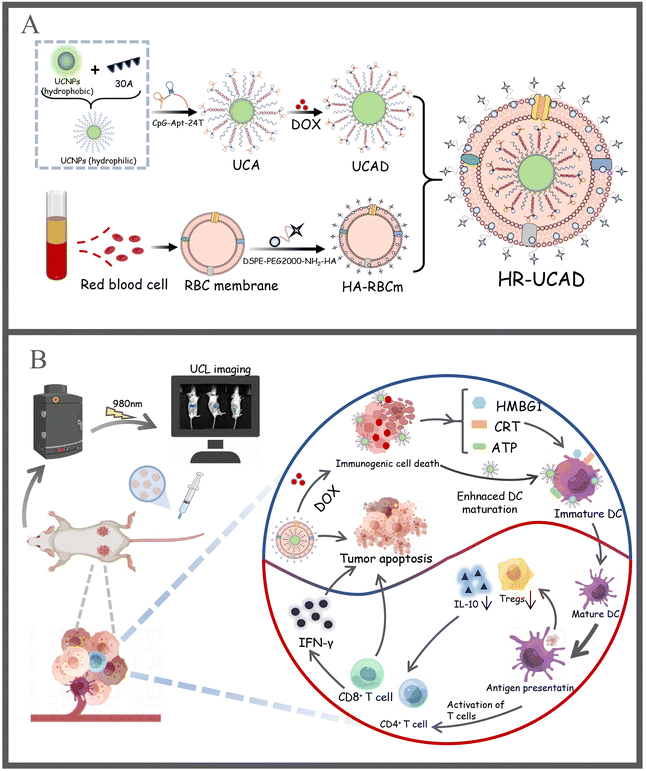
Erythrocyte membrane-camouflaged DNA-functionalized upconversion nanoparticles for tumor-targeted chemotherapy and immunotherapy†
Qinjie
Kou
 a,
Yufen
Huang
a,
Yanrong
Su
a,
Lu
Lu
b,
Xisheng
Li
a,
Haiye
Jiang
a,
Rong
Huang
b,
Jian
Li
*b and
Xinmin
Nie
a,
Yufen
Huang
a,
Yanrong
Su
a,
Lu
Lu
b,
Xisheng
Li
a,
Haiye
Jiang
a,
Rong
Huang
b,
Jian
Li
*b and
Xinmin
Nie
 *ac
*ac
aDepartment of Laboratory Medicine, The Third Xiangya Hospital, Central South University, Changsha, Hunan Province, China
bDepartment of Blood Transfusion, The Third Xiangya Hospital, Central South University, Changsha, 410013, Hunan, China. E-mail: 603196@csu.edu.cn
cHunan Engineering Technology Research Center of Optoelectronic Health Detection, Changsha, 410000, Hunan, China. E-mail: niexinmin@csu.edu.cn
First published on 25th April 2023
Abstract
A synergistic combination of treatment with immunogenic cell death (ICD) inducers and immunoadjuvants may be a practical way to boost the anticancer response and successfully induce an immune response. The use of HR@UCNPs/CpG-Apt/DOX, new biomimetic drug delivery nanoparticles generated to combat breast cancer, is reported here as a unique strategy to produce immunogenicity and boost cancer immunotherapy. HR@UCNPs/CpG-Apt/DOX (HR-UCAD) consists of two parts. The core is composed of an immunoadjuvant CpG (a toll-like receptor 9 agonist) fused with a dendritic cell-specific aptamer sequence (CpG-Apt) to decorate upconversion nanoparticles (UCNPs) with the successful intercalation of doxorubicin (DOX) into the consecutive base pairs of Apt-CpG to construct an immune nanodrug UCNPs@CpG-Apt/DOX. The targeting molecule hyaluronic acid (HA) was inserted into a red blood cell membrane (RBCm) to form the shell (HR). HR-UCAD possessed a strong capacity to specifically induce ICD. Following DOX-induced ICD of cancer cells, sufficient exposure to tumor antigens and UCNPs@CpG-Apt (UCA) activated the tumor-specific immune response and reversed the immunosuppressive tumor microenvironment. In addition, HR-UCAD has good biocompatibility and increases the active tumor-targeting effect. Furthermore, HR-UCAD exhibits excellent near-infrared upconversion luminescence emission at 804 nm under irradiation with a 980 nm laser, which has great potential in biomedical imaging. Thus, the RBCm-camouflaged drug delivery system is a promising targeted chemotherapy and immunotherapy nanocomplex that could be used for effective targeted breast cancer treatment.
1 Introduction
Breast cancer produces aggressive malignant tumors and has a high mortality rate in women.1 Current treatments are far from satisfactory due to their high costs, significant interindividual differences and severe side effects.2 Therefore, the development of new strategies with high efficiency and low toxicity to eradicate breast cancer is urgently needed. Recently, immunotherapy has made significant advances in the field of antitumor therapy, such as immune checkpoint blockade (ICB)3 and chimeric antigen receptor T-cell (CAR-T) therapy,4 but there are still some obstacles, such as weak immunogenicity, insufficient cytotoxic T lymphocyte (CTL) infiltration, and poor maturation of dendritic cells (DCs).5–7 Thus, enhancing antitumor immunogenicity and reversing the immunosuppressive tumor microenvironment (ITM) are crucially important.8 Recent studies have revealed that doxorubicin (DOX), a commonly used clinical chemotherapeutic drug, can elicit immunogenic cell death (ICD).9 Tumor cells undergoing ICD can generate a large number of tumor fragments in situ and release damage-associated molecular patterns (DAMPs) into the surrounding environment, including calreticulin (CRT), adenosine triphosphate (ATP), and high mobility group box 1 (HMGB1).10 CRT is an “eat me” signal that encourages the phagocytic engulfment of cancer cells by DCs.11 ATP serves as a “find me” signal to recruit antigen-presenting cells (APCs).12 HMGB1 binds to toll-like receptor 4 (TLR4) on DCs and presents antigens to CTLs.13 Therefore, ICD may provide a new strategy to combat tumors through immune cell recruitment. However, it has been reported that ICD cannot generate a sufficient antitumor immune response because of the ITM.14,15Cytosine-phosphate-guanosine oligonucleotides (CpG ODNs),16 as toll-like receptor 9 (TLR9) agonists of DCs, can trigger strong immunostimulatory activity via MyD88-dependent NF-κB and MAPK signaling and subsequently promote the release of proinflammatory cytokines such as interleukin 6 (IL-6), IL-12, and tumor necrosis factor-α (TNF-α), ultimately inducing a Th1-like innate immune response and adaptive immunity.17,18 The antigens generated by ICD can be efficiently delivered to APCs with the aid of the immunoadjuvant CpG. Therefore, ICD inducers combined with immunoadjuvants may be a promising tactic to combat cancer through a robust immune response.
The use of naked CpG ODNs as therapeutic agents is limited. TLR9 is mainly distributed on the endoplasmic reticulum (ER) of DCs rather than on the cell surface.19 The negatively charged CpG ODNs fail to penetrate the cell membrane and are easily digested by exonucleases in the serum or cytoplasm.20 In recent years, lanthanide-based upconversion nanoparticles (UCNPs) have come into focus due to their distinctive chemical and optical properties.21 According to research, UCNPs that carry and protect single-stranded DNA have been employed in cancer treatment and bioimaging.22 On the one hand, the fast coordination of the phosphate groups on the DNA backbone with Ln3+ exposed on the UCNPs causes DNA absorption onto the UCNP surface.23 This method does not require chemical modification of oligonucleotides and is time-saving and highly efficient. The DNA–UCNP composite synthesized by this method is stable, and the DNA attached onto the surface of UCNPs is capable of crossing the cell membrane without the need for transfection agents.24 On the other hand, unlike traditional fluorescent agents, UCNPs can convert near-infrared (NIR) radiation into tunable shorter wavelength luminescence, thus avoiding background signal noise from samples and the surroundings.25 NIR light penetrates deeper than visible light, and it can induce UCNPs to produce luminescence from UCNPs in deeper tissues, showing significant potential for in vivo imaging.26 As a result, UCNPs hold great promise for multifunctional drug delivery and biomedical imaging applications. In addition, we conjugated CpG ODNs with aptamers targeting DCs to form a novel DNA nanostructure (CpG-Apt), which endows the DNA nanostructure with high binding affinity for DCs and further stimulates DCs maturation.27,28 Notably, DNA nanostructures are also effective drug delivery nanocarriers, and DOX can intercalate into adjacent base pairs.29 In this study, UCNPs were surface modified using engineered DNA molecules (CpG-Apt/DOX) to create DNA–UCNP complexes. For the first time, DNA-functionalized UCNPs were used to deliver DOX, which has never been reported yet.
Nevertheless, UCNPs also have some biocompatibility concerns, as they can easily be regarded as exogenous substances and removed from the blood circulation by the reticuloendothelial system (RES).30 Moreover, these nanocarriers lack targeting ability, allowing the drug to prematurely leak from the bare material, thereby resulting in serious side effects.31 Fortunately, the use of the biomimetic cell membrane camouflage strategy can help nanoparticles cloak the encapsulated contents to evade RES elimination.32–34 Among natural biological carriers, erythrocyte membranes are accessible and frequently used to prepare biomimetic nanomaterials. In particular, the self-recognition protein CD47 on the erythrocyte membrane surface can inhibit erythrocyte phagocytosis by immune cells, which gives nanocarriers a superior half-life in systemic circulation.35 CD44 is highly expressed in breast cancer cells and can selectively bind to hyaluronic acid (HA).36–38 Thus, HA modification of the erythrocyte membrane can enhance the tumor-specific accumulation of nanodrugs.
Herein, we fabricated a novel multifunctional delivery system, erythrocyte membrane-camouflaged DNA-functionalized UCNPs, for targeted breast cancer chemotherapy and immunotherapy. UCNPs capped with oleic acid are normally hydrophobic. Related investigations have demonstrated that if CpG-Apt directly replaces the original capped ligands on UCNPs, the capped DNAs could collapse on the particle surface, which is not conducive to its ability to carry out its corresponding function and its further assembly.39 Additionally, it has been observed that inhibiting the 5′ end of CpG may lower immunological activity because this portion is necessary for TLR9-mediated immune responses.40,41 On this basis, we first modified UCNPs with a 30-mer polyA oligonucleotide to change the UCNPs from hydrophobic to hydrophilic. Moreover, a 24-mer polyT oligonucleotide was fused with the 3′ end of CpG-Apt to form a 5′-CpG-Apt-24T-3′ sequence. According to Watson Crick's matching method, polyA on the UCNPs and polyT of CpG-Apt-24T will hybridize to form a duplex that resembles a satellite-like structure (Fig. 1A). This novel strategy, the 3′ end of CpG fused with a DC-targeted aptamer sequence, has not yet been reported in the literature. Subsequently, DOX was introduced into base pairs to construct UCNPs@CpG-Apt/DOX, which was encapsulated within the erythrocyte membrane. This novel nanocomplex (HR-UCAD) exhibits good biocompatibility and immune escape. With the aid of HA and the enhanced permeability and retention (EPR) effect, HR-UCAD can actively accumulate at the tumor site, while UCNPs@CpG-Apt (UCA) can precisely target DCs, thereby activating antitumor immune responses. Moreover, DNA modification and membrane encapsulation do not affect the upconversion optical properties of the UCNPs. Therefore, this novel biomimetic nanoplatform is also useful for efficient tumor imaging. Our work lays the foundation for UCNPs in multifunctional drug delivery and biomedical imaging applications, providing a new way to treat breast cancer.
