Open Access Article
Open Access ArticleA novel nanocomposite drug delivery system for SARS-CoV-2 infections†
Uday
Chintapula‡
a,
Shazeed-Ul
Karim‡
b,
Priyanka Raghunathan
Iyer‡
a,
Haritha
Asokan-Sheeja
c,
Biswas
Neupane
b,
Farzana
Nazneen
 b,
He
Dong
b,
He
Dong
 c,
Fengwei
Bai
*b and
Kytai T.
Nguyen
*a
c,
Fengwei
Bai
*b and
Kytai T.
Nguyen
*a
aDepartment of Bioengineering, University of Texas at Arlington, Arlington, TX 76010, USA. E-mail: knguyen@uta.edu
bDepartment of Cell and Molecular Biology, University of Southern Mississippi, Hattiesburg, MS 39406, USA. E-mail: fengwei.bai@usm.edu
cDepartment of Chemistry and Biochemistry, University of Texas at Arlington, Arlington, TX 76010, USA
First published on 17th June 2024
Abstract
To develop an inhalable drug delivery system, we synthesized poly (lactic-co-glycolic acid) nanoparticles with Remdesivir (RDV NPs) as an antiviral agent against SARS-CoV-2 replication and formulated Remdesivir-loaded nanocomposites (RDV NCs) via coating of RDV NPs with novel supramolecular cell-penetrating peptide nanofibers (NFs) to enhance cellular uptake and intracellular drug delivery. RDV NPs and RDV NCs were characterized using variou techniques, including Transmission Electron Microscopy (TEM), Dynamic Light Scattering (DLS), and fluorescent microscopy. The cytotoxicity of RDV NCs was assessed in Vero E6 cells and primary human lung epithelial cells, with no significant cytotoxicity observed up to 1000 μg mL−1 and 48 h. RDV NCs were spherically shaped with a size range of 200–300 nm and a zeta potential of ∼+31 mV as well as indicating the presence of coated nanofibers. Reverse Transcription-quantitative Polymerase Chain Reaction (RT-qPCR), immunofluorescence and plaque assays of SARS-CoV-2 infected Vero E6 treated with RDV NCs showed significantly higher antiviral activities compared to those of free drug and uncoated RDV NPs. RDV NCs exhibited high antiviral activity against SARS-CoV-2, and the nanocomposite platform has the potential to be developed into an inhalable drug delivery system for other viral infections in the lungs.
1. Introduction
Coronaviruses can cause significant illnesses such as respiratory tract infections in humans and other mammals.1 December 2019 marked the outbreak of the new coronavirus (SARS-CoV-2), which was first detected in Wuhan, China and now almost all the nations of the world are victims of this pandemic. Scientists are making great efforts to find treatments or vaccines, but still, there is no permanent remedy or cure for the patients suffering from COVID-19 and long-term COVID-19 complications. Although the pandemic has come to an end, the emergence of a pandemic-potential virus is probable due to the growing urbanization of societies and global connectivity. Currently, vaccines are the only tools to prevent the spread, while the scientific community is engaged in developing treatment strategies for the diseases caused by these viruses.Everyone, irrespective of age, can get infected with SARS-CoV-2 but its complications are of major concern for older people and people with diabetes, where increased glucose levels in airway secretion significantly increase influenza virus replication and other complications including inflammation and hypertension, which can be life-threatening.2–4 Drugs such as Remdesivir, Tocilizumab, and molnupiravir, among others, are currently under clinical trials with the aim of either interfering with viral replication or reducing complications in the lungs.5–9 Very few drugs such as paxlovoid, sotrovimab and molnupiravir were approved by the FDA, which address only mild to typical COVID-19. Out of all these drugs, Remdesivir and Nirmatrelvir (paxlovid) showed promising results in inhibiting viral reproduction.10 However, free drugs are more vulnerable and susceptible to enzymatic degradation, uptake by macrophages, and clearance by the immune system. In addition, the free drug is not entirely available at the targeted site due to its non-specificity, which leads to the requirement of multiple drug dosages, leading to increased cost and other side effects. Therefore, it is necessary to develop a drug delivery system that can increase drug delivery efficiency and targeting specificity.
Nanotechnology has demonstrated great promise in the medical field. The major factors contributing to its popularity are increased bioavailability, enhanced drug targeting, improved drug solubility and stability, controlled drug release, and facilitated patient adherence. These properties of nanotechnology make it beneficial to effectively treat various lung diseases.11 The concerns associated with free drugs are addressed by using a nanoparticle system that can have sustained drug release and can efficiently deliver the drug to the target site. In a study done by Jurek et al.,12 edema in acute lung injury was prevented for 3 days via inhalation delivery of ruthenium red loaded polylactic-co-glycolic acid (PLGA) nanoparticles over free ruthenium red for 60 minutes in high-pressure mechanical ventilation (HPMV)-mediated mice ex vivo perfusion studies, showing the potential significance of nanoparticle-mediated drug delivery for treating patients with respiratory failure. Similarly, a free anti-inflammatory drug delivered to treat lung injury-related inflammation in mice was almost undetectable in the lungs after 4 hours compared to targeted nanoparticles, which were seen for over 24 hours, in a study by Zhang et al.13 In other studies, pre-treatment with an anti-inflammatory molecule α-bisabolol loaded lipid-core nanocapsule (LNC) in LPS-induced lung injury showed a higher concentration of the anti-inflammatory molecule and significant reduction of injury in the lungs compared to free α-bisabolol.14 Various nanoparticle-mediated drug delivery approaches have been successfully utilized previously to treat acute lung diseases.15
Although nanoparticles have shown their superiority in delivering drugs to the lungs, some viral infections have been shown to reduce the cell's ability to take up nanoparticles.16 Recently, technologies utilizing cell-penetrating peptides (CPPs) have exhibited an enhanced uptake of drugs in cells via higher binding affinity to cell membranes. Towards that approach, we apply our novel drug delivery system, consisting of CPP nanofiber-coated PLGA NPs to deliver antiviral drugs for the treatment of SARS-CoV-2 infections (Fig. 1). Cellular uptake and viral load in SARS-CoV-2 infected Vero E6 cells were examined to assess the efficacy of RDV NCs in inhibiting SARS-CoV-2 infection in vitro. We have also demonstrated the stability of nanocomposites in inhalable formulations in our previous study, where our nanocomposites improved cell uptake compared to nanoparticles when delivered via a nebulizer to the cells in vitro.17 Our novel drug delivery system has the potential to deliver drugs via inhalation to the lungs for the treatment of lung diseases, especially, lung infections such as COVID-19.
