Recent advances in nanomaterial-enhanced enzyme-linked immunosorbent assays
Lu
Gao
a,
Qianfan
Yang
 *a,
Peng
Wu
*a,
Peng
Wu
 a and
Feng
Li
a and
Feng
Li
 *ab
*ab
aKey laboratory of Green Chemistry & Technology of Ministry of Education, College of chemistry, Analytical & Testing Center, Sichuan University, 29 Wangjiang Road, Chengdu, Sichuan, China 610064. E-mail: fli@brocku.ca
bDepartment of Chemistry, Brock University, 1812 Sir Isaac Brock Way, St. Catharines, Ontario, Canada L3S 3A1
First published on 4th May 2020
Abstract
Despite serving as a gold standard for protein analysis, the classic enzyme-linked immunosorbent assay (ELISA) is currently challenged by the ever-increasing needs of sensitivity and simplicity. Towards the ongoing needs, recent advances in nanomaterials have offered numerous promising tools for enhancing the performance and broadening the applicability of ELISA. In this review, we highlight nanomaterial-enabled strategies that drastically improve the assay performance without significantly altering the classic ELISA format. Particular attention will be focused on the functional roles of nanomaterials as novel readout systems in ELISA, including those serving as substrate-alternatives, enzyme-alternatives, and non-enzymatic signal amplifiers.
1. Introduction
Since first introduced in 1971,1 enzyme-linked immunosorbent assay (ELISA) has been widely accepted as a gold standard for protein detection and quantification in disease diagnosis,2 food safety testing,3 and environmental monitoring.4 A typical ELISA assay involves four main steps,5 including (1) immobilization of a target protein on a solid support either through a capture antibody or nonspecific adsorption, (2) target recognition using detection antibodies, (3) signal amplification via an enzymatic reaction, and (4) signal readout generated by a chromogenic or fluorogenic substrate. Combining antibody-mediated target recognition and enzyme-driven signal transduction, ELISA allows the specific detection of as low as picomolar level proteins.Although sufficient for routine protein analysis, better diagnostic and/or prognostic uses of ELISA demand improved sensitivity.6,7 For example, post-surgical monitoring prostate specific antigen (PSA) at femtomolar level has shown better predictive values for the reoccurrence of prostate cancer.8 As such, many research efforts have to improve the sensitivity of ELISA, revealing at least two viable solutions. The first solution involves the modification or replacement of conventional enzyme-based signal readout with more sensitive ones. A classic example is immuno-PCR,9 where a synthetic DNA strand and polymerase chain reaction (PCR) are used to facilitate the signal amplification for ELISA. The second solution focuses on converting the classic microplate-based ELISA into digital ELISA,10 where each assay is aliquoted into thousands of microwells or droplets and thus generates digital signal readouts. Besides sensitivity, advances in novel immunoassays are also driven by their applicability and user-friendship in infrastructure-limited settings. Miniaturization of ELISA into rapid, portable, point-of-care tests (POCT) represents a new trend towards personalized medical care and disease diagnosis in resource-limited countries or regions.11,12 Along with this trend, numerous efforts have been made to develop novel readout systems for ELISA, which are both sensitive and user-friendly.13,14
Towards the ongoing need of improved sensitivity and/or user-friendship, diverse biohybrid nanomaterials have been designed and employed as novel readouts in immunoassays.15 Immunoassays harnessing unique optical, electrochemical, catalytic, or structural properties of nanomaterials have been reviewed extensively in the past years.16–20 The previous reviews discussed nanomaterial-enhanced immunoassays based primarily on the types of materials or signal readouts. The functional roles of nanomaterials in immunoassays have not been reviewed extensively. Herein, we focus this review on their functional roles as novel signal readouts for enhancing ELISA, including those serve as substrate-alternatives, enzyme-alternatives, or enzyme-free amplifiers (Fig. 1). This review will help guide the design and translation of novel nanomaterial-based readout systems for improving ELISA with minimal alterations of the classic assay format.
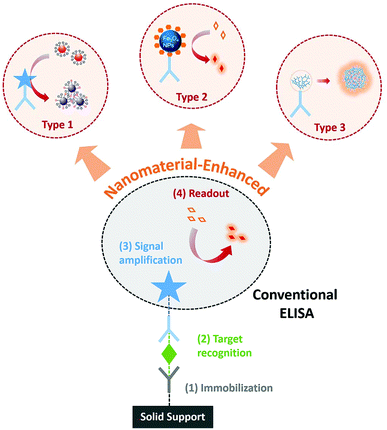 | ||
| Fig. 1 Schematic diagram showing the enhancement of conventional ELISA by using nanomaterials as substrate-alternatives (Type 1), enzyme-alternatives (Type 2), or enzyme-free amplifiers (Type 3). | ||
2. Immunoassays using nanomaterials as substrates
Detection signals of classic ELISA are generated through the enzymatic conversion of chromogenic or fluorogenic substrates, such as 3,3′,5,5′-tetramethylbenzidine (TMB), 2,2′-azino-bis-(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) and o-Phenylenediamine (OPD). One limiting factor to the sensitivity of ELISA is the low extinction coefficients of these small molecular substrates (at 104 M−1 cm−1 level).21,22 On the other hand, extinction coefficients for plasmonic nanoparticles, such as gold nanoparticles (AuNPs), can reach as high as 108 to 109 M−1 cm−1 because of the unique localized surface plasmon resonance (LSPR).23 According to Beer–Lambert law, the solution absorbance is linearly correlated to the extinction coefficient. The higher extinction coefficient would be beneficial for higher detection sensitivity. As such, replacing commonly used small molecular substrates with plasmonic nanoparticles can lead to a drastic improvement of the conventional ELISA in terms of the assay sensitivity. Thus inspired, numerous strategies have been explored to integrate ELISA with plasmonic readouts.24–27 These types of novel immunoassays are generally accepted as plasmonic ELISA.The idea of plasmonic ELISA was first introduced by Stevens and coworkers in 2012 (Fig. 2A).28 The assay workflow is adapted from ELISA, except that plasmonic AuNPs were used instead of conventional small molecular substrates. In the absence of the analyte, gold ions were rapidly reduced by hydrogen peroxide (H2O2) to form quasi-spherical, non-aggregated AuNPs, and the color of the reaction solution is red. In the presence of the target protein, however, catalase is introduced to the reaction mixture through the target-specific immunocomplexes. As catalase consumes H2O2, the newly formed AuNPs are of ill-defined morphology with aggregates. The surface plasmon resonance is caused by the collective oscillation of the conduction electrons across the nanoparticle. Therefore, the aggregates of AuNPs results in significant red-shifting (from ∼520 to ∼650 nm) due to the interparticle plasmon coupling, leading to a sharp red-to-blue color transition. Using this principle, the plasmonic ELISA enabled ultra-sensitive detection of PSA and HIV-1 capsid antigen p24 in serum samples with LODs as low as 1 × 10−18 g mL−1. In addition to catalase, other enzymes, such as glucose oxidase (GOx),29 alcohol dehydrogenase,30 have been used to catalyze the growth of metal nanoparticles to achieve ultra-sensitive naked-eye detection (Table 1).