2 Methods and materials
2.1 Materials
Lanthanide-based UCNPs (XF201-3) were purchased from Nanjing XFNANO Materials Tech Co., Ltd. (Nanjing, China). DSPE-PEG2000-NH2 (PS2-NDE-1K) and DSPE-PEG2000-FITC (PS2-DEFT) were produced by Ponsure Biotechnology (China). All nonmethylated oligodeoxynucleotides were synthesized by Shanghai Sangon Biological Engineering Technology & Services (Shanghai, China). The sequences are as follows (Fig. S1†): DNA for the preparation of A30-UCNPs (5′-AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA-3′); CpG 1826 (5′-TCCATGACGTTCCTGACGTT-3′); the aptamer sequence targeting DCs (5′-GGCTGGCACTGGTCGAGGTATGTTGGGGCAGCT-3′); and the CpG-Apt-24T sequence (5′-TCCATGACGTTCCTGACGTTTTTTTGGCTGGCACTGGTCGAGGTATGTTGGGGC AGCTTTTTTTTTTTTTTTTTTTTTTTTT-3′). A penicillin–streptomycin cocktail (PB180120), fetal bovine serum (FBS; 164210-50), DMEM (PM150210), and RPMI 1640 (PM150110) were purchased from Procell Biotech (Wuhan, China). Calcein-AM/propidium iodide (PI) (C2015S), cell counting kit-8 (C0037) ATP detection (S0027) and Annexin V-FITC apoptosis detection kits (C1062S) were purchased from Beyotime Biotechnology (China). Hematoxylin and eosin (H&E), DAPI, and crystal violet were produced by Servicebio Technology (China). Anti-HMGB1 (66525-1-Ig), anti-CRT (27298-1-AP), and anti-CD86 (13395-1-AP) antibodies were manufactured by Proteintech (Wuhan, China). Anti-CD11c (AB_2841076), anti-IL-10 (AB_2838853), anti-Foxp3 (AB_2847268), and anti-IFN-γ (AB_2838015) antibodies were manufactured by Affinity Biosciences (USA). Anti-CD80 (104707), anti-CD86 (105011), anti-CD3 (100235), anti-CD4 (100405), and anti-CD8 (155007) antibodies and the Zombie Aqua™ Fixable Viability Kit (423101) were obtained from BioLegend (USA). Transwell plates (3422) were purchased from Corning Incorporated (USA). The enzyme-linked immunosorbent assay (ELISA) kits (IL-6 and TNF-α) and the micro BCA™ protein assay reagent kit (AR1110) were obtained from Boster Biological Technology (China). DOX, the dialysis membrane (2 kDa), the mouse tumor-infiltrating lymphocyte isolation kit (P9000), hyaluronidase (H8030), and DNase I (D-8071) were purchased from Solarbio (China). Collagenase IV (2091) was purchased from BioFroxx.2.2 Cells
All cells were purchased from Central South University's Advanced Research Center. DC2.4 is a murine dendritic cell line. RAW264.7 is a macrophage that is used in immune escape experiments. 4T1, a breast cancer cell line, is the primary validation cell for this study. HeLa is a cervical cancer cell line used as a control tumor cell in HA targeting experiments. MCF-10A is the normal breast epithelial cell line. HEK-293T cells are human embryonic kidney cells. MCF-10A and HEK-293T cells were used as controls. RAW264.7, DC2.4, and 4T1 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. HeLa, MCF-10A, and HEK-293T cells were cultured in DMEM/high glucose medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin.2.3 Preparation of HR-UCAD
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. The pellets were washed repeatedly until a reddish color was observed to obtain erythrocyte membranes. Finally, erythrocyte vesicles were prepared by ultrasonication at a frequency of 42 kHz and 100 W for 5 min, and the extrusion process was continued. PBS (3 ml) was used to dissolve HA (3.75 mg), DSPE-PEG2000-NH2 (15 mg), 1-ethyl-3-(dimethylaminopropyl) carbodiimide (EDC; 1.68 mg), and N-hydroxysuccinimide (NHS; 1 mg), and the mixture was agitated for 6 h at 37 °C. The solution was then dialyzed in a dialysis bag for 48 h to obtain DSPE-PEG2000-NH2-HA. HR was synthesized after DSPE-PEG2000-NH2-HA was incubated with the prepared RBCms at 37 °C for 1 h.
000 rpm for 5 min. The pellets were washed repeatedly until a reddish color was observed to obtain erythrocyte membranes. Finally, erythrocyte vesicles were prepared by ultrasonication at a frequency of 42 kHz and 100 W for 5 min, and the extrusion process was continued. PBS (3 ml) was used to dissolve HA (3.75 mg), DSPE-PEG2000-NH2 (15 mg), 1-ethyl-3-(dimethylaminopropyl) carbodiimide (EDC; 1.68 mg), and N-hydroxysuccinimide (NHS; 1 mg), and the mixture was agitated for 6 h at 37 °C. The solution was then dialyzed in a dialysis bag for 48 h to obtain DSPE-PEG2000-NH2-HA. HR was synthesized after DSPE-PEG2000-NH2-HA was incubated with the prepared RBCms at 37 °C for 1 h.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–8
1–8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were sonicated (3 min, 42 kHz, 100 W) and then filtered 20 times through a 200 nm porous membrane syringe filter. Excess HR (3000 rpm, 10 min) was separated by centrifugation to obtain HR-UCAD. In order to evaluate the membrane-to-nucleus mass ratio with the highest RBC membrane coating efficiency, the amount of membrane proteins on HR-UCAD (1 ml) was measured using a BCA protein assay reagent kit.
1 were sonicated (3 min, 42 kHz, 100 W) and then filtered 20 times through a 200 nm porous membrane syringe filter. Excess HR (3000 rpm, 10 min) was separated by centrifugation to obtain HR-UCAD. In order to evaluate the membrane-to-nucleus mass ratio with the highest RBC membrane coating efficiency, the amount of membrane proteins on HR-UCAD (1 ml) was measured using a BCA protein assay reagent kit.
2.4 Characterization of HR-UCAD
The particle size and shape of the nanoparticles were assessed by transmission electron microscopy (TEM; Tecnai F20, FEI, USA) to confirm the successful encapsulation of UCA into the RBCm. A Zetasizer Nano ZS (Malvern Nano series, Malvern, UK) was used to assess the particle size distribution and zeta potential/surface charge. The absorption peaks were observed by UV-vis spectroscopy (Malvern Nano series, Malvern, UK). RBCm proteins were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) after staining with Coomassie Brilliant Blue staining reagent (Beyotime, China). The upconversion luminescence (UCL) spectra of UCNPs (2 mg ml−1) and HR-UCA (UCNP concentration: 2 mg ml−1) were recorded and characterized using a steady state and transient state fluorescence spectrometer (Edinburgh FLS-920) with an external adjustable CW laser (power ≈ 800 mW) at 980 nm (Connet Fiber Optics, China). The optimal ratio of A30 to UCNP attachment and CpG-Apt to A30-UCNP was determined using an agarose gel retardation assay. The gels were visualized using a gel imaging analysis system (Bio-Rad ChemiDoc XRS+ System). The X-ray diffraction (XRD) pattern of UCNPs was measured using X-ray diffraction instrument (XRD, Rigaku Ultima IV, Japan) at a scanning rate of 2° min−1 in the 2θ range of 10–90°. The elemental composition of UCNPs was evaluated by X-ray photoelectron spectroscopy (XPS, Thermo Scientific K-Alpha, USA).2.5 Release of DOX from HR-UCAD and the intracellular localization of nanoparticles
To determine whether DOX could be released from HR-UCAD in a pH-dependent manner, drug release tests were conducted. Dialysis bags were filled with 1 ml each of R-UCAD (DOX, 1 mg ml−1) and UCAD (1 mg ml−1) and then placed in PBS at pH = 5.0 and pH = 7.4. At 5 h, 10 h, 15 h, 20 h, 25 h, and 30 h, dialysate samples were collected, and their absorbance values at 480 nm were measured (EnSpire 2300, PerkinElmer, USA). The DOX content in the dialysate was calculated, and drug release curves were plotted.4T1 cells were seeded into 6-well plates at a density of 2 × 105 per well and cultured at 37 °C overnight. 4T1 cells were incubated with HR-UCAD (10 μg ml−1) for 0 h, 1 h, 2 h, 3 h, 4 h, 5 h, and 6 h. Then, the culture medium was removed and the cells were washed three times with PBS. Nuclei were stained with Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342. Finally, the fluorescence of the cells was measured by CLSM.
342. Finally, the fluorescence of the cells was measured by CLSM.
The element Y is the characteristic element of UCNPs, so Y is used to quantify the uptake efficiency of nanoparticles by cancer cells. We used inductively coupled plasma (ICP) to quantify the mass percentage of Y in UCNPs, which is approximately 32.58% in UCNPs. Then, 4T1 cells were inoculated into 6-well plates (2 × 105 cells per well) and treated with 3 preparations including UCNPs, R@UCNPs and HR@UCNPs at the same dose of UCNPs for 24 h. After being trypsinized, the cells were then centrifuged at 5000 rpm for 10 min and collected in a 1.5 mL Eppendorf tube. The cells were dissolved in an aqua regia solution and then quantified using an Agilent 7500 cs/ce Quadrupole ICP-MS. For each mouse administered with all the three preparations (UCNPs, R@UCNPs and HR@UCNPs, 0.05 mg ml−1 UCNPs per group), the harvested tumor tissues were weighed, subsequently cut into 1–2 mm2 pieces and dissolved in an aqua regia solution. Furthermore, the biodistribution of Y was quantified by ICP-MS.
2.6 Hemocompatibility and immune escape ability assay
The biocompatibility of HR-UCAD was examined by determining the hemolysis rate. Five different concentrations of nanoparticles were incubated with 5% erythrocytes at 37 °C for 4 h, followed by centrifugation at 3500 rpm for 5 min. Multifunctional enzyme labeling instrument (PerkinElmer EnSpire, USA) was employed to measure the absorbance of the supernatant at 540 nm. Distilled water (A100%) and PBS (A0%) were used as positive and negative controls, respectively. The formula used to calculate the hemolysis rate is as follows: hemolysis (%) = (Asample − A0%)/(A100% − A0%) × 100%. To detect the immune escape ability of the nanoparticles, RAW264.7 cells were added to a 6-well plate (2 × 105 cells per well) and treated with UCAD (DOX, 5 μg ml−1), HR-UCAD (DOX, 5 μg ml−1), and R-UCAD (DOX, 5 μg ml−1) for 3 h at 37 °C. The nuclei of the macrophages were stained with DAPI, and the fluorescence signals of DOX in the macrophages after nanoparticle phagocytosis were observed under a fluorescence microscope (ZEISS Axio Vert.A1, Germany).2.7 Evaluation of HR-UCAD targeting in vitro
To observe the ability of HA to target breast cancer cells, 4T1, HeLa, HEK-293T and MCF-10A cells (as controls) were seeded in 6-well plates (2 × 105 cells per well) and subsequently incubated with DSPE-FITC-labeled HA-RBCm vesicles (2 mg) for 6 h. The nuclei were stained blue using DAPI. The cells were then washed three times with PBS and observed under a fluorescence microscope. Furthermore, the uptake of UCNPs@CpG-Apt by DC2.4 cells was analyzed by fluorescence microscopy. DC2.4 cells were seeded into 6-well plates at a density of 2 × 105 cells per well and maintained for 24 h. Then, the cells were incubated with free CpG (Cy5-CpG ODN, 10 μmol ml−1), UCNPs@CpG (Cy5-CpG ODN, 10 μmol ml−1), and UCNPs@CpG-Apt (Cy5-CpG ODN, 10 μmol ml−1) for 6 h. CpG ODNs were modified with a fluorophore (Cy5.5) at the 5′ end. After incubation, the cells were washed and fixed with 4% paraformaldehyde and stained with DAPI for 20 min. A fluorescence microscope (ZEISS Axio Vert.A1, Germany) was employed to probe the cellular uptake of the different CpG formulations.2.8 In vitro uptake of DOX and cytotoxic effects of HR-UCAD
4T1 cells (6-well plates, 2 × 105 cells per well) were treated with free DOX, UCAD, or HR-UCAD at the same dose of DOX (3 μg ml−1) for 6 h to determine DOX uptake. A flow cytometer (FACS CantoTM II, BD, USA) was utilized to measure the DOX fluorescence intensity (3 × 104 cells).The cytotoxicity of HR-UCAD was measured by the CCK-8 assay according to the manufacturer's instructions. 4T1 cells were inoculated into 96-well plates (7 × 103 cells per well) and treated with 6 preparations including PBS, UCNPs, DOX (3 μg ml−1 DOX), CpG-Apt + DOX (CAD) (3 μg ml−1 DOX mixed with free CpG-Apt), UCAD (3 μg ml−1 DOX), and HR-UCAD (3 μg ml−1 DOX) for 24 h. Then, 10 μl of CCK-8 solution was added to each well for 2 h incubation, and the absorbance at 450 nm was measured using a microplate reader (EnSpire 2300, PerkinElmer, USA). After treatment of the above six groups, live and dead cells were stained with calcein-AM and PI and then photographed under a fluorescence microscope.
2.9 Immunogenic cell death (ICD) in vitro
4T1 cells were seeded into a 6-well plate (2 × 105 cells per well) for incubation overnight. After that, the cells were exposed to PBS, UCNPs, DOX, CAD, UCAD, and HR-UCAD for 24 h at the same concentration of DOX (3 μg ml−1). CRT was detected by immunofluorescence. After fixing with 4% paraformaldehyde and washing twice with PBS, the cells were exposed to an anti-CRT antibody for 1 h. Then, the cells were treated with a secondary antibody conjugated to Alexa Fluor 488 for an additional 30 min. Subsequently, the cells were labeled with DAPI for 20 min, and a fluorescence microscope was used for viewing the cells. The supernatants were collected to determine HMGB1 release by western blotting and ATP release with an ATP luminescence assay system. To evaluate the migration rate of immune cells recruited by the nanodelivery system, a transwell system was constructed with 8 μm polycarbonate porous membranes in 6-well plates. In this system, the upper chamber contained DC2.4 cells (2 × 104 cells per well) in serum-free media, whereas the lower chamber contained 4T1 cells (1 × 105 cells per well).44 To allow the immune function of DCs to be prominent, we set the concentration of DOX to 0.5 μg ml−1 to guarantee that the DC2.4 cells viability remained above 80% after incubation with HR-UCAD. After the 4T1 cells were treated with PBS, UCNPs, DOX, CAD, UCAD, and HR-UCAD (at the same concentration of DOX, 0.5 μg ml−1) for 12 h, the cells in the upper wells were transferred and coincubated with 4T1 cells for 12 h. Then, the migrated DC2.4 cells were collected, fixed with 4% paraformaldehyde, and stained with crystal violet. The numbers of migrating cells from 3 randomly selected fields were counted using a microscope, and the migration rates were calculated.2.10 Coculture of immune cells and cancer cells in vitro
The coculture system used in this study was adopted to investigate the antitumor immune effect of HR-UCAD in vitro as described previously.45 After coincubation, DC2.4 cells (3 × 104) were collected to evaluate the expressions of CD86 and CD80 by flow cytometry (FACS CantoTM II, BD, USA). The cytokines in the supernatant, including IL-6 and TNF-α, were analyzed using ELISA kits. 4T1 cells (3 × 104 cells) were subjected to flow cytometry (FACS CantoTM II, BD, USA) for apoptosis analysis with an apoptosis detection kit.2.11 In vivo animal xenograft models
BALB/c mice (female, 8 weeks old) were purchased from Hunan SJA Laboratory Animal Co., Ltd. (China). All animal experiments were performed in compliance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and according to a protocol authorized by the Central South University Ethics Committee. The subcutaneous space of BALB/c mice was injected with 6 × 107 4T1 cells in 100 μL of PBS to construct a 4T1 tumor-bearing xenograft animal model. The tumor model was considered to be effectively established when the tumor volume reached 100 mm3 (volume = length × width2/2).2.12 In vivo UCL imaging
For in vivo UCL imaging, nine BALB/c mice bearing 4T1 tumor grafts were randomly assigned into 3 groups (n = 3 per group) and subsequently injected with 200 μl of PBS containing different nanoparticle preparations (UCNPs, R-UCAD, and HR-UCAD) at a concentration of 3 mg ml−1 UCNPs via the tail vein. At 6, 24, and 48 h after injection, in vivo UCL images were obtained using an ex/in vivo imaging system (IVIS Lumina XRMS Series III, PerkinElmer Inc.) (excitation/emission = 980/805 nm). The CW laser power density of the 980 nm laser used in the mice experiment was set as 600 mW cm−2 and the field of view was 12.5 cm in diameter. Images were acquired for 30 s and further analyzed with Living Image Software (PerkinElmer, USA). Next, the mice were euthanized to collect the major organs (heart, liver, spleen, lungs, and kidneys) and tumors to conduct ex vivo UCL signal analysis.2.13 HR-UCAD treatment of breast cancer-bearing mice
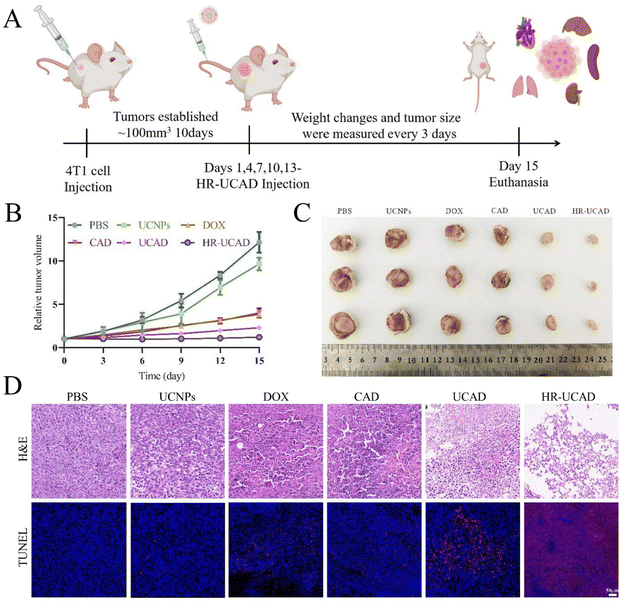
When the tumor volume reached approximately 100 mm3, the BALB/c mice were randomly divided into 6 groups, with 3 mice in each group. 100 μl of PBS, UCNPs (380 μg), DOX (200 μg), CAD (200 μg DOX and 230 nmol CpG), UCAD (200 μg DOX and 230 nmol CpG), and HR-UCAD (200 μg DOX and 230 nmol CpG) were administered to the mice via the tail vein once every three days for a total of five injections. Every 3 days, the length and width of each tumor were measured using an electronic Vernier caliper. After the tumor volume was calculated, a tumor volume–time curve was drawn. Each mouse was weighed once every three days, and a time curve showing mouse weight fluctuation was also plotted. All of the animals were sacrificed after 15 days of therapy, and whole blood was collected for hematology (blood routine instrument, BC-5390; Mindray, China) and blood biochemical index (7100 automatic biochemical analyzer, Hitachi, Japan) examination. The primary organs (tumor, heart, liver, spleen, lungs, and kidneys) were collected, fixed with 4% paraformaldehyde for 24 h, embedded in paraffin, and cut into sections for H&E staining to observe any histological abnormalities.2.14 Flow cytometry
Helper T cells (Th cells, CD4+) and cytotoxic T cells (Tc, CD8+) were analyzed in the tumors and spleens by flow cytometry. After euthanasia, the tumors and spleens of the mice were removed to extract single cells. The spleen was crushed using the plunger of the syringe and then the spleen fragments were repeatedly washed with RPMI 1640 medium in a 40-μm cell strainer to obtain a single splenocyte. Red blood cell lysis buffer was used to remove all of the red blood cells (RBCs) from a single splenocyte. A mouse lymphocyte isolation kit was used to obtain splenic lymphocytes. Surgical scissors were used to cut the tumors into small pieces, which were then digested for 90 min at 37 °C in RPMI 1640 medium supplemented with 2% FBS, hyaluronidase, DNase I, and collagenase IV. A mouse tumor-infiltrating lymphocyte isolation kit was used to remove the cancer cells and enrich the lymphocytes. The cells (3 × 104) were incubated with fluorescence-conjugated antibodies for 30 min at 4 °C in accordance with the manufacturer's instructions for flow cytometry sorting (FACS CantoTM II, BD, USA). A Zombie Aqua™ Fixable Viability Kit was used to exclude dead cells. Flow cytometry data were plotted and quantified with FlowJo software.2.15 Immunohistochemical analysis
The excised tumors were used for immunofluorescence staining (CRT, CD11c CD86, and Foxp3) and immunohistochemical analysis (IL-10 and IFN-γ). The apoptotic tumor cells were also evaluated with a TUNEL apoptosis assay. Briefly, paraffin-embedded tumor samples were used for detachment and antigen retrieval. Then, anti-CRT, anti-CD11c, anti-CD86, anti-Foxp3, and TDT in situ apoptosis kits were used for immunofluorescence staining, and the nuclei were stained with DAPI. Anti-IL-10 and anti-IFN-γ were used for immunohistochemical staining. Images of the tissue sections were observed under a fluorescence microscope.2.16 Statistical analysis