 | ||
| Fig. 1 Graphical abstract of nanocomposite-based intracellular delivery of Remdesivir to treat SARS-CoV-2 infection. | ||
2. Methods
2.1. Ethics statement and biosafety
All the experiments involving live SARS-CoV-2 were carried out by trained and certified personnel in the USDA-certified Biosafety Level 3 (BSL3) facilities at the University of Southern Mississippi by following the biosafety protocol approved by the USM Institutional Biosafety Committee.2.2. Viruses
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2, strain USA_WA1/2020, GenBank accession MT020880) was provided by the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch. A single passage of parental viruses was propagated in Vero E6 cells (ATCC® CRL-1586™) and then collected as viral stock for this study. The viral titer of the stocks was determined by performing plaque assays in plaque forming units (PFU) per millilitre.2.3. Cells
Vero E6 cells (ATCC® CRL-1586™) were maintained in Minimum Essential Medium (Gibco™ MEM, Life Technologies) containing 1% L-glutamine, 1% penicillin/streptomycin, and 10% fetal bovine serum (FBS). Alveolar Type I (AT1) cells (ABM, T4003) were maintained in Iscove's Modified Dulbecco's Medium (IMDM, Sigma Life Science) containing 1% L-glutamine, 1% penicillin/streptomycin, and 10% FBS.2.4. Synthesis of PLGA nanoparticles and nanocomposites
Poly (lactic-co-glycolic acid) based nanoparticles (blank NPs, RDV-NPs, and RDV-NCs) were synthesized via a modified single emulsion (O/W) technique as described previously.17 Briefly, 10 mg of Remdesivir dissolved in DMSO (Sigma-Aldrich, St. Louis, USA) was added to 90 mg of PLGA 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (Polyscitech, West Lafayette, USA) in 3 mL of DCM (Sigma-Aldrich, St. Louis, USA) dropwise and sonicated at 30 W for 1 minute to allow dispersion of PLGA and Remdesivir in the solvent. The resulting solution was added dropwise to 20 mL of filtered 5% (w/v) poly(vinyl) alcohol (PVA, 13 kDa) solution under stirring conditions. The suspension was then sonicated at 30 W for 2 minutes and then allowed to stir overnight to evaporate the organic solvent. The obtained nanoparticle suspension was centrifuged at 15
50 (Polyscitech, West Lafayette, USA) in 3 mL of DCM (Sigma-Aldrich, St. Louis, USA) dropwise and sonicated at 30 W for 1 minute to allow dispersion of PLGA and Remdesivir in the solvent. The resulting solution was added dropwise to 20 mL of filtered 5% (w/v) poly(vinyl) alcohol (PVA, 13 kDa) solution under stirring conditions. The suspension was then sonicated at 30 W for 2 minutes and then allowed to stir overnight to evaporate the organic solvent. The obtained nanoparticle suspension was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm/27
000 rpm/27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 g for 30 minutes. The supernatant was saved for the drug loading evaluation, and the PLGA NP pellet was re-suspended in 3 mL of DI water followed by freeze-drying for 24 hours. Nanoparticles for imaging techniques were synthesized by a similar procedure with Rhodamine dyes instead of Remdesivir using a double emulsion technique, and blank nanoparticles were made with no drugs or dyes encapsulated in the polymer.
200 g for 30 minutes. The supernatant was saved for the drug loading evaluation, and the PLGA NP pellet was re-suspended in 3 mL of DI water followed by freeze-drying for 24 hours. Nanoparticles for imaging techniques were synthesized by a similar procedure with Rhodamine dyes instead of Remdesivir using a double emulsion technique, and blank nanoparticles were made with no drugs or dyes encapsulated in the polymer.
NFs were synthesized as previously described (refer to the ESI section for the method)17,18 (ESI Fig. S1†). Labelled and non-labelled NF peptides were dissolved in tris(hydroxymethyl)-aminomethane (Tris) buffer (pH 7.4, 20 mM) at 20 mM. After lyophilization of NPs, 2 mg of RDV-NPs were dissolved in Tris buffer as separate groups, and 0.5 mg of NF in suspension was added to the NP suspensions to prepare RDV NCs. The mixture was left to react electrostatically by rotating the solution for an hour at room temperature. Later, the sample was centrifuged at 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200×g for 8 minutes to remove free NFs and collect the samples, which contained RDV NCs. Rhodamine dye-loaded nanocomposites were also prepared in the same manner.
200×g for 8 minutes to remove free NFs and collect the samples, which contained RDV NCs. Rhodamine dye-loaded nanocomposites were also prepared in the same manner.
2.5. DLS measurements
A ZETAPALS90 dynamic light scattering (DLS) detector (Brookhaven Instrument, Holtsville, NY) was used to determine the size, charge, and polydispersity of the NPs. For DLS measurements, 15 μL of 1 mg mL−1 NP suspension was mixed with 3 mL of DI water in a transparent cuvette and placed in the instrument to measure size, while a DLS probe was used to measure the zeta potential of the NF-coated NPs.2.6. Fluorescence microscopy
Fluorescein-terminated peptides were synthesized as previously described.18 5(6)-Carboxyl fluorescein (FITC)-tagged peptides were mixed with Rho B PLGA NPs. Green colour tagged NFs were incubated with NPs loaded with Rhodamine B (red colour). The NCs formed were washed three times to remove any unbound NFs. Another set of NPs was similarly washed and imaged without any NF. A fluorescence microscope (ECHO, San Diego, CA) with FITC (for NFs) and Texas Red channels (for Rho B NPs) at 40× magnification was used to image the NF coating on the NPs.2.7. Transmission electron microscopy (TEM)
To generate TEM images of NF-coated NPs, 10 μl of 2 mg mL−1 NP or NC suspension was added to plasma-treated formvar square mesh copper grids and air-dried after incubating with uranyl acetate for negative staining. An H-7500 TEM (Hitachi) transmission electron microscope was used to visualize the particle morphologies.2.8. Drug loading and drug release kinetics of RDV NCs
The drug/dye loading efficiency was calculated by an indirect method, where the drug present in the supernatant collected from the nanoparticle synthesis process was measured with the help of HPLC, and the loading efficiency was calculated using eqn (1).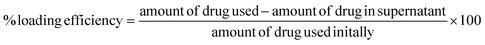 | (1) |
The Remdesivir release study was carried out for 10 days. Briefly, 1 mg of RDV-NPs was taken at a concentration of 1 mg mL−1 and incubated at 37 °C. At each predetermined time point, the samples were centrifuged at 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200×g for 30 minutes, and supernatants were collected and stored at −20 °C for later analysis. The pellets were re-suspended in fresh 1× PBS and incubated for further time points. Each of the drug-release aliquots was analysed using the HPLC method. The amount of drug released was determined against a standard curve for Remdesivir.
200×g for 30 minutes, and supernatants were collected and stored at −20 °C for later analysis. The pellets were re-suspended in fresh 1× PBS and incubated for further time points. Each of the drug-release aliquots was analysed using the HPLC method. The amount of drug released was determined against a standard curve for Remdesivir.
2.9. HPLC method for nanocomposite drug loading and release study
Chromatographic analysis was performed on a liquid chromatograph (Agilent 1260) with a UV-visible detector. RDV was analysed at a flow rate of 1.2 mL min−1 using a mobile phase composed of 20 mM potassium dihydrogen phosphate solution and acetonitrile (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v). Before use, the mobile phase was filtered and degassed through a 0.22 μm membrane filter. An Agilent Extend C18 (4.6 mm × 250 mm, 5.0 μm particle size) column was used and operated at 25 °C. RDV was detected with the UV detector at 247 nm. The run time under these conditions was 10 minutes.
50, v/v). Before use, the mobile phase was filtered and degassed through a 0.22 μm membrane filter. An Agilent Extend C18 (4.6 mm × 250 mm, 5.0 μm particle size) column was used and operated at 25 °C. RDV was detected with the UV detector at 247 nm. The run time under these conditions was 10 minutes.
2.10. Cytocompatibility of nanoparticles and nanocomposites
In this study, primary lung epithelial cells and kidney epithelial cells (Vero E6) were used to assess the toxicity of NPs. NCs were prepared as described above, where 4000 cells per well of primary Alveolar Type I epithelial cells (AT1) and Vero E6 cells were seeded in 96-well plates. After overnight culture, RDV-NCs were added to the cells in triplicate at various concentrations ranging from 0 to 1 mg mL−1. An optimized NF to NP ratio of 0.25 was used for the study. After 48 hours, the cells were washed three times with PBS, and MTS reagent was given to the cells to assess the cell viability following the company's instructions.2.11. Cell treatment and viral infection
Vero E6 cells were seeded at a concentration of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 12-well plates and incubated overnight at 37 °C. Following overnight attachment, the cells were infected with 0.5 MOI of SARS-CoV-2 for 2 hours. After infection, Remdesivir (200 nM), blank NPs (100 μg mL−1), and various concentrations (10, 100, and 1000 μg mL−1) of RDV NPs and RDV NCs were added to the cells separately for 24 and 48 hours. After 24 and 48 hours, the cell morphology and viability were observed under a microscope. Subsequently, supernatants were collected for plaque assay, and cells were collected in TRI-reagent for RT-qPCR analysis to evaluate viral inhibition.
000 cells per well in 12-well plates and incubated overnight at 37 °C. Following overnight attachment, the cells were infected with 0.5 MOI of SARS-CoV-2 for 2 hours. After infection, Remdesivir (200 nM), blank NPs (100 μg mL−1), and various concentrations (10, 100, and 1000 μg mL−1) of RDV NPs and RDV NCs were added to the cells separately for 24 and 48 hours. After 24 and 48 hours, the cell morphology and viability were observed under a microscope. Subsequently, supernatants were collected for plaque assay, and cells were collected in TRI-reagent for RT-qPCR analysis to evaluate viral inhibition.
AT1 cells were seeded at a density of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 12-well plates and incubated at 37 °C until reaching approximately 80% confluence. Subsequently, the cells were treated with Remdesivir (200 nM), blank NPs (100 μg mL−1), RDV NPs (100 μg mL−1), and RDV NCs (100 μg mL−1) for 24 hours without viral infection. After the treatment, cells were collected in TRI-reagent for qPCR analysis of inflammatory cytokines.
000 cells per well in 12-well plates and incubated at 37 °C until reaching approximately 80% confluence. Subsequently, the cells were treated with Remdesivir (200 nM), blank NPs (100 μg mL−1), RDV NPs (100 μg mL−1), and RDV NCs (100 μg mL−1) for 24 hours without viral infection. After the treatment, cells were collected in TRI-reagent for qPCR analysis of inflammatory cytokines.
2.12. Cell uptake of nanoparticles
Vero E6 cells were seeded in a 24-well glass bottom plate at a density of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and incubated overnight. The cells were infected with 0.5 MOI (Multiplicity of Infection) SARS-CoV-2 for 2 hours. The cells were treated with Rhodamine B fluorescently labelled NPs and NCs at different concentrations (0, 50, and 100 μg mL−1) for 2 hours. The infected cells were fixed with 4% paraformaldehyde (PFA) for 30 minutes at room temperature (RT). The cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (300 nM, Invitrogen) for 5 minutes at RT. The plates were observed and imaged under a Stellaris STED confocal microscope (Leica) to see the uptake level of NPs and NCs. Random sections of the fluorescent images were analysed for mean fluorescence intensity following previous studies.17,19
000 cells per well and incubated overnight. The cells were infected with 0.5 MOI (Multiplicity of Infection) SARS-CoV-2 for 2 hours. The cells were treated with Rhodamine B fluorescently labelled NPs and NCs at different concentrations (0, 50, and 100 μg mL−1) for 2 hours. The infected cells were fixed with 4% paraformaldehyde (PFA) for 30 minutes at room temperature (RT). The cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (300 nM, Invitrogen) for 5 minutes at RT. The plates were observed and imaged under a Stellaris STED confocal microscope (Leica) to see the uptake level of NPs and NCs. Random sections of the fluorescent images were analysed for mean fluorescence intensity following previous studies.17,19
2.13. Reverse transcriptase quantitative real-time PCR (RT-qPCR)
The infected Vero E6 cells were collected in TRI-reagent, and RNA was extracted using chloroform and isopropanol reagents. RNA concentration was quantified using a NanoDrop spectrometer (Thermo Scientific™). First-strand complementary DNA (cDNA) was synthesized from total RNA using an iSCRIPT cDNA synthesis kit (Bio-Rad). The QPCR was performed in a CFX Connect Real-Time System (Bio-Rad) using an iTaq Universal Probes Supermix (Bio-Rad) for the detection of 2019-novel Coronavirus Nucleocapsid N1 (2019-nCoV_N1) and cellular β-actin. Viral RNA copy numbers were expressed as the ratio of nCoV-N1 to β-actin. Relative fold change (RFC) to the control was done using the comparative threshold cycle ΔΔCT method after normalizing to cellular β-actin. nCoV_N1 and cellular actin gene primer and probe sequences were adapted according to previous publications: 2019nCoV_N1 (ref. 20) and β-actin.21–23 AT1 cells were used to evaluate the expression of inflammatory cytokines including interleukin-6 (IL-6), tumor necrosis factor alpha (TNF-α), interleukin-1 beta (IL-1β), and interleukin-10 (IL-10) after treatment with blank NPs, RDV NPs, and RDV NCs. The primer sequences were obtained using NCBI's primer-designing tool and synthesized by Integrated DNA Technologies or Invitrogen (Table S3†).2.14. Immunofluorescence assays (IFAs)
Vero E6 cells were seeded in a 24-well glass bottom plate with a concentration of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well. The cells were infected with 0.5 MOI SARS-CoV-2 followed by various drug-loaded nanoparticle and nanocomposite treatments for 24 hours. The infected cells were fixed with 4% paraformaldehyde (PFA) for 30 minutes at RT. They were permeabilized with 0.1% Triton X for 20 minutes at RT and blocked in antibody dilution buffer (ADB) for 1 hour at RT. The cells were stained with a primary SARS-CoV-2 nucleocapsid monoclonal antibody (2 μg mL−1, 1
000 cells per well. The cells were infected with 0.5 MOI SARS-CoV-2 followed by various drug-loaded nanoparticle and nanocomposite treatments for 24 hours. The infected cells were fixed with 4% paraformaldehyde (PFA) for 30 minutes at RT. They were permeabilized with 0.1% Triton X for 20 minutes at RT and blocked in antibody dilution buffer (ADB) for 1 hour at RT. The cells were stained with a primary SARS-CoV-2 nucleocapsid monoclonal antibody (2 μg mL−1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in ADB, 200 μl per well, Invitrogen) overnight at 4 °C covered in foil, and then stained with a FITC conjugated goat anti-rabbit IgG (H + L) cross-adsorbed secondary antibody (2 μg mL−1, 1.3
500 in ADB, 200 μl per well, Invitrogen) overnight at 4 °C covered in foil, and then stained with a FITC conjugated goat anti-rabbit IgG (H + L) cross-adsorbed secondary antibody (2 μg mL−1, 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in ADB, 200 μl per well, Invitrogen) on a shaker for 1 hour at RT covered in foil. Their nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (300 nM, Invitrogen) for 5 minutes at RT. Their images were captured using a Stellaris STED confocal microscope (Leica).
1000 in ADB, 200 μl per well, Invitrogen) on a shaker for 1 hour at RT covered in foil. Their nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (300 nM, Invitrogen) for 5 minutes at RT. Their images were captured using a Stellaris STED confocal microscope (Leica).
