, S. Ye, X. Liu, B. Reinius, S. J. Elsasser, T. Nyman, G. Sandh, X. Yin and V. Pelechano, Sci. Rep., 2021, 11, 1820 Search PubMed.
- P. Novo, V. Chu and J. P. Conde, Lab Chip, 2014, 14, 1991–1995 Search PubMed.
- R. J. Meagher, A. Priye, Y. K. Light, C. Huang and E. Wang, Analyst, 2018, 143, 1924–1933 Search PubMed.
- C. S. Ball, Y. K. Light, C. Y. Koh, S. S. Wheeler, L. L. Coffey and R. J. Meagher, Anal. Chem., 2016, 88, 3562–3568 Search PubMed.
- L. Becherer, N. Borst, M. Bakheit, S. Frischmann, R. Zengerle and F. von Stetten, Anal. Methods, 2020, 12, 717–746 Search PubMed.
- I. F. Pinto, M. R. Aires-Barros and A. M. Azevedo, Pharm. Bioprocess., 2015, 3, 263–279 Search PubMed.
- T. Matos, J. A. Queiroz and L. Bülow, J. Mol. Recognit., 2014, 27, 184–189 Search PubMed.
- S. Carturan, A. Quaranta, M. Bonafini, A. Vomiero, G. Maggioni, G. Mattei, C. de Julián Fernández, M. Bersani, P. Mazzoldi and G. Della Mea, Eur. Phys. J. D, 2007, 42, 243–251 Search PubMed.
- V. L. Fowler, B. Armson, J. L. Gonzales, E. L. Wise, E. L. A. Howson, Z. Vincent-Mistiaen, S. Fouch, C. J. Maltby, S. Grippon, S. Munro, L. Jones, T. Holmes, C. Tillyer, J. Elwell, A. Sowood, O. de Peyer, S. Dixon, T. Hatcher, H. Patrick, S. Laxman, C. Walsh, M. Andreou, N. Morant, D. Clark, N. Moore, R. Houghton, N. J. Cortes and S. P. Kidd, J. Infect., 2021, 82, 117–125 Search PubMed.
- F. Wolters, J. van de Bovenkamp, B. van den Bosch, S. van den Brink, M. Broeders, N. H. Chung, B. Favie, G. Goderski, J. Kuijpers, I. Overdevest, J. Rahamat-Langedoen, L. Wijsman, W. J. Melchers and A. Meijer, J. Clin. Virol., 2020, 128, 104426 Search PubMed.
- A. Ganguli, A. Mostafa, J. Berger, M. Y. Aydin, F. Sun, S. A. S. Ramirez, E. Valera, B. T. Cunningham, W. P. King and R. Bashir, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 22727–22735 Search PubMed.
- M. P. McRae, G. W. Simmons, N. J. Christodoulides, Z. Lu, S. K. Kang, D. Fenyo, T. Alcorn, I. P. Dapkins, I. Sharif, D. Vurmaz, S. S. Modak, K. Srinivasan, S. Warhadpande, R. Shrivastav and J. T. McDevitt, Lab Chip, 2020, 20, 2075–2085 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1lc00266j |
| This journal is © The Royal Society of Chemistry 2021 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536) divided by 1000.
536) divided by 1000.