Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Extracting DNA from ocean microplastics: a method comparison study†
Pavla
Debeljak
a,
Maria
Pinto
b,
Maira
Proietti
 *ac,
Julia
Reisser
a,
Francesco F.
Ferrari
*ac,
Julia
Reisser
a,
Francesco F.
Ferrari
 a,
Ben
Abbas
d,
Mark C. M.
van Loosdrecht
a,
Ben
Abbas
d,
Mark C. M.
van Loosdrecht
 d,
Boyan
Slat
a and
Gerhard J.
Herndl
d,
Boyan
Slat
a and
Gerhard J.
Herndl
 be
be
aThe Ocean Cleanup Foundation, Martinus Nijhofflaan 2, 18th Floor, 2624 ES Delft, The Netherlands. E-mail: mairaproietti@gmail.com
bDepartment of Bio-Oceanography and Limnology, University of Vienna, Althanstraβe 14, A-1090 Vienna, Austria
cInstituto de Oceanografia, Universidade Federal do Rio Grande, Avenida Itália Km 08, 96203-900, Rio Grande, Brazil
dDepartment of Biotechnology, Delft University of Technology, van der Maasweg 9, 2629 HZ Delft, The Netherlands
eNIOZ Netherlands Institute for Sea Research, Department of Marine Microbiology and Biogeochemistry, Utrecht University, PO Box 59, 1790 AB Den Burg, The Netherlands
First published on 21st November 2016
Abstract
The ubiquity of plastics in oceans worldwide raises concerns about their ecological implications. Suspended microplastics (<5 mm) can be ingested by a wide range of marine organisms and may accumulate up the food web along with associated chemicals. Additionally, plastics provide a stable substrate to a wide range of organisms and, owing to their widespread dispersal, may function as vectors for harmful and invasive species. Despite the growing application of molecular techniques to study ocean microplastic colonizers, to date there is no comparative study on DNA extraction methods for ocean plastic biofilms. The present study aims to fill this gap by comparing DNA yield, amplification efficiency, costs and processing time of different DNA extraction techniques applied to oceanic microplastics. DNA was extracted with five methods (four extraction kits, and standard phenol:chloroform purification) using two mechanical lysis techniques (bead beating and cryogenic grinding with liquid nitrogen) applied to three plastic quantities (1, 15, and 50 fragments per extraction) and size classes (0.05–0.15 and 0.15–0.5 mm). All methods resulted in DNA suitable for downstream applications and were successfully amplified. Overall, the Qiagen Puregene Tissue kit yielded relatively high DNA concentrations for most sizes and amounts of plastics at relatively low costs and short processing time. This study provides a detailed evaluation of DNA extraction methods from ocean plastics, and may assist future research using molecular techniques to study ocean plastic biofilms.
Introduction
Buoyant ocean plastics harbor a wide range of rafting organisms on their surfaces that can have potentially negative ecological impacts, e.g. when plastics serve as vectors for harmful microorganisms and/or invasive species.1–3 Organisms living on plastics in the North Pacific4 and waters around Australia5 have been studied using scanning electron microscopy, with the identification of a large number of diatoms, bacteria, coccolithophorids, and even some invertebrate groups. The complexity of fragmented microplastics (<5 mm in length), which display irregular shapes resulting in high surface to volume ratios, could favour colonization by marine microorganisms.1,2,5,6Molecular techniques are being increasingly used to gain better insights into the composition of ‘epiplastic’ communities from different aquatic environments, as well as particle types and sizes (Table 1). Techniques are based on the extraction of nucleic acids from plastic biofilms, generally followed by amplification of selected genes and amplicon sequencing. These genetic studies have consistently revealed a wide range of epiplastic groups,1,7–11 including potential pathogens and organisms that could play a role in the fate of plastics, such as hydrocarbon-degrading bacteria and fungi.2,12 Reported genera of microorganisms with potential pathogenic strains include Vibrio, Aeromonas, Enterobacter, Halomonas, Mycobacterium, Photobacterium, Pseudomonas, Rhodococcus, and Shigella.1,2,12 Potential hydrocarbon degraders include Alcanivorax, Marinobacter, Pseudomonas, Acinetobacter and Rhodobacteraceae,13–15 as well as fungi of the genus Pestalotiopsis.16 The plastic-degrading capabilities of microorganisms remain to be assessed, though a microbial enzyme has recently been shown to affect the degradation of plastics.17 First identifications of the eukaryotic organisms on plastic particles through metagenomics and amplicon sequencing have also been reported and included diatom groups Coscinodiscophytina and Bacillariophytina, the brown algae Phaeophyceae, the ciliate group Conthreep and the green algae Chlorophyta,18 as well as Hydrozoa, Maxillopoda and Aphragmophora.19