2.15. Plaque assays
Vero E6 cells were seeded in 6-well plates at a density of 600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and incubated overnight. Supernatants that were collected from the pre-infection followed by drug-treated cells for the PCR were serially diluted tenfold and used to inoculate monolayers of Vero E6 cells. After 1 hour of incubation at 37 °C with 5% CO2, the virus inoculum was removed and covered with an overlay medium containing 1% SeaPlaque agarose (Lonza). The plates were incubated for 24 to 48 hours until plaques were formed. To determine the virus titer, the plaques were stained with neutral red for 3 hours before counting. The titer of the virus was calculated in plaque forming units per millilitre (PFU mL−1) using the formula given below in eqn (2)
000 cells per well and incubated overnight. Supernatants that were collected from the pre-infection followed by drug-treated cells for the PCR were serially diluted tenfold and used to inoculate monolayers of Vero E6 cells. After 1 hour of incubation at 37 °C with 5% CO2, the virus inoculum was removed and covered with an overlay medium containing 1% SeaPlaque agarose (Lonza). The plates were incubated for 24 to 48 hours until plaques were formed. To determine the virus titer, the plaques were stained with neutral red for 3 hours before counting. The titer of the virus was calculated in plaque forming units per millilitre (PFU mL−1) using the formula given below in eqn (2) | (2) |
2.16. Statistical analysis
All data were generated using triplicates if not mentioned. Data were processed using GraphPad Prism software with one-way ANOVA analysis followed by Tukey's test for multiple comparisons.3. Results and discussion
3.1. Synthesis and characterization of Remdesivir-loaded PLGA nanocomposites
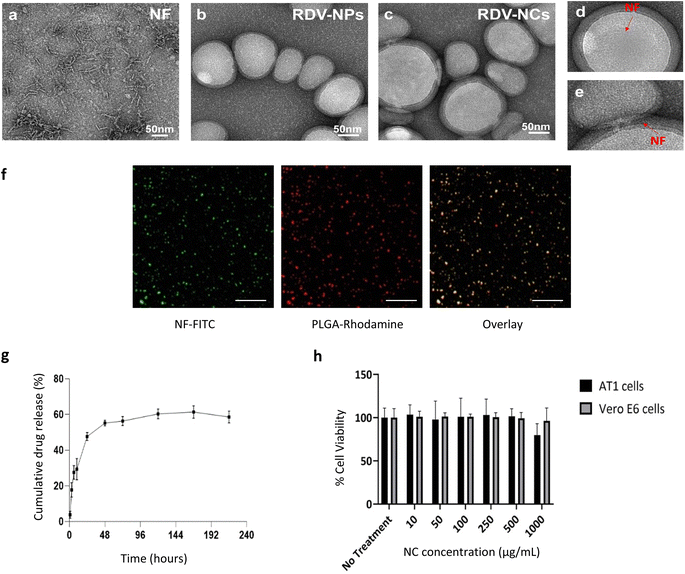
Based on recent developments in drug discoveries to treat COVID-19 and promising results in a randomized clinical trial,10 we have chosen Remdesivir (RDV) as the anti-viral drug for developing a drug delivery system against COVID-19. Here, Remdesivir is chosen as a model drug based on recent potential activity, but various other potential antiviral drugs such as Ritonavir, Lopinavir, and Nirmatrelvir7,24 can also be optimally loaded into our drug delivery system.Drug-loaded nanoparticles and nanocomposites were synthesized using solvent evaporation and physical adsorption methods.17 The electrostatic interaction of positively charged nanofibers and negatively charged drug loaded PLGA nanoparticles was applied for coating (Fig. S2†). The prepared materials were subjected to several characterization procedures to confirm the loading of Remdesivir and coating of cell penetration nanofiber peptides onto the PLGA nanoparticles. Transmission electron microscopy (TEM) was employed to characterize NFs, RDV NPs, and RDV NCs. TEM analysis of NFs revealed the presence of short fibers (Fig. 2a). The TEM images of RDV-NPs exhibited a spherical morphology with an approximate size of 100 nm (Fig. 2b). In the case of RDV-NCs, TEM analysis indicated the coexistence of both NPs and NFs (Fig. 2c–e). However, the presence of NFs was observed to be minimal, likely due to their masking by staining agents and the drying process during sample preparation. Nevertheless, some fibers were observed surrounding certain NPs, highlighting morphological distinctions between the two structures. Nanofiber coating was performed as per previously established methods from our group.17 Nanofiber coating of Remdesivir-loaded NPs increased the average size of nanoparticles from 154 ± 18 nm to 210 ± 70 nm. FITC-labelled nanofibers were coated onto PLGA NPs via physical adsorption exploiting the electrostatic interaction between positively charged nanofibers and the negatively charged PLGA NP surface. Fluorescence microscopy revealed the presence of nanoparticle and nanofiber overlap in the composites (Fig. 2f). The zeta potential values of uncoated RDV NPs increased from −26 mV to +31 mV after incubation with nanofibers, indicating the presence of a positively charged nanofiber coating (Table S1†). Similar physiological trends in peptide coatings were observed in other studies, especially in the study done by Ucar et al.,25 where model antiviral drug-loaded NPs coated with peptides showed an increase in size from 162 nm to 226 nm and an increase in surface potential from −23.9 to −4.59 mV.26 Our results show the successful coating of nanofibers onto PLGA NPs to make nanocomposites. Additionally, MALDI was utilized to assess the integrity of the cell-penetrating peptide within the nanocomposite. The peak observed at M/Z = 3614 was evident in RDV NCs, while it was entirely absent in RDV NPs. This observation indicates that the peak originates from the cell-penetrating peptide, which remains intact within the nanocomposite (Fig. S1†).
Remdesivir loading was analysed using liquid chromatography. Drug loading efficiency calculated via the indirect loading method was found to be ∼25%. Various nanoparticle drug release solutions were collected via dialysis under physiological conditions including 37 °C and pH 7.4 PBS buffer as a sink. Drug release of Remdesivir from RDV NPs showed a biphasic behaviour with an initial burst release of 47% over 24 hours followed by a sustained release until day 10 with 55% of total drug release. The drug release profiles of Remdesivir from PLGA show a burst release suitable for faster kinetics in inhibiting viral replication, which is followed by sustained release, maintaining the therapeutic levels of the drug. Antivirals loaded into PLGA have previously shown similar drug release behaviours to those seen in our study (Fig. 2g).17,18 Yang et al. compared the drug release of antivirals from PLGA NPs with similar results of biphasic drug release behaviour with burst release followed by sustained release.26 Overall, our nanoparticle-based drug delivery can improve antiviral drug bioavailability, provide controlled drug release, and reduce side effects from multiple doses needed to maintain the therapeutic levels of the drug in situ.
Conventional cell membrane penetrating peptides with their cationic nature show cytotoxicity at higher concentrations.27,28 Here, we show that our nanofiber coated RDV NPs show no significant cytotoxicity up to 1000 μg mL−1 and 48 h (Fig. 2h). Tolerance to high concentrations of nanocomposites in cells can be explained by the coating/physical adsorption approach, which, unlike chemical linking, has higher motility and low temporal activity. The nanofiber coated onto the nanoparticle improves the binding affinity of the nanoparticle during uptake, leading to enhanced intracellular drug delivery.
The encapsulation of Remdesivir in nanoparticles with a nanofiber coating was assessed for its influence on drug release during the manufacturing process. We quantified the release of the drug after each washing step following the application of the nanofiber coating to the Remdesivir-loaded nanoparticles. Our data indicate that the initial wash resulted in a release of approximately <10% of the total drug content, with minimal release observed in subsequent washes up to five times (Table S2†). These findings suggest that the nanofiber coating on PLGA nanoparticles does not significantly impact the release of the encapsulated Remdesivir drug.
3.2. Nanofiber-coated PLGA NPs show improved uptake in SARS-CoV-2 infected Vero E6 cells
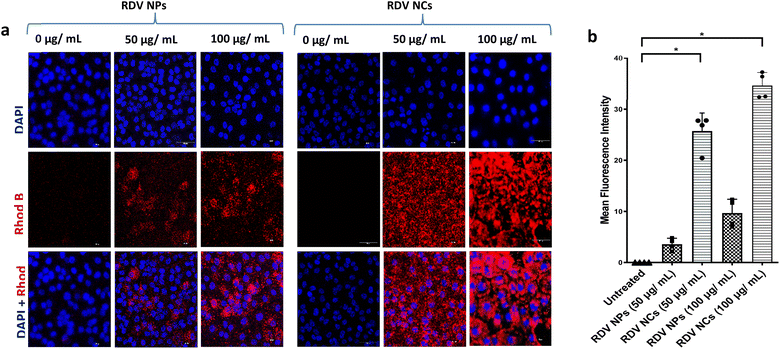
Enhancing NP uptake in cells can improve the therapeutic index of antiviral drugs by increasing drug availability intracellularly, especially in the case of inhibiting viral replication, which occurs mostly inside the cytoplasm of the cell. Our previous work showed that coating PLGA NPs with cell-penetrating nanofibers can improve the uptake of NPs in primary lung cells and is suitable for pulmonary drug delivery.17 Accordingly, various other cell-penetrating peptides have also been employed to improve the drug delivery of antivirals but concerns about toxicity remain.29 Here we explored nanocomposites with better safety profiles employed in enhancing intracellular drug delivery into SARS-CoV-2-infected cells. PLGA NPs were labelled by loading Rhodamine B dye to visualize their uptake in cells. Nanocomposites with nanofiber coating showed a dose-dependent increase in cellular uptake similar to plain nanoparticles up to 250 μg mL−1 of particles (Fig. 3). Note that we observed a significantly enhanced uptake of nanocomposites compared to plain particles without nanofiber coating. This enhancement can be explained by the highly efficient membrane penetrating nanofiber binding activity during the cell membrane uptake of nanoparticles. Increasing the residence time of nanoparticles near the cell membrane can help improve the endocytosis of the particles. Previous studies show an alteration in the nanoparticle uptake of virally infected cells, with some viral infections reducing the cell uptake of nanoparticles.16 Further exploration is needed to understand whether SARS-CoV-2 has a significant effect on the alteration of the nanoparticle uptake of the cell. Nevertheless, the developed nanocomposites with nanofiber coating have exhibited a superior cell uptake compared to plain PLGA nanoparticles in SARS-CoV-2 infected cells. Based on our previous study, macropinocytosis is a major endocytic pathway utilized by primary mammalian cells for the uptake of these nanocomposites.17 In outlook, nanocomposites with enhanced uptake in cells can be ideal drug carriers to deliver antivirals with an intracellular mode of action such as inhibition of viral replication, especially among pulmonary pathologies.3.3. Remdesivir-loaded nanocomposites inhibit SARS-CoV-2 infections in vitro
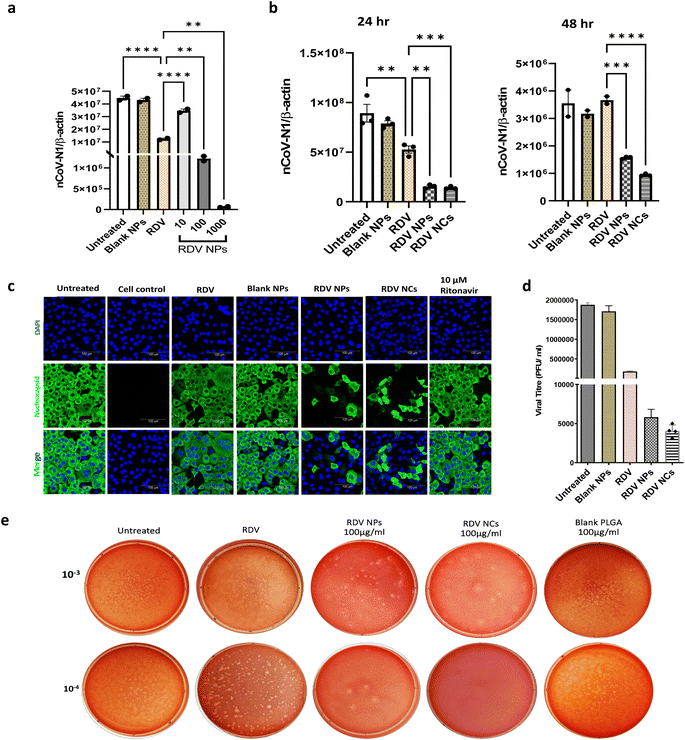
Remdesivir, which binds to the viral RNA-dependent RNA polymerase, has been reported to be effective against SARS-CoV-2 infection both in vivo and in vitro.30 However, the instability and lack of specificity of free Remdesivir within the body pose challenges in achieving an efficient treatment at lower concentrations, and the requirement of multiple dosages often leads to varying degrees of side effects experienced by patients.31–33 Nanotechnology has emerged as a highly promising technology for effectively addressing viral detection, prevention, and treatment. Biocompatible nanoparticle platforms, such as polymer- and lipid-based nanocarriers with drugs, offer better stability, release, uptake, and bioavailability.34 In a study by Mandel et al., PLGA NPs were used to encapsulate the antiretroviral drug (Emtricitabine) to treat HIV-1 infection. This PLGA-based nanocarrier system showed prolonged inhibition of HIV infection in in vitro studies.35 Similarly, PLGA nanoparticles loaded with Bictegravir showcased extended drug release and prolonged intracellular retention, which ultimately improved protection against HIV-1.36 Nanotechnology has also brought great advances to overcome different barriers to combat SARS-CoV-2, and several nanoparticle-based drug delivery approaches have already shown improved antiviral efficacy against the novel coronavirus.37–42 In that context, PLGA polymer-based nanoparticles have demonstrated their utility in developing anti-viral drug delivery systems.39,43 In the same vein, our nanofiber-coated NPs (RDV NCs) have exhibited enhanced uptake inside SARS-CoV-2 infected mammalian cells compared to uncoated RDV NPs.In this study, the antiviral activity of RDV NPs or RDV NCs was evaluated in Vero E6 cells. Vero E6 cells were employed for our study due to their abundant expression of ACE2 crucial for SARS-CoV-2 entry.44 Human cell lines are comparatively slower to become infected compared to Vero E6 cells, especially seen in airway epithelial cells.45 Additionally, Vero E6 cells are well-established cell lines for studying coronaviruses. Vero E6 cells were infected with SARS-CoV-2 and treated with the RDV NPs at various concentrations (10, 100, and 1000 μg mL−1). RT-qPCR data with nCoV-N1/beta-actin gene analysis show a reduction in viral load above 10 μg mL−1, while 100 and 1000 μg mL−1 exhibited approximately 5- and 70-fold higher antiviral activity respectively compared to 200 nM Remdesivir (Fig. 4a).
Note that 100 μg mL−1 RDV NPs have an equivalent drug concentration of 200 nM Remdesivir drug and 100 μg mL−1 NPs showed 5 times higher inhibition than 200 nM Remdesivir. A time-dependent study was also performed to evaluate the antiviral efficacy of RDV NPs and RDV NCs. We observed an enhanced reduction of viral load in RDV NC and RDV NP treated cells compared to the free Remdesivir drug and untreated control. As seen, after 48 hours, Remdesivir did not show any significant reduction in viral reduction plausibly due to the degradation or reduced uptake in cells, while NCs and NPs loaded with Remdesivir showed increased viral inhibition (Fig. 4b). Following drug release profiles, 24 hours showed a drastic inhibition of the virus compared to 48 hours in agreement with the burst drug release profile of nanoparticles. Note that nanocomposites showed higher viral inhibition compared to nanoparticles after 48 hours, indicating higher bioavailability of antiviral drugs via increased uptake ability of nanocomposites by SARS-CoV-2 infected Vero E6 cells. The improved antiviral properties against SARS-CoV-2 have also been reported in other nanofiber-based drug delivery studies at different levels of infection.39,46 However, there has been no research comparing the efficacy of nanofiber-based nanoparticles combined with a widely used drug like Remdesivir for the treatment of SARS-CoV-2, and our novel nanofiber-based delivery approach exhibits greater promise in reducing the transmission of the virus, supporting other nanofiber-based drug delivery strategies with effective inhibition of SARS-CoV-2 at different levels of pathogenesis.39,46
To visualize the inhibition of SARS-CoV-2, an immunofluorescence assay was performed in accordance with protocols outlined in previous studies to detect SARS-CoV-2 nucleocapsid proteins.43,47 Nucleocapsid antibody staining on SARS-CoV-2 infected Vero E6 cells enabled visualizing the viral load in various treatment groups. Clearly, both RDV NPs and RDV NCs have shown more than 80% viral inhibition without inducing cytotoxicity as seen by nuclei staining (Fig. 4c). An immunofluorescence study shows the ability of RDV NCs to successfully deliver Remdesivir to the cytoplasm, thus improving their biological activity in inhibiting viral replication intracellularly compared to Remdesivir (200 nM) and Ritonavir (10 μM). The results of RT-qPCR and Immunofluorescence analysis (IFA) were further validated by viral plaque assays. Plaque assays have been considered one of the most precise methods for the direct quantification of viruses through counting the isolated plaques in cell culture.48–50 The plaque assay was performed following the guidelines of previous studies, to estimate the viral titer from the supernatants that were collected after 24 hours of drug treatment in Vero E6 cells pre-infected with SARS-CoV-2.49,51 The virus titer from different wells was calculated in PFU mL−1, and differences in the viral titer were compared to the untreated virus control (Fig. 4d). The viral titer in RDV NC and RDV NP treated supernatants showed a 463- and 322-fold reduction, respectively, compared to the untreated control. The plaque assay showed a reduced number of plaques in RDV NP and RDV NC treated cells, which indicated an increased level of inhibition of SARS-CoV-2 compared to the untreated and RDV control (Fig. 4e). The inhibition pattern of SARS-CoV-2 found in plaque assays confirmed the results of RT-qPCR and IFA. Our novel nanocomposites increase the bioavailability of antivirals via enhanced intracellular delivery and improve therapeutic efficacy.
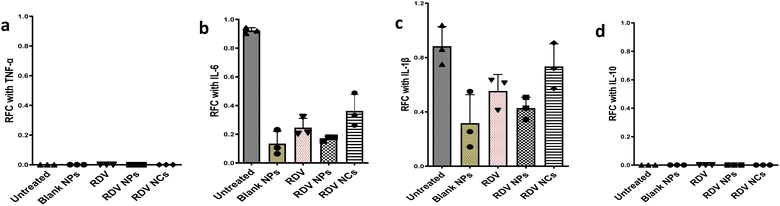
During SARS-CoV-2 infection in humans, significant quantities of proinflammatory cytokines are released including IL-6, TNF-α, and IL-1β.52,53 IL-6 is an important cytokine, which is correlated with inflammation and its expression is high in SARS-CoV-2 infection leading to extensive lung damage.54 IL-10, an anti-inflammatory cytokine, functions in a negative feedback mechanism to suppress inflammation, and contributes to the pathological severity of COVID-19.55 Here, our study investigated whether the nanocomposite material changes the expression of inflammatory markers in the absence of SARS-CoV-2 infection (Fig. 5). Human alveolar type 1 (AT1) cells, which cover the large surface in lungs, were employed to evaluate the expression of proinflammatory and anti-inflammatory cytokines with and without nanocomposite treatment. The RT-qPCR results demonstrate that our formulations don't alter the expression of these cytokines in AT1 cells compared to the control group (Fig. 5a–d). Noticeably, Remdesivir and our nanocomposite material show low expression of IL-6 indicating low inflammatory response (Fig. 5c). These results further confirm that there are negligible changes in the inflammatory cytokines compared to untreated controls. Hence, our nanocomposite materials can be applied to deliver drugs without altering the immune response. Overall, our evidence suggests that RDV NCs show potent effects against SARS-CoV-2 due to their enhanced anti-viral drug delivery.
Translating our RDV NCs into in vivo models via inhalation represents a promising strategy to enhance the penetration and distribution in lung tissue, which are often inadequate with intravenous delivery.56 Aerosolized nanoparticles have demonstrated improvements in the delivery of the antiviral drug Remdesivir to the lungs, including both aggregate Remdesivir and liposomal Remdesivir formulations.57,58 These studies underscore the effectiveness of inhalation delivery in increasing Remdesivir bioavailability in the lungs. Given our previous success in delivering NCs via nebulization,17 our NCs are strong candidates for the inhalation delivery of antiviral drugs to the lungs.
4. Conclusion
A nanofiber coating onto PLGA NPs enabled their improved uptake in SARS-CoV-2 infected Vero E6 cells. Remdesivir-loaded nanoparticles showed a sustained drug release of Remdesivir under physiological conditions. The results of RT-qPCR, IFA, and plaque assays showed significant SARS-CoV-2 inhibition with nanocomposites compared to uncoated nanoparticles, indicating their superior ability of intracellular drug delivery and their potential as a drug carrier for anti-viral therapy in pulmonary infections. Additionally, our nanocomposite material does not induce inflammatory cytokine expression revealing its biocompatible nature in SARS-CoV-2 infected cells. Further investigation in primary mammalian cells, along with an in vivo animal study, is needed to optimize the nanocomposite drug delivery system. Our nanocomposites may be applied as an inhalable drug delivery system with their optimized size and drug release, which is ideal for pulmonary infections.Author contributions
Uday Chintapula: conceptualisation, methodology, software, visualisation, formal analysis, writing, investigation, and original draft preparation. Shazeed-Ul Karim: methodology, formal analysis, investigation, and writing. Priyanka Iyer Raghunathan – methodology, investigation, and writing. Biswas Neupane – methodology and investigation. Farzana Nazneen – methodology and investigation. Haritha Asokan Shreeja – methodology and resources. He Dong – funding acquisition, resources, and writing – review editing. Fengwei Bai & Kytai Nguyen – conceptualisation, funding acquisition, methodology, and writing – review editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Graphical images were made using Biorender.com. We would like to thank the Shimadzu Center for Advanced Analytical Chemistry at UTA for their support in liquid chromatography experiments. This work is supported by NSF DMR1824614 (H.D.) and the Alfred R. and Janet H. Potvin Bioengineering Endowed Professorship in the Department of Bioengineering, College of Engineering, University of Texas at Arlington (K. T. N.).References
- Organization WH, COVID-19 statistics worldwide, 2020, Accessed May 27, https://covid19.who.int/.
- W. Guo, M. Li and Y. Dong, et al., Diabetes is a risk factor for the progression and prognosis of COVID-19, Diabetes/Metab. Res. Rev., 2020, e3319, DOI:10.1002/dmrr.3319.
- C. Cristelo, C. Azevedo, J. Moreira Marques, R. Nunes and B. Sarmento., SARS-CoV-2 and Diabetes: New Challenges for the Disease, Diabetes Res. Clin. Pract., 2020, 164, 108228 CrossRef CAS PubMed.
- K. D. Hulme, L. A. Gallo and K. R. Short, Influenza Virus and Glycemic Variability in Diabetes: A Killer Combination?, Front. Microbiol., 2017, 8, 861, DOI:10.3389/fmicb.2017.00861.
- C. Liu, Q. Zhou and Y. Li, et al., Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases, ACS Cent. Sci., 2020, 6(3), 315–331, DOI:10.1021/acscentsci.0c00272.
- J. S. Faust, A. Kumar and J. Shah, et al., Oral Nirmatrelvir and Ritonavir for Covid-19 in Vaccinated, Non-Hospitalized Adults, Ages 18-50 Years, Clin. Infect. Dis., 2023, 77, 1257–1264, DOI:10.1093/cid/ciad400.
- M. Marzi, M. K. Vakil, M. Bahmanyar and E. Zarenezhad, Paxlovid: Mechanism of Action, Synthesis, and, BioMed Res. Int., 2022, 2022, 7341493, DOI:10.1155/2022/7341493.
- A. Hossein Heydari, S. Ghaffari, Z. Khani, S. Heydari, Z. Eskandari and M. Esmaeil Heidari, MiR-21 and Tocilizumab interactions improve COVID-19 myocarditis outcomes, Ther. Adv. Cardiovasc. Dis., 2023, 17, 17539447231182548, DOI:10.1177/17539447231182548.
- M. Sun, H. Lai, J. Huang, J. Liu, Y. Li, J. Tian, C. Zhang, J. Estill, Z. Zhang and L. Ge, Molnupiravir for the treatment of non-severe COVID-19: a systematic review and meta-analysis of 14 randomized trials with 34
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 570 patients, J. Antimicrob. Chemother., 2023, 78(9), 2131–2139, DOI:10.1093/jac/dkad216.
570 patients, J. Antimicrob. Chemother., 2023, 78(9), 2131–2139, DOI:10.1093/jac/dkad216. - Gilead Announces Results From Phase 3 Trial of Investigational Antiviral Remdesivir in Patients with Severe COVID-19, acessed April 29.
- M. Doroudian, R. MacLoughlin, F. Poynton, A. Prina-Mello and S. C. Donnelly, Nanotechnology based therapeutics for lung disease, Thorax, 2019, 74(10), 965, DOI:10.1136/thoraxjnl-2019-213037.
- S. C. Jurek, M. Hirano-Kobayashi, H. Chiang, D. S. Kohane and B. D. Matthews, Prevention of ventilator-induced lung edema by inhalation of nanoparticles releasing ruthenium red, Am. J. Respir. Cell Mol. Biol., 2014, 50(6), 1107–1117, DOI:10.1165/rcmb.2013-0163OC.
- C. Y. Zhang, W. Lin and J. Gao, et al., pH-Responsive Nanoparticles Targeted to Lungs for Improved Therapy of Acute Lung Inflammation/Injury, ACS Appl. Mater. Interfaces, 2019, 11(18), 16380–16390, DOI:10.1021/acsami.9b04051.
- A. P. L. D’Almeida, M. T. Pacheco de Oliveira and É. T. de Souza, α-bisabolol-loaded lipid-core nanocapsules reduce lipopolysaccharide-induced pulmonary inflammation in mice, Int. J. Nanomed., 2017, 12, 4479–4491, DOI:10.2147/IJN.S130798.
- H. X. Nguyen, Targeted Delivery of Surface-Modified Nanoparticles: Modulation of Inflammation for Acute Lung Injury, Surface Modification of Nanoparticles for Targeted Drug Delivery, 2019, pp. 331–353, DOI:10.1007/978-3-030-06115-9_17.
- Y. Abo-Zeid, G. R. Williams, L. Touabi and G. R. McLean, An investigation of rhinovirus infection on cellular uptake of poly (glycerol-adipate) nanoparticles, Int. J. Pharm., 2020, 589, 119826, DOI:10.1016/j.ijpharm.2020.119826.
- U. Chintapula, S. Yang and T. Nguyen, et al., Supramolecular Peptide Nanofiber/PLGA Nanocomposites for Enhancing Pulmonary Drug Delivery, ACS Appl. Mater. Interfaces, 2022, 14(51), 56498–56509, DOI:10.1021/acsami.2c15204.
- Y. Su and H. Dong, Modular design and self-assembly of multidomain peptides towards cytocompatible supramolecular cell penetrating nanofibers, RSC Adv., 2020, 10, 29469–29474 RSC.
- S. Dhakal, M. Mondal and A. Mirzazadeh, et al., α-Synuclein emulsifies TDP-43 prion-like domain—RNA liquid droplets to promote heterotypic amyloid fibrils, Commun. Biol., 2023, 6, 1227, DOI:10.1038/s42003-023-05608-1.
- A. Anantharajah, R. Helaers and J. P. Defour, et al., How to choose the right real-time RT-PCR primer sets for the SARS-CoV-2 genome detection?, J. Virol. Methods, 2021, 295, 114197, DOI:10.1016/j.jviromet.2021.114197.
- F. Bai, T. Wang, U. Pal, F. Bao, L. H. Gould and E. Fikrig, Use of RNA interference to prevent lethal murine west nile virus infection, J. Infect. Dis., 2005, 191(7), 1148–1154, DOI:10.1086/428507.
- F. Bai, K.-F. Kong and J. Dai, et al., A Paradoxical Role for Neutrophils in the Pathogenesis of West Nile Virus, J. Infect. Dis., 2010, 202(12), 1804–1812, DOI:10.1086/657416.
- A. M. Paul, D. Acharya and L. Duty, et al., Osteopontin facilitates West Nile virus neuroinvasion via neutrophil "Trojan horse" transport, Sci. Rep., 2017, 7(1), 4722, DOI:10.1038/s41598-017-04839-7.
- M. S. Mousavi Maleki, S. Sardari, A. Ghandehari Alavijeh and H. Madanchi, Recent Patents and FDA-Approved Drugs Based on Antiviral Peptides and Other Peptide-Related Antivirals, Int. J. Pept. Res. Ther., 2023, 29(1), 5, DOI:10.1007/s10989-022-10477-z.
- B. Ucar, T. Acar, P. P. Arayici and S. Derman, A nanotechnological approach in the current therapy of COVID-19: model drug oseltamivir-phosphate loaded PLGA nanoparticles targeted with spike protein binder peptide of SARS-CoV-2, Nanotechnology, 2021, 32(48), 485601, DOI:10.1088/1361-6528/ac1c22.
- H. Yang, J. Li, S. K. Patel, K. E. Palmer, B. Devlin and L. C. Rohan, esign of Poly(lactic-co-glycolic Acid) (PLGA) Nanoparticles for Vaginal Co-Delivery of Griffithsin and Dapivirine and Their Synergistic Effect for HIV Prophylaxis, Pharmaceutics, 2019, 11(4), 184, DOI:10.3390/pharmaceutics11040184.
- H. Derakhshankhah and S. Jafari, Cell penetrating peptides: A concise review with emphasis on biomedical applications, Biomed. Pharmacother., 2018, 108, 1090–1096, DOI:10.1016/j.biopha.2018.09.097.
- J. Xie, Y. Bi and H. Zhang, et al., Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application, Front. Pharmacol, 2020, 11, 697, DOI:10.3389/fphar.2020.00697.
- I. Sadeghian, R. Heidari, S. Sadeghian, M. J. Raee and M. Negahdaripour, Potential of cell-penetrating peptides (CPPs) in delivery of antiviral therapeutics and vaccines, Eur. J. Pharm. Sci., 2022, 169, 106094, DOI:10.1016/j.ejps.2021.106094.
- M. Wang, R. Cao and L. Zhang, et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res., 2020, 30(3), 269–271, DOI:10.1038/s41422-020-0282-0.
- Remdesivir (Veklury) and Food and Drug Administration, https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/214787Orig1s015lbl.pdf.
- S. Gandhi, J. Klein and A. J. Robertson, et al., De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report, Nat. Commun., 2022, 13(1), 1547, DOI:10.1038/s41467-022-29104-y.
- M. Cheng, R. Fowler, S. Murthy, R. Pinto, N. L. Sheehan and A. Tseng, Remdesivir in Patients With Severe Kidney Dysfunction: A Secondary Analysis of the CATCO Randomized Trial, JAMA Netw. Open, 2022, 5(8), e2229236, DOI:10.1001/jamanetworkopen.2022.29236.
- A. Puri, K. Loomis and B. Smith, et al., Lipid-based nanoparticles as pharmaceutical drug carriers: from concepts to clinic, Crit. Rev. Ther. Drug Carrier Syst., 2009, 26(6), 523–580, DOI:10.1615/critrevtherdrugcarriersyst.v26.i6.10.
- S. Mandal, M. Belshan, A. Holec, Y. Zhou and C. J. Destache, An Enhanced Emtricitabine-Loaded Long-Acting Nanoformulation for Prevention or Treatment of HIV Infection, Antimicrob. Agents Chemother., 2017, 61(1), e01475, DOI:10.1128/AAC.01475-16.
- S. Mandal, P. K. Prathipati, M. Belshan and C. J. Destache, A potential long-acting bictegravir loaded nano-drug delivery system for HIV-1 infection: A proof-of-concept study, Antiviral Res., 2019, 167, 83–88, DOI:10.1016/j.antiviral.2019.04.007.
- R. M. Hathout, S. G. Abdelhamid and A. A. Metwally, Chloroquine and hydroxychloroquine for combating COVID-19: Investigating efficacy and hypothesizing new formulations using Bio/chemoinformatics tools, Inform. Med. Unlocked, 2020, 21, 100446, DOI:10.1016/j.imu.2020.100446.
- M. Karmacharya, S. Kumar, O. Gulenko and Y. K. Cho, Advances in Facemasks during the COVID-19 Pandemic Era, ACS Appl. Bio Mater., 2021, 4(5), 3891–3908, DOI:10.1021/acsabm.0c01329.
- M. H. Emam, R. S. Elezaby, S. A. Swidan, S. A. Loutfy and R. M. Hathout, Cerium Oxide Nanoparticles/Polyacrylonitrile Nanofibers as Impervious Barrier against Viral Infections, Pharmaceutics, 2023, 15(5), 1494, DOI:10.3390/pharmaceutics15051494.
- X. Cai, M. Chen and A. Prominski, et al., A Multifunctional Neutralizing Antibody-Conjugated Nanoparticle Inhibits and Inactivates SARS-CoV-2, Adv. Sci., 2022, 9(2), e2103240, DOI:10.1002/advs.202103240.
- A. Ejsmont, A. Warowicka, J. Broniarczyk and J. Goscianska, The synergistic effect of Cu-MOF nanoparticles and immunomodulatory agent on SARS-CoV-2 inhibition, Chem. Commun., 2023, 59(33), 4907–4910, 10.1039/d3cc00894k.
- V. Sanna, S. Satta, T. Hsiai and M. Sechi, Development of targeted nanoparticles loaded with antiviral drugs for SARS-CoV-2 inhibition, Eur. J. Med. Chem., 2022, 231, 114121, DOI:10.1016/j.ejmech.2022.114121.
- M. Mohammadi, P. Dehghani, A. Mohseninia, M. Roozbehani, A. Hemphill and K. Hesamizadeh, Incorporation of the Tat cell-penetrating peptide into nanofibers improves the respective antitumor immune response, J. Cell. Physiol., 2021, 236(2), 1401–1417, DOI:10.1002/jcp.29946.
- N. S. Ogando, T. J. Dalebout and J. C. Zevenhoven-Dobbe, et al., SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology, J. Gen. Virol., 2020, 101(9), 925–940, DOI:10.1099/jgv.0.001453.
- M. Essaidi-Laziosi, F. J. Perez Rodriguez, N. Hulo, F. Jacquerioz, L. Kaiser and I. Eckerle, Estimating clinical SARS-CoV-2 infectiousness in Vero E6 and primary airway epithelial cells, Lancet Microbe, 2021, 2(11), e571, DOI:10.1016/S2666-5247(21)00216-0.
- M. H. Emam, H. Nageh and F. Ali, et al., Inhibition of SARS-CoV-2 spike protein entry using biologically modified polyacrylonitrile nanofibers, RSC Adv., 2022, 12(25), 16184–16193, 10.1039/d2ra01321e.
- A. K. Zhu, S. S. Li and S. P. Yu, et al., A pair of SARS-CoV-2 nucleocapsid protein monoclonal antibodies shows high specificity and sensitivity for diagnosis, Virol. Sin., 2022, 37(6), 942–945, DOI:10.1016/j.virs.2022.10.003.
- E. J. Mendoza, K. Manguiat, H. Wood and M. Drebot, Two Detailed Plaque Assay Protocols for the Quantification of Infectious SARS-CoV-2, Curr. Protoc. Microbiol., 2020, 57(1), ecpmc105, DOI:10.1002/cpmc.105.
- B. Neupane and F. Bai, Quantification of West Nile Virus by Plaque-Forming Assay, Methods Mol. Biol., 2023, 2585, 9–14, DOI:10.1007/978-1-0716-2760-0_2.
- S. U. Karim and F. Bai, Introduction to West Nile Virus, Methods Mol. Biol., 2023, 2585, 1–7, DOI:10.1007/978-1-0716-2760-0_1.
- F. Nazneen, E. A. Thompson, C. Blackwell, J. S. Bai, F. Huang and F. Bai, An effective live-attenuated Zika vaccine candidate with a modified 5' untranslated region, npj Vaccines, 2023, 8(1), 50, DOI:10.1038/s41541-023-00650-w.
- C. J. Neufeldt, B. Cerikan, M. Cortese, J. Frankish, J. Y. Lee, A. Plociennikowska, F. Heigwer, V. Prasad, S. Joecks, S. S. Burkart, D. Y. Zander, B. Subramanian, R. Gimi, S. Padmanabhan, R. Iyer, M. Gendarme, B. El Debs, N. Halama, U. Merle, M. Boutros, M. Binder and R. Bartenschlager, SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB, Commun. Biol., 2022, 5(1), 45, DOI:10.1038/s42003-021-02983-5.
- N. Taki, J. M. Tatro, R. Lowe, V. M. Goldberg and E. M. Greenfield, Comparison of the roles of IL-1, IL-6, and TNFalpha in cell culture and murine models of aseptic loosening, Bone, 2007, 40(5), 1276–1283, DOI:10.1016/j.bone.2006.12.053.
- X. Wang, G. Tang and Y. Liu, et al., The role of IL-6 in coronavirus, especially in COVID-19, Front. Pharmacol, 2022, 13, 1033674, DOI:10.3389/fphar.2022.1033674.
- L. Lu, H. Zhang, D. J. Dauphars and Y. W. He, A Potential Role of Interleukin 10 in COVID-19 Pathogenesis, Trends Immunol., 2021, 42(1), 3–5, DOI:10.1016/j.it.2020.10.012.
- C. Y. Loo, W. H. Lee and Q. T. Zhou, Recent Advances in Inhaled Nanoformulations of Vaccines and Therapeutics Targeting Respiratory Viral Infections, Pharmacal Res., 2023, 40(5), 1015–1036, DOI:10.1007/s11095-023-03520-1.
- R. Vartak, S. M. Patil, A. Saraswat, M. Patki, N. K. Kunda and K. Patel, Aerosolized nanoliposomal carrier of remdesivir: an effective alternative for COVID-19 treatment in vitro, Nanomedicine, 2021, 16(14), 1187–1202, DOI:10.2217/nnm-2020-0475.
- S. Sahakijpijarn, C. Moon and Z. N. Warnken, et al., In vivo pharmacokinetic study of remdesivir dry powder for inhalation in hamsters, Int. J. Pharm.: X, 2021, 3, 100073, DOI:10.1016/j.ijpx.2021.100073.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00361f |
| ‡ Chintapula U., Shazeed-Ul Karim and Priyanka Raghunathan Iyer have contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |