Temperature and particles interact to affect human norovirus and MS2 persistence in surface water†
Lauren C.
Kennedy
 ,
Sarah A.
Lowry
,
Sarah A.
Lowry
 and
Alexandria B.
Boehm
and
Alexandria B.
Boehm
 *
*
Department of Civil and Environmental Engineering, Stanford University, Y2E2 Room 189, Stanford, CA 94305, USA. E-mail: aboehm@stanford.edu
First published on 28th November 2023
Abstract
Modeling the fate and transport of viruses and their genetic material in surface water is necessary to assess risks associated with contaminated surface waters and to inform environmental surveillance efforts. Temperature has been identified as a key variable affecting virus persistence in surface waters, but the effects of the presence of biological and inert particles and of their interaction with temperature have not been well characterized. We assessed these effects on the persistence of human norovirus (HuNoV) genotype II.4 purified from stool and MS2 in surface water. Raw or filter-sterilized creek water microcosms were inoculated and incubated in the dark at 10 °C, 15 °C, and 20 °C. HuNoV (i.e., genome segments and intact capsids) and MS2 (i.e., infectious MS2, genome segments, and intact capsids) concentrations were followed over 36 days. The range in positive, significant first-order decay rate constants for HuNoV in this study was 0.14 to 0.69 day−1 compared with 0.026 to 0.71 day−1 for that of MS2. Decay rate constants for HuNoV genome segments and infectious MS2 were largest in creek water that included biological and inert particles and incubated at higher temperatures. In addition, for HuNoV and MS2 incubated in raw or filter-sterilized creek water at 15 °C, capsid damage was not identified as a dominant inactivation mechanism. Environmental processes and events that affect surface water biological and inert particles, temperature, or both could lead to variable virus decay rate constants. Incorporating the effects of particles, temperature, and their interaction could enhance models of virus fate and transport in surface water.
Environmental significanceHuman norovirus is a leading cause of gastroenteritis that can be transmitted via contact with contaminated surface water. However, its fate in surface water is not well characterized. This research fills an important knowledge gap by quantifying human norovirus persistence in environmental surface water and providing parameters for fate and transport models. |
1 Introduction
Surface waters are valuable environmental resources that can be used for recreation and as sources of potable water. Surface water can be contaminated with pathogens from point and nonpoint sources including untreated wastewater discharge, sewer pipe leakage, wastewater effluent, and land-based runoff.1,2 Gastrointestinal illnesses from exposure to contaminated surface waters are often attributed to viral pathogens in particular,3–5 and modeling the fate and transport of viruses in surface water is necessary to assess risks associated with contaminated surface waters. In addition, for some unsewered communities, virus occurrence in surface water that receives raw wastewater can be used to assess the disease burden in a community (i.e., through environmental surveillance6). For environmental surveillance, it is also important to understand the impact of environmental conditions on the virus signal.Virus persistence in surface water is dependent on the local environmental conditions. Temperature has been identified as a key factor that affects virus persistence in water,2,7–9 but the effect of biological and inert particles is equivocal.8,9 For example, a systematic review and meta-analysis of non-enveloped virus persistence found that presence of biological and inert particles in surface waters significantly affected first-order decay rate constants of viruses in half of the virus families included in the review (i.e., in Herpesviridae, Orthomyxoviridae, and Togaviridae9). Olive et al.10 found that the effect of biological and inert particles on the virus decay rate constants in water was dependent on the type of virus (i.e., echovirus 11, human adenovirus type 2, or bacteriophage H6) and the incubation temperature. Additional research is needed to better understand the effects of the presence of biological and inert particles (or “particle content”) on virus persistence in environmental waters.
The majority of virus persistence studies in environmental matrices have been completed with culture-based methods to enumerate viruses,7–9 but use of molecular methods (e.g., quantitative polymerase chain reaction (qPCR) or reverse transcription qPCR (RT-qPCR)) can be advantageous in some cases. Molecular methods generally have faster turnaround and are more sensitive compared to culture-based methods.11 However, molecular methods enumerate active viruses as well as naked genomic RNA and DNA, which are persistent in environmental matrices under certain conditions.12,13 Persistent genetic material could affect the decay rate constants observed for viruses quantified via molecular methods when compared to culture-based methods.7,9 In the case of environmental surveillance in surface waters, sensitivity and quick turnaround times are desired, so molecular methods are particularly useful. In addition, for viruses that are difficult to culture, such as human norovirus (HuNoV)14 the decay rate constants determined for genome segments are conservative compared to those for infectious virus (i.e., virus inactivation).15,16 For HuNoV, additional methods have been developed to quantify intact capsids and to assess genome damage specifically.16,17
HuNoV is a leading cause of gastroenteritis that has been associated with 18% of global acute gastroenteritis cases18 and with 22% of outbreaks from recreational environmental water exposures in the United States.19 The persistence of HuNoV in environmental water has been assessed via a human intestinal enteroid assay15,16,20 as well as molecular methods.16,21–23 However, its persistence in surface water is not well characterized. Two studies of HuNoV incubated in surface water in dark conditions observed larger first-order decay rate constants at warmer compared to cooler incubation temperatures.22,23 In contrast, in a recent study, we observed larger HuNoV first-order decay rate constants in filter-sterilized surface water incubated at 15 °C compared to 20 °C.16 To our knowledge, the combined effect of biological and inert particles with incubation temperature on HuNoV decay rate constants in surface water has not been investigated.
We aimed to quantify the combined effects of particle content with incubation temperature on HuNoV persistence in surface water. We followed intact HuNoV capsids as well as genome segments over time in raw and filter-sterilized creek water microcosms incubated at three temperatures (10, 15, and 20 °C). We included the bacteriophage MS2 as an internal biological control and followed the loss of MS2 infectivity, intact capsids and genome segments over time. The HuNoV decay rate constants determined in this study are compared to inactivation rate constants from a previous study of infectious HuNoV incubated in the same sample of filter-sterilized creek water.16 To our knowledge, this study is the first to assess the effects of particle content on HuNoV decay rate constants in surface water. The 33 decay rate constants provided herein will be useful parameters in fate and transport models.
2 Methods
2.1 Overview
To determine the effect of particle content on HuNoV decay rate constants, HuNoV was seeded into raw (i.e., including natural biological and inert particles) and filter-sterilized (i.e., without particles larger than 0.22 μm) creek water microcosms along with MS2 as an internal biological control. The microcosms were incubated at a range of temperatures that are environmentally relevant to California coastal waters in the United States and that reflect the average surface seawater temperatures of many coastal communities globally (10, 15, and 20 °C).24,25 To isolate the effects of particle content and temperature on virus decay rate constants, the reactors were kept in the dark throughout the experiments. The persistence of HuNoV and MS2 intact viral capsids and genome segments as well as the infectivity of MS2 were followed over time. Molecular methods were adapted to quantify long genome segments that were >300 nucleotides (nt) as well as intact viral capsids by pretreating samples with RNase through enzyme pretreatment RT-qPCR (ET-RT-qPCR). RNase was chosen as the method to assess capsid integrity because it was found to work most consistently of the methods tested during heat inactivation of HuNoV and human rotavirus.26 In addition, ET-RT-qPCR has been used successfully to assess sunlight inactivation of HuNoV and MS2.17 This research was approved by the Stanford Administrative Panel on Biosafety and was conducted with biosafety level 2 precautions.2.2 Preparation of HuNoV and MS2 inoculants
A stool positive for HuNoV (GII.4, MK762570) was used to prepare HuNoV inoculants. The stool was diluted into autoclaved phosphate buffered saline (PBS; Thermo Fisher Scientific, Waltham, MA, USA), centrifuged, and sequentially filtered as described previously.16 The HuNoV inoculant was stored at −80 °C until use.MS2 was propagated as described in United States Environmental Protection Agency method 1602,27 and stored in 15% glycerol at −80 °C. To prepare the MS2 inoculant for the experiments, an aliquot of MS2 glycerol stock was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in autoclave-sterilized PBS and stored at −80 °C until the experiments. Additional details of the preparation of the inoculants are provided in the ESI.†
10 in autoclave-sterilized PBS and stored at −80 °C until the experiments. Additional details of the preparation of the inoculants are provided in the ESI.†
2.3 Creek water collection
San Pedro Creek in Pacifica, CA, USA (37.59 N, −122.5 W) was sampled on 11/08/2021 using sterile technique as described previously.16 The temperature and salinity of the raw water were measured upon collection. An aliquot of the raw water was filter sterilized using a 0.22 μm pore size filter. The turbidity, pH, UV-vis absorbance, intracellular and extracellular ATP concentrations were measured for the raw and filter-sterilized creek water. No attempt was made to further characterize inert or biological particles. Experiments commenced immediately following collection (day 0).2.4 Experimental setup
Fourteen autoclave-sterilized 100 mL pyrex bottles covered in aluminum foil served as reactors. Reactors were filled with 19.5 mL: six with raw creek water, six with filter-sterilized creek water, and two with autoclave-sterilized PBS. Waters of all but the two negative control reactors (autoclave-sterilized PBS) were inoculated with HuNoV and MS2 such that the concentrations at the start of the experiments were approximately 106 gene copies per mL (gc mL−1) HuNoV and 108 gc mL−1 MS2. The reaction volume was chosen to ensure that at least 10 mL would remain in the reactor at the end of the experiment; given the limited availability of HuNoV seed virus and the needed high concentration at time 0 to conduct the experiment, using a larger volume of reactor was not feasible.Two raw creek water and two filter-sterilized creek water reactors were stored at each of three temperatures (10, 15, and 20 °C). The two negative control reactors were incubated at 15 and 20 °C, respectively. All reactors oscillated at 100 revolutions per minute in the dark. Samples were collected from the reactors at the following time points: 0, 1, 3, 5, 7, 10, 14, 22, 29, 36 days. The exception was the negative control reactor incubated at 20 °C from which samples were collected at the following time points: 0, 1, 7, 14, 21, 28 days (logistical constraints precluded sampling this reactor on the same days as the others). To collect a sample at each time point, the oscillator was paused, and 1 mL of sample was withdrawn from the reactors using sterile technique. 400 μL was used immediately to cultivate MS2 and the remaining 600 μL was stored at −80 °C until analysis (within five months).
At each time point, MS2 infectivity was assessed by a plaque assay. The concentrations of a short segment (89 nt) of HuNoV genome (targeting the ORF1-ORF2 junction), a long segment (506 nt) of the HuNoV genome, and a long segment (313 nt) of the MS2 genome, were measured using both RT-qPCR and, for a subset of reactors, in conjunction with enzymatic pretreatment (ET-RT-qPCR). Samples collected from the same reactor were pretreated, extracted, and quantified in the same batch. We assumed that the recovery efficiency of samples within the same batch would be similar and cancel out in eqn (1) ln(N(t)/N0) (see data analysis section below). Details of these methods are provided below.
2.5 MS2 infectivity
MS2 infectivity was assessed by following method 1602.27 Fourteen technical negative controls of tryptic soy broth and the seven technical positive controls of MS2 inoculant were included. Additional methodological details are in the ESI.†2.6 Enzymatic pretreatment
A subset of samples from the experiments included paired enzymatic pretreatment to digest RNA not protected by an intact capsid as described previously.16,17 250 μL aliquots with 0.1 U μL−1 of RNase I (Thermo Fisher Scientific, Waltham, MA, USA) were incubated at 37 °C for 15 minutes, and RNA extractions immediately followed the incubation period.2.7 RNA extraction and cDNA synthesis
The Qiagen AllPrep DNA/RNA kit (Qiagen, Germantown, MD, USA) without bead beating was used to extract RNA from 200 μL aliquots of samples and eluted into 60 μL of nuclease-free water as described previously.16,17 An iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) with 20 μL reactions and 15 μL of template RNA was used to immediately reverse transcribe a portion of the RNA extract to cDNA, as per manufacturer specifications. The cDNA and the remaining RNA extract were stored at −80 °C until RT-qPCR. Four extraction negative controls (nuclease-free water), three MS2 positive controls (MS2 inoculant diluted in nuclease free water), and one HuNoV positive control (HuNoV inoculant diluted in the filter-sterilized creek water) were included as extraction negative and positive controls.2.8 RT-qPCR assays
The Environmental Microbiology Minimum Information guidelines were followed.28 To quantify HuNoV and MS2, three RT-qPCR assays were used (Tables S1–S3†): A probe-based assay in a one-step format using RNA as template targeting a short (89 nt) region of the ORF1-ORF2 junction of the HuNoV GII genome (ORF)29 and two intercalating dye-based assays each in a two-step format with cDNA as a template targeting long (>300 nt) segments of HuNoV (NR1) and MS2 (MP1) genomes.17 qPCR plates included standard curves of gBlocks Gene Fragments (Integrated DNA Technologies, Coralville, IA, USA; Table S4†) and no template controls (NTCs) of nuclease-free water. Master standard curves were generated from individual standard curves to convert experimental values from threshold cycle (Ct) to quantity (Table S5†). Values were deemed quantifiable if they had a Ct value less than the Ct value of the lowest quantity standard (Table S4†). For each RT-qPCR assay, NTCs were deemed negative if they were not quantifiable. Additional details are provided in the ESI (Supplemental Methods, Fig. S1 and Tables S1–S5†).2.9 Data analysis
R (v4.1.3) was used for all data analysis. Observed first-order decay rate constants (“k”; day−1) were calculated using eqn (1). N0 is the concentration of HuNoV or MS2 immediately following reactor inoculation (day 0), Nt is the concentration of the viruses at other time points (“t”) in days, β0 is the intercept, and ε is the error term. The arithmetic mean of experimental duplicates for ln(N(t)/N0), the “log fraction of remaining virus,” at each time point with at least one quantifiable experimental replicate was plotted versus time. The slope (k), its standard error, and its p value were calculated using the lm() function in R. In a previous study, HuNoV was incubated in filter-sterilized creek water collected from the same creek on the same date (i.e., San Pedro Creek Water collected on 11/8/2021), and HuNoV inactivation was assessed a human intestinal enteroid assay.16 Decay rate constants for HuNoV from the previous study were included for comparison, and the data analysis code was adapted for use in this study. Data analysis code and dataset are available through the Stanford Digital Repository [https://doi.org/10.25740/hk584kf2556].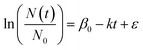 | (1) |
A multiple linear regression was used to assess the effect of incubation temperature, particle content, and their interaction on the persistence of HuNoV short genome segments and infectious MS2 (eqn (2)). For this multiple linear regression, we included only virus targets most commonly measured in environmental waters (i.e., the MS2 plaque assay and the RT-qPCR assay targeting the ORF1-ORF2 junction of HuNoV). Temperature (“T”) was modeled as a continuous variable, where 15 °C was subtracted from all incubation temperatures (in Celsius) to set the reference condition at 15 °C.7 A dummy variable was generated for the filter-sterilized water type (“F”). The coefficients (β0, β1, β2, β12, β3, β13, β23, and β123), their standard errors, and their p values were calculated using the lm() function in R. For eqn (2), the reference conditions were the raw water type incubated at 15 °C.
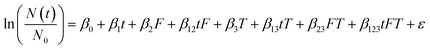 | (2) |
The persistence of most virus targets were measured for all conditions in the study (i.e., infectious MS2, long genome segments, and short genome segments), but intact capsids were measured for a subset of conditions. The subset of conditions with a complete dataset for all virus targets for both MS2 and HuNoV was raw and filter-sterilized water incubated at 15 °C. Thus, to compare the decay of various virus targets, and thereby gain insight into potential inactivation mechanisms, a multiple linear regression was used that included all measured virus targets for HuNoV and MS2 incubated at 15 °C (eqn (3)). Dummy variables were generated for the filter-sterilized water type (“F”), long genome segments (“L”) and intact capsids (“I”). The coefficients (β0, β1, β2, β12, β3, β13β4, and β14), their standard errors, and their p values were calculated using the lm() function in R. The reference conditions are the persistence measured for short genome segments (HuNoV) or infectious virus (MS2) in raw water.
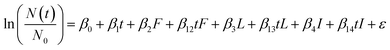 | (3) |
3 Results
Upon collection, the San Pedro Creek water sample had a salinity of 0.3 ppt and temperature of 11.3 °C (Table S6†). Raw and filter-sterilized creek water had turbidity of <1 NTU and similar absorption spectra (Fig. S2†). The biological activity in the raw creek water was higher than that of the filter-sterilized creek water, with intracellular ATP of 4.71 × 10−2 ± 3.15 × 10−3 compared to 8.97 × 10−4 ± 3.69 × 10−4 nM respectively (error is the standard deviation of triplicates; Table S6†). Uninoculated raw and filter-sterilized creek water had no amplification of genomic RNA targets from MS2 or HuNoV (MP1 and NR1 RT-qPCR assays). Additional details are in the ESI.†3.1 Quality assurance and quality control
We included experimental negative controls as well as extraction, infectivity, and RT-qPCR negative and positive controls. All positive controls were positive and negative controls were negative. Two reactors containing only PBS and incubated at 15 and 20 °C had no amplification for RT-qPCR assays (MP1 and NR1) and no plaques (MS2 plaque assay) at all time points. The extraction negative controls had no amplification (MP1 and NR1), the MS2 extraction positive controls ranged from 3.9 × 106 to 1.1 × 1010 gene copies per mL (gc mL−1; MP1), and the HuNoV extraction positive control had 1.9 × 105 gc mL−1 (ORF). For the MS2 plaque assay, the fourteen technical negative controls of tryptic soy broth had no plaques and the seven positive controls of MS2 inoculant ranged from 1.4 × 108 to 2.3 × 1010 plaque forming units per mL (PFU mL−1). The RT-qPCR no template controls were negative. Additional details are in the ESI.†3.2 Effect of temperature and particles on HuNoV persistence
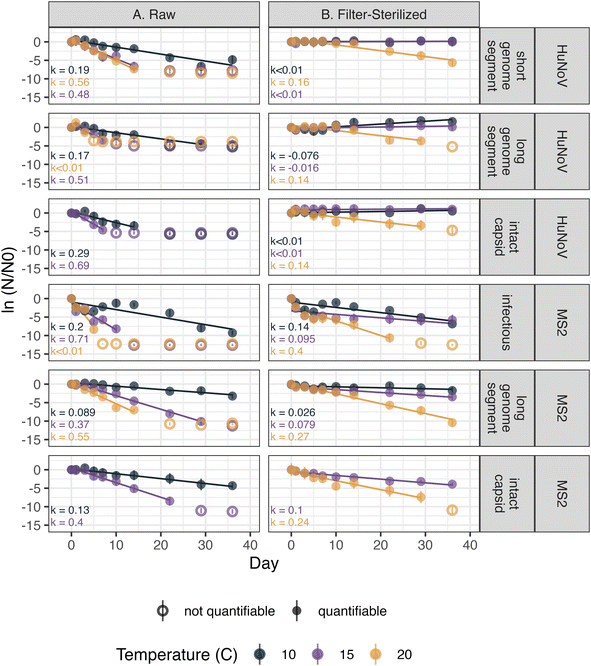
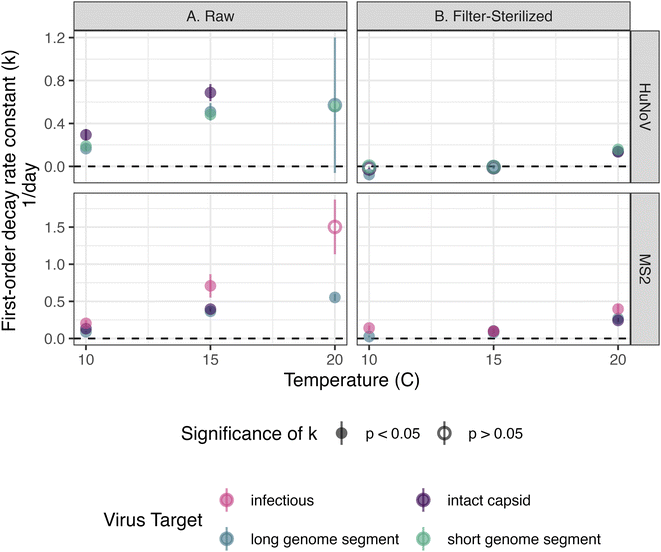
The persistence of HuNoV incubated in raw and filter-sterilized creek water at 10, 15, and 20 °C in dark conditions was assessed by following the concentration of short HuNoV genome segments, long HuNoV genome segments or intact HuNoV capsids over 36 days. The decay rate constants (“k values”) were generally larger in raw creek water and at higher incubation temperatures (Fig. 1). The positive, significant (p < 0.05) k values ranged from 0.17 ± 0.020 to 0.69 ± 0.080 day−1 (errors are standard errors of the slope in eqn (1), k) in raw creek water compared to 0.14 ± 0.025 to 0.16 ± 0.015 day−1 in filter-sterilized creek water (Table S7†). The effect of incubation temperature on the k values was more prominent in raw creek water compared to filter-sterilized creek water (Fig. 2). Five of the incubation conditions tested had no significant decay (Table S7†), with four of them from filter-sterilized creek water. Two k values were significant and negative, and both were long genome segments incubated in filter-sterilized creek water (k = −0.076 ± 0.017 and −0.016 ± 0.0065). This result reflects experimental error (not captured by the standard error and p value of k) as there is no mechanism whereby concentrations of HuNoV genome segments may increase in the experiments.To test the effect of incubation temperature and particle content on the decay rate constants of HuNoV short genome segments, a multiple linear regression was used (eqn (2)). Incubation temperature had a significant negative effect (Table S8;†β13 = −0.042 day−1; p < 0.0001) while particle content (i.e., filter-sterilization) had a significant positive effect (Table S8;†β12 = 0.34 day−1; p < 0.0001) on the log fraction of remaining HuNoV over time. These results suggest that significantly larger decay rate constants were observed for HuNoV at higher temperatures and that significantly smaller decay rate constants were observed in filter-sterilized water. In addition, a significant interaction effect between temperature and particle content was observed (Table S8;†β123 = 0.026 day−1; p < 0.001). For example, in raw water, the log fraction of remaining HuNoV decreased by 0.60 units each day incubated at 20 °C and 0.39 units at 15 °C. In comparison, in filter-sterilized water, the log fraction of remaining HuNoV decreased by 0.13 units each day incubated at 20 °C and 0.05 units at 15 °C. These results suggest that the decay rate constants for HuNoV were largest in water with the higher particle content (raw water) and combined with higher incubation temperature (20 °C).
3.3 Effect of temperature and particles on MS2 persistence
The persistence of MS2 incubated in raw and filter-sterilized creek water at 10, 15, and 20 °C in dark conditions was assessed by following the concentrations of infectious MS2, long MS2 genome segments, and intact MS2 capsids over 36 days. The k values of long MS2 genome segments and infectious MS2 were larger in magnitude in raw creek water and at higher incubation temperatures (Fig. 1). The k values observed ranged from 0.089 ± 0.0094 (p < 0.0001) to 1.5 ± 0.37 (p = 0.055) day−1 in raw creek water compared to 0.026 ± 0.011 (p < 0.05) to 0.40 ± 0.063 (p < 0.001) day−1 in filter-sterilized creek water (Table S7†). The effect of incubation temperature on the k values was more prominent in raw creek water compared to filter-sterilized creek water, especially for infectious MS2 (Fig. 2).A multiple linear regression was used to test the effect of incubation temperature and particle content on the decay rate constants of infectious MS2 (eqn (2)). Similar to the findings for HuNoV, incubation temperature had a significant negative effect (Table S9;†β13 = −0.12 day−1; p < 0.0001) and particle content had a significant positive effect (Table S9;†β12 = 0.62 day−1; p < 0.001) on the log fraction of remaining MS2 over time, with a significant interaction effect between these variables (Table S9;†β123 = 0.10 day−1; p < 0.0001). For example, the log fraction of remaining MS2 in raw water decreased by 1.39 units each day incubated at 20 °C and 0.79 units at 15 °C. In comparison, in filter-sterilized water, the log fraction of remaining MS2 decreased by 0.25 units each day incubated at 20 °C and 0.18 units at 15 °C. These results suggest that the decay rate constants for MS2 were largest in water with higher particle content (raw water) and combined with higher incubation temperature (20 °C).
3.4 HuNoV target persistence
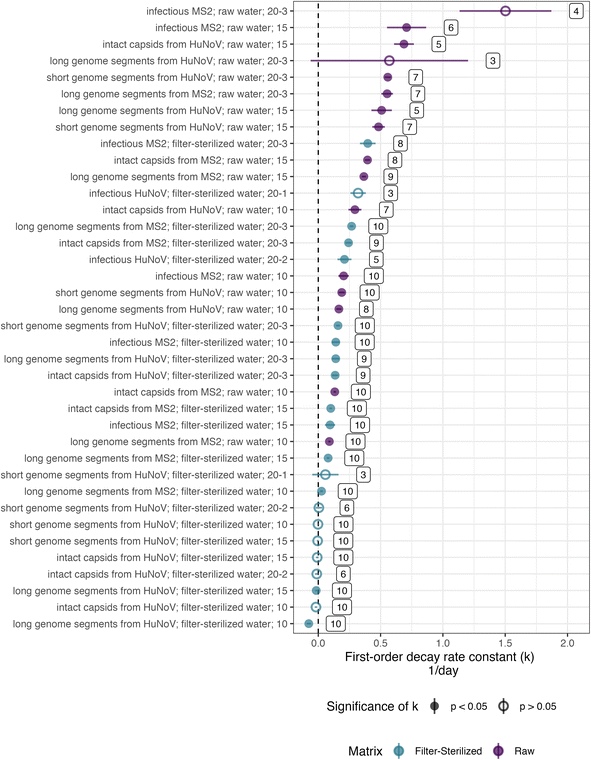
The persistence of HuNoV was assessed by following the concentrations of different virus targets (e.g., short genome segments compared to long genome segments or intact capsids) during the experiment. k values for long or short genome segments were similar in magnitude when measured for the same incubation conditions and virus (Fig. 2). However, positive, significant k values for HuNoV intact capsids were larger in magnitude than those of genome segments incubated in raw water at the temperatures tested: 10 °C (0.29 ± 0.052 compared to 0.17 ± 0.020 day−1 respectively) and 15 °C (0.69 ± 0.080 compared to 0.51 ± 0.082 day−1 respectively). For HuNoV incubated in raw or filter-sterilized water at 15 °C, a multiple linear regression (eqn (3)) was used to assess the effect of virus target on HuNoV persistence. Intact capsids and long genome segments had similar log fraction of remaining HuNoV over time compared to short genome segments (Table S10†). This finding suggests that k values for HuNoV virus targets incubated at 15 °C were not significantly different when incubated in water with the same particle content.To compare k values from HuNoV genome segments and intact HuNoV capsids to those of infectious HuNoV, k values from our previous study were compared to all k values in this study (Fig. 3).16 In the previous study, HuNoV (GII.4, OL913976) was incubated at 20 °C in filter-sterilized creek water collected from the same creek on the same day as was used herein. It should be noted that this study used a different specimen of HuNoV (GII.4, MK762570). The k values from infectious HuNoV (in filter-sterilized water incubated at 20 °C) were the largest k values in magnitude of all HuNoV targets in filter-sterilized water (Fig. 3). However, when compared with all of the k values assessed, the eight largest k values were from viruses incubated in raw creek water, regardless of virus target (Fig. 3).
3.5 MS2 target persistence
The persistence of MS2 was assessed by following the concentrations of different virus targets (e.g., infectious virus compared to long genome segments or intact capsids) during the experiment. k values for intact viral capsids were similar to those of genome segments within the same incubation conditions for all incubation conditions tested (Fig. 2). For MS2 incubated in raw creek water, k values observed for infectious MS2 were larger in magnitude compared to long genome segments when incubated at 10 °C (0.20 ± 0.042 compared to 0.089 ± 0.0094 day−1 respectively) and 15 °C (0.71 ± 0.16 compared to 0.37 ± 0.016 day−1 respectively). A multiple linear regression (eqn (3)) was applied to assess the effect of virus target on persistence of MS2 incubated in raw or filter-sterilized water at 15 °C specifically. No significant effect on the log fraction of remaining MS2 over time was observed for capsids and long genome segments compared to infectious MS2 (Table S11†). This finding suggests that k values for MS2 virus targets incubated at 15 °C were not significantly different when incubated in water with the same particle content, as was found for HuNoV.4 Discussion
The persistence of HuNoV and MS2 was assessed in surface water incubated with varied particle content (i.e., raw or filter-sterilized creek water) and temperature (10, 15, and 20 °C) conditions. Surface water temperature fluctuates with season and is a key variable affecting virus decay rate constants.7,8 Biological and inert particles in surface water also fluctuate with environmental events. For example, a four year study of a coastal sea identified estuary freshets, rainstorms, phytoplankton blooms, and jellyfish blooms as environmental events that affected particle flux.30 However, to our knowledge, the effect of particle content on the persistence of HuNoV in surface water has not been studied. This study filled that important knowledge gap, and we found that decay rate constants (“k values”) were larger in water with higher particle content. Furthermore, an interaction between particle content and temperature was observed such that the largest decay rate constants for MS2 and HuNoV were from water incubated at a higher incubation temperature and particle content. These findings demonstrate the importance of quantifying interactions between environmental conditions on the fate and transport of viruses in surface water. The k values reported herein provide parameters that can be incorporated to enhance fate and transport models.In our study, HuNoV and MS2 had k value ranges that were comparable to other studies of infectious HuNoV and MS2 persistence in environmental waters. The range in positive, significant k values for HuNoV in this study was 0.14 ± 0.025 to 0.69 ± 0.080 day−1, which corresponds to the time to 90% reduction in concentration (“T90”) of 3.3 to 16 days. Previous studies of infectious HuNoV found a similar T90 range when significant decay was observed during the study period (1.0 to 29 days).15,16 In one previous study, HuNoV remained infectious following inoculation in groundwater and incubation at room temperature in dark conditions for up to 61 days.31 More work has been done with MS2 and the range in significant k values for infectious MS2 in this study (0.095 ± 0.040 to 0.71 ± 0.016 day−1) and T90 (3.2 to 24 days) are within the range identified in the systematic review by Boehm et al.7 Boehm et al.7 identified 20 k values (median k = 0.39 day−1) for MS2 incubated in surface water in dark conditions and at temperatures from 4 °C to 25 °C (k ranged from 0.00001 to 5.1 day−1).7,32,33 Of these studies, three included infectious MS2 incubated in raw freshwater in the dark at temperatures included in this study: 10 °C (k = 0.41 day−1), 15 °C (k = 0.44 day−1), and 20 °C (k = 0.16 day−1).7,32,34,35 For these three studies, the k values notably did not correlate with temperature. In addition, the k values from these studies were smaller than those reported herein for infectious MS2 incubated in raw water at 15 °C (k = 0.71 day−1, p < 0.05) and 20 °C (k = 1.5, p = 0.055) and larger than our findings of that at 10 °C (k = 0.20 day−1, p < 0.01).
We observed larger decay rate constants for HuNoV and MS2 at higher incubation temperatures in raw and filter-sterilized freshwater. Incubation temperature has been shown to affect virus persistence in surface waters,7,8 including for six of eight viruses included in a systematic literature review (i.e., Enterovirus, Norwalk virus, Rotavirus A, Mastadenovirus, somatic coliphage, and F+ coliphage).7 However, recent findings from individual studies of the effects of incubation temperature on the persistence of HuNoV and the surrogates murine norovirus and F+ coliphage are equivocal. A study that included HuNoV and F+ coliphage incubated in freshwater in dark conditions found that HuNoV persisted longer at 4 °C (T90 = 12.1 days) compared to 15 °C (T90 = 5.5 days) in dark conditions, but the effect was not as pronounced for F+ coliphage (4 °C T90 = 2.6 days compared to 15 °C T90 = 1.7 days).36 We previously observed larger decay rate constants at 20 °C compared to 15 °C for infectious HuNoV incubated in filter-sterilized creek water.16 In addition, one study found largest decay rate constant at an incubation temperature of 24 °C (T90 = 7.6 days) as opposed to the highest temperature tested (37 °C; T90 = 25.7 days) for murine norovirus incubated in autoclaved freshwater.37 Environmental conditions beyond the type of surface water (i.e., marine, estuarine, or fresh) and temperature may obfuscate relationships between incubation conditions and decay rate constants. Indeed, we previously observed large differences in k values for HuNoV incubated at 15 °C in freshwater collected from the same creek on different dates.16
Raw water includes a microbial community that could contribute to virus inactivation through complex microbial interactions, including predation,38 as well as organic and inorganic inert particles that could affect virus fate and transport through adsorption.39–41 In this study, particle content in raw water contributed to the variation in decay rate constants observed for HuNoV and MS2, with the effect sizes for particle content larger than that of incubation temperature (Tables S8 and S10†): HuNoV and MS2 incubated in raw water had larger decay rate constants than when incubated in filter-sterilized creek water. These findings are consistent with previous studies of enterococcal bacteriophages42 and echovirus 11 (ref. 10) in raw and filter-sterilized fresh and marine waters. In contrast, organic substances (>0.45 μm in diameter) had a shielding effect on F+ coliphage in autoclaved freshwater.34 In another study, the persistence of adenovirus 40/41 and of crAssphage in raw compared to filter-sterilized freshwater were similar.43 However, the experimental design included spiking sewage into the samples after the filter-sterilization, which likely introduced biological and inert particles. In general, the effect of particle content on virus persistence in environmental waters is not well characterized,8 and, to our knowledge, has not been assessed for HuNoV in environmental water prior to this study.
We found that particle content and incubation temperature interacted to produce larger decay rate constants in water with higher particle content and incubation temperatures. These findings are consistent with others who have studied the effect of particle content and incubation temperature for enterococcal bacteriophages and echovirus 11 in environmental waters.10,42 Predation could contribute to this interaction effect: others have observed temperature dependence in predation response for protists consuming viruses.10 More research is needed testing interaction effects between particle content and incubation temperature on virus decay rate constants as well as to develop a mechanistic understanding of the effect.
Infectious virus, intact capsid, and genome segment decay rate constants can be compared to assess virus inactivation mechanisms. For MS2, these targets were compared via a multiple linear regression for raw and filter-sterilized water incubated at 15 °C. The decay rate constants from intact capsids and genome segments were similar, which suggests that capsid damage was not a major contributor to the overall decay of infectious virus (Table S11†). In addition, the decay rate constants for infectious virus and genome segments were similar, which suggests genome damage was a major contributor to the inactivation of MS2 for the conditions tested. Genome damage was previously identified as a likely inactivation mechanism for HuNoV incubated in filter-sterilized water from the same creek at 15 °C.16 However, it should be noted that k values for infectious MS2 were generally larger in magnitude than that of genome segments for all incubation conditions, particularly in raw water. Thus, damage to viral proteins could have contributed to MS2 inactivation. Indeed, we observed non-linear decay behavior for infectious MS2 incubated at 10 °C in raw creek water and at all temperatures in filter-sterilized creek water (Fig. 1), and others have hypothesized that nonlinear decay in phage could be from multiple inactivation mechanisms combined with varied phenotypic states.44 We were not able to directly compare the persistence of infectious HuNoV to that of intact capsids and genome segments. In addition, the capsid integrity methods applied in this study cannot distinguish active from inactive viruses, but instead only distinguish intact viruses from naked genomic RNA. However, for raw and filter-sterilized water incubated at 15 °C, the decay rate constants from intact capsids and genome segments were similar, which suggests that capsid damage was not a major contributor to HuNoV decay (Table S10†).
5 Conclusion
Our findings suggest that environmental processes and events that affect particle content in surface waters could also affect the persistence of viruses; such processes and events include stormwater runoff,45 earthquakes,46 and particle deposition of dust,47 black carbon,48 and soot.49 Variable virus decay rate constants could be observed following these processes and events, particularly following those wherein both particle content and temperature of surface waters are affected (e.g., droughts exacerbated by high ambient temperatures). Incorporating the effects of particle content, incubation temperature, and their interactions could enhance virus fate and transport models. For example, Li et al.50 found that incorporating the effects of environmental stressors on rotavirus and enterovirus persistence in a lake reduced the estimated risk of infection while swimming by 1000 times. In addition to infectious virus persistence, HuNoV genome segment persistence is useful when data for infectious viruses are not available as well as for environmental surveillance efforts. The global burden of HuNoV is likely underreported due to factors like healthcare-seeking behaviors and limited resources,51,52 and, thus, environmental surveillance of HuNoV in surface water could be a useful tool in unsewered communities.6 The decay rate constants presented herein will be useful to estimate HuNoV signal decay in surface water.Author contributions
LCK: conceptualization, data curation, formal analysis, investigation, methodology, project administration, software, validation, visualization, writing – original draft, writing – review and editing. SAL: data curation, investigation, validation, writing – review and editing. ABB: conceptualization, funding acquisition, methodology, project administration, validation, resources, supervision, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Drs Jan Vinje and Veronica Costantini, and the CDC CaliciNet lab for providing the clinical stool for the experiments. This work was supported by the NSF CBET-1804169 and a grant from the Woods Institute for the Environment at Stanford University. SAL was supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1656518.References
- K. E. Gibson, Viral pathogens in water: occurrence, public health impact, and available control strategies, Curr. Opin. Virol., 2014, 4, 50–57 CrossRef.
- C. Ferguson, A. M. R. de Husman, N. Altavilla, D. Deere and N. Ashbolt, Fate and Transport of Surface Water Pathogens in Watersheds, Crit. Rev. Environ. Sci. Technol., 2003, 33, 299–361 CrossRef.
- J. A. Soller, T. Bartrand, N. J. Ashbolt, J. Ravenscroft and T. J. Wade, Estimating the primary etiologic agents in recreational freshwaters impacted by human sources of faecal contamination, Water Res., 2010, 44, 4736–4747 CrossRef CAS PubMed.
- E. J. Viau, D. Lee and A. B. Boehm, Swimmer Risk of Gastrointestinal Illness from Exposure to Tropical Coastal Waters Impacted by Terrestrial Dry-Weather Runoff, Environ. Sci. Technol., 2011, 45, 7158–7165 CrossRef CAS PubMed.
- G. B. McBride, R. Stott, W. Miller, D. Bambic and S. Wuertz, Discharge-based QMRA for estimation of public health risks from exposure to stormwater-borne pathogens in recreational waters in the United States, Water Res., 2013, 47, 5282–5297 CrossRef PubMed.
- World Health Organization, Environmental Surveillance for SARS-COV-2 to Complement Public Health Surveillance, 2022 Search PubMed.
- A. B. Boehm, A. I. Silverman, A. Schriewer and K. Goodwin, Systematic review and meta-analysis of decay rates of waterborne mammalian viruses and coliphages in surface waters, Water Res., 2019, 164, 114898 CrossRef PubMed.
- K. Dean and J. Mitchell, Identifying water quality and environmental factors that influence indicator and pathogen decay in natural surface waters, Water Res., 2022, 211, 118051 CrossRef PubMed.
- A. I. Silverman and A. B. Boehm, Systematic Review and Meta-Analysis of the Persistence of Enveloped Viruses in Environmental Waters and Wastewater in the Absence of Disinfectants, Environ. Sci. Technol., 2021, 55, 14480–14493 CrossRef PubMed.
- M. Olive, C. Gan, A. Carratalà and T. Kohn, Control of Waterborne Human Viruses by Indigenous Bacteria and Protists Is Influenced by Temperature, Virus Type, and Microbial Species, Appl. Environ. Microbiol., 2020, 86, e01992 CrossRef.
- J. W.-F. Law, N.-S. Ab Mutalib, K.-G. Chan and L.-H. Lee, Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations, Front. Microbiol., 2015, 5, 770 Search PubMed.
- Y. L. Tsai, B. Tran and C. J. Palmer, Analysis of viral RNA persistence in seawater by reverse transcriptase-PCR, Appl. Environ. Microbiol., 1995, 61, 363–366 CrossRef PubMed.
- D. Dancer, R. E. Rangdale, J. A. Lowther and D. N. Lees, Human Norovirus RNA Persists in Seawater under Simulated Winter Conditions but Does Not Bioaccumulate Efficiently in Pacific Oysters (Crassostrea gigas), J. Food Prot., 2010, 73, 2123–2127 CrossRef.
- K. Ettayebi, V. R. Tenge, N. W. Cortes-Penfield, S. E. Crawford, F. H. Neill, X.-L. Zeng, X. Yu, B. V. Ayyar, D. Burrin, S. Ramani, R. L. Atmar and M. K. Estes, New Insights and Enhanced Human Norovirus Cultivation in Human Intestinal Enteroids, mSphere, 2021, 6, e01136 CrossRef PubMed.
- M. Shaffer, K. Huynh, V. Costantini, K. Bibby and J. Vinjé, Viable Norovirus Persistence in Water Microcosms, Environ. Sci. Technol. Lett., 2022, 9(10), 851–855 CrossRef PubMed.
- L. C. Kennedy, V. P. Costantini, K. A. Huynh, S. K. Loeb, W. C. Jennings, S. Lowry, M. C. Mattioli, J. Vinjé and A. B. Boehm, Persistence of Human Norovirus (GII) in Surface Water: Decay Rate Constants and Inactivation Mechanisms, Environ. Sci. Technol., 2023, 57, 3671–3679 CrossRef PubMed.
- S. K. Loeb, W. C. Jennings, K. R. Wigginton and A. B. Boehm, Sunlight Inactivation of Human Norovirus and Bacteriophage MS2 Using a Genome-Wide PCR-Based Approach and Enzyme Pretreatment, Environ. Sci. Technol., 2021, 55, 8783–8792 CrossRef CAS PubMed.
- S. M. Ahmed, A. J. Hall, A. E. Robinson, L. Verhoef, P. Premkumar, U. D. Parashar, M. Koopmans and B. A. Lopman, Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis, Lancet Infect. Dis., 2014, 14, 725–730 CrossRef.
- K. L. Vanden Esschert, M. C. Mattioli, E. D. Hilborn, V. A. Roberts, A. T. Yu, K. Lamba, G. Arzaga, M. Zahn, Z. Marsh, S. M. Combes, E. S. Smith, T. J. Robinson, S. R. Gretsch, J. P. Laco, M. E. Wikswo, A. D. Miller, D. M. Tack, T. J. Wade and M. C. Hlavsa, Outbreaks Associated with Untreated Recreational Water — California, Maine, and Minnesota, 2018–2019, Morb. Mortal. Wkly. Rep., 2020, 69, 781–783 CrossRef.
- M. Desdouits, D. Polo, C. Le Mennec, S. Strubbia, X.-L. Zeng, K. Ettayebi, R. L. Atmar, M. K. Estes and F. S. Le Guyader, Use of Human Intestinal Enteroids to Evaluate Persistence of Infectious Human Norovirus in Seawater, Emerging Infect. Dis., 2022, 28, 1475–1479 CrossRef CAS PubMed.
- E. s. Ngazoa, I. Fliss and J. Jean, Quantitative study of persistence of human norovirus genome in water using TaqMan real-time RT-PCR, J. Appl. Microbiol., 2008, 104, 707–715 CrossRef CAS.
- J. Bae and K. J. Schwab, Evaluation of Murine Norovirus, Feline Calicivirus, Poliovirus, and MS2 as Surrogates for Human Norovirus in a Model of Viral Persistence in Surface Water and Groundwater, Appl. Environ. Microbiol., 2008, 74, 477–484 CrossRef CAS.
- J. Flannery, P. Rajko-Nenow, S. Keaveney, V. O'Flaherty and W. Doré, Simulated sunlight inactivation of norovirus and FRNA bacteriophage in seawater, J. Appl. Microbiol., 2013, 115, 915–922 CrossRef CAS PubMed.
- National Oceanic and Atmospheric Administration - National Centers for Environmental Information, Coastal Water Temperature Guide - All Coastal Regions Table, https://www.ncei.noaa.gov/access/coastal-water-temperature-guide/all_table.html, accessed 6 November 2023 Search PubMed.
- S. Earle, Physical Geology, BCcampus, Victoria, B.C., 2nd edn, 2019 Search PubMed.
- S. Oristo, H. J. Lee and L. Manula, Performance of pre-RT-qPCR treatments to discriminate infectious human rotaviruses and noroviruses from heat-inactivated viruses: applications of PMA/PMAxx, benzonase and RNase, J. Appl. Microbiol., 2018, 124, 1008–1016 CAS.
- Environmental Protection Agency, Method 1602: Male-specific (F+) and Somatic Coliphage in Water by Single Agar Layer (SAL) Procedure, 2001 Search PubMed.
- M. A. Borchardt, A. B. Boehm, M. Salit, S. K. Spencer, K. R. Wigginton and R. T. Noble, The Environmental Microbiology Minimum Information (EMMI) Guidelines: qPCR and dPCR Quality and Reporting for Environmental Microbiology, Environ. Sci. Technol., 2021, 55, 10210–10223 CrossRef CAS PubMed.
- F. Loisy, R. L. Atmar, P. Guillon, P. Le Cann, M. Pommepuy and F. S. Le Guyader, Real-time RT-PCR for norovirus screening in shellfish, J. Virol. Methods, 2005, 123, 1–7 CrossRef CAS PubMed.
- S. C. Johannessen, R. W. Macdonald, C. A. Wright and D. J. Spear, Short-term variability in particle flux: Storms, blooms and river discharge in a coastal sea, Cont. Shelf Res., 2017, 143, 29–42 CrossRef.
- S. R. Seitz, J. S. Leon, K. J. Schwab, G. M. Lyon, M. Dowd, M. McDaniels, G. Abdulhafid, M. L. Fernandez, L. C. Lindesmith, R. S. Baric and C. L. Moe, Norovirus Infectivity in Humans and Persistence in Water, Appl. Environ. Microbiol., 2011, 77, 6884–6888 CrossRef CAS.
- S. V. Ravva and C. Z. Sarreal, Persistence of F-Specific RNA Coliphages in Surface Waters from a Produce Production Region along the Central Coast of California, PLoS One, 2016, 11, e0146623 CrossRef.
- F. E. Eregno, I. Tryland, M. Myrmel, A. Wennberg, A. Oliinyk, M. Khatri and A. Heistad, Decay rate of virus and faecal indicator bacteria (FIB) in seawater and the concentration of FIBs in different wastewater systems, Microb. Risk Anal., 2018, 8, 14–21 CrossRef.
- Y. Yang and M. W. Griffiths, Comparative Persistence of Subgroups of F-Specific RNA Phages in River Water, Appl. Environ. Microbiol., 2013, 79, 4564–4567 CrossRef.
- S. C. Long and M. D. Sobsey, A comparison of the survival of F+RNA and F+DNA coliphages in lake water microcosms, J. Water Health, 2004, 2, 15–22 CrossRef.
- A. Tiwari, A. Kauppinen, P. Räsänen, J. Salonen, L. Wessels, J. Juntunen, I. T. Miettinen and T. Pitkänen, Effects of temperature and light exposure on the decay characteristics of fecal indicators, norovirus, and Legionella in mesocosms simulating subarctic river water, Sci. Total Environ., 2023, 859, 160340 CrossRef PubMed.
- E. M. E. Ibrahim, M. A. El-Liethy, A. L. K. Abia, B. A. Hemdan and M. N. Shaheen, Survival of E. coli O157:H7, Salmonella Typhimurium, HAdV2 and MNV-1 in river water under dark conditions and varying storage temperatures, Sci. Total Environ., 2019, 648, 1297–1304 CrossRef PubMed.
- M. Zhang, N. Altan-Bonnet, Y. Shen and D. Shuai, Waterborne Human Pathogenic Viruses in Complex Microbial Communities: Environmental Implication on Virus Infectivity, Persistence, and Disinfection, Environ. Sci. Technol., 2022, 56, 5381–5389 CrossRef PubMed.
- M. G. Weinbauer, Y. Bettarel, R. Cattaneo, B. Luef, C. Maier, C. Motegi, P. Peduzzi and X. Mari, Viral ecology of organic and inorganic particles in aquatic systems: avenues for further research, Aquat. Microb. Ecol., 2009, 57, 321–341 CrossRef PubMed.
- Y. Yamada, R. Guillemette, A.-C. Baudoux, N. Patel and F. Azam, Viral Attachment to Biotic and Abiotic Surfaces in Seawater, Appl. Environ. Microbiol., 2020, 86, e01687 CrossRef PubMed.
- L. Kernegger, I. Zweimüller and P. Peduzzi, Effects of suspended matter quality and virus abundance on microbial parameters: experimental evidence from a large European river, Aquat. Microb. Ecol., 2009, 57, 161–173 CrossRef PubMed.
- N. Booncharoen, S. Mongkolsuk and K. Sirikanchana, Comparative persistence of human sewage-specific enterococcal bacteriophages in freshwater and seawater, Appl. Microbiol. Biotechnol., 2018, 102, 6235–6246 CrossRef PubMed.
- W. Ahmed, S. Toze, C. Veal, P. Fisher, Q. Zhang, Z. Zhu, C. Staley and M. J. Sadowsky, Comparative decay of culturable faecal indicator bacteria, microbial source tracking marker genes, and enteric pathogens in laboratory microcosms that mimic a sub-tropical environment, Sci. Total Environ., 2021, 751, 141475 CrossRef PubMed.
- R. H. Heineman and S. P. Brown, Experimental Evolution of a Bacteriophage Virus Reveals the Trajectory of Adaptation across a Fecundity/Longevity Trade-Off, PLoS One, 2012, 7, e46322 CrossRef.
- K. Williamson, J. Harris, J. Green, F. Rahman and R. Chambers, Stormwater runoff drives viral community composition changes in inland freshwaters, Front. Microbiol., 2014, 5, 105 CrossRef PubMed.
- T. Nagata, Y. Yang and H. Fukuda, Disturbed virus–bacteria dynamics in Otsuchi Bay (Japan) after the mega-earthquake and tsunami in March 2011, Aquat. Microb. Ecol., 2022, 88, 31–41 CrossRef.
- J. Dinasquet, E. Bigeard, F. Gazeau, F. Azam, C. Guieu, E. Marañón, C. Ridame, F. Van Wambeke, I. Obernosterer and A.-C. Baudoux, Impact of dust addition on the microbial food web under present and future conditions of pH and temperature, Biogeosciences, 2022, 19, 1303–1319 CrossRef CAS.
- R. Cattaneo, C. Rouviere, F. Rassoulzadegan and M. G. Weinbauer, Association of marine viral and bacterial communities with reference black carbon particles under experimental conditions: an analysis with scanning electron, epifluorescence and confocal laser scanning microscopy, FEMS Microbiol. Ecol., 2010, 74, 382–396 CrossRef CAS.
- X. Mari, J. Lefèvre, J.-P. Torréton, Y. Bettarel, O. Pringault, E. Rochelle-Newall, P. Marchesiello, C. Menkes, M. Rodier, C. Migon, C. Motegi, M. G. Weinbauer and L. Legendre, Effects of soot deposition on particle dynamics and microbial processes in marine surface waters, Global Biogeochem. Cycles, 2014, 28, 662–678 CrossRef CAS.
- C. Li, É. Sylvestre, X. Fernandez-Cassi, T. R. Julian and T. Kohn, Waterborne virus transport and the associated risks in a large lake, Water Res., 2023, 229, 119437 CrossRef CAS.
- Centers for Disease Control and Prevention, Reporting and Surveillance for Norovirus, https://www.cdc.gov/norovirus/reporting/index.html, accessed 26 July 2023 Search PubMed.
- World Health Organization, WHO Estimates of the Global Burden of Foodborne Diseases, 2015 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3em00357d |
| This journal is © The Royal Society of Chemistry 2024 |