| Publication | Extraction method | Pieces per extraction | Number of extractions | DNA yield (ng μl−1) | Plastic length | Identification method | Plastic type | Plastic origin |
|---|---|---|---|---|---|---|---|---|
| a Personal communication. | ||||||||
| Zettler et al. (2013) | Puregene | 1 | 6 | <5a | 2–18 mm | 16S amplicon sequencing | PE/PP fragments | North Atlantic subtropical gyre water |
| Oberbeckmann et al. (2014) | Lyse-and-Go reagent | 1 | 131 | <5a | 0.5–10 mm | 16S DDGE and sequencing | Fragments | North Sea, Baltic Sea and English Channel water |
| McCormick et al. (2014) | Powersoil | 5–10a | — | — | 2–5 mma | 16S amplicon sequencing | Fragments and pellets | US River water |
| Harrison et al. (2014) | Powersoil | 6 | 63 | — | 5 mm | 16S CARD-FISH | LDPE pellets | Purchased then incubated with UK estuary sediment |
| De Tender et al. (2015) | Powersoil | 1 | 26 | <5 | >25 mm/<5 mm | 16S amplicon sequencing | Fragments/pellets | Belgium ocean sediment/beach sediment |
| Amaral-Zettler et al. (2015) | Puregene | 1 | 346 | — | <5 mm | 16S amplicon sequencing | PE/PP/PS/PET fragments | North Pacific and Atlantic subtropical gyre water |
| Bryant et al. (2016) | DNeasy blood and tissue | 1 | 12 | — | 0.2–2 mm, 2–5 mm, >5 mm | Metagenomic sequencing | Fragments | North Pacific Subtropical Gyre water |
| Oberbeckmann et al. (2016) | Phenol:chloroform | 1 | 27 | <5a | 10 cm2 (0.5 g) | 16S & 18S amplicon sequencing | PET fragments | Purchased then incubated in North Sea off the U.K. coast |
Despite the growing application of molecular techniques to identify microplastic colonizers, there is currently no standard protocol on the extraction of DNA from ocean plastic biofilms, and the available literature does not detail methods or resulting DNA yields. In this study, we compare DNA yields and amplification success obtained with five extraction methods using two mechanical lysis techniques, applied to different sizes (0.5–1.5 mm and 1.5–5 mm) and amounts (1, 15, 50 particles) of oceanic plastics. Furthermore, we compare costs and processing time of these different extraction methods, which are also relevant for future research involving the characterization of epiplastic communities through genetic analyses.
Materials and methods
1. Sampling and sorting of microplastics
DNA extractions were done on plastic fragments collected with Manta nets (frame dimensions 90 × 15 cm, 500 μm mesh size) in August 2015 aboard the RV Ocean Starr. Five paired net tows were conducted at surface waters around 29° N latitude and 140–142° W longitude. This area is within the North Pacific accumulation zone.20 After each net tow, the net was rinsed with seawater, and its cod-end was removed and placed in zip-lock bags, and immediately frozen at −2 °C for storage and transportation. We acknowledge that the ideal temperature for storage of samples for molecular analyses is −20 °C or lower,21 but due to logistical reasons, samples were transported at −2 °C. Once in the laboratory, the zip-lock bags were opened, and contents were thawed and washed with filtered artificial seawater (salinity 35) into sieves (Giuliani, micron: 500, 1500, 5000) that separated the material into two size classes: 0.5–1.5 and 1.5–5 mm.Hard plastic fragments of each size class were randomly selected with forceps. In order to standardise our samples in terms of polymer type, we placed the particles into a 0.94 g ml−1 solution composed of sterile seawater and analytical-grade ethanol, and separated those that sank for further use. According to density data presented by Morét-Ferguson et al.,22 particles that completely sink in this solution are composed of HDPE. To validate this density separation, we used Raman spectroscopy to determine polymer type of five plastic pieces that floated and five that sunk in the above-described solution, and confirmed that the latter were HDPE. We acknowledge however that this is only a partial validation of the separation method due to the small number of particles analysed by Raman spectroscopy. Additionally, processes like biofouling may alter the density of polymers over time; nonetheless, the microplastics used here did not have a visible amount of biofouling, and therefore most likely did not suffer alterations in their density due to this process. The plastic pieces were then grouped into samples according to the experimental design described in the following section. All materials used in our experiments were autoclaved and/or cleaned with 96% ethanol and heated at 150 °C.
2. DNA extraction
Five DNA extraction methods were used to determine DNA yield and amplification efficiency: four commercial extraction kits – Gentra Puregene Tissue kit (Qiagen, Venlo, The Netherlands), MOBIO Powersoil and Powerbiofilm (MOBIO LABORATORIES, INC., Carlsbad, USA), MPBio Fast DNA (MP Biomedicals, LLC., Santa Ana, USA) – and standard phenol:chloroform purification. These methods were chosen according to DNA extraction techniques previously used for ocean plastic biofilms (see Table 1). For all treatments, Ready-Lyse™ lysozyme (10 μl of 1000 units per μl stock; Epicentre, Madison, WI) was added to the samples and incubated at 37 °C for 30 min to improve nucleic acid extraction efficiency. Extractions using kits were conducted following the manufacturers' instructions, and phenol:chloroform extraction was done using phenol and phenol:chloroform:isoamyl alcohol, with ethanol precipitation. Detailed descriptions of protocols are available in ESI.† For all methods, a standard volume of 40 μl elution buffer was used for DNA re-suspension. Extracted DNA was checked on a 1% agarose gel stained for 30 min in a freshly prepared 250 ml 1× TAE (Tris-acetate–EDTA) buffer containing SYBR Gold (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). All extracts were kept at 4 °C during experimental procedures and later stored at −80 °C.
000). All extracts were kept at 4 °C during experimental procedures and later stored at −80 °C.
To evaluate the influence of plastic particle size and quantity on resulting DNA yield, we applied the five DNA extraction methods to 1, 15, and 50 pieces of 0.5–1.5 mm microplastics, and to 1 and 15 pieces of 1.5–5 mm microplastics. We also evaluated whether initial mechanical lysis methods – grinding with liquid nitrogen or bead beating – influence the quantity of the resulting DNA. For bead beating, if provided by the kit, beads were used according to the manufacturer's protocol; if not provided, zirconium beads (0.1 mm diameter, BioSpec Products) were added. For cryogenic grinding, particles were placed in a sterile mortar, flash-frozen in liquid nitrogen, and then grounded with a sterile pestle. Each combination of variables was performed in triplicate, amounting to a total of 150 extraction tests and 2460 plastic particles.
The costs of each extraction method including all required reagents were calculated with prices retrieved from the manufacturers' online order pages (https://mobio.com; https://www.mpbio.com, https://www.qiagen.com) and suppliers (phenol, chloroform: https://www.sigmaaldrich.com; Ready-Lyse™ lysozyme: https://www.epibio.com/enzymes/lysozymes/ready-lyse-lysozyme-solution).
3. DNA yield quantity and quality
To assess the amount of DNA obtained with the different methods, a PicoGreen assay of all extracted samples was performed using a Quant-iT™ PicoGreen® dsDNA Assay kit (ThermoFisher). A standard curve ranging from 0 to 300 ng ml−1 was prepared using the standard provided by the kit (100 μl ml−1). 1× TE (Tris–EDTA) buffer was pipetted into each well of a black 96-well MICROLON® 200 microplate, to which the standards and 1 μl of each sample were added. Fluorescence was measured at 485 nm excitation and 530 nm emission wavelength on a microplate reader (Tecan Infinite), and the DNA concentration of each sample was determined using the standard curve. Extraction quality was also assessed by measuring the absorbance at 260 and 280 nm wavelengths, using a NanoDrop spectrometer.4. 16S rRNA amplification
Full-length 16S bacterial rRNA genes were amplified through Polymerase Chain Reactions (PCR) on a Mastercycler (Eppendorf) with primers 27-F (5′ AGAGTTTGATCCTGGCTCAG 3′) and 1492-R (5′ GGTTACCTTGTTACGACTT 3′) under the following conditions: initial denaturation of 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, and a final extension at 72 °C for 10 min. Each reaction contained 2.5 μl dNTPs, 0.1 μl Taq polymerase (Thermo Scientific), 2.5 μl of the corresponding buffer, 2 μl MgCl2, 0.2 μl of each primer and DNA sample. PCR reactions were set up to a final volume of 25 μl with sterile H2O. Amplicons were checked on a 1% agarose gel stained for 30 min in freshly prepared 250 ml 1× TAE buffer containing SYBR Gold (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000).
000).
5. Statistical analysis
To assess the efficiency of the different lysis methods, extraction protocols and number of microplastic pieces, we fitted multiple linear regression models to our DNA concentration dataset for each size class, with the above-mentioned extraction treatments as categorical variables. These analyses were performed with the ‘stats’ package in R.23Results & discussion
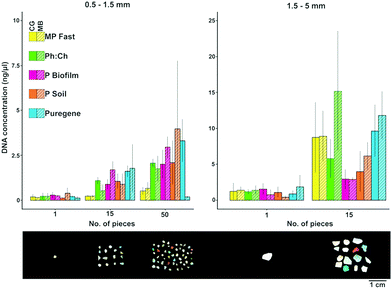
Concentrations of DNA extracted from ocean microplastic biofilms ranged from 0.06 to 25.86 ng μl−1 (Fig. 1). The overall low DNA concentrations obtained (most values <5 ng μl−1) were similar to those obtained by previous studies (see Table 1). This might be due to (1) a generally low abundance of microorganisms present on the individual plastic pieces, and/or (2) a low efficiency of the methods examined due to the highly irregular surfaces of marine microplastics (see images in Zettler et al.1 and Reisser et al.5). Under all the experimental conditions, DNA yields were higher for microplastics in the 1.5–5 mm size range (mean ± SD = 4.37 ± 4.86 ng μl−1) than for 0.5–1.5 mm particles (mean ± SD = 1.06 ± 1.33 ng μl−1).When extracting DNA from microplastics smaller than 1.5 mm in diameter, most applied extraction methods resulted in similar DNA concentrations, but the MP Fast Spin kit yielded consistently lower values (Fig. 1). For microplastics ranging from 1.5 to 5 mm, the Qiagen Puregene and MP Fast Spin kits, as well as the phenol:chloroform method, resulted in relatively high DNA yields (Fig. 1). Despite the overall low quality of extracted DNA (see Table 1 for A260/280 and A260/230 values), the five extraction methods led to the successful amplification of the full-length fragment of 16S rRNA, indicating that all tested methods are suitable for downstream applications for bacterial community analysis.
The amount of microplastics required for molecular analyses highly depends on the desired downstream procedure, and this should be considered when deciding the number of particles per extraction used. In our extractions, 15 particles of 0.5–1.5 mm sized plastics yielded on average 1.16 ng μl−1 (SD = 0.86 ng μl−1), a similar amount of DNA as one particle of 1.5–5 mm sized plastics (mean ± SD = 0.99 ± 0.77 ng μl−1; see Fig. 1). Compared to one larger particle, 15 smaller microplastics likely have a larger surface area available for microbial colonization. This indicates that the abundance of the plastic-associated microorganisms is directly proportional to the size of the particles. Alternatively, the complex surface structure of these weathered smaller particles might make the extraction of cells more difficult, resulting in a similar amount of extracted DNA in the large versus small (but multiple) plastics.
The extraction method and number of pieces had a significant influence on the extraction efficiency for both size 0.5–1.5 mm (p = 0.02 and p = 3.0 × 10−7 respectively; n = 89) and size 1.5–5 mm (p = 0.01 and p = 1.1 × 10−9 respectively; n = 60), while the lysis method did not influence efficiency (size 0.5–1.5 mm, p = 0.94; size 1.5–5 mm, p = 0.18; Fig. 1). Despite the fact that increasing the number of pieces per extraction led to higher DNA yields, analysing single plastic pieces can be valuable if the research question at hand is related to specific particle properties; for instance, Zettler et al.10 used single plastic pieces to analyse epiplastic communities and evaluate whether they reflected factors such as polymer type and biogeographic origins.
DNA concentration variance between extractions was explained by the fitted model in 38% (R2 = 0.38) for plastics of size 0.5–1.5 mm, and 57% (R2 = 0.57) for size 1.5–5 mm. These relatively low R2 values suggest that there is a high variability between individual plastic pieces due to their inherent characteristics, such as time spent in the ocean and fragmentation processes, which could influence biomass and community composition.
Although all tested extraction and lysis techniques led to successful 16S amplification, it is likely that different methods favour acquisition of DNA from different groups. This influence is shown in McCarthy et al.,24 who report that the type of extraction protocol affects perceived bacterial community composition of water samples. In the case of our extraction tests, we believe that, when compared to bead beating, cryogenic grinding could more thoroughly remove bioeroding organisms embedded in the microplastics. This type of influence should be considered when planning a molecular study of epiplastic communities.
The cost of each extraction method including all required reagents ranged from € 1.39 per sample for phenol:chloroform to € 7.08 for MOBIO Powerbiofilm kit (Table 2). However, we highlight that these costs can vary substantially depending on the purchasing conditions of research institutions, as well as customs and tax charges in different countries. In terms of time, extractions with kits ranged from three to five hours per 15 samples, while phenol:chloroform was the most labour-intensive method with 36–37 hours (including overnight incubation of around 14 hours) required per 15 samples. Additionally, the latter is the only method that includes highly toxic substances. Labour costs were not considered in the final calculations as these are highly variable, but if taken into account, the costs of the phenol:chloroform extraction would increase substantially. Cryogenic grinding increased extraction time by one hour per 15 samples when compared to bead beating. Since no significant difference in resulting DNA yields was observed between the two methods, we recommend bead beating as the mechanical lysis method.
| Qiagen Puregene | MPBio Fast DNA | MOBIO Powersoil® | MOBIO Powerbiofilm® | Phenol:chloroform | |
|---|---|---|---|---|---|
| DNA yields from particles <1.5 mm (mean ± SE) | 1.20 ± 0.33 | 0.32 ± 0.06 | 1.49 ± 0.53 | 1.34 ± 0.25 | 0.98 ± 0.19 |
| DNA yield from particles >1.5 mm (mean ± SE) | 6.03 ± 1.58 | 5.05 ± 1.37 | 2.90 ± 0.82 | 2.03 ± 0.35 | 5.87 ± 2.09 |
| A 260/A280 | 2.43 ± 1.14 | 2.34 ± 0.40 | 1.85 ± 1.14 | 1.36 ± 0.69 | 1.45 ± 0.24 |
| A 260/A230 | 0.23 ± 0.12 | 0.04 ± 0.06 | 0.54 ± 0.20 | 0.13 ± 0.11 | 1.18 ± 0.08 |
| 16S amplification successful | Yes | Yes | Yes | Yes | Yes |
| Cost per sample (in €) | 1.79 | 4.29 | 4.98 | 7.08 | 1.39 |
| Extraction time per 15 samples with CG (h) | 5 | 4 | 4.5 | 4.5 | 37 |
| Extraction time per 15 samples with BB (h) | 4 | 3 | 3.5 | 3.5 | 36 (including overnight incubation) |
| Toxicity | Low | Low | Low | Low | High |
Conclusions
This study provides a guide for DNA extraction from different sizes and amounts of marine microplastics. The choice of the extraction method depends on the desired DNA yield, which is dependent on the size and amount of microplastics, and should be pondered along with an evaluation of cost and time efficiency. Based on our comparisons, the most cost-effective method was bead beating followed by purification with the Qiagen Puregene Tissue kit. However, the other methods also yielded suitable and amplifiable DNA, and researchers should consider their individual scenarios when selecting an extraction technique for marine microplastic biofilms. Furthermore, we attempted to focus only on HDPE hard microplastics due to their wide distribution in oceanic waters,25 and highlight that DNA yields could differ when extracting from other polymers and particle types (e.g. soft plastics, fibres). Our comparison of extraction methods provides guidance for researchers aiming to further characterize marine ‘epiplastic’ communities, which may include pathogenic, invasive and polymer-degrading groups.Acknowledgements
We thank The Ocean Cleanup supporters and the Mega Expedition project team. We also acknowledge Christian Baranyi (University of Vienna) for advice on nucleic acid purification and assistance in the lab, as well as colleagues at the TU Delft Biotechnology for support during extractions. We thank Charles de Boer for assistance with Raman spectroscopy at the Kavli Nanolab at TU Delft. This study was supported by a DOC grant of the Austrian Academy of Sciences (Grant number: 24446) to M. P. and by the Austrian Science Fund (FWF; ARTEMIS project P28781-B21) to G. J. H.Notes and references
- E. R. Zettler, T. J. Mincer and L. A. Amaral-Zettler, Environ. Sci. Technol., 2013, 47, 7137–7146 CAS.
- S. Oberbeckmann, M. G. J. Löder and M. Labrenz, Environ. Chem., 2015, 12, 551–562 CrossRef CAS.
- T. J. Mincer, E. R. Zettler and L. A. Amaral-Zettler, in The Handbook of Environmental Chemistry. Volume: Hazardous Chemicals Associated with Plastics in the Marine Environment, ed. H. Takada and H. K. Karapanagioti, Springer International Publishing, Switzerland, 1st edn, 2016, DOI:10.1007/698_2016_12.