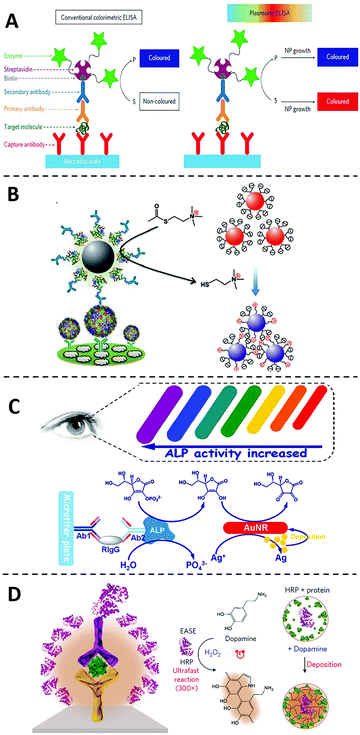 | ||
| Fig. 2 Immunoassays using nanomaterials as substrate-alternatives. (A) Schematic diagram of the plasmonic ELISA by the growth of plasmonic nanoparticles and the comparation to the conventional one. In conventional colorimetric ELISA (left), enzymatic biocatalysis generates a coloured compound. While in plasmonic ELISA (right) the biocatalytic cycle of the enzyme generates coloured nanoparticle solutions of characteristic tonality. Reprinted from ref. 28, Copyright, 2012, Springer Nature. (B) Schematic diagram of the ultra-sensitive visual Immunoassay based on the direct aggregations of AuNPs. Reprinted from ref. 31, Copyright, 2013, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (C) Schematic diagram of the multicolour plasmonic ELISA based on the size changes of AuNRs. Reprinted from ref. 40, Copyright, 2014, American Chemical Society. (D) Schematic diagram of the ultra-sensitive immunoassay based on polymeric nanoparticles. Reprinted from ref. 43, Copyright, 2017, Springer Nature. | ||
| The role of nanomaterials | Target protein | Detection limit | Readout | Ref. |
|---|---|---|---|---|
| Substrate alternatives | Prostate specific antigen (PSA) and HIV-1 capsid antigen p24 | 1 ag mL−1 | Colorimetric | 28 |
| PSA | 3.1 fg mL−1 | Colorimetric | 29 | |
| Hepatitis B surface antigen and α-fetoprotein | 1 pg mL−1 | Colorimetric | 30 | |
| PSA | 3 fg mL−1 | Colorimetric | 38 | |
| Alpha fetal protein | 0.2 ng mL−1 | Colorimetric | 24 | |
| Enterovirus 71 | 104 copies per mL | Colorimetric | 31 | |
| Treponema pallidum antibodies | 1 pg mL−1 | Colorimetric | 32 | |
| Rabbit antihuman IgG; mycoplasma; pneumonia | 2 ng mL−1 | Colorimetric | 33 | |
| Human IgG | 1 ng mL−1 | Colorimetric | 40 | |
| E. coli O157:H7 | 50 CFU mL−1 | Colorimetric | 27 | |
| PSA | 75 pg ml−1 | Colorimetric | 41 | |
| Aflatoxin B1 (AFB1) | 12.5 pg ml−1 | Colorimetric | 39 | |
| HIV antigen | 3 fg ml−1 | Colorimetric | 42 | |
| Enzyme alternatives | Hepatitis B virus surface antigen | Not reported | Colorimetric | 47 |
| Mouse interleukin 2 (IL-2) | Not reported | Colorimetric | 44 | |
| Ebola virus | 1 ng mL−1 | Colorimetric | 48 | |
| PSA | 31 fg mL−1 | Colorimetric | 53 | |
| C-Reactive protein (CRP) | 0.67 pg mL−1 | Fluorescent | 54 | |
| HIV-1 capsid antigen p24 | 0.8 pg mL−1 | Colorimetric | 55 | |
| CEA | 9 pg mL−1 | Colorimetric | 56 | |
| Apolipoprotein A1 (ApoA1) | 20 pg mL−1 | Colorimetric | 57 | |
| Nanomaterials as nanocarriers | PSA | 32 fg mL−1 | Fluorescent | 65 |
| AFP | 0.74 ng mL−1 | Fluorescent | 68 | |
| PSA | 30 amol L−1 | Grayscale | 70 | |
| Rabbit IgG, Salmonella, Listeria, and E. coli O157 | 30 amol L−1 | Colorimetric | 71 | |
| HIV-1 gp41 antigen | 0.1 ng mL−1 | Colorimetric | 72 | |
| DNA nanotechnology | Cancerous exosomes | 2.1 × 104 mL−1 | Electrochemical | 73 |
| Golgi protein 73 (GP73) | 15 pg mL−1 | Electrochemical | 74 | |
| immunoglobulin G (IgG) | 2.8 pg mL−1 | Electrochemical | 84 | |
| PSA | 40 pg mL−1 | Fluorescent | 84 | |
| Cytokine | <1 ng mL−1 | Fluorescent | 93 | |
| HeLa cells | 4400 cells per mL | Fluorescent | 98 | |
Besides controlling the growth of plasmonic nanoparticles, directly inducing aggregations of metal nanoparticles is another commonly used strategy to enhance the sensitivity of ELISA. For example, Chen and coworkers31 designed a novel plasmonic ELISA (Fig. 2B), where acetylcholinesterase (AChE) was used to produce thiocholine. Thiocholine induced the subsequent aggregation of citrate-capped AuNPs and thus resulting in a strong red-to-purple color transition. This method allows the visual detection of enterovirus 71 with a LOD (∼104 copies per mL) comparable to that obtained by quantitative PCR. Strategies involving aggregation of plasmonic nanoparticles can be easily adapted into the currently available ELISA platforms and have been successively applied to the detection of various targets, including Treponema pallidum antibodies,32 mycoplasma,33 and HCV.34
As LSPR are highly shape- and size-dependent, plasmonic ELISA with multicolored readout have also been achieved by inducing changes in the shape and/or size of plasmonic nanomaterials through etching or deposition.35–39 For example, Tang and coworkers40 have developed an ultra-sensitive plasmonic ELISA based on the size changes of gold nanorods (AuNRs) (Fig. 2C). In this assay, ALP was utilized to catalyze the hydrolysis of ascorbic acid 2-phosphate to produce ascorbic acid, which is capable of reducing silver ion to metallic silver. The metal silver then deposited on AuNRs, resulting in a rainbow-like color transition from red, to orange, to yellow, and finally to green. The multicolour transition has also been achieved by etching the anisotropic AuNRs. For example, Lin and coworkers41 recently developed a multicolour plasmonic ELISA by using TMB to etch AuNRs, allowing the visual detection of carcinoembryonic antigen (CEA) in human serum with a LOD of 2.5 ng mL−1.42 Comparing with monochromatic intensity changes, the multicolour transition is an ideal readout for naked-eye detection and quantification in POCT settings.