Statistical analysis was performed using SPSS 16.0, and the data are presented as the mean ± SD. Differences between groups were assessed using one-way ANOVA, followed by Tukey's post hoc test (*p < 0.05, **p < 0.01, and ***p < 0.001).3 Results and discussion
3.1 Preparation and characterization of HR-UCAD
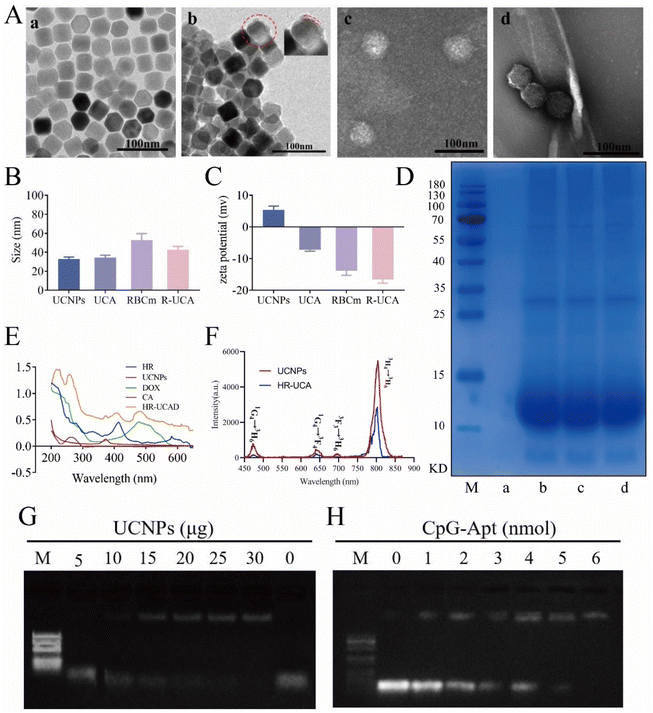
Upconversion nanoparticles, an emerging kind of nano-luminescent material, are composed of inorganic nanocrystals doped with rare earth ions. Intense upconversion luminescence was successfully acquired by developing a hydrophobic core–shell nanocomposite with NaYF4:Yb/Tm as the core and NaYF4 as the shell by the thermal decomposition method. We used ICP to quantify the mass percentages of Yb and Tm in UCNPs, which were about 20% and 1%, respectively. Yb was the sensitizer and Tm was the activator. The excitation wavelength was 975 nm and the emission wavelength was 804 nm. The hydrophobic core–shell upconversion nanoparticles were homogeneous in size and displayed polygonal structures with a size of 32.7 ± 2.2 nm (Fig. 2Aa and Fig. S2D†). The HRTEM image of UCNPs in Fig. S2A† shows that the lattice distance was 0.52 nm, corresponding to the typical plane of the hexagonal NaYF4 structure. The shell thickness of the UCNPs was about 1.9 nm (Fig. S2B†). Fig. S2C† illustrates the result of XRD examination to determine the crystal structure of the prepared UCNPs. All of the diffraction peaks associated with the pure hexagonal phase identified in β-NaYF4 (JCPDS No. 064-0156) could be observed, according to the XRD results of UCNPs. XPS analysis was performed to confirm the chemical composition of the prepared UCNPs. In the XPS spectrum (Fig. S2C†), the strong peaks located at 1071.7, 158.8, 684.9, 184.3, 182.2, 286.6, and 529.7 eV were attributed to the binding energies of Na 1s, Y 3d, F 1s, Yb 4d, Tm 4d, C 1s, and O 1s, respectively.As illustrated in Fig. 1A, the preparation of HR-UCAD mainly consisted of four steps: (1) synthesis of A30-UCNPs using a 30-mer polyA oligonucleotide; (2) conjugation of CpG-Apt-24T to the A30-UCNPs; (3) DOX loading into UCA to obtain UCAD; and (4) wrapping UCAD with HA-RBCm vesicles via co-extrusion. The TEM images of UCA illustrated that they were evenly distributed in water without changing the form, indicating that these DNA-functionalized UCNPs are hydrophilic (Fig. 2Ab). As previously reported,46 the UCNPs were surrounded by a uniform DNA layer (2.99 ± 0.3 nm). After negative staining with 1% phosphotungstic acid, the RBCms displayed an oval shape with a diameter of 52.8 ± 6.4 nm (Fig. 2Ac), and R-UCA presented a typical core–shell structure with a size of 42.4 ± 3.6 nm (Fig. 2Ad). The sizes of UCNPs, UCA, RBCm and R-UCA were 32.7 ± 2.2 nm, 34.3 ± 2.4 nm, 52.8 ± 6.4 nm and 42.3 ± 3.6 nm, respectively (Fig. 2B). The thickness of the RBCm on HR-UCADs was approximately 10 nm, which was approximately equal to the thickness of one natural RBCm (7.8 nm).47 The zeta potential changed from 5.35 ± 1.03 mV (UCNPs) to approximately −7.23 ± 0.36 mV (UCA) because the nucleic acid aptamer has a negative charge (Fig. 2C). The zeta potential of R-UCA was −16.67 ± 0.88 mV, similar to the zeta potential of the RBCm (−13.83 ± 1.20 mV). This change in the charge might be a result of charge shielding caused by RBCm encapsulation, indicating the successful encapsulation of R-UCA. The SDS-PAGE results (Fig. 2D) demonstrated that the protein components in the RBCms were well preserved in HR-UCAD. As shown in the UV−vis spectrum (Fig. 2E), HR-UCAD had absorption peaks at approximately 413 nm, 373 nm, 480 nm, and 260 nm, which were consistent with the peaks of RBCms, UCNPs, DOX, and nucleic acids, respectively. These data further confirmed the successful construction of HR-UCAD. After 980 nm NIR irradiation (Fig. 2F), the corresponding UCL spectrum of HR-UCA in water resembled that of UCNPs in cyclohexane, indicating that the upconversion optical characteristics were conserved. As shown in Fig. 2F, the emission peaks at 475, 644, 694 and 804 nm of the prepared UCNP and HR-UCA can be indexed to the 1G4 → 3H6, 1G4 → 3F4, 3F3 → 3H6 and 3H4 → 3H6 transitions of Tm3+, respectively. The optimal ratio of A30 and CpG-Apt to UCNP attachment was quantified by gel electrophoresis. As shown in Fig. 2G, the band of free 30A DNA (10 nmol) was completely retarded when anchored to 15 μg UCNPs, indicating that A30-mer polyA oligonucleotides were totally decorated onto the surface of the UCNPs according to a previous strategy. Similarly, 6 nmol of CpG-Apt-24T was able to completely hybridize with 30A-UCNPs from the previous step without the appearance of a free band (Fig. 2H). The electrophoretic mobility shift assay results demonstrated that the optimal amounts of reagents needed to construct UCNPs@CpG-Apt were 10 nmol of A30-mer polyA oligonucleotides, 15 μg of UCNPs, and 6 nmol of Apt-CpG. Fig. S3† shows the results of the amount of proteins on HR-UCAD. There was a good correlation between the bionanoparticle surface membrane protein content and the HR-UCAD mass ratio when the ratio was below 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. When this ratio exceeded 5
1. When this ratio exceeded 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the erythrocyte membranes on the bionanoparticle surface were gradually saturated. Thus, the optimal ratio for erythrocyte RBC membranes to completely encapsulate UCAD is 5
1, the erythrocyte membranes on the bionanoparticle surface were gradually saturated. Thus, the optimal ratio for erythrocyte RBC membranes to completely encapsulate UCAD is 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
3.2 Drug loading efficiency and the release rate
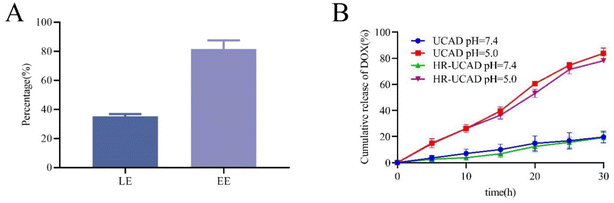
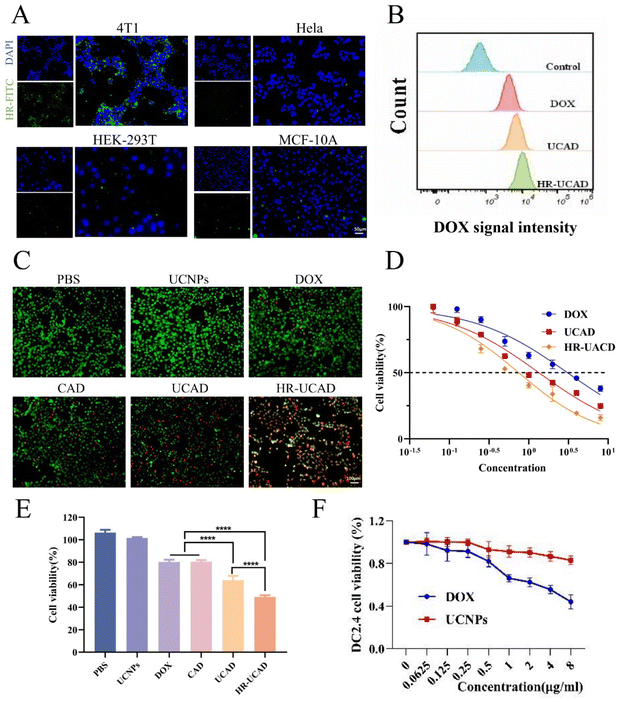
In this system, CpG-Apt modified the surface of the UCNPs well according to phosphate group coordination and base pairing rules, and DOX was inserted into CpG-Apt via noncovalent interactions; hence, UCNPs might be an ideal carrier. The LE and EE of DOX in the UCAD nanocomplexes were 35.2 ± 1.4% and 81.5 ± 4.9%, respectively (Fig. 3A). Thus, the concentration of DOX encapsulated in UCAD was 8.148 μg of DOX per 15 μg of UCNPs. To explore the release characteristics of DOX in an acidic tumor microenvironment (TME), we investigated the release behavior of DOX in PBS (pH 7.4 and 5.0) at 37 °C (Fig. 3B). When the pH was 7.4, DOX was released from HR-UCAD and UCAD at rates of 19.3 ± 3.1% and 19.6.0 ± 3.7%, respectively, whereas at pH 5.0, these rates were 78.1 ± 0.9% and 83.8 ± 3.4%, respectively. In conclusion, DOX may be more readily released from this system at pH 5.0 than at pH 7.4, indicating that the acidic milieu of tumor tissues (pH 5.0) may stimulate the release of DOX. An intracellular localization experiment of the nanoparticles was carried out to better understand the process of HR-UCAD uptake and release. As shown in Fig. S5,† the characteristic red fluorescence of DOX in tumor cells began to increase at 4 h of incubation. Subsequently, the fluorescence intensity did not change significantly, indicating that the nanocomplexes were able to deliver most of the DOX into the cells at around 5 h. The cellular uptake of nanodelivery systems by 4T1 cells was quantified using an inductively coupled plasma (ICP) technique. As shown in Fig. S4,† the uptake percentages of UCNPs, R@UCNPs and HR@UCNPs in 4T1 cells were 22%, 29% and 65%, respectively. The uptake percentages of UCNPs, R@UCNPs and HR@UCNPs in tumor tissues were 18%, 34% and 79%, respectively. HR@UCNPs have the strongest tumor targeting ability because HA can bind to the CD44 molecules of tumor cells.3.3 Biocompatibility and immune escape of HR-UCAD
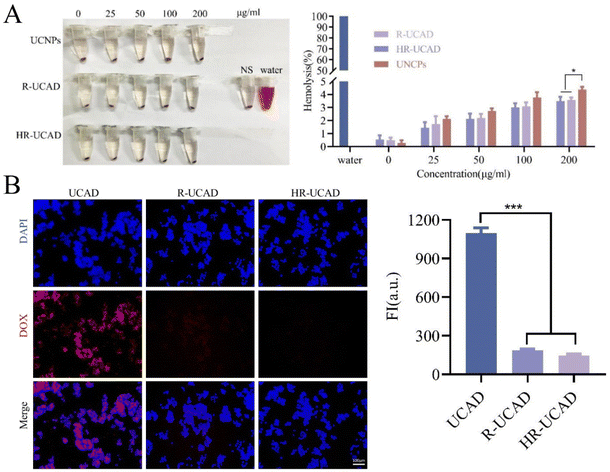
First, a hemolysis test was performed to evaluate the blood compatibility of HR-UCAD. Different concentrations of UCNPs, R-UCAD, and HR-UCAD (0, 25, 50, 100, and 200 μg ml−1) were incubated with 5% RBCs for 4 h. As indicated in Fig. 4A, the rates of hemolysis produced by various concentrations of UCNPs, R-UCAD, and HR-UCAD were all less than 5%, suggesting that HR-UCAD has good blood compatibility and is safe for intravenous delivery. Second, the phagocytosis effect was examined to investigate the immunological escape ability of HR-UCAD. As shown in Fig. 4B, the red DOX signal in the UCAD group was significantly higher than that in the R-UCAD and HR-UCAD groups because the camouflage provided by the erythrocyte membrane inhibited phagocytosis by macrophages.48 These findings supported the notion that the camouflaging RBCm coating on these nanoparticles can reduce self-recognition and clearance by the RES and prolong their half-life in circulation in vivo.423.4 In vitro cytotoxicity of HR@UCNPs/CpG-Apt/DOX
CD44+CD24− cells with stem cell-like features are the most common cell type in breast cancer,49 which is associated with tumor recurrence, metastasis and drug resistance.50 The CD44 antigen is a multifunctional transmembrane glycoprotein associated with cell adhesion, activation, migration and differentiation and is considered a promising tumor marker.51 CD44 is a specific marker of breast cancer cells to which HA can specifically bind.52 To validate the ability of HA-modified RBC vesicles to target breast cancer cells, HA-RBCms were labeled with DSPE-PEG2000-FITC (green fluorescence). As shown in Fig. 5A, compared with HeLa, HEK-293T, and MCF-10A cells, the green fluorescence around the 4T1 cells was the most notable, indicating that the RBC vesicles modified with HA significantly adhered to 4T1 cells and had an active targeting ability.Prior to investigating the toxicity of HR-UCAD to tumor cells, it was necessary to evaluate its uptake by tumor cells. The fluorescence intensity of DOX was measured by flow cytometry analysis. Fig. 5B demonstrates that the tumor cells in the HR-UCAD treatment group exhibited the best uptake of DOX, illustrating that UCA is a powerful nanocarrier that may improve DOX uptake.
Next, the toxicity of HR-UCAD was investigated in 4T1 cells by using a CCK-8 assay. From the results in Fig. 5E, HR-UCAD showed the most intense and highest toxicity to 4T1 cells. The half-maximal inhibitory concentration (IC50) of HR-UCAD was 0.7484 μg ml−1, which was significantly lower than that of UCAD (1.343 μg ml−1) and DOX (3.076 μg ml−1) (Fig. 5D). The live/dead staining images were consistent with the CCK-8 results (Fig. 5C). In addition, UCNPs were not only minimally toxic to tumor cells but also almost nontoxic to DCs. DCs were relatively unharmed when the dose of DOX was below 0.5 μg ml−1 (Fig. 5F).
3.5 HR-UCAD induces ICD in 4T1 cells in vitro
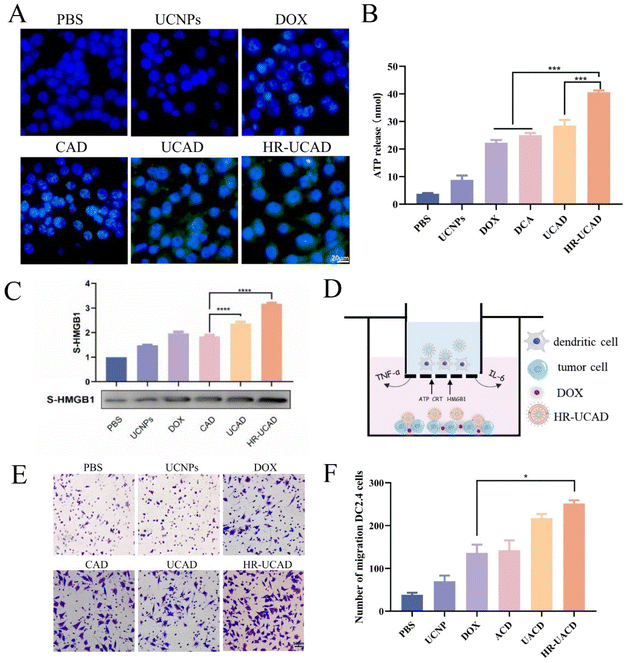
To verify the effect of ICD caused by HR-UCAD, we detected the ICD markers: CRT exposure on the cell surface, HMGB-1 release in the cell medium, and ATP secretion in the cell medium.53 The cells treated with HR-UCAD and UCAD, but not PBS or UCNPs, showed considerable CRT expression on the cell surface according to the immunofluorescence data (Fig. 6A). Moreover, the results in Fig. 6B and C illustrate that the amounts of ATP and S-HMGB1 secreted by HR-UCAD-treated cells were the highest compared to the cells treated with DOX and UCAD. Our findings revealed that treating 4T1 cells with HR-UCAD produced a greater ICD effect than free DOX, owing to the enhanced cellular uptake by the nanodelivery system and the good tumor cell targeting ability provided by HA. ICD is essential for the development of antitumor immunity. During the progression of ICD, dying cancer cells are able to release DAMP-based signals and chemical mediators, which represent the connection between cell death induction and the immune response.54 These released factors efficiently recruit and activate DCs, which can engulf fractions of cancer cells and deliver antigens to naïve T lymphocytes for prompting differentiation in CD8+cytotoxic and CD4+ helper T cells.55Finally, a transwell system44 mimicking the in vivo TME was constructed to evaluate the migration rate of immune cells recruited by the nanodelivery system (Fig. 6D). 4T1 cells in the bottom wells were first incubated for 12 hours with various formulations. Following that, DC2.4 cells seeded in the upper wells were cocultured with bottom 4T1 cells for another 12 hours. The results of the migration assay shown in Fig. 6E indicate that HR-UCAD treatment promoted the most significant migration of DC2.4 cells. The migration rate of the cells in the HR-UCAD group was 1.8 times higher than that in the DOX group. This was the result of ICD-evoked DAMPs recruiting APCs to reach the TME.56 Taken together, these findings implied that HR-UCAD caused increased ICD to initiate a robust immune response compared to DOX treatment.
3.6 HR-UCAD-mediated immune activation in vitro
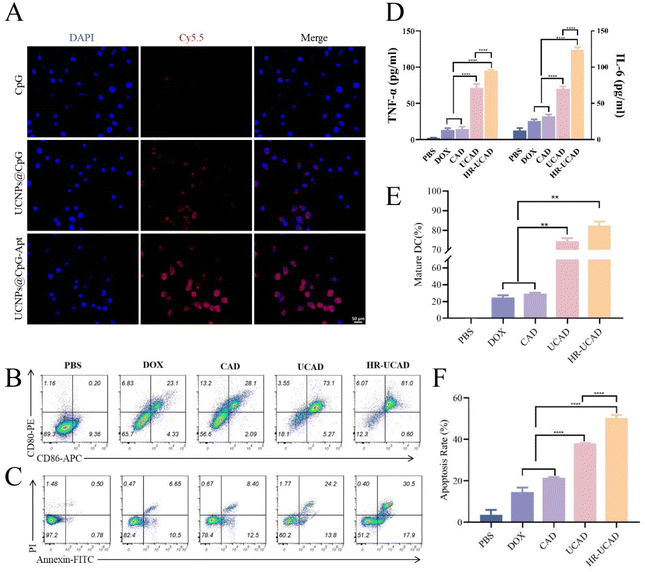
Following the DOX chemotherapy-triggered ICD of tumor cells, the produced tumor pieces can act as tumor-associated antigens, which, with the aid of CpG ODNs as immunoadjuvants, induce potent antitumor immune responses.57 In our strategy, the UCNPs protected the CpG ODNs from nuclease degradation, as they were capable of crossing the cell membrane without the need for transfection agents, and the DNA aptamer-conjugated UCNPs readily targeted DCs. First, the cellular uptake efficiency of UCA was assessed by fluorescence microscopy. A fluorophore (Cy5) was added to the 5′ end of the CpG ODNs. As illustrated in Fig. 7A, no visible fluorescence was seen in cells incubated with free CpG ODNs. In cells treated with UCA, the strongest fluorescence signal was observed throughout the cytoplasm. As a result, the combination of UCNPs and an aptamer significantly increased the efficiency of CpG ODN cellular uptake. The DC-targeted aptamer was produced by Moghadam et al. via cell-SELEX58 and has been successfully applied in the field of nanomedicine.27,28To further confirm whether HR-UCAD could promote DCs maturation and activation, which are crucial for maintaining the T-cell-mediated immune response, we cocultured HR-UCAD-treated 4T1 cells with DC2.4 cells (Fig. 6D). After culture with various formulations, the expression levels of the costimulatory molecules CD86 and CD80 were examined using flow cytometry to assess the degree of DCs maturation. As shown in Fig. 7B, HR-UCAD treatment significantly increased the maturity of DCs (81%) compared with the control group (0.2%). CAD treatment caused only 28.1% DCs activation, which was consistent with the fact that bare CpG has difficulty crossing the cytoplasmic membrane.59 DOX treatment alone also had little effect on activating DCs (23%), suggesting that ICD failed to induce a sufficient antitumor immune response.60 Furthermore, the production of various cytokines, including TNF-α and IL-6, was assessed in the coculture system. Both TNF-α and IL-6 are DCs activation-related cytokines that are essential indicators of cellular and humoral immune system activation.61–63 The ELISA results showed that the secretion of TNF-α and IL-6 was significantly enhanced in the HR-UCAD group, which was consistent with the observed DCs maturation (Fig. 7D). Together, these results demonstrated that HR-UCAD, with excellent adjuvant performance, has the potential to induce a powerful antitumor immune response. Moreover, tumor cells in the coculture system were also collected to evaluate apoptosis induction by HR-UCAD via annexin V-FITC/PI double staining (Fig. 7C). Both DOX and CpG-Apt from HR-UCAD were able to remarkably trigger the apoptosis of 4T1 cells with a total apoptotic ratio of 48.5%, which may be a result of the combined effects of HA-RBCm targeting to increase DOX accumulation and cytokine induction. Collectively, these results suggested that HR-UCAD has outstanding antitumor efficacy and could be a prospective chemoimmunotherapeutic drug.
3.7 Biodistribution
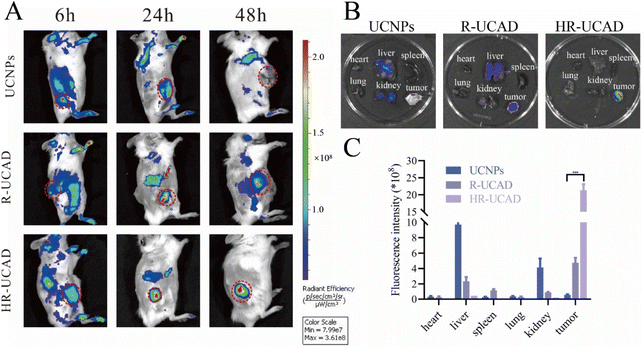
UCNPs possess superior optical properties for visualizing tumors through UCL.64 The optical features of UCNPs in conjunction with surface modification chemistry to link adjuvants and chemotherapy drugs for immunotherapy may offer a promising and practical technique.65,66 To detect the characteristic in vivo, the tumor accumulations of different formulations were investigated and monitored in 4T1 tumor-bearing BALB/c mice using a noninvasive NIR optical imaging system (IVIS Lumina XRMS Series III, PerkinElmer Inc, equipped with fluorescence filter sets, excitation = 980 nm, 600 mW cm−2, emission = 805 nm). As depicted in Fig. 8A, the HR-UCAD group displayed the strongest UCL signal at the tumor site due to CD44 targeting of the HA-RBCm,36 indicating the ability of the HA-RBCm to target 4T1 tumors. The R-UCAD and UCNPs could have a small amount of accumulation at tumor tissues through enhanced permeability and retention effects.67 The fluorescence signals in the R-UCAD and UCNPs groups were weaker than that in the HR-UCAD group. Forty-eight hours after injection, the mice were euthanized, and the organs were harvested for ex vivo imaging. The results in Fig. 8B show that the signal in the liver was significantly stronger in the UCNPs group than in the R-UCAD group, suggesting that the RBCm coating could indeed reduce RES uptake. Thus, the quick accumulation and long-term retention of HR-UCAD at the tumor site was evidence of its good active targeting.3.8 In vivo antitumor efficacy
On the basis of the outstanding efficacy of the biodistribution and accumulation of HR-UCAD verified above, the antitumor effects of HR-UCAD were examined in 4T1 tumor-bearing mice. Fig. 9A shows the design of the in vivo experiment. The BALB/c mice bearing 4T1 breast tumor were randomly divided into six groups (n = 3) and treated with PBS, UCNPs, DOX (2 mg ml−1), DOX + CpG (2 mg ml−1 DOX and 8 μM CpG), UCAD (2 mg ml−1 DOX and 8 μM CpG), and HR-UCAD (2 mg ml−1 DOX and 8 μM CpG) every 3 days during 15 days of treatment. As shown in Fig. 9B, after 15 days of treatment with CAD (DOX mixed with free CpG-Apt), the tumor growth suppression effect was similar to that of DOX treatment alone, which may be because CpG-Apt was cleared by the systemic circulation. UCAD further inhibited the development of tumors compared to CAD, confirming that DOX combined with UCA could augment the antitumor effect. Notably, HR-UCAD treatment most significantly inhibited the growth of tumor tissue. It was therefore concluded that the DNA-functionalized UCNP nanostructure provided systemic drug biostability and site-specific drug release for precise immunochemotherapy. Furthermore, the camouflage provided by the HA-RBCm showed a natural tumor targeting ability and played a protective role. Moreover, to identify tumor tissue necrosis and apoptosis, H&E and TUNEL stains were applied. The HR-UCAD group had more necrotic cells in the tumor tissues than those in the other groups according to H&E staining (Fig. 9D). TUNEL analysis demonstrated that the proportion of apoptotic cells (red fluorescence) caused by HR-UCAD treatment was significantly higher than that in the other groups (Fig. 9D). The TUNEL results were consistent with the apoptosis induction effects produced by HR-UCAD in vitro.3.9 HR-UCAD enhances cancer immunotherapy in vivo
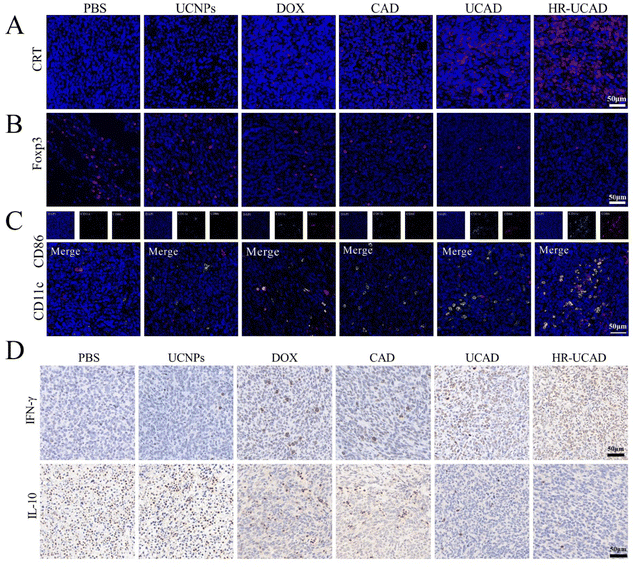
Given the potent effects of HR-UCAD on DAMP exposure in vitro, we assessed whether different formulations may trigger ICD in the tumor tissues of 4T1 tumor-bearing mice. As shown in Fig. 10A, CRT exposure was significantly increased by HR-UCAD administration compared with UCAD and free DOX treatment. The results suggested that ICD is markedly enhanced because of the targeted delivery of DOX by HR-UCAD. Consistent with previous reports, loading ICD inducers in a nanodelivery system can enhance ICD by releasing more antigens and DAMPs than free chemotherapeutic agents, which can enhance antigenicity and adjuvanticity in situ, resulting in strong innate and adaptive immune responses.68To assess whether the CpG-based multifunctional nanoplatform might activate a tumor-specific immune response in vivo, we studied the proportions of tumor-infiltrating CD86+ DCs and Foxp3+ regulatory T cells (Tregs) in tumor tissues by immunofluorescence staining (Fig. 10B). CD86 is regarded as a DC maturation marker. Mature DCs play a pivotal role in engulfing and presenting antigens from dying tumor cells to T lymphocytes, thus promoting intratumoral infiltration of CD8+ CTLs.69 Immunosuppressive Tregs are often enriched in the tumor to protect tumor cells from attack by the immune system.70 As shown in Fig. 10C, HR-UCAD treatment induced significantly more DCs maturity (CD86+ in CD11c+ DCs) in the TME than the other treatments, while the infiltration of Tregs was remarkably decreased. These results indicated that our constructed nanosystem could improve DCs maturation and alleviate the suppression of the TME to some extent.
Interferon-γ (IFN-γ) is secreted by immune cells and has immunomodulatory and antitumor properties.71 In contrast, IL-10, as an immunosuppressive cytokine, promotes tumor immune escape by diminishing antitumor immune responses in the TME.72 To deeply evaluate the antitumor immunity evoked by HR-UCAD, immunohistochemical staining was used to detect the production of IFN-γ and IL-10 in tumors. There were more IFN-γ-positive cells (brownish-yellow) in the HR-UCAD group than in the other groups, whereas relatively low IL-10 secretion was observed (Fig. 10D). These results suggested that the HR-UCAD nanocomposite is an effective immunomodulatory agent.
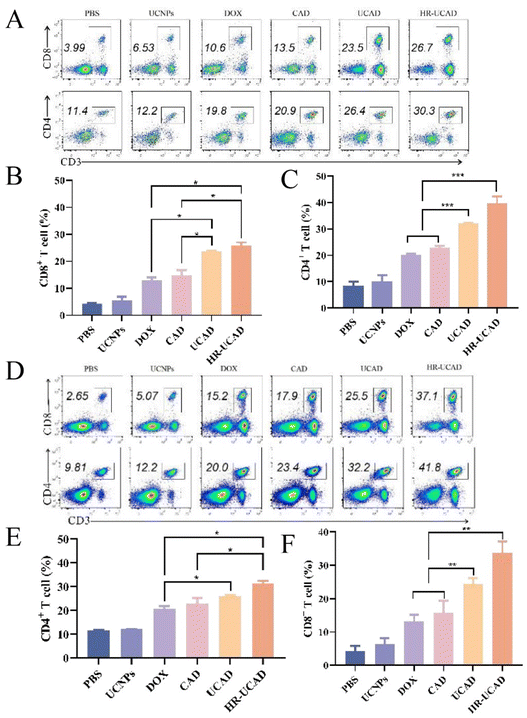
Next, the levels of CD4+ and CD8+ T cells in isolated tumor tissues from each group were further evaluated via flow cytometry. CTLs (CD8+ T cells) can directly attack tumor cells by secreting cytotoxins such as perforin and granulysin, while Th cells (CD4+ T cells) are of vital importance in the process of immunoregulation.73,74 The results of flow cytometry in Fig. 11A reveal that the proportions of both CD4+ T cells and CD8+ T cells (gated on CD3+ T cells) recruited to tumors dramatically increased after treatment with HR-UCAD compared with the other treatments. Furthermore, we explored the systemic immune effects triggered by HR-UCAD and examined the quantity and phenotype of T lymphocytes in the spleen, the body's largest immune organ (Fig. 11D). Notably, the percentages of CD8+ T cells and CD4+ T cells in the spleens clearly increased after HR-UCAD administration. In conclusion, our data strongly demonstrated that HR-UCAD enhanced tumor-specific ICD and remodeled the TME, promoting powerful immune responses.
