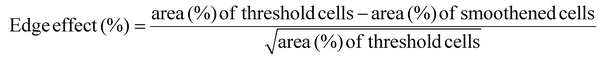Open Access Article
Open Access ArticleBicyclic RGD peptides enhance nerve growth in synthetic PEG-based Anisogels†
Sitara
Vedaraman
 ab,
Dominik
Bernhagen
c,
Tamas
Haraszti
ab,
Dominik
Bernhagen
c,
Tamas
Haraszti
 ab,
Christopher
Licht
ab,
Arturo
Castro Nava
ab,
Christopher
Licht
ab,
Arturo
Castro Nava
 ab,
Abdolrahman
Omidinia Anarkoli
ab,
Peter
Timmerman
ab,
Abdolrahman
Omidinia Anarkoli
ab,
Peter
Timmerman
 cd and
Laura
De Laporte
cd and
Laura
De Laporte
 *abe
*abe
aDWI Leibniz Institute for Interactive Materials, Forckenbeckstrasse 50, 52074 Aachen, Germany
bInstitute for Technical and Macromolecular Chemistry, RWTH Aachen, Worringerweg 1-2, 52074 Aachen, Germany. E-mail: delaporte@dwi.rwth-aachen.de
cPepscan Therapeutics, Zuidersluisweg 2, 8243 RC Lelystad, the Netherlands
dVan't Hoff Institute for Molecular Sciences, University of Amsterdam, Science Park 904, 1098 XH Amsterdam, the Netherlands
eInstitute of Applied Medical Engineering, RWTH University, Pauwelsstraße 20, 52074 Aachen, Germany
First published on 16th March 2021
Abstract
Nerve regeneration scaffolds often consist of soft hydrogels modified with extracellular matrix (ECM) proteins or fragments, as well as linear and cyclic peptides. One of the commonly used integrin-mediated cell adhesive peptide sequences is Arg-Gly-Asp (RGD). Despite its straightforward coupling mechanisms to artificial extracellular matrix (aECM) constructs, linear RGD peptides suffer from low stability towards degradation and lack integrin selectivity. Cyclization of RGD improves the affinity towards integrin subtypes but lacks selectivity. In this study, a new class of short bicyclic peptides with RGD in a cyclic loop and ‘random screened’ tri-amino acid peptide sequences in the second loop is investigated as a biochemical cue for cell growth inside three-dimensional (3D) synthetic poly(ethylene glycol) (PEG)-based Anisogels. These peptides impart high integrin affinity and selectivity towards either αvβ3 or α5β1 integrin subunits. Enzymatic conjugation of such bicyclic peptides to the PEG backbone enables the formulation of an aECM hydrogel that supports nerve growth. Furthermore, different proteolytic cleavable moieties are incorporated and compared to promote cell migration and proliferation, resulting in enhanced cell growth with different degradable peptide crosslinkers. Mouse fibroblasts and primary nerve cells from embryonic chick dorsal root ganglions (DRGs) show superior growth in bicyclic RGD peptide conjugated gels selective towards αvβ3 or α5β1, compared to monocyclic or linear RGD peptides, with a slight preference to αvβ3 selective bicyclic peptides in the case of nerve growth. Synthetic Anisogels, modified with bicyclic RGD peptides and containing short aligned, magneto-responsive fibers, show oriented DRG outgrowth parallel to the fibers. This report shows the potential of PEG hydrogels coupled with bicyclic RGD peptides as an aECM model and paves the way for a new class of integrin selective biomolecules for cell growth and nerve regeneration.
Introduction
The native neuronal tissue is soft (0.1–1 kPa) and consists of a highly ordered microarchitecture to enable connections between neurons and their supporting cells to form a functional neural network.1 Efficient nerve regeneration in an artificial extracellular matrix (aECM) is achieved by mimicking the extracellular matrices (ECM) with three-dimensional (3D) scaffolds where the cells inside the scaffold experience a combination of chemical, physical, and architectural cues.2,3Ex vivo mimicry of biological systems is established to create relevant models and micro-environments that enable cells to adhere, proliferate and migrate either by squeezing through the aECM (amoeboid) or by degrading their local environment by secreting proteases (mesenchymal).4 Most of the studied ECM mimicking neural niches and 3D scaffolds are based on endogenous protein sources, such as collagen,5 fibrin,6,7 hyaluronic acid,8,9 or a combination of these.10,11 Although proteinaceous gels or scaffolds offer a natural environment for cell growth, their batch to batch variability and lack of controlled properties are limiting factors. On the other hand, scaffolds made from synthetic polymers or other macromolecules are reproducible, more resistant to immunogenic reactions, and easy to vary in properties, such as stiffness, degradability, architecture, biomolecule modification, etc.12 To combine the advantages of both natural and synthetic building blocks, hybrid materials are often employed. Arginylglycylaspartic acid (Arg-Gly-Asp or RGD) is the smallest integrin recognizable ligand present in vivo, which is widely used to induce cell adhesion in synthetic scaffolds.13 For example, elastin-like proteins (ELP) with controlled RGD ligand density and optimized scaffold stiffness enabled neurite outgrowth of chick dorsal root ganglions (DRGs).14 Semi-synthetic scaffolds, prepared from PEGylated albumin or PEGylated fibrinogen enabled nerve growth at low concentrations of PEG (1.6 w/v% and 2.3 w/v%, respectively) with 0.8 w/v% of the respective proteins.15 Besides RGD, the laminin-derived adhesion peptide IKVAV is often employed to induce vascularization and innervation, for example in composite hydrogels consisting of elastin-like polypeptides and PEG, creating a microporous environment with controlled mechanical properties.16 In the case of non-endogenous sources, proteinaceous gels, such as silk17 and alginate18-based hydrogels provide a promising environment as a 3D neuronal scaffold.In the case of polymeric hydrogel scaffolds, the mechanical properties can be varied by the composition, structure, molecular weight, reaction mechanism, and concentration of the pre-polymer, and optimized to support nerve growth and axon protrusion.19,20 Inside these constructs, spatial distribution14,21 of integrin-binding ligands and the internal structure are crucial factors in bio-mimicking the native cell microenvironment. Designing hydrogels with large pores that facilitate perfusion of growth factors and nutrients,22 while at the same time permitting temporally controlled degradation is primordial to promote cell proliferation and enable cell motility and scaffold invasion.23 Among the several reported polymeric hydrogel systems, PEG-based networks offer a biocompatible microenvironment for cells with a low risk of immune reaction. By using degradable crosslinks, the degradation products can be eliminated from the body via natural biochemical mechanisms.24–26 The PEG system used in this report employs an enzymatic crosslinking and bio-conjugation method, inspired by fibrin clotting,27 while degradation moieties are included in the backbone to enable proteolytic degradation by matrix metalloproteases (MMPs).24,28 These MMP sensitive domains in the crosslinker degrade when cells produce MMPs, resulting in gel degradation on cell-demand and providing space for growing cell populations. Such a system has been reported by our group to promote 3D nerve growth when a specially designed fibronectin fragment FNIII9*-10/12-14 was coupled inside the gel.20 Compared to linear RGD, much lower concentrations of the fragment were sufficient to support cell growth. FNIII9*-10/12-14 has previously demonstrated to enhance wound and bone regeneration when bound to a fibrin gel, likely due to the synergistic effect of providing integrin binding domains adjacent to growth factor binding domains.29 In the case of RGD coupling to a synthetic fibrin-mimicking PEG gel, Ehrbar et al. determined the binding efficiency via the transglutaminase reaction using release studies by reverse phase chromatography and mass spectroscopy, resulting in ∼72% RGD binding efficiency for 5 wt/v% gels in the case of 4.3% RGD peptides of total PEG-K arms.30 We previously translated these gels with FNIII9*-10/12-14 to Anisogels to create anisotropy imparting gels by incorporating magneto-responsive rod-shaped microgels or short fibers and aligning them along external magnetic field lines to promote unidirectional neurite extension.20
Integrin binding domains, incorporated inside aECMs are involved in bidirectional transmembrane signalling between cells and their environment and play an important role in cell growth.31 Integrins are involved in the regulation of physiological processes such as migration, proliferation, survival, and apoptosis. The focal adhesion mechanisms between the ECM ligand and the integrin receptor heterodimers, consisting of α and β integrin subunits, have been extensively studied.32,33 Linear RGD peptides, cyclic RGD peptides, and peptidomimetic ligands have evolved progressively for binding integrin subunits.34 Flanking linear RGD peptides with amino acids, such as glycine (G) at the N terminus and serine (S) at the C terminus, results in a decrease in half-maximum inhibitory concentration (IC50) values by 3 and 4 folds for αvβ3 and α5β1 integrin subunits, respectively.35 Lower IC50 values indicate a higher affinity to integrin subunits and the lowest IC50 value is observed for heptapeptide GRGDSPK, which is 7 folds lower than for tripeptide RGD in the case of αvβ3.35 Similarly, the cyclization of linear RGD peptides (RGDfK) and incorporation of one D-amino acid residue between Asp (D) and Lys (K) improves affinity to integrin subunits and increases stability towards enzymatic degradation.35,36 Monocyclic RGD peptides show moderate to low IC50 values towards multiple integrin subunits, such as αvβ3, αvβ5, αvβ6, and α5β1. The lowest IC50 values are observed for αvβ3 subunits and cyclization improves integrin αvβ3 binding affinity in the range of 1.5–6.0 nM and integrin α5β1 binding affinity in the range of 141–236 nM.35 Similar to flanking in linear RGD, flanking in cyclic RGD peptides with linear polyproline improves cell spreading and focal adhesions in rat embryo fibroblasts in comparison to other spacers like PEG.37 Another class of high affinity and selective integrin-binding ligands are the peptidomimetics, which can distinguish between different integrin subtypes in vitro and in vivo.38,39
It is important to understand the role of integrin-mediated adhesion to its corresponding ligands in the ECM for achieving directed cell fate in tissue engineering applications. For example, the ability of cells to sense the mechanical properties of its fibronectin based ECM is mediated by the synergy between the α5β1 and αv-class integrin subunits.40 Similarly, the inter-dependence of integrins (α5β1, αvβ3) employed during focal adhesions is explored on 2D platforms with integrin specific RGD-coupled gold nanoparticles on PEG gels. The study revealed that during focal adhesion, the recruitment of αvβ3 integrins occurs on patterned substrates that are exclusive for α5β1, but the vice versa is not true.41 In addition, 3D hyaluronic acid hydrogels with specific integrin activation (α3/α5β1) induced sprouting in an endothelial cell angiogenic model in vitro and enabled the formation of non-leaky blood vessels in stroke models.8 Such examples show the importance of integrin-mediated cell adhesion and migration, which in turn determines cell fate.
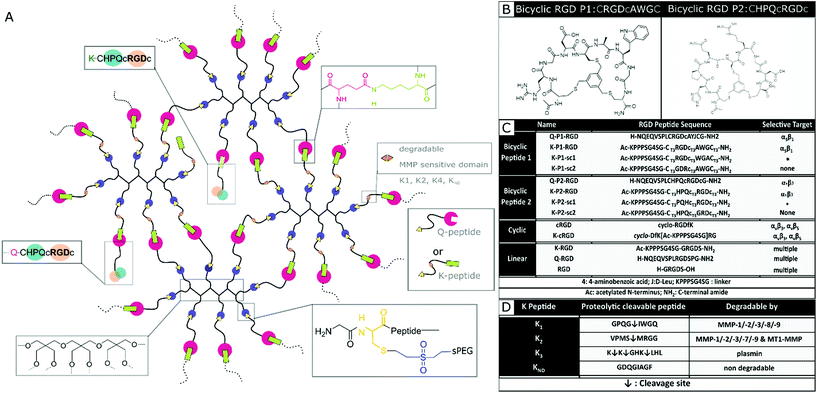
In this report, a new class of integrin selective engineered bicyclic peptides with one loop containing the integrin-binding RGD peptide is employed to study neuronal growth in 3D PEG-based hydrogels and Anisogels. These peptides have demonstrated greater conformational rigidity, metabolic stability, and most importantly, high integrin selectivity. The primary loop consists of an RGD peptide flanked with cysteine residues that bind to a tri-covalent scaffold (1,3,5-tris(bromomethyl)benzene). The secondary loop consists of a tri amino-acid sequence, which is screened using a ‘random approach’ and imparts the potential to affect the RGD selectivity towards different integrin subunits (αvβ3 or α5β1).42,43 Elastin-like recombinamers (ELR), produced with recombinant proteins and coupled with bicyclic RGD peptides selective for α5β1 show an increase in HUVEC proliferation at early periods of culture.44 Here, we describe for the first time the effect of bicyclic RGD peptides for nerve growth in synthetic 3D PEG hydrogels to study the affinity towards selective integrin sites. Soft, synthetic, degradable multi-arm PEG hydrogels are enzymatically crosslinked using transglutaminase and conjugated with different bicyclic RGD peptides. Domains cleavable by MMP 1, MMP 2, and plasmin are incorporated in the PEG precursors to enable on cell-demand degradation at the crosslinking arms. The MMP 1 and MMP 2 sensitive domains enable extensive cell growth inside the hydrogel. Expansion of both mouse fibroblasts and primary nerve cells from chick DRGs is investigated and compared with linear and monocyclic RGD and a specific fibronectin fragment FNIII9*-10/12-1429 (FN*). Bicyclic RGD conjugated gels show superior nerve and fibroblast growth. Also, cell growth and innervation in PEG gels are dependent on the type of proteolytic cleavable MMP-sensitive peptide present in the crosslinker. Finally, fully synthetic Anisogels are prepared by mixing and orienting short, magneto-responsive polycaprolactone (PCL) fibers (diameter ∼5 μm, length ∼50 μm, 1 vol%) before crosslinking of the surrounding PEG hydrogels, modified with the bicyclic peptides and optimal degradation domain VPMS↓MRGG (K2). Embedded DRGs inside Anisogels, tethered with bicyclic RGD peptides (K-P2 RGD), linear RGD (K-RGD) and fibronectin fragment (FN*) demonstrate unidirectional nerve growth.
Results and discussion
Designing RGD conjugated PEG hydrogels
Nerve and fibroblast cell growth in synthetic PEG hydrogels is compared among bicyclic peptides and standard RGD peptides, commonly used for biomaterial development. The RGD peptides are tethered to the PEG hydrogels (Fig. 1a) either by its amine in the lysine (K), its amide in glutamine (Q), or via the amine from the N terminus of any unmodified RGD peptide. The RGD peptides with K or Q residues bind to the arms PEG-Q or PEG-K, respectively. Transglutaminase reaction between the Q and the K residues is initiated in the presence of an enzyme (activated FXIII), resulting in the formation of an iso-peptide bond.45 The multiple arms in PEG facilitate crosslinking and bio-conjugation simultaneously. PEG polymer is used at 1 wt/v% to fabricate hydrogels with a storage modulus of ∼10 Pa,20 to which a fixed concentration (100 μM) of RGD peptide is conjugated. In a previous report, we demonstrate superior cell growth using this RGD concentration.20 The proteolytic cleavable sites are incorporated in the arms of PEG-K; PEG-K1: FKGG-GPQG↓IWGQ-PEG (K1), PEG-K2: FKGG-VPMS↓MRGG-PEG (K2), PEG-K3: FKGG-K↓K↓GHK↓LHL-PEG (K3), PEG-KND: FKGG-GDQGIAGF-PEG (KND) (Fig. 1d), where KND is an 8 amino acid sequence not degraded by MMPs. These MMP sensitive domains are cleaved by proteases.24,46Mixing the precursor solution with the cells and controlling the crosslinking rate by the concentration of activated Factor XIII (FXIIIa) enables 3D encapsulation of cells. Bicyclic RGD peptides, engineered by Bernhagen et al., contain two loops, consisting of an RGD sequence and integrin selectivity imparting sequence, where Ala-Trp-Gly (AWG) results in selectivity towards α5β1 and loop His-Pro-Gln (HPQ) leads to selectivity towards αvβ3 (Fig. 1b). Bicyclic RGD peptides (Fig. 1c) (K-linker-CT3RGDcT3AWGCT3,43 NQEQVSPL-CT3RGDcT3AYJCT3G, K-linker-CT3HPQcT3RGDcT3,42 NQEQVSPLCT3HPQcT3RGDcT3G,), cyclic RGD peptides (cyclo-RGDfK,35 cyclo-DfK[K-linker]RG), and linear RGD peptides (K-linker-GRGDS, NQEQVSPLRGDSPG, GRGDS35) are used as cell adhesive domains in this study with controls, such as scrambled second loops (K-linker-CT3RGDcT3WGACT3, K-linker-CT3PQHcT3RGDcT3) and scrambled RGD peptides (K-linker-CT3GDRcT3AWGCT3, K-linker-CT3HPQcT3GRDcT3). Fibronectin fragment FNIII 9*-10/12-14 is used as positive control and PEG gels without RGD as another negative control.