In addition to plasmonic nanoparticles, polymeric nanoparticles have also been introduced as substrate alternatives to conventional ELISA. For example, Gao and coworkers42 developed an ultra-sensitive immunoassay by combining the polymerization of dopamine with horseradish peroxidase (HRP)-based conventional ELISA in 2017 (Fig. 2D). In the system, HRP catalyzes the localized deposition of polydopamine to form the polydopamine layer, and then the polydopamine layer accumulates more HRP enzymes in turn, which leads to a much stronger color transition. Using this principle, as low as 3 fg mL−1 of HIV antigen could be detected in blood samples, which is over 1000-fold more sensitive than the conventional ELISA.
The uses of plasmonic or polymeric nanoparticles as substrate alternatives in ELISA allows the improvement of both sensitivity and user-friendship without significantly altering the assay format. Such strategies can be employed directly as “add-on” units to current ELISA platforms, which may help reduce the cost and infrastructural barriers and accelerate their uses in real-world applications.
3. Immunoassays using nanozymes
Enzyme is a core component in ELISA to transduce and amplify the specific affinity recognition of target proteins into sensitive detectable signals. Despite the high catalytic efficiency, commonly used enzymes, such as HRP, ALP, and GOx, are proteins in their chemical nature and thus are susceptible to environmental influences (e.g., pH or temperature). The massive production of such enzymes is also tedious and expensive. As such, nanomaterials have also been investigated as chemical alternatives of naturally existing enzymes to improve the assay robustness and reduce the cost of ELISA.43–45 These enzyme-like nanomaterials capable of catalyzing chromogenic or fluorogenic reactions in aqueous solutions are generally known as nanozymes.46The enzyme-liked nanozyme was first discovered by Yan and coworkers in 2007,47 where Fe3O4 magnetite nanoparticles (Fe3O4 MNPs) were found to possess intrinsic catalytic activity similar to natural peroxidase (Fig. 3A). Comparing with HRP, Fe3O4 MNPs have shown higher affinity and faster catalytic rate to the substrate TMB, and are more tolerant to environmental changes. This nanozyme was further integrated into a standard ELISA method as an emzyme-alternative to detect hepatitis B virus surface antigen preS1. The same nanozyme was also integrated by the same lab to an immunochromatographic strip, which allows the visual detection of glycoprotein of Ebola virus with a LOD as low as 1 ng mL−1.48 Remarkably, this method is ∼100 times more sensitive than the conventional lateral flow strips operated by ELISA. Following these pioneering works, many nanozymes with diverse chemical compositions have been discovered, including cobalt-based,49 platinum-based,50 copper-based,51 and manganese-based,52 the most of which have been successfully integrated into ELISA assays for ultrasensitive protein detection.53–57
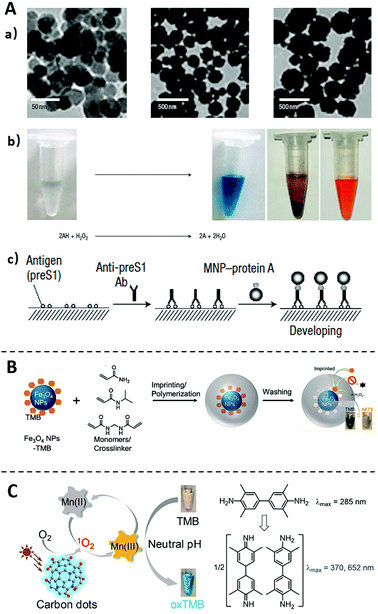 | ||
| Fig. 3 Immunoassays using nanozymes (A) and the method for improving nanozymes properties (B and C). (A) TEM images of nanozyme Fe3O4 MNPs, and the Fe3O4 MNPs catalyse oxidation of various peroxidase substrates in the presence of H2O2 to produce different colour reactions. Scheme of the Immunoassay based on the catalysis of Fe3O4 MNPs. Reprinted from ref. 47, Copyright, 2007, Springer Nature. (B) Schematic diagram of imprinting TMB on Fe3O4 NPs to improve their substate-specificity. Reprinted from ref. 59, Copyright, 2017, American Chemical Society. (C) Schematic diagram of expanding the pH limitation of Carbon dots nanozyme by using Mn(II) as a mediator. Reprinted from ref. 61, Copyright, 2019, American Chemical Society. | ||
Despite the exciting advancement of nanozymes in recent years, their wide applications to immunoassays remain limited by several challenges.58 For example, unlike natural enzymes with well-defined binding pockets, nanozymes do not contain specific substrate-binding sites and thus are generally lack of substate-specificity. To address this issue, Liu and coworkers59 have successfully created substrate (TMB and ABTS) recognition sites on Fe3O4 MNPs using molecular imprinting technology (Fig. 3B). It was found that the imprinted material not only improved the specificity to TMB by nearly 100-times, but also improved the catalytic activity. This study points a possible solution to improve the catalytic specificity of nanozymes, which will expand the application of nanozymes in ELISA.60
Another challenge for using nanozymes is their poor catalytic activities at neutral pH. For most nanozymes, their optimal catalytic activities are achieved at acidic conditions rather than physiological pH ranges. Liu and coworkers61 recently addressed this pH limitation by using Mn(II) as a mediator (Fig. 3C). It was established on the previous observation that the inhibition of the catalytic activity of nanozymes was related to the difficulty of the oxidation of the substrate TMB in neutral conditions.62 When Mn(II) is added to the reaction, it can be photooxidized to Mn(III) by the nanozymes under neutral conditions. Mn(III) then oxidizes TMB to generate the colorless-to-blue transition. Once integrated to ELISA, nanozymes of extended workable pH range will further improve the assay robustness against environmental factors and thus expand their applicability to POCT and field-based tests.