3.10 Evaluation of the safety of HR-UCAD in vivo
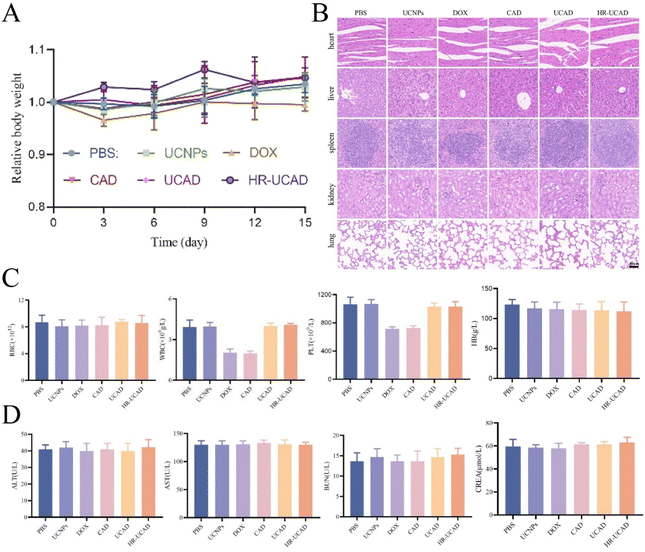
There is always considerable concern regarding the potential in vivo toxicity of nanomaterials, particularly given that UCNPs are nonbiodegradable.75 Moreover, this issue appears particularly vital for nanoparticles that are camouflaged by RBCms, as blood group mismatches may sometimes lead to transfusion reactions or even death.76 Fluctuation in the body weight is a sensitive indicator of the in vivo toxicity of nanomaterials. During the treatment period, no apparent changes in the mouse body weight were observed (Fig. 12A), indicating the low or modest toxicity and minimal side effects of the nanodelivery system. On the 15th day after injection, all mice were euthanized, and their blood and main organs (heart, liver, spleen, lungs, and kidneys) were collected for blood tests and histological analysis. Due to the myelosuppressive effects of DOX, white blood cell (WBC) and platelet (PLT) counts were notably decreased in the DOX and CAD groups. The blood biochemistry indicators ALT, AST, ALP, BUN, and CREA and hematological indices (RBCs, hemoglobin (Hb), WBCs, and PLTs) did not vary significantly among the treatment and PBS groups (Fig. 12C and D). Furthermore, H&E staining of the heart, liver, spleen, lungs, and kidneys showed that the nanodrugs did not change the histological structures of these major organs (Fig. 12B). All of the above results revealed the low in vivo toxicity and good biocompatibility of HR-UCAD.4 Conclusion
In conclusion, we successfully developed an erythrocyte membrane-cloaked upconversion nanocomplex (HR-UCAD) as a multifunctional nanoplatform to generate a robust antitumor immune response for combating breast cancer. This nanoplatform is composed of erythrocyte membrane-camouflaged UCNPs loaded with DOX for ICD induction and the immunoadjuvant CpG-Apt for TLR9 stimulation. HR-UCAD exhibited many outstanding abilities, such as immune escape, specific drug delivery, and response to a weakly acidic environment. Given the remarkable NIR fluorescence emission performance of UCNPs, HR-UCAD was employed for highly specific in vivo tumor imaging. HR-UCAD can effectively deliver DOX to tumor cells and enhance their immunogenicity. Synergistically, the released UCA provided the possibility of activating endosomal TLR9 in infiltrating APCs. Moreover, our findings demonstrated that HR-UCAD improved the activities of CD8+ T cells and abrogated the immunosuppressive activity of Tregs. As a result, we believe that this nanocomplex will not only be used as a promising tool for the treatment of breast cancer but also pave the way for applying synergistic chemoimmunotherapy to boost cancer immunotherapy against other tumor types.Abbreviations
| UCNPs | Upconversion nanoparticles |
| UCA | UCNPs@CpG-Apt |
| CAD | CpG-Apt + DOX |
| UCAD | UCNPs@CpG-Apt/DOX |
| R-UCAD | R@UCNPs/CpG-Apt/DOX |
| HR-UCAD | HR@UCNPs/CpG-Apt/DOX |
| ICD | Immunogenic cell death |
| DC | Dendritic cell |
| HA | Hyaluronic acid |
| RBCm | Red blood cell membrane |
| DOX | Doxorubicin |
| CTL | Cytotoxic T lymphocyte |
| DAMPs | Damage-associated molecular patterns |
| CRT | Calreticulin |
| HMGB1 | High mobility group box 1 |
| APCs | Antigen-presenting cells |
| CpG ODNs | Cytosine-phosphate-guanosine oligonucleotides |
| IL-6 | Interleukin 6 |
| TNF-α | Tumor necrosis factor-α |
| NIR | Near-infrared |
| RES | Reticuloendothelial system |
| EE | Encapsulation efficiency |
| LE | Drug loading efficiency |
| SDS-PAGE | Sulfate-polyacrylamide gel electrophoresis |
| UCL | Upconversion luminescence |
| XRD | X-ray diffraction |
| XPS | X-ray photoelectron spectroscopy |
| TME | Tumor microenvironment |
| IC50 | Half-maximal inhibitory concentration |
| WBC | White blood cell |
| PLT | Platelet |
| ALT | Aspartate aminotransferase |
| AST | Alanine aminotransferase |
| ALP | Alkaline phosphatase |
| BUN | Blood urea nitrogen |
| CREA | Creatinine |
| Hb | Hemoglobin |
Ethical statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Central South University and approved by the Animal Ethics Committee of Central South University.Author contributions
XN and JL conceived and designed the experiments. QK carried out the experiments. RH, HJ, XL, YH, LL and YS contributed to analyzing the experimental results. QK wrote the manuscript. All authors read and approved the final manuscript.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81971748 and 82001165), the Hunan Provincial Natural Science Foundation of China (Grant No. 2020JJ4131, 2021JJ40926, and 2021JJ40923), the Key Research and Development Program of Hunan Province (Grant No. 2022SK2004), the Graduate Self-Exploration and Innovation Project of the Central South University of China (Grant no. 2021zzts1084 and 2022ZZTS0890), the Wisdom Accumulation and Talent Cultivation Project of the Third Xiangya Hospital of Central South University (Grant No. YX202206) and the Science and Technology Program of Changsha (Grant No. kh2201057).References
- K. L. Britt, J. Cuzick and K. A. Phillips, Nat. Rev. Cancer, 2020, 20, 417–436 CrossRef CAS PubMed.
- Y. Liang, H. Zhang, X. Song and Q. Yang, Semin. Cancer Biol., 2020, 60, 14–27 CrossRef CAS PubMed.
- R. Ma, Z. Li, E. A. Chiocca, M. A. Caligiuri and J. Yu, Trends Cancer, 2022, 9, 122–139 CrossRef PubMed.
- Y. Hu, J. Feng, T. Gu, L. Wang, Y. Wang, L. Zhou, R. Hong, E. Tan Su Yin, M. Zhang, P. Lu and H. Huang, Lancet Haematol., 2022, 9, e930–e941 CrossRef CAS PubMed.
- L. Qin, H. Zhang, Y. Zhou, C. S. Umeshappa and H. Gao, Small, 2021, 17, e2006000 CrossRef PubMed.
- S. Ma, X. Liang, N. Yang, J. Yang, J. Zhang, X. Pan, Y. Wei, Z. Liu and Q. Shen, Biomater. Sci., 2022, 11, 518–532 RSC.
- Y. Chen, L. Zeng, H. Zhu, Q. Wu, R. Liu, Q. Liang, B. Chen, H. Dai, K. Tang, C. Liao, Y. Huang, X. Yan, K. Fan, J. Z. Du, R. Lin and J. Wang, Small Methods, 2022, e2201086 CrossRef PubMed.
- Q. Yang, T. Liu, H. Zheng, Z. Zhou, Y. Huang, H. Jia, S. Fu, X. Zhang, H. Zhang, Y. Liu, X. Chen and W. Shan, Biomaterials, 2023, 292, 121936 CrossRef CAS PubMed.
- H. Wu, G. Wei, L. Luo, L. Li, Y. Gao, X. Tan, S. Wang, H. Chang, Y. Liu, Y. Wei, J. Song, Z. Zhang and J. Huo, Biomater Res., 2022, 26, 77 CrossRef CAS PubMed.
- Y. Yang, J. Huang, M. Liu, Y. Qiu, Q. Chen, T. Zhao, Z. Xiao, Y. Yang, Y. Jiang, Q. Huang and K. Ai, Adv Sci., 2022, 10, e2204365 CrossRef PubMed.
- J. Wan, X. Zhang, Z. Li, F. Mo, D. Tang, H. Xiao, J. Wang, G. Rong and T. Liu, Adv. Healthc. Mater., 2022, e2202710, DOI:10.1002/adhm.202202710.
- F. Mpekris, M. Panagi, C. Michael, C. Voutouri, M. Tsuchiya, C. Wagatsuma, H. Kinoh, A. Osada, S. Akinaga, S. Yoshida, J. D. Martin and T. Stylianopoulos, J. Controlled Release, 2023, 353, 956–964 CrossRef CAS PubMed.
- J. Hu, M. Liang, M. Ye, J. Xu, H. Liu, X. Zhang, W. Sun, P. Xue, Y. Kang and Z. Xu, Carbohydr. Polym., 2023, 301, 120365 CrossRef CAS PubMed.
- Z. Zhao, S. Dong, Y. Liu, J. Wang, L. Ba, C. Zhang, X. Cao, C. Wu and P. Yang, ACS Nano, 2022, 16, 20400–20418 CrossRef CAS PubMed.
- L. Li, M. Zhang, T. Liu, J. Li, S. Sun, J. Chen, Z. Liu, Z. Zhang and L. Zhang, Acta Biomater., 2022, 154, 454–466 CrossRef CAS PubMed.
- S. Takano, Y. Miyashima, S. Fujii and K. Sakurai, Biomacromolecules, 2023, 24, 1299–1309 CrossRef CAS PubMed.
- D. Huang, T. Wu, S. Lan, C. Liu, Z. Guo and W. Zhang, Biomaterials, 2022, 289, 121808 CrossRef CAS PubMed.
- Z. Fan, Y. Wang, L. Li, F. Zeng, Q. Shang, Y. Liao, C. Liang and L. Nie, ACS Nano, 2022, 16, 16177–16190 CrossRef CAS PubMed.
- Y. Jia, K. Shi, L. Dai, X. He, H. Deng, R. Han, F. Yang, B. Chu, J. Liao, X. Wei and Z. Qian, Small Methods, 2022, e2201087 CrossRef PubMed.
- Y. Zhang, H. Xu, L. Jiang, Z. Liu, C. Lian, X. Ding, C. Wan, N. Liu, Y. Wang, Z. Yu, L. Zhu, F. Yin and Z. Li, ACS Nano, 2022, 16, 19509–19522 CrossRef CAS PubMed.
- S. Bi, Z. Deng, J. Huang, X. Wen and S. Zeng, Adv. Mater., 2022, 35, e2207038 CrossRef PubMed.
- C. Yao, J. Ou, J. Tang and D. Yang, Acc. Chem. Res., 2022, 55, 2043–2054 CrossRef CAS PubMed.
- D. Zhang, R. Peng, W. Liu, M. J. Donovan, L. Wang, I. Ismail, J. Li, J. Li, F. Qu and W. Tan, ACS Nano, 2021, 15, 17257–17274 CrossRef CAS PubMed.
- W. Ren, S. Wen, S. A. Tawfik, Q. P. Su, G. Lin, L. A. Ju, M. J. Ford, H. Ghodke, A. M. van Oijen and D. Jin, Chem. Sci., 2018, 9, 4352–4358 RSC.
- P. K. Chintamaneni, D. Nagasen, K. C. Babu, A. Mourya, J. Madan, D. A. Srinivasarao, R. K. Ramachandra, P. M. Santhoshi and S. Pindiprolu, J. Controlled Release, 2022, 352, 652–672 CrossRef CAS PubMed.
- D. Zhang, R. Peng, W. Liu, M. J. Donovan, L. Wang, I. Ismail, J. Li, J. Li, F. Qu and W. Tan, ACS Nano, 2021, 15, 17257–17274 CrossRef CAS PubMed.
- S. K. Shahbaz, A. R. Varasteh, K. Koushki, S. H. Ayati, K. Mashayekhi, M. Sadeghi, M. Moghadam and M. Sankian, Int. Immunopharmacol., 2020, 85, 106603 CrossRef PubMed.
- M. Sadeghi, K. Koushki, K. Mashayekhi, S. H. Ayati, S. K. Shahbaz, M. Moghadam and M. Sankian, Int. Immunopharmacol., 2020, 86, 106690 CrossRef CAS PubMed.
- D. Gu, Y. Qiao, H. Fu, H. Zhao, X. Yue, S. Wang, Y. Yin, R. Xi, X. Fu, X. Zhao and M. Meng, ACS Appl. Mater. Interfaces, 2022, 14, 38048–38055 CrossRef CAS PubMed.
- J. Yin, P. Wang, Y. Yin, Y. Hou and X. Song, Drug Delivery, 2017, 24, 1891–1897 CrossRef CAS PubMed.
- M. Zhang, W. Wang, F. Wu, K. Graveran, J. Zhang and C. Wu, Chemistry, 2018, 24, 12890–12901 CrossRef CAS PubMed.
- M. Imran, V. Gowd, P. Saha, S. Rashid, A. A. Chaudhary, M. Y. A. Mohamed, A. S. Alawam and R. Khan, Int. J. Pharm., 2023, 631, 122407 CrossRef CAS PubMed.
- N. Khosravi, E. Pishavar, B. Baradaran, F. Oroojalian and A. Mokhtarzadeh, J. Controlled Release, 2022, 348, 706–722 CrossRef CAS PubMed.
- A. Naskar, H. Cho, S. Lee and K. S. Kim, Pharmaceutics, 2021, 13, 1887 CrossRef CAS PubMed.
- M. Song, S. Dong, X. An, W. Zhang, N. Shen, Y. Li, C. Guo, C. Liu, X. Li and S. Chen, J. Controlled Release, 2022, 345, 744–754 CrossRef CAS PubMed.
- X. Kang, F. Bu, W. Feng, F. Liu, X. Yang, H. Li, Y. Yu, G. Li, H. Xiao and X. Wang, Adv. Mater., 2022, 34, e2206765 CrossRef PubMed.
- Q. Ma, S. Wu, L. Yang, Y. Wei, C. He, W. Wang, Y. Zhao, Z. Wang, S. Yang, D. Shi, Y. Liu, Z. Zhou, J. Sun and Y. Zhou, Adv Sci., 2022, 10, e2202416 CrossRef PubMed.
- A. M. Carvalho, R. L. Reis and I. Pashkuleva, Adv. Healthc. Mater., 2022, 12, e2202118 CrossRef PubMed.
- H. Ge, D. Wang, Y. Pan, Y. Guo, H. Li, F. Zhang, X. Zhu, Y. Li, C. Zhang and L. Huang, Angew. Chem., Int. Ed., 2020, 59, 8133–8137 CrossRef CAS PubMed.
- A. H. Najafabadi, Z. I. N. Abadi, M. E. Aikins, K. E. Foulds, M. M. Donaldson, W. Yuan, E. B. Okeke, J. Nam, Y. Xu, P. Weerappuli, T. Hetrick, D. Adams, P. A. Lester, A. M. Salazar, D. H. Barouch, A. Schwendeman, R. A. Seder and J. J. Moon, J. Controlled Release, 2021, 337, 168–178 CrossRef CAS PubMed.
- E. Fehri, E. Ennaifer, R. Bel Haj Rhouma, M. Ardhaoui and S. Boubaker, Cells, 2022, 12, 152 CrossRef PubMed.
- L. Ran, B. Lu, H. Qiu, G. Zhou, J. Jiang, E. Hu, F. Dai and G. Lan, Bioact Mater, 2021, 6, 2956–2968 CrossRef CAS PubMed.
- P. A. Jeshvaghani, M. Pourmadadi, F. Yazdian, H. Rashedi, K. Khoshmaram and M. N. Nigjeh, Int. J. Biol. Macromol., 2023, 226, 1100–1115 CrossRef CAS PubMed.
- Q. Jiang, K. Wang, X. Zhang, B. Ouyang, H. Liu, Z. Pang and W. Yang, Small, 2020, 16, e2001704 CrossRef PubMed.
- Y. Han, B. Ding, Z. Zhao, H. Zhang, B. Sun, Y. Zhao, L. Jiang, J. Zhou and Y. Ding, Biomaterials, 2018, 185, 205–218 CrossRef CAS PubMed.
- Y. Lu, L. Wang and H. Chen, Spectrochim. Acta, Part A, 2019, 223, 117345 CrossRef CAS PubMed.
- I. Kim, D. Lee, S. W. Lee, J. H. Lee, G. Lee and D. S. Yoon, ACS Nano, 2021, 15, 6386–6394 CrossRef CAS PubMed.
- M. M. T. Jansman, C. Coll-Satue, X. Liu, P. J. Kempen, T. L. Andresen, P. W. Thulstrup and L. Hosta-Rigau, Biomater. Adv., 2022, 134, 112691 CrossRef PubMed.
- C. Zhou, D. Wang, J. Li, Q. Wang, L. Wo, X. Zhang, Z. Hu, Z. Wang, M. Zhan, M. He, G. Hu, X. Chen, K. Shen, G. Q. Chen and Q. Zhao, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2117988119 CrossRef CAS PubMed.
- C. Liu, Y. Zhang, J. Gao, Q. Zhang, L. Sun, Q. Ma, X. Qiao, X. Li, J. Liu, J. Bu, Z. Zhang, L. Han, D. Zhao and Y. Yang, Drug Resistance Updates, 2022, 66, 100903 CrossRef PubMed.
- N. Kumar, S. Yadav, M. A. Sadique and R. Khan, Biosensors, 2022, 12, 966 CrossRef CAS PubMed.
- J. H. Kim, J. M. Park, E. Jung, J. Lee, J. Han, Y. J. Kim, J. Y. Kim, J. H. Seo and J. S. Kim, Biomaterials, 2022, 289, 121781 CrossRef CAS PubMed.
- M. Zhan, J. Qiu, Y. Fan, L. Chen, Y. Guo, Z. Wang, J. Li, J. P. Majoral and X. Shi, Adv. Mater., 2022, 35, e2208277 CrossRef PubMed.
- C. Sansone, A. Bruno, C. Piscitelli, D. Baci, A. Fontana, C. Brunet, D. M. Noonan and A. Albini, Cells, 2021, 10, 231 CrossRef CAS PubMed.
- G. Huang, L. Liu, H. Pan and L. Cai, Small Methods, 2022, e2201412 CrossRef PubMed.
- F. Zhou, B. Feng, H. Yu, D. Wang, T. Wang, Y. Ma, S. Wang and Y. Li, Adv. Mater., 2019, 31, e1805888 CrossRef PubMed.
- Q. Chen, J. Chen, Z. Yang, J. Xu, L. Xu, C. Liang, X. Han and Z. Liu, Adv. Mater., 2019, 31, e1802228 CrossRef PubMed.
- M. Moghadam, M. Sankian, K. Abnous, A. Varasteh, S. M. Taghdisi, M. Mahmoudi, M. Ramezani, Z. Gholizadeh and A. Ganji, Int. Immunopharmacol., 2016, 36, 324–332 CrossRef CAS PubMed.
- D. Qiao, L. Li, L. Liu and Y. Chen, ACS Appl. Mater. Interfaces, 2022, 14, 50592–50600 CrossRef CAS PubMed.
- X. Wen, X. Xiong, G. Yang, W. Xiao, J. Hou, T. Pan, Y. Hu and S. Zhou, J. Controlled Release, 2022, 353, 535–548 CrossRef PubMed.
- S. Anirudh, A. Rosenberger, E. Schwarzenberger, C. Schaefer, H. Strobl, A. Zebisch, H. Sill and A. Wölfler, Cells, 2020, 9, 1223 CrossRef CAS PubMed.
- A. Psarras, A. Antanaviciute, A. Alase, I. Carr, M. Wittmann, P. Emery, G. C. Tsokos and E. M. Vital, J. Immunol., 2021, 206, 785–796 CrossRef CAS PubMed.
- A. N. Wilkinson, K. Chang, R. D. Kuns, A. S. Henden, S. A. Minnie, K. S. Ensbey, A. D. Clouston, P. Zhang, M. Koyama, J. Hidalgo, S. Rose-John, A. Varelias, S. Vuckovic, K. H. Gartlan and G. R. Hill, Blood, 2019, 134, 2092–2106 CrossRef PubMed.
- H. Fang, M. Li, Q. Liu, Y. Gai, L. Yuan, S. Wang, X. Zhang, M. Ye, Y. Zhang, M. Gao, Y. Hou and X. Lan, Nanomicro Lett., 2020, 12, 62 CAS.
- B. Sun, S. S. Mullapudi, Y. Zhang and K. G. Neoh, Mikrochim. Acta, 2022, 189, 349 CrossRef CAS PubMed.
- Z. Yu, O. Vepris, C. Eich, Y. Feng, I. Que, M. G. M. Camps, H. Zhang, F. A. Ossendorp and L. J. Cruz, Mikrochim. Acta, 2022, 189, 368 CrossRef CAS PubMed.
- H. Zhang, W. Chen, J. Wang, W. Du, B. Wang, L. Song, Y. Hu and X. Ma, Biomaterials, 2022, 293, 121954 CrossRef PubMed.
- Z. Li, X. Lai, S. Fu, L. Ren, H. Cai, H. Zhang, Z. Gu, X. Ma and K. Luo, Adv. Sci., 2022, 9, e2201734 CrossRef PubMed.
- T. Katopodi, S. Petanidis, C. Charalampidis, I. Chatziprodromidou, P. Eskitzis, D. Tsavlis, P. Zarogoulidis, C. Kosmidis, D. Matthaios and K. Porpodis, Cells, 2022, 11, 3183 CrossRef CAS PubMed.
- A. S. Bulygin, J. N. Khantakova, N. S. Shkaruba, H. Shiku and S. S. Sennikov, Front. Immunol., 2022, 13, 1016670 CrossRef CAS PubMed.
- D. Jorgovanovic, M. Song, L. Wang and Y. Zhang, Biomark Res., 2020, 8, 49 CrossRef PubMed.
- Q. Zeng, R. Wang, Y. Hua, H. Wu, X. Chen, Y. C. Xiao, Q. Ao, X. Zhu and X. Zhang, Nano Res., 2022, 15, 9286–9297 CrossRef CAS PubMed.
- E. Renaude, M. Kroemer, C. Borg, P. Peixoto, E. Hervouet, R. Loyon and O. Adotévi, Front. Immunol., 2021, 12, 669992 CrossRef CAS PubMed.
- Y. Huang, A. Jia, Y. Wang and G. Liu, Immunology, 2023, 168, 30–48 CrossRef CAS PubMed.
- F. Jin, J. Qi, D. Liu, Y. You, G. Shu, Y. Du, J. Wang, X. Xu, X. Ying, J. Ji and Y. Du, J. Controlled Release, 2021, 337, 90–104 CrossRef CAS PubMed.
- S. Wang, Y. Wang, K. Jin, B. Zhang, S. Peng, A. K. Nayak and Z. Pang, Expert Opin. Drug Delivery, 2022, 19, 965–984 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr00542a |
| This journal is © The Royal Society of Chemistry 2023 |