- H. S. Carson, M. S. Nerheim, K. Carroll and M. Eriksen, Mar. Pollut. Bull., 2013, 75, 126–132 CrossRef CAS PubMed.
- J. Reisser, J. Shaw, G. Hallegraeff, M. Proietti, D. K. Barnes and M. Thums, PLoS One, 2014, 9, 1–11 Search PubMed.
- J. P. Harrison, M. Sapp, M. Schratzberger and A. M. Osborn, Mar. Technol. Soc. J., 2011, 45, 12–20 CrossRef.
- J. P. Harrison, M. Schratzberger, M. Sapp and A. Osborn, BMC Microbiol., 2014, 14, 1–15 CrossRef PubMed.
- A. McCormick, T. J. Hoellein, S. A. Mason, J. Schluep and J. J. Kelly, Environ. Sci. Technol., 2014, 48, 11863–11871 CrossRef CAS PubMed.
- S. Oberbeckmann, M. G. J. Löder, G. Gerdts and M. Osborn, FEMS Microbiol. Ecol., 2014, 49, 478–492 Search PubMed.
- L. A. Amaral-Zettler, E. R. Zettler, B. Slikas, G. D. Boyd, D. W. Melvin and C. E. Morrall, Front. Ecol. Environ., 2015, 13, 541–546 CrossRef.
- C. A. De Tender, L. I. Devriese, A. Haegeman, S. Maes, T. Ruttink and P. Dawyndt, Environ. Sci. Technol., 2015, 49, 9629–9638 CrossRef CAS PubMed.
- I. V. Kirstein, S. Kirmizi, A. Wichels, A. Garin-Fernandez, R. Erler, M. Löder and G. Gerdts, Mar. Environ. Res., 2016, 120, 1–8 CrossRef CAS PubMed.
- J. G. Leahy and R. R. Colwell, Microbiol. Rev., 1990, 54, 305–315 CAS.
- M. Shimao, Curr. Opin. Biotechnol., 2001, 12, 242–247 CrossRef CAS PubMed.
- J. E. Kostka, O. Prakash, W. A. Overholt, S. J. Green, G. Freyer, A. Canion, J. Delgardio, N. Norton, T. C. Hazen and M. Huettel, Appl. Environ. Microbiol., 2011, 77, 7962–7974 CrossRef CAS PubMed.
- J. R. Russell, J. Huang, P. Anand, K. Kucera, A. G. Sandoval, K. W. Dantzler, D. Hickman, J. Jee, F. M. Kimovec, D. Koppstein and D. H. Marks, Appl. Environ. Microbiol., 2011, 77, 6076–6084 CrossRef CAS PubMed.
- S. Yoshida, K. Hiraga, T. Takehana, I. Taniguchi, H. Yamaji, Y. Maeda, K. Toyohara, K. Miyamoto, Y. Kimura and K. Oda, Science, 2016, 351, 1196–1199 CrossRef CAS PubMed.
- S. Oberbeckmann, A. M. Osborn and M. B. Duhaime, PLoS One, 2016, 11, e0159289 Search PubMed.
- J. A. Bryant, T. M. Clemente, D. A. Viviani, A. A. Fong, K. A. Thomas, P. Kemp, D. M. Karl, A. E. White and E. F. DeLong, mSystems, 2016, 1, e00024 CrossRef PubMed.
- N. Maximenko, J. Hafner and P. Niiler, Mar. Pollut. Bull., 2012, 65, 51–62 CrossRef CAS PubMed.
- L. Prendini, R. Hanner and R. DeSalle, in Techniques in molecular evolution and systematics, ed. R. DeSalle, G. Giribet and W. C. Wheeler, Birkhaeuser Verlag AG., Basel, 2002, pp. 176–248 Search PubMed.
- S. Morét-Ferguson, K. L. Law, G. Proskurowski, E. K. Murphy, E. E. Peacock and C. M. Reddy, Mar. Pollut. Bull., 2010, 60, 1873–1878 CrossRef PubMed.
- R Core Team, R (3.2.4 Revised Version), R Foundation for Statistical Computing, Vienna, Austria, 2016, http://www.R-project.org Search PubMed.
- A. McCarthy, E. Chiang, M. L. Scmidt and V. J. Denef, PloS One, 2015, 10(3), e0121659 Search PubMed.
- V. Hidalgo-Ruz, L. Gutow, R. C. Thompson and M. Thiel, Environ. Sci. Technol., 2012, 46, 3060–3075 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ay03119f |
| This journal is © The Royal Society of Chemistry 2017 |