4. Immunoassays using nanomaterials as non-enzymatic signal amplifiers
While the enhancement of enzymatic reactions by nanomaterials has proven to be an effective solution to improve the sensitivity and user-friendship of ELISA, the assay performance may still be limited by the choice of either enzymes (in the case of substrate alternatives) or substrates (in the case of nanozymes). As such, many research efforts have also been emphasized on the development of non-enzymatic signal amplifiers that are compatible with ELISA. Despite no enzymatic reaction is involved, similar operational protocols are applied and newly developed signal amplifiers can be adapted with low technological barriers.63–68 Nanomaterials have been used exclusively to fulfill this goal, as they may either serve as nanocarriers capable of releasing hundreds to thousands of signal reporters all at once or be programmed into unique DNA nanostructures to amplify optical or electrochemical signals.4.1 Non-enzymatic signal amplifiers using nanomaterials as nanocarriers
One of the most frequently adopted mechanisms for designing non-enzymatic signal amplifiers for ELISA is to use nanomaterials as nanocarriers that can accommodate and release large amounts of signal reporters.69 One classical example of such is the biobarcode assay introduced by Mirkin and coworkers in 2003 (Fig. 4A),70 where detection antibody and hundreds of DNA barcodes were conjugated onto the same AuNP. Using this nanobarcode, the detection of trace amounts of target protein is translated into the detection of large numbers of DNA barcodes. When combined with PCR, the LOD of this biobarcode assay reached as low as 3 aM. The clinical usefulness of this assay was further demonstrated by monitoring the level of PSA for patients undergone radical prostatectomy.8 Because of the ultrahigh sensitivity, this assay redefined the undetectable level of PSA and thus effectively improve the predictive value of serum PSA for prostate cancer recurrence.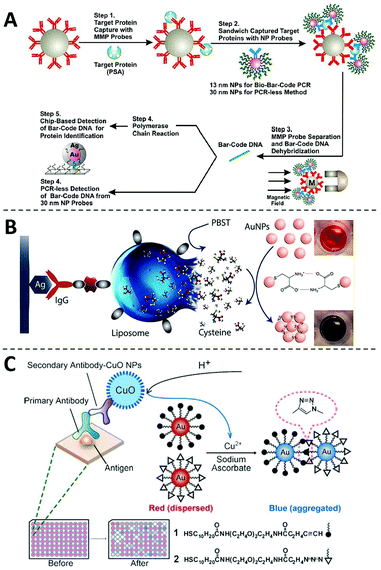 | ||
| Fig. 4 The non-enzymatic signal amplifiers using nanomaterials as nanocarriers. (A) Schematic diagram of the probe design, preparation and PSA detection of the biobarcode assay, in which the detection of trace amounts of target protein is translated into the detection of large numbers of DNA barcodes. Reprinted from ref. 70, Copyright, 2003, Springer Nature. (B) Schematic diagram of the liposome-assisted plasmonic ELISA, in which liposomes can load large amounts of cysteine to amplify the detection signals. Reprinted from ref. 71, Copyright, 2015, American Chemical Society. (C) Schematic diagram of the immunoassay based on CuO-labeled antibody and click chemistry. Reprinted from ref. 72, Copyright, 2011, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
Besides surfaces of nanomaterials, signal reporters can also be encapsulated inside, which will be released during the detection step. Abbas and coworkers developed a liposome-assisted plasmonic ELISA capable of visual detection of specific antibody with a LOD of 6.7 aM (Fig. 4B).71 In this assay, large amounts of cysteine molecules were loaded into a single liposome. In the presence of the target, the liposome was captured through a detection antibody and then hydrolyzed to release cysteines those can induce the rapid aggregation of AuNPs. Through this non-enzymatic amplification mechanism, this assay improves the sensitivity of conventional ELISA by 6 orders of magnitude. This assay has also been successfully applied to the detection of pathogens, such as Salmonella, Listeria, and E. coli O157.71
The chemical composition of nanoparticles can also be used as signal amplifiers for designing ELISA with non-enzymatic readouts. For example, Jiang and coworkers have successfully harnessed copper monoxide nanoparticles (CuO NPs) as signal amplifiers for ultrasensitive protein analysis (Fig. 4C).72 As Cu(II) is the main chemical composition of CuO NPs, millions of Cu(II) can be released upon acid hydrolysis. Cu(II) is then reduced to Cu(I) in the presence of sodium ascorbate, which accelerates a click reaction between alkyn-modified AuNPs and azid-modified AuNPs and induces their aggregation. This method enabled rapid and equipment-free detection of HIV antibodies, showing its potential to be used as a POCT diagnostic test.
4.2 Non-enzymatic signal amplifiers using DNA nanotechnology
Beyond their roles as nanocarriers, nanomaterials can also be programmed into rationally designed structures or reaction networks for signal amplification. Particularly, recent advances in DNA nanotechnology offer numerous structural and dynamic approaches enabling programmable signal amplification for ELISA.73–75Since first introduced by Seeman and coworkers in 1982,76 numerous DNA nanostructures and dynamic devices have been created and found diverse applications in sensing, drug delivery, and biocomputing.77–82 When integrated with an ELISA protocol and electrochemistry for protein analysis, Fan and coworkers found that the three-dimensional (3D) DNA tetrahedron could effectively control the spatial distribution of antibodies on the electrode and thus effectively eliminate entanglement and masking between adjacent probes (Fig. 5A).83 As a result, target proteins could be captured with much higher efficiency and the assay sensitivity was much improved with a low LOD of 2.8 pg mL−1. Inspired by this work, the DNA tetrahedra and other 3D DNA nanostructures, such as DNA nanostars, have also been explored to enhance immunoassays.84–87
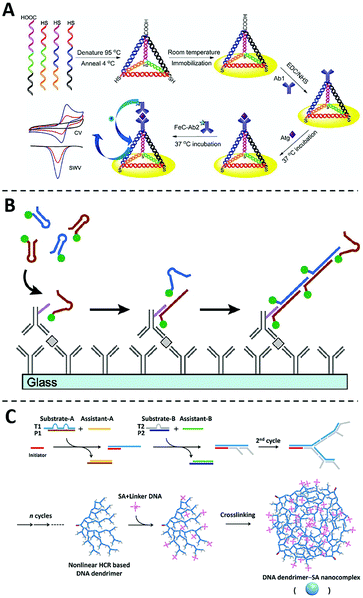 | ||
| Fig. 5 The non-enzymatic signal amplifiers using DNA nanotechnology. (A) Schematic diagram of the sandwich immunoassay using the DNA tetrahedron modified Au electrode, in which entanglement and masking between adjacent probes can be effectively eliminated and target proteins could be captured with much higher efficiency. Reprinted from ref. 83, Copyright, 2014, Springer Nature. (B) Schematic diagram of the immuno-HCR, in which the target recognition can trigger the subsequent formation of long HCR products containing hundreds to thousands of fluorophores. Reprinted from ref. 93, Copyright, 2011, American Chemical Society. (C) Schematic diagram of the formation of DNA dendrimer-SA nanocomplex via a nonlinear HCR reaction, which can be integrated with conventional ELISA to amplify detection signals. Reprinted from ref. 98, Copyright, 2017, American Chemical Society. | ||
DNA nanotechnology also offers a series of dynamic DNA reactions capable of signal amplification in an isothermal and enzyme-free manner.88–91 Because of the ability for in situ signal amplification, hybridization chain reaction (HCR) has emerged as one of the most widely used DNA amplifiers for protein analysis and imaging.92 The integration of HCR with ELISA was first explored by Love and coworkers in 2011, where an HCR initiator was conjugated to the detection antibody (Fig. 5B).93 Once captured by the target protein through the sandwiched binding complex, the initiator triggers the subsequent hybridization of two fluorescently labeled DNA hairpin probes. A long HCR product is then created containing hundreds to thousands of fluorophores. This technology, termed immuno-HCR, allows the quantification of multiple cytokines from human peripheral blood mononuclear cells with LODs at sub-pM levels. Using a similar design, Luo and coworkers further developed a multiplexed immuno-HCR for the simultaneous detection/imaging of multiple protein markers in clinical samples and cells.94 Immuno-HCR have also been successfully designed for the ultra-sensitive detection of IgG,95 CEA,96 and matrix metalloproteinase-797 through electrochemical signal readouts.
In addition to linear DNA structures, dendrimeric DNA structures created by hyperbranched HCR reactions have also been integrated with ELISA as non-enzymatic signal amplifiers.99–102 For example, Liu and coworkers developed a DNA dendrimer-streptavidin nanocomplex via a nonlinear HCR reaction (Fig. 5C).98 This dendrimeric DNA nanostructure was further intercalated with large numbers of SYBR Green I dye and thus significantly amplified fluorescent signals. By integrating this fluorescent DNA dendrimer with ELISA, this assay can detect HeLa cells with a detection limit of 4.4 × 103 cells per mL.
5. Conclusion and perspectives
Over the past years, techniques for protein detection and quantification have been driven by the ever-increasing needs of ultra-high sensitivity and assay simplicity.11–14 Advances in nanotechnology have offered even finer tools for designing better strategies for protein analysis.102 Techniques of nano-/micro- fabrication drive the development of digital ELISA,10 where proteins can be encapsulated and analyzed at a single molecular level. Protein-responsive DNA nanotechnology, such as proximity ligation assays103,104 and binding-induced DNA assembly,105,106 allows the development of homogeneous assays for ultrasensitive protein detection without the need for tedious washing steps. Novel biohybrid nanomaterials, with their unique structural, chemical, catalytic, and optical properties, have facilitated the design of better signal transduction or amplification strategies for protein detection and quantification.16–20In this review, we highlighted the emerging roles of nanomaterials for enhancing or replacing conventional enzyme-based signal readout systems in ELISA. Plasmonic or polymeric nanomaterials have been used as substrate-alternatives in ELISA, enabling ultrasensitive visual readouts that are ideal for POCT applications. Diverse nanozymes have been created, which demonstrate comparable or even better enzymatic activities than natural enzymes but are chemically more stable and less sensitive to environmental factors. Non-enzymatic readout systems making use of nanocarriers and DNA nanotechnology have also been adapted to conventional ELISA systems, allowing sensitive and rapid signal amplification with flexible assay designs. These nanomaterial-enhanced immunoassays drastically improve the analytical performance of conventional ELISA without significantly altering the assay format. As such, these technical advances can be rapidly adopted to existing ELISA detection platforms for diverse biological and clinical applications.
To further expand nanomaterial-enhanced immunoassays for real-world applications, several challenges remain to be addressed. First, antibody-based immunoassays are known to subject to nonspecific adsorption and cross-reactions.107 Better ligands need to designed and integrated to nanomaterials for improving assay performance. Synthetic ligands, such as aptamers and molecularly imprinted polymers,108 are ideal candidates for such applications, as they can be evolved and/or designed for more specific biorecognition than antibodies. Recent advances in structural DNA nanotechnology have enabled the design of multivalent ligands with well-defined spatial distribution.77–80 Second, most nanomaterial-enhanced readout systems are designed for the detection of a single or limit number of proteins. Multiplexed detection of a panel of protein markers is often necessary for making better clinical decisions. Combining nanomaterials with advanced microfluidic ELISA systems may help address the throughput issue.109–111 Finally, similar to conventional ELISA, most nanomaterial-enhanced immunoassays are currently developed and validated in the microplate systems. The movement of these assays from laboratory setting to POCT and field-based applications, it is necessary to combine nanomaterial-based readout with miniaturized ELISA platforms, such as those achieved using paper-based microfluidics and portable device fabrication.112–115
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work is supported by the Fundamental Research Funds for the Central Universities, the National Sciences and Engineering Research Council of Canada, and the Ontario Ministry of Research, Innovation and Science.Notes and references
- E. Engvall and P. Perlmann, Immunochemistry, 1971, 8, 871–874 CrossRef CAS PubMed.
- S. S. Pierangeli and E. N. Harris, Nat. Protoc., 2008, 3, 840–848 CrossRef CAS PubMed.
- L. Asensio, I. González, T. García and R. Martín, Food Control, 2008, 19, 1–8 CrossRef CAS.
- B. M. Beatriz, et al. , Environ. Sci. Technol., 2005, 39, 3896–3903 CrossRef PubMed.
- M. F. Clark, R. M. Lister and M. B. Joseph, Methods Enzymol., 1986, 118, 742–766 CAS.
- W. Jongbloed, M. I. Kester, W. M. Flier, R. Veerhuis, P. Scheltens, M. A. Blankenstein and C. E. Teunissen, Alzheimers Dement., 2013, 9, 276–283 CrossRef PubMed.
- V. H. Flood, J. C. Gill, P. A. Morateck, P. A. Christopherson, K. D. Friedman, S. L. Haberichter, R. G. Hoffmann and R. R. Montgomery, Blood, 2011, 117, e67–e74 CrossRef CAS PubMed.
- C. S. Thaxton, R. Elghanian, A. D. Thomas, S. I. Stoeva, J.-S. Lee, N. D. Smith, A. J. Schaeffer, H. Klocker, W. Horninger, G. Bartsch and C. A. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 18437–18442 CrossRef CAS PubMed.
- T. Sano, C. L. Smith and C. R. Cantor, Science, 1992, 258, 120–122 CrossRef CAS PubMed.
- D. M. Rissin, et al. , Nat. Biotechnol., 2010, 28, 595–600 CrossRef CAS PubMed.
- B. Berg, B. Cortazar, D. Tseng, H. Ozkan, S. Feng, Q. Wei, O. B. Garner and A. Ozcan, ACS Nano, 2015, 9, 7857–7866 CrossRef CAS PubMed.
- T. Laksanasopin, T. W. Guo, S. Nayak, A. A. Sridhara, J. E. Justman, S. Nsanzimana and S. K. Sia, Sci. Transl. Med., 2015, 7, 273re1 CrossRef PubMed.
- X. Wu, M. K. K. Oo, K. Reddy, Q. Chen, Y. Sun and X. Fan, Nat. Commun., 2014, 5, 3779 CrossRef PubMed.
- Y. Song, Y. Huang, X. Liu, X. Zhang, M. Ferrari and L. Qin, Trends Biotechnol., 2014, 32, 132–139 CrossRef CAS PubMed.
- D. Q. González and A. Merkoçi, Chem. Soc. Rev., 2018, 47, 4697–4709 RSC.
- S. Song, Y. Qin, Y. He, Q. Huang, C. Fan and H. Y. Chen, Chem. Soc. Rev., 2010, 39, 4234–4243 RSC.
- X. Pei, B. Zhang, J. Tang, B. Liu, W. Lai and D. Tang, Anal. Chim. Acta, 2013, 758, 1–18 CrossRef CAS PubMed.
- Y. Zhang, Y. Guo, Y. Xianyu, W. Chen, Y. Zhao and X. Jiang, Adv. Mater., 2013, 25, 3802–3819 CrossRef CAS PubMed.
- X. Yang, Y. Tang, R. R. Alt, X. Xie and F. Li, Analyst, 2016, 141, 3473–3481 RSC.
- Z. Farka, T. Juřík, D. Kovář, L. Trnková and P. Skládal, Chem. Rev., 2017, 117, 9973–10042 CrossRef CAS PubMed.
- P. K. Jain, K. S. Lee, I. H. El-Sayed and M. A. El-Sayed, J. Phys. Chem. B, 2006, 110, 7238–7248 CrossRef CAS PubMed.
- B. Jiang, D. Duan, L. Gao, G. Nie, M. Liang and X. Yan, Nat. Protoc., 2018, 13, 1506–1520 CrossRef CAS PubMed.
- P. D. Josephy, T. Eling and R. P. Mason, J. Biol. Chem., 1982, 257, 3669–3675 CAS.
- Z. Xuan, M. Li, P. Rong, W. Wang, Y. Li and D. Liu, Nanoscale, 2016, 8, 17271–17277 RSC.
- J. Satija, N. Punjabi, D. Mishraac and S. Mukherji, RSC Adv., 2016, 6, 85440–85456 RSC.
- M. Salomón and M. N. Eden, Front. Bioeng. Biotechnol., 2019, 7, 1–10 CrossRef PubMed.
- L. Zheng, G. Cai, S. Wang, M. Liao, Y. Li and J. Lin, Biosens. Bioelectron., 2019, 124–125, 143–149 CrossRef CAS PubMed.
- R. de la Rica and M. M. Stevens, Nat. Nanotechnol., 2012, 7, 821–824 CrossRef CAS PubMed.
- D. Liu, J. Yang, H.-F. Wang, Z. Wang, X. Huang, Z. Wang, G. Niu, A. R. H. Walker and X. Chen, Anal. Chem., 2014, 86, 5800–5806 CrossRef CAS PubMed.
- M. Peng, W. Ma and Y.-T. Long, Anal. Chem., 2015, 87, 5891–5896 CrossRef CAS PubMed.
- D. Liu, Z. Wang, A. Jin, X. Huang, X. Sun, F. Wang, Q. Yan, S. Ge, Ni. Xia, G. Niu, G. Liu, A. R. H. Walker and Xi. Chen, Angew. Chem., Int. Ed., 2013, 52, 14065–14069 CrossRef CAS PubMed.
- X. M. Nie, R. Huang, C. X. Dong, L. J. Tang, R. Gui and J. H. Jiang, Biosens. Bioelectron., 2014, 58, 314–319 CrossRef CAS PubMed.
- Y. Xianyu, Z. Wang and X. Jiang, ACS Nano, 2014, 8, 12741–12747 CrossRef CAS PubMed.
- Y. Xianyu, Y. Chen and X. Jiang, Anal. Chem., 2015, 87, 10688–10692 CrossRef CAS PubMed.
- H. Chen, L. Shao, Q. Li and J. Wang, Chem. Soc. Rev., 2013, 42, 2679–2724 RSC.
- H. Liao and J. H. Hafner, Chem. Mater., 2005, 17, 4636–4641 CrossRef CAS.
- S. A. Alex, N. Chandrasekaran and A. Mukherjee, Anal. Methods, 2016, 8, 2131–2137 RSC.
- Y. Li, X. Ma, Z. Xu, M. Liu, Z. Lin, B. Qiu, L. Guo and G. Chena, Analyst, 2016, 141, 2970–2976 RSC.
- Y. Lin, S. Xu, J. Yang, Y. Huang, Z. Chen, B. Qiu, Z. Lin, G. Chen and L. Guo, Sens. Actuators, B, 2018, 267, 502–509 CrossRef CAS.
- Z. Gao, K. Deng, X.-D. Wang, M. Miró and D. Tang, ACS Appl. Mater. Interfaces, 2014, 6, 18243–18250 CrossRef CAS PubMed.
- X. Ma, Y. Lin, L. Guo, B. Qiu, G. Chen, H. Yang and Z. Lin, Biosens. Bioelectron., 2017, 87, 122–128 CrossRef CAS PubMed.
- J. Li, M. A. Baird, M. A. Davis, W. Tai, L. S. Zweifel, K. M. A. Waldorf, M. Jr, L. Rajagopal, R. H. Pierce and X. Gao, Nat. Biomed. Eng., 2017, 1, 1–12 CrossRef PubMed.
- A. Asati, S. Santra, C. Kaittanis, S. Nath and J. M. Perez, Angew. Chem., Int. Ed., 2009, 48, 2308–2312 CrossRef CAS PubMed.
- W. He, Y. Liu, J. Yuan, J. J. Yin, X. Wu, X. Hu, K. Zhang, J. Liu, C. Chen, Y. Ji and Y. Guo, Biomaterials, 2011, 32, 1139–1147 CrossRef CAS PubMed.
- Y. Wana, P. Qia, D. Zhang, J. Wu and Y. Wang, Biosens. Bioelectron., 2012, 33, 69–74 CrossRef PubMed.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- L. Z. Gao, J. Zhuang, L. Nie, J. B. Zhang, Y. Zhang, N. Gu, T. H. Wang, J. Feng, D. L. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- D. Duan, K. Fan, D. Zhang, S. Tan, W. Liu, X. Qiu, G. P. Kobinger, G. F. Gao and X. Yan, Biosens. Bioelectron., 2015, 74, 134–141 CrossRef CAS PubMed.
- J. Mu, Y. Wang, M. Zhao and L. Zhang, Chem. Commun., 2012, 48, 2540–2542 RSC.
- C. Zheng, A.-X. Zheng, B. Liu, X.-L. Zhang, Y. He, J. Li, H. H. Yang and G. Chen, Chem. Commun., 2014, 50, 13103–13106 RSC.
- W. Chen, J. Chen, A. L. Liu, L. M. Wang, G. W. Li and X. H. Lin, ChemCatChem, 2011, 3, 1151–1154 CrossRef CAS.
- N. Singh, M. A. Savanur, S. Srivastava, P. D'Silva and G. Mugesh, Angew. Chem., 2017, 129, 14455–14459 CrossRef.
- H. Ye, K. Yang, J. Tao, Y. Liu, Q. Zhang, S. Habibi, Z. Nie and X. Xia, ACS Nano, 2017, 11, 2052–2059 CrossRef CAS PubMed.
- L. Jiao, L. Zhang, W. Du, H. Li, D. Yang and C. Zhu, Nanoscale, 2018, 10, 21893–21897 RSC.
- C. N. Loynachan, M. R. Thomas, E. R. Gray, D. A. Richards, J. Kim, B. S. Miller, J. C. Brookes, S. Agarwal, V. Chudasama, R. A. McKendry and M. M. Stevens, ACS Nano, 2018, 12, 279–288 CrossRef CAS PubMed.
- X. Zhang, L. Deng, C. Huang, J. Zhang, X. Hou, P. Wu and J. Liu, Chem. – Eur. J., 2018, 24, 2602–2608 CrossRef CAS PubMed.
- C. Penga, M. Hua, N. Li, Y. Hsu, Y. Chen, C. Chuang, S. Pang and H. Yang, Biosens. Bioelectron., 2019, 126, 581–589 CrossRef PubMed.
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119, 4357–4412 CrossRef CAS PubMed.
- Z. Zhang, X. Zhang, B. Liu and J. Liu, J. Am. Chem. Soc., 2017, 139, 5412–5419 CrossRef CAS PubMed.
- Z. Zhang, Y. Li, X. Zhang and J. Liu, Nanoscale, 2019, 11, 4854–4863 RSC.
- J. Zhang, S. Wu, X. Lu, P. Wu and J. Liu, Nano Lett., 2019, 19, 3214–3220 CrossRef CAS PubMed.
- W. He, Y. Liu, J. Yuan, J. J. Yin, X. Wu, X. Hu, K. Zhang, J. Liu, C. Chen, Y. Ji and Y. Guo, Biomaterials, 2011, 32, 1139–1147 CrossRef CAS PubMed.
- W. Zhao, R. Chen, P. Dai, X. Li, J. Xu and H. Chen, Anal. Chem., 2014, 86, 11513–11516 CrossRef CAS PubMed.
- L. Feng, Z. Bian, J. Peng, F. Jiang, G. Yang, Y. Zhu, D. Yang, L. Jiang and J. Zhu, Anal. Chem., 2012, 84, 7810–7815 CrossRef CAS PubMed.
- D. Liu, X. Huang, Z. Wang, A. Jin, X. Sun, L. Zhu, F. Wang, Y. Ma, G. Niu, A. R. H. Walker and X. Chen, ACS Nano, 2013, 7, 5568–5576 CrossRef CAS PubMed.
- G. Fu, S. T. Sanjay, M. Doua and X. Li, Nanoscale, 2016, 8, 5422–5427 RSC.
- M. Lee, H. Kim, B. Kim, J. Jung and T. Kang, ACS Appl. Mater. Interfaces, 2018, 10, 37829–37834 CrossRef CAS PubMed.
- Q. Liu, S. Cheng, R. Chen, J. Ke, Y. Liu, Y. Li, W. Feng and F. Li, ACS Appl. Mater. Interfaces, 2020, 12, 4358–4365 CrossRef CAS PubMed.
- J. M. Nam, S. J. Park and C. A. Mirkin, J. Am. Chem. Soc., 2002, 124, 3820–3821 CrossRef CAS PubMed.
- J. M. Nam, C. S. Thaxton and C. A. Mirkin, Science, 2003, 301, 1884–1886 CrossRef CAS PubMed.
- M.-P. N. Bui, S. Ahmed and A. Abbas, Nano Lett., 2015, 15, 6239–6246 CrossRef CAS PubMed.
- W. Qu, Y. Liu, D. Liu, Z. Wang and X. Jiang, Angew. Chem., Int. Ed., 2011, 50, 3442–3445 CrossRef CAS PubMed.
- S. Wang, L. Zhang, S. Wan, S. Cansiz, C. Cui, Y. Liu, R. Cai, C. Hong, I. T. Teng, M. Shi, Y. Wu, Y. Dong and W. Tan, ACS Nano, 2017, 11, 3943–3949 CrossRef CAS PubMed.
- Y. Lin, J. Jia, R. Yang, D. Chen, J. Wang, F. Luo, L. Guo, B. Qiu and Z. Lin, Anal. Chem., 2019, 91, 3717–3724 CrossRef CAS PubMed.
- J. Wang, C. Xia, L. Yang, Y. Li, C. Li and C. Huang, Anal. Chem., 2020, 92, 4046–4052 CrossRef CAS PubMed.
- N. C. Seeman, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed.
- A. R. Chandrasekaran and O. Levchenko, Chem. Mater., 2016, 28, 5569–5581 CrossRef CAS.
- P. Chidchob and H. F. Sleiman, Curr. Opin. Chem. Biol., 2018, 46, 63–70 CrossRef CAS PubMed.
- Z. Li, J. Wang, Y. Li, X. Liu and Q. Yuan, Mater. Chem. Front., 2018, 2, 423–436 RSC.
- Y. Zhang, J. Tu, D. Wang, H. Zhu, S. K. Maity, X. Qu, B. Bogaert, H. Pei and H. Zhang, Adv. Mater., 2018, 30, 1–44 Search PubMed.
- W. Wang, S. Yu, S. Huang, S. Bi, H. Han, J. Zhang, Y. Lu and J. Zhu, Chem. Soc. Rev., 2019, 48, 4892–4920 RSC.
- S. D. Mason, Y. Tang, Y. Li, X. Xie and F. Li, Trends Anal. Chem., 2018, 107, 212–221 CrossRef CAS.
- L. Yuan, M. Giovanni, J. Xie, C. Fan and D. T. Leong, NPG Asia Mater., 2014, 6, e1120 Search PubMed.
- Z. Li, B. Zhao, D. Wang, Y. Wen, G. Liu, H. Dong, S. Song and C. Fan, ACS Appl. Mater. Interfaces, 2014, 6, 17944–17953 CrossRef CAS PubMed.
- X. Liu, Y. Xu, T. Yu, C. Clifford, Y. Liu, H. Yan and Y. Chang, Nano Lett., 2012, 12, 4254–4259 CrossRef CAS PubMed.
- N. R. Sundah, N. R. Y. Ho, G. S. Lim, A. Natalia, C. W. Chan, T. P. Loh and H. Shao, Nat. Biomed. Eng., 2019, 3, 684–694 CrossRef CAS PubMed.
- P. S. Kwon, S. Ren, S. Kwon, R. J. Linhardt, J. Chao and X. Wang, Nat. Chem., 2020, 12, 26–35 CrossRef CAS PubMed.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- C. Jung and A. D. Ellington, Acc. Chem. Res., 2014, 47, 1825–1835 CrossRef CAS PubMed.
- Y. Tang, Y. Lin, X. Yang, Z. Wang, X. C. Le and F. Li, Anal. Chem., 2015, 87, 8063–8066 CrossRef CAS PubMed.
- H. Zhang, F. Li, B. Dever, X.-F. Li and X. C. Le, Chem. Rev., 2013, 113, 2812–2841 CrossRef CAS PubMed.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- J. Choi, K. R. Love, Y. Gong, T. M. Gierahn and J. C. Love, Anal. Chem., 2011, 83, 6890–6895 CrossRef CAS PubMed.
- R. Lin, Q. Feng, P. Li, P. Zhou, R. Wang, Z. Liu, Z. Wang, X. Qi, N. Tang, F. Shao and M. Luo, Nat. Methods, 2018, 15, 275–278 CrossRef CAS PubMed.
- B. Zhang, B. Liu, D. Tang, R. Niessner, G. Chen and D. Knopp, Anal. Chem., 2012, 84, 5392–5399 CrossRef CAS PubMed.
- L. Hou, X. Wu, G. Chen, H. Yang, M. Lu and D. Tang, Biosens. Bioelectron., 2015, 68, 487–493 CrossRef CAS PubMed.
- W. Zhuang, Y. Li, J. Chen, W. Liua and H. Huang, Anal. Methods, 2019, 11, 2597–2604 RSC.
- Y. Zhao, S. Hu, H. Wang, K. Yu, Y. Guan, X. Liu, N. Li and F. Liu, Anal. Chem., 2017, 89, 6907–6914 CrossRef CAS PubMed.
- G. Wang, L. Chen, X. He, Y. Zhu and X. Zhang, Analyst, 2014, 139, 3895–3900 RSC.
- Y. Lv, R. Peng, Y. Zhou, X. Zhang and W. Tan, Chem. Commun., 2016, 52, 1413–1415 RSC.
- M. Gao, F. He, B. Yin and B. Ye, Analyst, 2019, 144, 1995–2002 RSC.
- Z. Farka, T. Jurik, D. Kovar, L. Trnkova and P. Skladal, Chem. Rev., 2017, 117, 9973–10042 CrossRef CAS PubMed.
- S. Fredriksson, M. Gullberg, J. Jarvius, C. Olsson, K. Pietras, S. M. Gustafsdottir, A. Ostman and U. Landegren, Nat. Biotechnol., 2002, 20, 473–477 CrossRef CAS PubMed.
- F. Li, H. Zhang, C. Lai, X.-F. Li and X. C. Le, Angew. Chem., Int. Ed., 2012, 51, 9317–9320 CrossRef CAS PubMed.
- F. Li, H. Zhang, Z. Wang, X. Li, X.-F. Li and X. C. Le, J. Am. Chem. Soc., 2013, 135, 2443–2446 CrossRef CAS PubMed.
- F. Li, Y. Lin and X. C. Le, Anal. Chem., 2013, 85, 10835–10841 CrossRef CAS PubMed.
- D. A. Giljohann and C. A. Mirkin, Nature, 2009, 462, 461–464 CrossRef CAS PubMed.
- F. Li, H. Zhang, Z. Wang, M. A. Newbigging, S. M. Reid, X.-F. Li and X. C. Le, Anal. Chem., 2015, 87, 274–292 CrossRef CAS PubMed.
- A. H. C. Ng, R. Fobel, C. Fobel, J. Lamanna, D. G. Rackus, E. Lam and A. R. Wheeler, Sci. Transl. Med., 2018, 10, eaar6067 Search PubMed.
- C. Liu, X. Xu, B. Li, B. Situ, W. Pan, Y. Hu, T. An, S. Yao and L. Zheng, Nano Lett., 2018, 18, 4226–4232 CrossRef CAS PubMed.
- C. Dixon, A. H. C. Ng, R. Fobel, M. B. Miltenburg and A. R. Wheeler, Lab Chip, 2016, 16, 4560–4568 RSC.
- J. Liu, Z. Geng, Z. Fan, J. Liu and H. Chen, Biosens. Bioelectron., 2019, 132, 17–37 CrossRef CAS PubMed.
- M. M. Gong and D. Sinton, Chem. Rev., 2017, 117, 8447–8480 CrossRef CAS PubMed.
- L. Soleymani and F. Li, ACS Sens., 2017, 2, 458–467 CrossRef CAS PubMed.
- L. Syedmoradi, M. Daneshpour, M. Alvandipour, F. A. Gomez, H. Hajghassem and K. Omidfar, Biosens. Bioelectron., 2017, 87, 373–387 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |