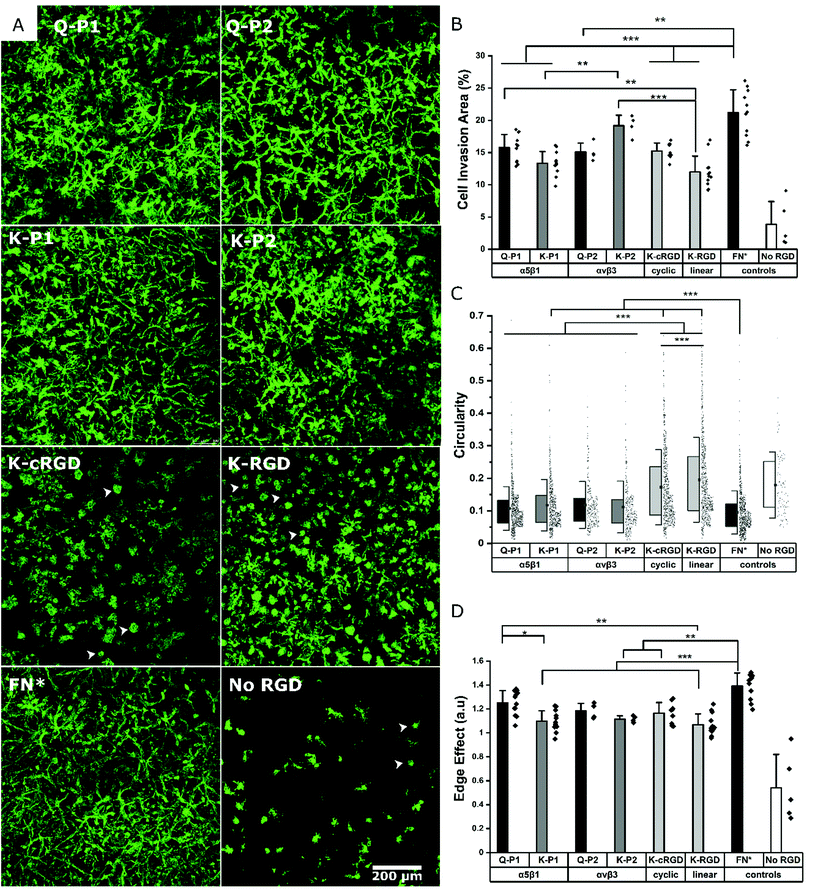
Influence of RGD peptides on fibroblast growth
Hydrogels conjugated with RGD peptides enable the spatial distribution of integrin-binding ligands for cell adhesion, spreading, and proliferation. These peptides are classified based on their structure as linear, cyclic, and bicyclic, reporting an increase in integrin affinity and selectivity based on IC50 from former to the latter.35,43,47 L929 mouse fibroblasts are encapsulated within the hydrogel at a fixed cell concentration. 3D cell growth in multiple planes is quantified after staining for actin filaments and imaging as z-stacks. A 200 μm z-stack image (Fig. S1†) of cells in a K-P2 conjugated hydrogel is represented by a color-coded depth profile, showing homogeneous cell distribution. Cells show different morphologies depending on the type of RGD peptide tethered to the hydrogels (Fig. 2a).The cells inside the hydrogels with bicyclic (K-P1, K-P2) and monocyclic (K-cRGD) RGD peptides spread and stretch to form network-like structures, in contrast to cells in gels with linear RGD (K-RGD) or no RGD. For hydrogels, modified with linear RGD, only some cells stretch, while others are more rounded with small protrusions (white arrows) resembling filipodia. In the control condition without RGD, most cells appear rounded, even though some cells are able to spread, likely due to their natural production of ECM proteins and proteases. The superimposed z stacks of the actin-stained images are compared for their cell invasion area (%) per field of view (Fig. 2b). K-P2 is the only peptide that statistically leads to similar cell invasion, compared to FN*, suggesting that the fibroblasts used in this culture may be more sensitive to αvβ3 binding domains. This is in agreement with the fact that the mono-cyclic peptide (more selective to αvβ3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35) leads to better cell invasion compared to the linear RGD peptide, which has 5× lower affinity to αvβ3 and binds to both αvβ3 and α5β1.35 In addition, none of the RGD peptides have the synergistic domain PHSRN, which is required for efficient α5β1 binding.48 This could explain the fact that the bicyclic peptide P2 shows a higher binding affinity towards αvβ3 (IC50: 30 nM
35) leads to better cell invasion compared to the linear RGD peptide, which has 5× lower affinity to αvβ3 and binds to both αvβ3 and α5β1.35 In addition, none of the RGD peptides have the synergistic domain PHSRN, which is required for efficient α5β1 binding.48 This could explain the fact that the bicyclic peptide P2 shows a higher binding affinity towards αvβ3 (IC50: 30 nM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 47) compared to peptide P1 towards α5β1 (IC50: 173 nM
47) compared to peptide P1 towards α5β1 (IC50: 173 nM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43).
43).
Among the bicyclic RGD peptides tethered via K, the peptide K-P2 show significantly higher cell invasion compared to K-P1, however, no such difference is observed in the case of bicyclic RGD tethered via Q. In addition, the K-P2 peptide leads to higher cell invasion compared to Q-P2, with the K-peptide having the shorter linker. Factors affecting the performance of a bicyclic peptide leading to the differences in cell behaviour can thus be attributed towards their Q or K residue peptide linkers, spatial availability of RGD peptide, and binding efficiency. When comparing the RGD peptides with fibronectin fragment FNIII9*-10/12-14 (5 μM, FN*),20 the fragment results in a more dense cell network. We reason that FN* provides a more natural ECM mimicking environment, compared to the RGD peptides, as the RGD sequence in FNIII10 can work together with its synergistic PHSRN sequence in FNIII9*,48 which is not present in any of the peptides used. In this experiment, linear K-RGD is employed to compare with the K-cRGD as both Q-RGD (100 μM) and K-RGD (100 μM) have a similar RGD potency when a proliferation assay is performed with L929 fibroblasts grown on thin PEG substrates (Fig. S2†). Further, a comparison of cell invasion in 3D gels, modified with Q-RGD (100 μM) or FN* (5 μM), leads to a similar Q-RGD vs. FN* ratio (ratio: 0.59) (Fig. S3†) as when comparing K-RGD (100 μM) with 5 μM FN* (ratio: 0.57) (Fig. 2b).
In parallel to the cell invasion area (%), which is a function of cell proliferation inside the hydrogels, the shape factors of the invading cells is investigated by quantifying the circularity and edge effect of the cells or cell clusters, which account for the shape, as well as the size of the cells. Since most of the hydrogel conditions support cell spreading, it is difficult to image single cells inside the network of fibroblasts. Hence, a circularity analysis tool (Fig. S4†) is employed to compute the perimeter of single cells or cell clusters. Circularity measures the irregular shape of spread cells and cell clusters, with a circularity of 1.0 indicating a rounded cell and a well-spread cell being represented by a circularity closer to 0. All bicyclic RGD peptides Q-P1, Q-P2, K-P1, and K-P2 show significantly lower circularity (0.10 to 0.12) compared to cyclic RGD (K-cRGD, 0.16) and linear (K-RGD, 0.19; Q-RGD, 0.15) (Fig. 2c, Fig. S3†). FN* still has the lowest circularity (0.09), validating the high network-like cell spreading observed in the fluorescent micrographs. No statistical difference in circularity is observed between the bicyclic peptides, inferring to the possible recruitment of multiple integrins during fibroblast cell adhesion and migration. Multiple integrin receptors contribute during focal adhesion kinase mediated fibroblast migration, such as α5β1 and α4β1 playing a major role and αvβ3, αvβ6, and αvβ8 a minor role on fibronectin.49 Similarly, the edge detection tool – edge effect – computes the ‘perimeter’ to ‘square root of surface area’ ratio, which is directly proportional to cell spreading (Fig. 2d, Fig. S3c†). A higher ratio reflects more spread cells. However, if multiple cells are clustered together or cells in multiple planes are overlapping with one another in the superimposed images, an increase in cell area is associated with a reduction in the edge effect and an increase in circularity due to hidden perimeters inside the clusters. While circularity considers the shape factor, the edge effect in addition considers how much the cells are expressing local protrusions (Fig. S4a†). FN* showed the largest edge effect followed by bicyclic RGD peptides, Q-P1, Q-P2, and monocyclic K-cRGD peptides. A significant increase in edge effect is observed for bicyclic Q-P1 compared to the linear K-RGD. Similarly, a significant increase in edge effect is observed for FN* compared to the bicyclic K-P1, K-P2, monocyclic K-cRGD, and linear K-RGD. Therefore, the combined reduction in circularity and increase in edge effect (Fig. S4b†) demonstrate the higher cell spreading for bicyclic RGD peptides compared to linear and monocyclic RGD peptides.
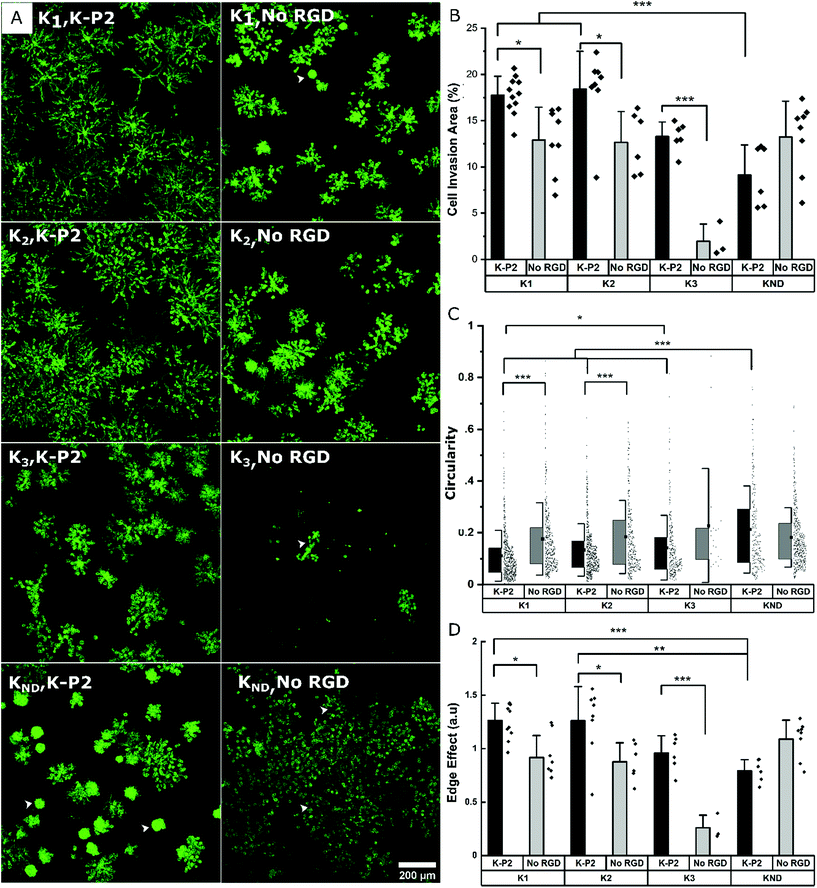
Influence of different degradable MMP sensitive domains on fibroblast growth
To render the hydrogel degradable, MMP sensitive domains are incorporated in the arms of the PEG-K precursors. This enables on cell-demand hydrogel degradation as the cells secrete the enzymes needed to cleave the crosslinks. This creates physical space for the cells to migrate and proliferate.25 PEG gels fabricated with di-cysteine K1 or K2 peptides as linear degradable crosslinkers previously reported an increase in cell invasion for K2 in comparison to K1 in the case of mouse myofibroblasts, owing to its high catalytic constant kcat for MMP1 and MMP2.24 Similarly, K3 derived from Secreted Protein Acidic and Rich in Cysteine (SPARC) has been demonstrated to be degraded by plasmin, while its high kcat enhanced cell invasion in PEG gels.46 Here, the growth of mouse fibroblasts is investigated in bicyclic K-P2 RGD-modified PEG hydrogels depending on these three proteolytic sensitive domains (K1, K2, K3), and compared to a non-degradable domain (KND) and no RGD. The fluorescent micrographs stained for actin (Fig. 3a) and their corresponding cell invasion areas (%) (Fig. 3b) indicate a significantly higher level of cell growth in PEG gels crosslinked with K1 or K2 (>17.5%) degradable domains and tethered with bicyclic K-P2 RGD peptide, when compared to their controls without RGD (<13%).The lower levels of cell growth in K1 and K2 gels in the absence of RGD indicate that the cells partly try to rescue themselves by secreting both ECM proteins and proteases. These observations are in agreement with a previous report from our research group where an increase in native fibronectin production by fibroblast cells was observed in the presence of bioinert microgels inside 3D Anisogel, compared to RGD-functionalized microgels.50 A low cell invasion area is observed for K3 gels with (∼13.5%) and without (∼2%) K-P2 RGD peptide, with the K3, no RGD showing exclusively rounded single cells in 3D. Although Patterson et al. reported improved cell invasion in gels made with K3 peptide, the results obtained here are not demonstrating enhanced cell growth. The PEG gels prepared with KND show a low cell invasion area (%) and a high circularity (Fig. 3c), indicating low cell spreading and moderate cell proliferation. However, in the presence of K-P2 RGD, the cells appear more clustered, which suggests that the presence of RGD does promote cell proliferation but due to lack of open space, spreading is limited. In the case of the edge effect (Fig. 3d), the hydrogels with degradable environment show higher edge effects for cells in gels modified with K-P2 compared to no RGD. Hydrogels with crosslinkers K1 and K2 have the highest edge effect offering a favourable environment for cell growth and spreading. Overall, the multi-parameter analysis reveals high fibroblast growth in hydrogels crosslinked with K1 or K2 degradable domains in the presence of bicyclic K-P2 RGD peptides, in comparison to K3 and KND, highlighting the importance of degradable domains in par with the cell adhesive ligands when developing artificial ECM scaffolds for functional cell growth.
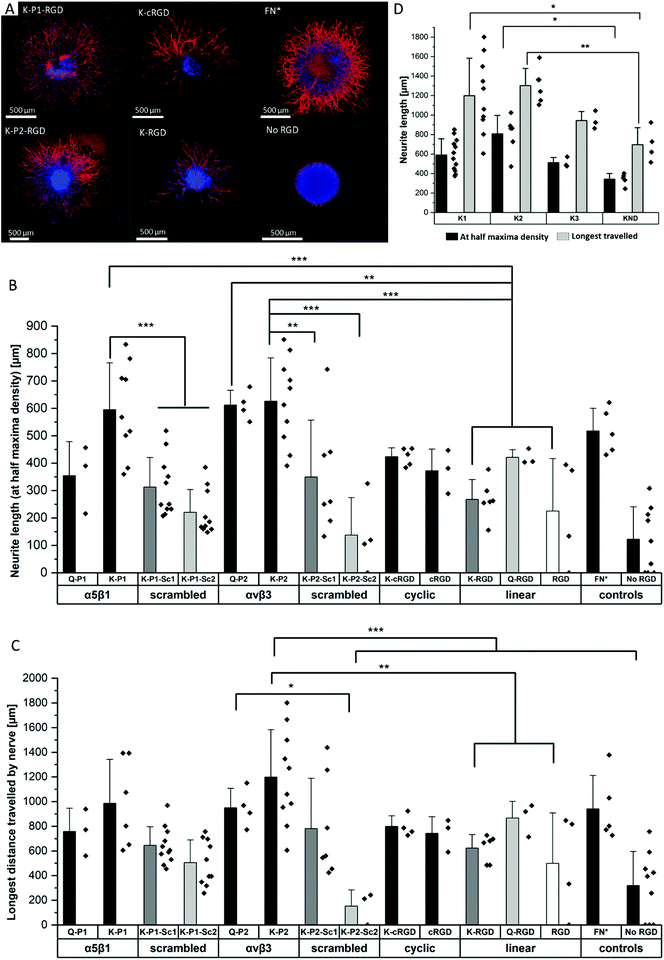
Influence of RGD peptides on nerve growth
To study the effect of RGD peptides on nerve growth, DRGs are encapsulated in PEG precursors with a fixed RGD concentration of 100 μM. The explants are cultured in PEG-based hydrogels, modified with bicyclic RGD (K-P1, K-P2, Q-P1, Q-P2), linear RGD (K-RGD, Q-RGD, RGD), or monocyclic RGD (K-cRGD, cRGD), and controls (no RGD, FN*) hydrogels. DRGs are cultured for 7 days, fixed, immunostained for β-tubulin fluorescent markers, and imaged with a minimum z-stack size of 200 μm (Fig. S5†). Neurite outgrowth (Fig. 4a) is analyzed using an image processing platform developed in-house to compute the nerve length at half-maximum density and the longest distance travelled by nerve. The fluorescent micrographs of bicyclic RGD peptide (K-P1, K-P2) show superior nerve growth in length (at half-maximum density) (∼600 μm) compared to monocyclic (K-cRGD, 420 μm) and linear (K-RGD, 270 μm), while comparable extension is observed among the bicyclic RGD peptides and FN* (520 μm). Consistent with the bright field images, the negative controls without RGD show minimal innervation of the PEG gels. This can be rationalized by the production of ECM proteins and proteases as discussed in the previous section. To quantify nerve growth, the distance of neurite extensions from the edge of the DRG is measured radially, until the distance where the neurite density drops to half of the maximum density (Fig. 4b). This reveals superior neurite growth in bicyclic RGD gels (K-P1, Q-P2, and K-P2) compared to the hydrogels, modified with linear RGD (K-RGD). Interestingly, neurite growth in gels, tethered with bicyclic RGD via K residues shows longer neurite extensions at half-maximum density compared to their corresponding bicyclic RGD peptides conjugated via Q residues. Interestingly, this difference is only observed for bicyclic peptide selective towards α5β1 whereas, no such major difference nerve growth is observed between bicyclic peptide selective for αvβ3. In addition, bicyclic RGD peptides with integrin selectivity towards α5β1 or αvβ3 show significantly better nerve growth compared to bicyclic peptides with a scrambled secondary loop, which imparts integrin selectivity, highlighting the influence of integrin selectivity in nerve growth. Scrambled RGD peptides (RDG) in the first loop also lead to worsened neurite outgrowth. The neurite growth at half maxima for Q-RGD (∼450 μm) is comparable to that of the monocyclic RGDs but is not significantly different than the two other linear RGD peptides. Enhanced neurite length at half-maximum density is observed for bicyclic RGD peptides (K-P1, Q-P2, K-P2) in comparison to our previously optimal condition FN* (5 μM),20 however, no statistical significance is observed. Such enhanced nerve growth is likely attributed towards the spatial cyclic conformation of the RGD loop in combination with the integrin selective secondary loop.Next to the neurite length at half-maximum density, the longest distance travelled by nerve inside the gels is compared among the different RGD-tethered hydrogels (Fig. 4c) with both parameters demonstrating similar trends. The longest neurite outgrowth distance (longest distance travelled by nerve) of ∼1200 μm is observed for bicyclic RGD peptide (K-P2), selective for αvβ3 integrin subunits, which is significantly longer than for linear RGD peptides K-RGD (∼600 μm) and RGD (∼500 μm). These results are in compliance with our previous observations of dissociated DRGs cultured on 2D sPEG hydrogels, where a larger decrease in nerve growth is observed when αv integrins are blocked relative to α5 integrins, revealing the importance of αv integrins in nerve growth. Similarly, integrin αvβ3 is reported to bind an RGD sequence in immunoglobulin Ig6 to promote neurite outgrowth in DRGs and retinal cells.51 Also, cellular migration of avian neural crest cells involves interaction between integrin αvβ3 and vitronectin,52 while an upregulation in αvβ3 is reported on astrocytes post-ischemia.53
Influence of degradable MMP sensitive domains on nerve growth
The different scaffold degradation domains are also investigated for neurite growth. Similar to the fibroblast culture, three proteolytic sensitive domains (K1, K2, K3) and one non-degradable domain (KND) are investigated in hydrogels modified with bicyclic RGD peptides (K-P2) (Fig. 4d). The neurite length at half-maximum density in K-P2 tethered gels with K1 and K2 crosslinkers is significantly longer in comparison to KND gels with K-P2. Among K1 and K2 with K-P2, longer neurite growth is observed for K2, however, no statistical significance between them is computed. Although K3 is degradable by plasmin, it shows comparable nerve length to the non-degradable KND. In addition to neurite length at half-maximum density, the longest distance travelled by nerve is quantified for these gels, revealing a similar trend with K2 gels conjugated with K-P2 RGD with a distance travelled by ∼1300 μm, followed by K1 gels with K-P2 RGD with ∼1200 μm (Fig. 4d). Both K3 and KND hydrogels, conjugated with K-P2 RGD, enable their longest travelled nerve up to ∼900 μm and ∼700 μm, respectively.Oriented nerve growth using anisometric magneto-responsive short PCL fibers
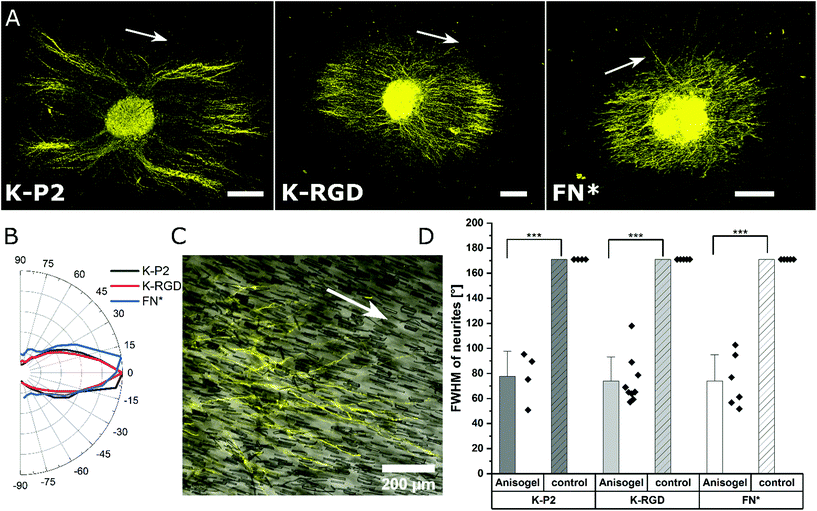
Among the investigated RGD bicyclic peptides and different proteolytically degradable domains, K-P2 RGD modified MMP based K2 PEG gels lead to the best nerve growth. Therefore, this combination is employed to prepare synthetic Anisogels with short magneto-responsive polycaprolactone (PCL) fibers (diameter of ∼5 μm and length of ∼50 μm), which are incorporated at a concentration of 1 v/v% and aligned in the presence of a low external magnetic field (50 mT) before crosslinking of the surrounding PEG gel.20 The short anisometric fibers act as directional guidance cues, function as physical barriers, and induce mechanical anisotropy in the hydrogel, to trigger nerves to grow parallel to their orientation, resulting in neurite alignment. K-P2 modified Anisogels are compared with linear (K-RGD) and fibronectin fragment (2 μM, FN*) and their respective controls without short fibers. Magnetically oriented fibers resulted in highly aligned neurites (Fig. 5a). The dimensions of the fibers and their innate surface properties are known to affect the cell adhesion and alignment of cells and nerves.54,55 Neurite orientation is analyzed with defined anisometric orientation kernels resulting in a range of orientation distributions between −90° to +90°. K-P2, K-RGD, and FN* all have a narrow distribution of aligned nerve growth with maxima around 0° (Fig. 5b). Bright-field images superimposed with tubulin stained (yellow) Anisogels show nerve growth along the direction of the fibers (Fig. 5c).All of the Anisogels studied showed superior nerve alignment compared to their respective controls without oriented short fibers (Fig. 5d). Anisogels tethered with bicyclic RGD (K-P2), linear RGD (K-RGD) and fibronectin fragment (2 μM, FN*) have comparable FWHM of ∼80° with no statistical significance between them. We define that nerves are highly aligned when the FWHM < 90°.56 These observations are consistent with our previous report on fully PEG-based Anisogels tethered with fibronectin fragment.20
Materials and methods
Fabrication of PEG gels
8-Arm 20 kDa PEG (Jenkem Technology, USA) is conjugated separately with two types of peptides H-NQEQVSPLERCG-NH2 (PEG-Q) and Ac-FKGG-GPQG↓IWGQ-ERCGNH2 (PEG-K1) or alternative MMP-sensitive domains (Pepscan, Netherlands) bearing conjugated Q and K amino acids, respectively. A stoichiometric ratio of 0.477 w/v% PEG-Q and 0.522 w/v% PEG-K is crosslinked in the presence of activated Factor XIII, (20 U mL−1, CSL Behring GmbH, 1250 U FXIII). Factor XIII is activated with thrombin (200 U mL−1) buffered with 10 mM Tris, 25 mM CaCl2, 150 mM NaCl for 30 min at 37 °C (briefly vortexing every 5 min). A buffer solution (10×, 0.1 M CaCl2, 0.5 M Tris, 1.1 M NaCl) is used to regulate the osmotic pressure of the gel precursor. All RGD-based bio-functional cell adhesive peptides are coupled to the gel at 100 μM concentration of the final gel volume, resulting in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24 between RGD peptides and total arms of star PEG molecules. In the case of complete peptide coupling, either 7.75% of the PEG-K arms would be tethered with RGD peptides or 8.46% of the PEG-Q arms, depending on the peptide domain used. The gels are incubated at 37 °C for 30 minutes for crosslinking and cultured in media.
24 between RGD peptides and total arms of star PEG molecules. In the case of complete peptide coupling, either 7.75% of the PEG-K arms would be tethered with RGD peptides or 8.46% of the PEG-Q arms, depending on the peptide domain used. The gels are incubated at 37 °C for 30 minutes for crosslinking and cultured in media.
Peptide synthesis
First, the linear RGD-sequences and other integrin-selectivity-promoting peptides are synthesized via Fmoc-based solid-phase peptide-synthesis using a fully automated peptide synthesizer as described by Bernhagen et al.43 Subsequently, these peptides are dissolved to 0.5 μM concentration in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 mixture of acetonitrile (ACN)/MilliQ-water and mixed with ∼1.1 equivalent of tris(bromomethyl) benzene (T3; dissolved in pure ACN at 10 mM concentration) until homogeneity, and left at room temperature for 1 h. Then, the reaction is quenched with 10% TFA/H2O to pH < 4 and the reaction mixture is purified via preparative HPLC, followed by lyophilization. All peptides were produced in PEPSCAN. B.V, the Netherlands.
3 mixture of acetonitrile (ACN)/MilliQ-water and mixed with ∼1.1 equivalent of tris(bromomethyl) benzene (T3; dissolved in pure ACN at 10 mM concentration) until homogeneity, and left at room temperature for 1 h. Then, the reaction is quenched with 10% TFA/H2O to pH < 4 and the reaction mixture is purified via preparative HPLC, followed by lyophilization. All peptides were produced in PEPSCAN. B.V, the Netherlands.
Encapsulation of cells in 3D hydrogel scaffolds and cell culture
L929 mouse-derived fibroblasts cells (ATCC, Germany) and DRGs from 10 days old embryonic chicks are harvested in RPMI and Hanks Balanced Salt Solution (HBSS, Gibco), respectively. In the case of single fibroblasts, the gel precursors are mixed with a pre-counted cell suspension in a defined volume, resulting in a cell concentration of 500 μL−1. Individual DRGs are embedded manually into the hydrogel precursors using tweezers. The cell encapsulated hydrogels or Anisogels are flipped every 10 minutes to ensure a homogeneous cell distribution and after 30 min, the gels are incubated with media at 37 °C to ensure complete precursor crosslinking before adding the media. The crosslinked gels are cultured for 7 days in RPMI basal medium, enriched with 10% Fetal Bovine Serum (FBS, Biowest) and 1% Antibiotic-Antimycotic (Gibco) for L929 cells and DMEM, enriched with 20 ng mL−1 nerve growth factor, 10% fetal bovine serum (FBS, Biowest) and 1% Antibiotic-Antimycotic (Gibco) in the case of DRGs. The media is exchanged every two days, and the cell-loaded gels are cultured at 37 °C, 5% CO2, and 95% humidity.Anisogel fabrication
Short magneto-responsive rod-shaped fibers are produced via solvent assisted spinning (SAS) based on previous reports.55 A polymer solution with 17 w/v% polycaprolactone (PCL) is prepared by dissolving PCL in 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 v/v chloroform
50 v/v chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetic acid, while superparamagnetic iron oxide nanoparticles (SPIONS) are dispersed into the solution before SAS.55 The fibers are collected on a cylindrical rotating drum and harvested for cryo-cutting at desirable lengths. The fiber concentration in the PEG hydrogel precursor solution is approximately 1 v/v%. The Anisogel precursor mixture is cast on a DRG between two magnets generating a field of ∼50 mT. This enables the fibers to align parallel to the magnetic field lines during crosslinking of the surrounding PEG-based hydrogel. The bulk matrix around the aligned fibers is allowed to crosslink for 30 minutes at 37 °C before adding more media.
acetic acid, while superparamagnetic iron oxide nanoparticles (SPIONS) are dispersed into the solution before SAS.55 The fibers are collected on a cylindrical rotating drum and harvested for cryo-cutting at desirable lengths. The fiber concentration in the PEG hydrogel precursor solution is approximately 1 v/v%. The Anisogel precursor mixture is cast on a DRG between two magnets generating a field of ∼50 mT. This enables the fibers to align parallel to the magnetic field lines during crosslinking of the surrounding PEG-based hydrogel. The bulk matrix around the aligned fibers is allowed to crosslink for 30 minutes at 37 °C before adding more media.
3D immunofluorescent staining
All the incubation steps in this staining protocol are performed on a plate shaker (at 30 rpm) unless stated otherwise. Encapsulated cells in 3D gels are washed twice with PBS for 30 minutes and are fixed with 4% paraformaldehyde (PFA) in PBS for 1 h on a plate shaker. After fixing, the gels are washed twice with PBS for 1 h and treated with 0.1% TritonX-100 for 40 minutes, and further washed twice with PBS for 1 h. For L929 fibroblasts, the gels are treated with iFluor-phalloidin 488 or 594 (Abcam, Germany) (1:1000) in PBS in combination with DAPI (1:100) and incubated for 4 h. The gels are washed twice with PBS for 1 h each and stored until imaging. The encapsulated DRGs in the gel are blocked with 5% bovine serum albumin (BSA) (Sigma, Germany) in PBS for 4 h, followed by the addition of primary beta-tubulin antibody TUJ1 (BioLegend, USA) (1:250) in 5% BSA solution for 18 h. The gels are washed twice with PBS for 1 h before adding the secondary antibody Alexa Fluor 633 goat anti-mouse (Invitrogen, Germany) (1:1000) in PBS and DAPI (1:100). The secondary antibody solution is incubated for 4 h and washed twice for 1 h each and stored in PBS until imaging with confocal microscopy.Imaging
Laser scanning confocal microscopy (SP8 Tandem Confocal, Leica Microsystems Inc.) is used to image the Z stacks of the cells encapsulated in the hydrogel. Air objective of 10×/0.3 N.A magnification is used to obtain fluorescent micrographs by exciting the fluorophores at different wavelengths and obtaining the emission signals at multiple channels.Analysis of fibroblast growth
The Z stack images are converted into a maximum projection image using FIJI image J after the background calculated with a rolling ball filter (size = 20) has been subtracted. The image is then converted into an 8-bit image with an auto-Otsu threshold to convert the images to binary. These images are referred to as threshold images and the percentage of cell invasion areas is computed. Cell invasion here is quantified based on % of actin pixels per field of view or per image. This data can be a combination of cells that are spread, as well as rounded cells, taking cell proliferation into account. To obtain a smoothened image, the threshold images are further treated with a shape smoothening plugin (relative proportion fds = 5, absolute number fds = 5). The edge effect is analyzed using the formula:In addition, each cell or cell cluster is analyzed for its circularity, after applying the ‘Otsu’ threshold, the images are processed using a particle analyzer tool (FIJI, Image J) for particle sizes >500 pixels. Hence, cells that are spreading well does not guarantee that they undergo similar proliferation and invasion rates. On the other hand, a large cell invasion does not always mean the cells are healthy and spreading. Cell invasion and the edge effect are both computed per field of view, while the circularity is measured for each cell or cell cluster. The data obtained from different gel conditions are compared and plotted in Origin.
Analysis of neurite outgrowth
The Z stack images are converted into a maximum projection image using FIJI image J. The explant/core of the DRG is manually fitted into a mask and the position of the centroid of the mask is used as the center for radial analysis. The tubulin stained images are Gauss smoothened with a window radius of 5.0 pixels and a width of 1.0 pixel. Power law dynamic compression/expansion is applied with power 0.5. The binary images subtracted with the explant mask is used for radial analysis. The number of non-zero pixels as a function of distance (radius) is obtained, which is further computed to get pixel density = pixels per unit area. Neurite growth at half-maximum density is computed by measuring the distance between the edge of the DRG explant and the distance at which the neurite density reaches half-maximum. The longest distance travelled by nerve is defined as the distance from the edge of the DRG explant until the pixel density < 0.001 pixels per μm2 (Fig. 6).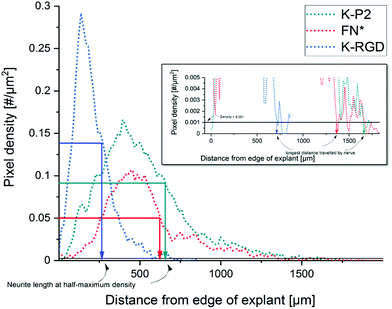 | ||
| Fig. 6 Analysis of neurite outgrowth using pixel density to compute the neurite length at half-maximum density and the longest distance travelled by nerve. | ||
The orientation of neurites inside a PEG-based Anisogel
Neurites stained for β-tubulin are imaged as z-stack. Neurite alignment is quantified inside the 3D hydrogel starting 10 μm above the first neurite in focus, starting from the bottom. A stack size of 100 μm is imaged and the maximum z-projection is used for orientation analysis. The DRG core is converted into a mask, to remove from the image during orientation analysis. Neurite orientation is quantified using an elliptical Mexican hat filter (a Laplace operator applied on an elliptic two-dimensional Gaussian function) as previously reported.20 The orientation kernels are designed to account for only the neurite data, excluding non-specific stains. The angles of the orientation kernels are plotted as a histogram and the maxima are set to zero to compare the relative distributions among other neurite orientations and thus using the full width at half-maximum (FWHM) as a measure of the degree of alignment of neurites in the Anisogels. Based on our previous reports, lower FWHM [FWHM < 90°] indicate higher neurite alignment.56Statistics
Statistical analysis is performed in Origin 2016 for Windows. A one-way ANOVA is applied with p-values below 0.05 being considered significant in combination with Bonferroni correction.Conclusions
PEG hydrogels crosslinked with proteolytically degradable domains and RGD bicyclic peptides selective towards integrin subunits are investigated to improve fibroblast growth and nerve extension inside synthetic PEG-based hydrogels. The short bicyclic RGD peptides are easily tethered to the PEG hydrogels via their amine or glutamine linkers. Fibroblast cells in 3D PEG gels show high network like formation compared to cells in gels tethered with linear and cyclic RGD peptides. The bicyclic peptides significantly enhance nerve growth (at half-maximum density) length (∼600 μm) and the longest distance travelled (∼1200 μm), when compared to linear and mono-cyclic RGD peptides. In the case of nerve growth, the αvβ3 selective P2 bicyclic peptides slightly outperformed the engineered fibronectin fragment FNIII9*-10/12-14, however not significantly. Degradable MMP cleavable crosslinkers incorporated in the gels with bicyclic RGD peptides improve cell invasion for K1 (GPQG↓IWGQ) and K2 (VPMS↓MRGG) MMP degradable crosslinkers and a marginal increase in cell growth for both fibroblasts and nerves in the case of K2. This report demonstrates for the first time the large potential of bicyclic RGD peptides inside PEG-based hydrogels, showing superior fibroblast and nerve growth compared to linear and monocyclic RGD peptides. Including oriented magneto-responsive short fibers inside bicyclic RGD-modified Anisogels demonstrates highly aligned nerve growth in the direction along the oriented fibers comparable to Anisogels modified with the fibronectin fragment.Author contributions
S.V carried out the experiments supervised by L.D.L based on the gels optimized by C.L and A.C.N using the bicyclic peptides designed by D.B and P.T. PCL fibers for the Anisogels were provided by A.O-A. The neurite growth and orientation were computationally analyzed by T.H. Manuscript was written by S.V with support from L.D.L. All the authors contributed in the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank J. Huffel for experimental assistance and C. Bergerbit for his support in writing. We acknowledge funding from the European Union's Horizon 2020 research and innovation programme under Marie Skłodowska-Curie grant agreement no. 642687 (Biogel). We gratefully acknowledge funding from the Leibniz Senate Competition Committee (SAW) under the Professorinnenprogramm (SAW-2017-PB62: BioMat). We acknowledge funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (ANISOGEL, grant agreement no. 637853). This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement no. 874586 (Organtrans).References
- S. Cheng, E. C. Clarke and L. E. Bilston, Med. Eng. Phys., 2008, 30, 1318–1337 CrossRef PubMed.
- M. J. Blewitt and R. K. Willits, Ann. Biomed. Eng., 2007, 35, 2159–2167 CrossRef PubMed.
- G. Huang, F. Li, X. Zhao, Y. Ma, Y. Li, M. Lin, G. Jin, T. J. Lu, G. M. Genin and F. Xu, Chem. Rev., 2017, 117, 12764–12850 CrossRef CAS PubMed.
- K. M. Yamada and M. Sixt, Nat. Rev. Mol. Cell Biol., 2019, 20, 738–752 CrossRef CAS PubMed.
- T. Nakaji-Hirabayashi, K. Kato and H. Iwata, Bioconjugate Chem., 2012, 23, 212–221 CrossRef CAS PubMed.
- D. Hodde, J. Gerardo-Nava, V. Wöhlk, S. Weinandy, S. Jockenhövel, A. Kriebel, H. Altinova, H. W. M. Steinbusch, M. Möller, J. Weis, J. Mey and G. A. Brook, Eur. J. Neurosci., 2016, 43, 376–387 CrossRef.
- A. R. Bento, P. Quelhas, M. J. Oliveira, A. P. Pêgo and I. F. Amaral, J. Tissue Eng. Regener. Med., 2017, 11, 3494–3507 CrossRef CAS PubMed.
- S. Li, L. R. Nih, H. Bachman, P. Fei, Y. Li, E. Nam, R. Dimatteo, S. T. Carmichael, T. H. Barker and T. Segura, Nat. Mater., 2017, 16, 953–961 CrossRef CAS PubMed.
- N. Broguiere, L. Isenmann and M. Zenobi-Wong, Biomaterials, 2016, 99, 47–55 CrossRef CAS PubMed.
- V. Carriel, J. Garrido-Gómez, P. Hernández-Cortés, I. Garzón, S. García-García, J. A. Sáez-Moreno, M. Del Carmen Sánchez-Quevedo, A. Campos and M. Alaminos, J. Neural Eng., 2013, 10, 026022 CrossRef PubMed.
- K. Chwalek, D. Sood, W. L. Cantley, J. D. White, M. Tang-Schomer and D. L. Kaplan, J. Visualized Exp., 2015, 2015, 1–7 Search PubMed.
- J. Zhu and R. E. Marchant, Expert Rev. Med. Devices, 2011, 8, 607–626 CrossRef CAS PubMed.
- E. F. Plow, T. A. Haas, L. Zhang, J. Loftus and J. W. Smith, J. Biol. Chem., 2000, 275, 21785–21788 CrossRef CAS PubMed.
- K. J. Lampe, A. L. Antaris and S. C. Heilshorn, Acta Biomater., 2013, 9, 5590–5599 CrossRef CAS.
- Y. Berkovitch and D. Seliktar, Int. J. Pharm., 2017, 523, 545–555 CrossRef CAS PubMed.
- B. P. dos Santos, B. Garbay, M. Fenelon, M. Rosselin, E. Garanger, S. Lecommandoux, H. Oliveira and J. Amédée, Acta Biomater., 2019, 99, 154–167 CrossRef CAS PubMed.
- A. M. Hopkins, L. De Laporte, F. Tortelli, E. Spedden, C. Staii, T. J. Atherton, J. A. Hubbell and D. L. Kaplan, Adv. Funct. Mater., 2013, 23, 5140–5149 CrossRef CAS.
- G. Palazzolo, N. Broguiere, O. Cenciarelli, H. Dermutz and M. Zenobi-Wong, Tissue Eng., Part A, 2015, 21, 2177–2185 CrossRef CAS PubMed.
- R. Boni, A. Ali, A. Shavandi and A. N. Clarkson, J. Biomed. Sci., 2018, 25, 1–21 CrossRef PubMed.
- C. Licht, J. C. Rose, A. O. Anarkoli, D. Blondel, M. Roccio, T. Haraszti, D. B. Gehlen, J. A. Hubbell, M. P. Lutolf and L. De Laporte, Biomacromolecules, 2019, 20, 4075–4087 CrossRef CAS PubMed.
- P. Naghdi, T. Tiraihi, F. Ganji, S. Darabi, T. Taheri and H. Kazemi, J. Tissue Eng. Regener. Med., 2016, 10, 199–208 CrossRef CAS.
- R.-S. Hsu, P.-Y. Chen, J.-H. Fang, Y.-Y. Chen, C.-W. Chang, Y.-J. Lu and S.-H. Hu, Adv. Sci., 2019, 6, 1900520 CrossRef PubMed.
- X. Li, E. Katsanevakis, X. Liu, N. Zhang and X. Wen, Prog. Polym. Sci., 2012, 37, 1105–1129 CrossRef CAS.
- J. Patterson and J. A. Hubbell, Biomaterials, 2010, 31, 7836–7845 CrossRef CAS PubMed.
- K. Bott, Z. Upton, K. Schrobback, M. Ehrbar, J. A. Hubbell, M. P. Lutolf and S. C. Rizzi, Biomaterials, 2010, 31, 8454–8464 CrossRef CAS PubMed.
- V. Truong, I. Blakey and A. K. Whittaker, Biomacromolecules, 2012, 13, 4012–4021 CrossRef CAS PubMed.
- M. Ehrbar, S. C. Rizzi, R. Hlushchuk, V. Djonov, A. H. Zisch, J. A. Hubbell, F. E. Weber and M. P. Lutolf, Biomaterials, 2007, 28, 3856–3866 CrossRef CAS PubMed.
- M. P. Lutolf, J. L. Lauer-Fields, H. G. Schmoekel, A. T. Metters, F. E. Weber, G. B. Fields and J. A. Hubbell, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 5413–5418 CrossRef CAS PubMed.
- M. M. Martino, F. Tortelli, M. Mochizuki, S. Traub, D. Ben-David, G. A. Kuhn, R. Müller, E. Livne, S. A. Eming and J. A. Hubbell, Sci. Transl. Med., 2011, 3(100), 100ra89 Search PubMed.
- M. Ehrbar, S. C. Rizzi, R. G. Schoenmakers, J. A. Hubbell, F. E. Weber and M. P. Lutolf, Biomacromolecules, 2007, 8, 3000–3007 CrossRef CAS PubMed.
- B. Geiger, J. P. Spatz and A. D. Bershadsky, Nat. Rev. Mol. Cell Biol., 2009, 10, 21–33 CrossRef CAS.
- F. G. Giancotti and E. Ruoslahti, Science, 1999, 285, 1028LP–1033 CrossRef.
- M. A. Dechantsreiter, E. Planker, B. Mathä, E. Lohof, G. Hölzemann, A. Jonczyk, S. L. Goodman and H. Kessler, J. Med. Chem., 1999, 42, 3033–3040 CrossRef CAS.
- C. Mas-Moruno, R. Fraioli, F. Rechenmacher, S. Neubauer, T. G. Kapp and H. Kessler, Angew. Chem., Int. Ed., 2016, 55, 7048 CrossRef CAS PubMed.
- T. G. Kapp, F. Rechenmacher, S. Neubauer, O. V. Maltsev, E. A. Cavalcanti-Adam, R. Zarka, U. Reuning, J. Notni, H. J. Wester, C. Mas-Moruno, J. Spatz, B. Geiger and H. Kessler, Sci. Rep., 2017, 7, 1–13 CrossRef.
- M. Kantlehner, P. Schaffner, D. Finsinger, J. Meyer, A. Jonczyk, B. Diefenbach, B. Nies, G. Hölzemann, S. L. Goodman and H. Kessler, Angew. Chem., Int. Ed., 2000, 39, 107–114 Search PubMed.
- D. Pallarola, A. Bochen, H. Boehm, F. Rechenmacher, T. R. Sobahi, J. P. Spatz and H. Kessler, Adv. Funct. Mater., 2014, 24, 943–956 CrossRef CAS PubMed.
- S. Neubauer, F. Rechenmacher, R. Brimioulle, F. S. Di Leva, A. Bochen, T. R. Sobahi, M. Schottelius, E. Novellino, C. Mas-Moruno, L. Marinelli and H. Kessler, J. Med. Chem., 2014, 57, 3410–3417 CrossRef CAS PubMed.
- D. Heckmann, A. Meyer, B. Laufer, G. Zahn, R. Stragies and H. Kessler, ChemBioChem, 2008, 9, 1397–1407 CrossRef CAS.
- H. B. Schiller, M. R. Hermann, J. Polleux, T. Vignaud, S. Zanivan, C. C. Friedel, Z. Sun, A. Raducanu, K. E. Gottschalk, M. Théry, M. Mann and R. Fässler, Nat. Cell Biol., 2013, 15, 625–636 CrossRef CAS PubMed.
- C. Diaz, S. Neubauer, F. Rechenmacher, H. Kessler and D. Missirlis, J. Cell Sci., 2020, 133, jcs232702 CAS.
- D. Bernhagen, V. Jungbluth, N. G. Quilis, J. Dostalek, P. B. White, K. Jalink and P. Timmerman, ACS Comb. Sci., 2019, 21, 198–206 CrossRef PubMed.
- D. Bernhagen, V. Jungbluth, N. G Quilis, J. Dostalek, P. B. White, K. Jalink and P. Timmerman, ACS Comb. Sci., 2019, 21, 598–607 CrossRef CAS PubMed.
- F. Cipriani, D. Bernhagen, C. Garcia-Arévalo, I. G. De Torre, P. Timmerman and J. C. Rodriguez-Cabello, Biomed. Mater., 2019, 14, 035009 CrossRef CAS PubMed.
- B. H. Hu and P. B. Messersmith, J. Am. Chem. Soc., 2003, 125, 14298–14299 CrossRef CAS PubMed.
- J. Patterson and J. A. Hubbell, Biomaterials, 2011, 32, 1301–1310 CrossRef CAS PubMed.
- D. Bernhagen, V. Jungbluth, N. G. Quilis, J. Dostalek, P. B. White, K. Jalink and P. Timmerman, ACS Comb. Sci., 2019, 21, 198–206 CrossRef PubMed.
- E. H. J. Danen, S. I. Aota, A. A. Van Kraats, K. M. Yamada, D. J. Ruiter and G. N. P. Van Muijen, J. Biol. Chem., 1995, 270, 21612–21618 CrossRef CAS PubMed.
- X. K. Zhao, Y. Cheng, M. Liang Cheng, L. Yu, M. Mu, H. Li, Y. Liu, B. Zhang, Y. Yao, H. Guo, R. Wang and Q. Zhang, Sci. Rep., 2016, 6, 19276 CrossRef CAS.
- J. C. Rose, D. B. Gehlen, T. Haraszti, J. Köhler, C. J. Licht and L. De Laporte, Biomaterials, 2018, 163, 128–141 CrossRef CAS PubMed.
- P. M. Yip, X. Zhao, A. M. P. Montgomery and C. H. Siu, Mol. Biol. Cell, 1998, 9, 277–290 CrossRef CAS PubMed.
- M. Delannet, F. Martin, B. Bossy, D. A. Cheresh, L. F. Reichardt and J. L. Duband, Development, 1994, 120, 2687–2702 CrossRef CAS.
- D. O. Clegg, K. L. Wingerd, S. T. Hikita and E. C. Tolhurst, Front. Biosci., 2003, 8, d723-50 CrossRef PubMed.
- A. Omidinia-anarkoli, R. Rimal, Y. Chandorkar, D. B. Gehlen, L. De Laporte, J. C. Rose and K. Rahimi, DOI:10.1021/acsami.8b17955.
- A. Omidinia-Anarkoli, S. Boesveld, U. Tuvshindorj, J. C. Rose, T. Haraszti and L. De Laporte, Small, 2017, 13, 1–8 CrossRef PubMed.
- J. C. Rose, D. B. Gehlen, A. Omidinia-Anarkoli, M. Fölster, T. Haraszti, E. E. Jaekel and L. De Laporte, Adv. Healthcare Mater., 2020, 2000886, 2000886 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional supplementary data on 3D homogeneous distribution of fibroblasts and DRGs in PEG hydrogels, circularity and edge effect quantification, proliferation assay comparing Q-RGD and K-RGD and comparative study of fibroblast cell growth in 3D PEG hydrogels with Q-RGD and FN*. See DOI: 10.1039/d0bm02051f |
| This journal is © The Royal Society of Chemistry 2021 |