Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cross-linking of aromatic phenolate groups by cytochrome P450 enzymes: a model for the biosynthesis of vancomycin by OxyB†
Hafiz Saqib
Ali
 ab,
Richard H.
Henchman
ab,
Richard H.
Henchman
 ab and
Sam P.
de Visser
ab and
Sam P.
de Visser
 *ac
*ac
aManchester Institute of Biotechnology, The University of Manchester, 131 Princess Street, Manchester M1 7DN, UK. E-mail: sam.devisser@manchester.ac.uk
bDepartment of Chemistry, The University of Manchester, Oxford Road, Manchester M13 9PL, UK
cDepartment of Chemical Engineering and Analytical Science, The University of Manchester, Oxford Road, Manchester M13 9PL, UK
First published on 3rd June 2020
Abstract
The cytochromes P450 are a versatile class of enzymes involved in many chemical reactions in biosystems and as such they take part in biodegradation as well as biosynthesis pathways in many organisms. These enzymes use molecular oxygen on a heme centre and often react as mono-oxygenases. Lesser known reactions catalyzed by the P450s include desaturation pathways and ring-closure reactions. In this work we study the aromatic cross-linking of glycopeptide units as, for instance, performed by the P450 isozyme OxyB as part of vancomycin biosynthesis. A series of density functional theory studies are reported on a large active site cluster model of 258 atoms containing the heme with its coordinated ligands, a representative substrate and its interacting protein residues. We show that the catalytic cycle intermediates Compound I and Compound II of P450 can rapidly and successively abstract a phenolic hydrogen atom from adjacent peptide groups to give a biradical intermediate with small reaction barriers. The latter can form the ether cross-link between the two aromatic residues, which is the rate-determining step in the reaction mechanism and involves a simultaneous proton transfer from the ipso-position to the ketone. A thermochemical analysis reveals that weak phenolic O–H bonds lead to hydrogen atom abstraction easily by Compound I and Compound II, enabling a selective aromatic cross-linking reaction.
Introduction
The cytochromes P450 are important heme enzymes found in most forms of life, where they typically catalyse mono-oxygenation reactions.1 These processes are highly versatile and processes leading to substrate hydroxylation (aliphatic and aromatic), epoxidation and heteroatom oxidation have been observed.2 Lesser known and studied reactions by the P450s relate to substrate desaturation and ring-closure reactions, whereby molecular oxygen on the heme active site is reduced to two water molecules.3 The versatility of the P450s, therefore, has given them key functions in the liver for the metabolism of a broad range of xenobiotics and drug molecules.4 Generally, this means the P450 enzymes are not highly selective and give a large number of reaction products. On the other hand, a range of P450 isozymes are involved in more selective biosynthesis reactions, for instance, in several steps in the biosynthesis of the hormone and signalling molecules oestrogen and serotonin.An unusual ring-closure reaction catalysed by the P450s is observed in bacteria, namely the aromatic cross-linking reaction between phenolic amino acid residues. For instance, the biosynthesis of glycopeptide antibiotics, such as vancomycin, includes a side-chain cross-linking between phenolic and aromatic residues that is believed to give the natural products their characteristic three-dimensional structure.5 Generally, these glycopeptide structures contain a protein chain of several Tyr amino acid residues, whereby the central one in vancomycin has a Cl-substitution on the ortho-position with respect to the phenol. Glycopeptide natural products are common in chemical biology and have been well studied;6 however, details on how the P450s activate glycopeptide structures to initiate these cross-linked structures are unknown.
The P450 isozymes involved in the aromatic amino acid cross-linking are OxyB and OxyA that perform consecutive cross-linking reactions to a central phenolate group of the substrate in both ortho-positions from the central hydroxo group.7 The first of those reactions is performed by OxyB and the overall reaction mechanism catalysed by OxyB enzymes includes the aromatic cross-linking of two Tyr residues in a protein chain (Fig. 1). Thus, the phenolate of the Cl-substituted aromatic ring (ring A) forms an ether bond with another aromatic ring further down the chain (ring B) that gives the natural product its three-dimensional structure and rigidity. As little is known about these types of aromatic cross-linking processes, we undertook a computational study into this unusual reaction mechanism.
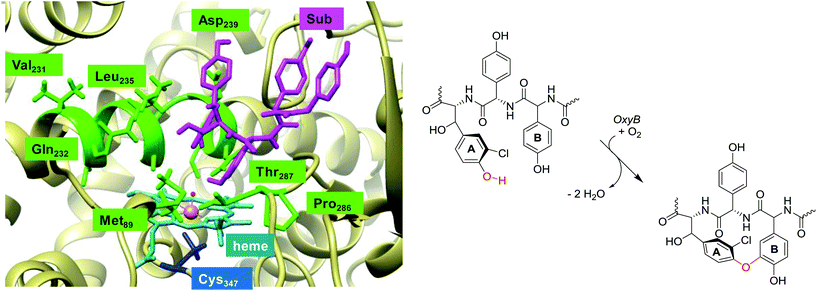 | ||
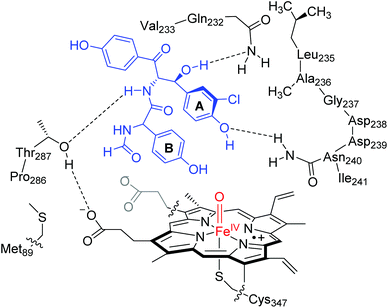
| Fig. 1 Lowest energy binding conformation of substrate (Sub) into the active site of P450 OxyB as taken from the 1LG9 pdb file and the general reaction mechanism catalysed. | ||
A resting state crystal structure of OxyB was characterized and shows the typical features of P450 isozymes with a heme group deeply buried inside the protein.8Fig. 1 shows an extract of the active site of OxyB from the 1LG9 protein databank (pdb) file,8 where we added a model substrate representing a glycopeptide unit (in purple) inside the substrate binding pocket. The central heme group (highlighted in light blue) is buried inside the protein and tightly positioned through covalent interactions and salt bridges. Like many P450s, the heme iron (in amber) binds to a sulphur atom of a cysteinate group in the axial position (Cys347), which is believed to induce a push-effect and affect its oxidative properties.9 The substrate binding pocket is tight and lined with a series of aliphatic residues, e.g. Val231, Leu235 and Pro286, as well as several hydrogen bonding donor groups of the side chains of Gln232, Asn240 and Thr287. As such, substrate will bind in a tight conformation for catalysis.
To gain insight into the glycopeptide cross-linking mechanism catalysed by P450 enzymes, we decided to create a large active site model of the OxyB isozyme with a glycopeptide model substrate included, the heme and a large part of the substrate binding pocket. The calculations show that weak phenolic O–H bonds are easily abstracted by compound I and two consecutive hydrogen atom abstractions then enables a cross-linking reactions while reducing dioxygen to two water molecules.
Results and discussion
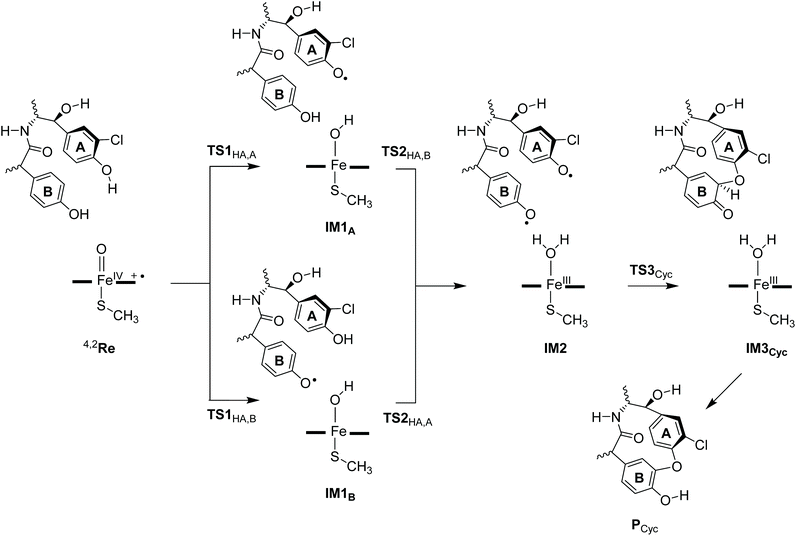
Based on the crystal structure coordinates deposited under the 1LG9 protein databank file,8 we created an active site model complex and studied the mechanism of aromatic cross-linking in OxyB enzymes. As the crystal structure is a resting state geometry without substrate bound, we docked a tripeptide substrate of three aromatic residues into the binding pocket using the AutoDock Vina software package and took the lowest energy binding pose with the phenol groups of ring A and B pointing towards the heme.10 Following previous experience in the field,11 an active site cluster model was created from the substrate-bound structure that includes the heme, the substrate and key interactions of the oxidant and substrate with the protein. Scheme 1 shows the 258 atom cluster model created for this study. As one of the propionate side chains of the heme forms hydrogen-bonding interactions with peptide groups of the substrate, we kept it in the model, while all other side chains of the heme were abbreviated to hydrogen atoms. The axial cysteinate was abbreviated to methylmercaptate and the distal water molecule replaced by an oxo group to create a high-valent iron(IV)-oxo heme cation radical species commonly labelled as Compound I (CpdI). This species was experimentally characterized for P450cam with spectroscopic methods,12 and computational studies identified it as the active oxidant in substrate activation for many P450-type reaction mechanisms.13 The substrate binding pocket was included in the model by taking the three small protein chains Gln232-Val233, Leu235-Ala236-Gly237-Asp238-Asp239-Asn240-Ile241 and Pro286-Thr287. Since the Asp238 side-chain points away from the substrate, it was abbreviated to a Gly residue. The protein chains were cut after the terminal Cα carbon of the chain and a hydrogen atom was added to complete its valency. In addition, the Met89 side chain was included as a dimethylsulfide group.We investigated several reaction pathways for the aromatic cross-linking reaction in glycopeptide substrate that started from the reactant complex (Re) as shown in Scheme 1. Thereafter, the reaction was studied for either initial hydrogen atom abstraction from the phenol of ring A or ring B, see Scheme 2. These pathways include a mechanism proceeding with a hydrogen atom abstraction from a phenolic O–H group via a transition state TS1HA,A to form a radical intermediate IM1A or a hydrogen atom transition state TS1HA,B to form the radical intermediate IM1B, both of those representing an iron-hydroxo species, also known as Compound II, with a nearby radical. The aromatic ring (A or B) that is activated in each step is given in subscript after the label. Next, a second hydrogen atom abstraction takes place from the other phenolic group to convert the iron-hydroxo species (IM1A and IM1B) into the iron(III)–water complex (IM2) via transition states TS2HA,B and TS2HA,A, respectively. Thereafter, the phenoxyl group of ring A attacks the ortho-position in ring B to form the aromatic cross-linked product (IM3) via transition state TS3Cyc. In a final step the ipso-proton is transferred to the phenolate oxygen of ring B to produce the product complex (PCyc) for aromatic ring cross-linked products. The full reaction profile for the mechanisms shown in Scheme 2 was calculated for all low-lying doublet and quartet spin states and various electromers for each intermediate were tested.
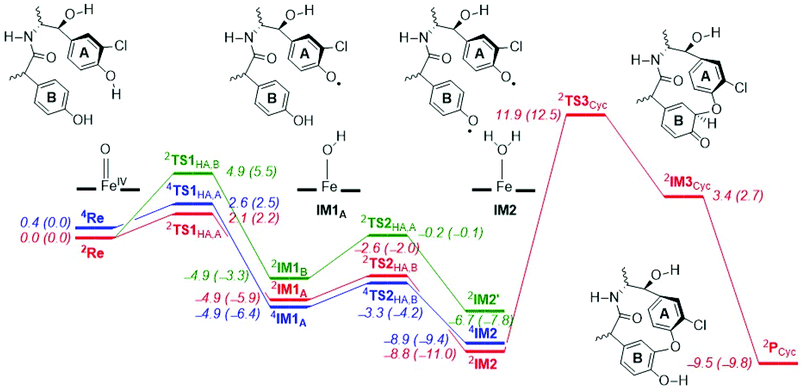
The potential energy landscape for the reaction mechanism of aromatic cross-linking of aromatic residues by P450 CpdI is described in Fig. 2, whereby the initial hydrogen atom abstraction comes from phenol ring A and the second one from phenol ring B or vice versa. The reaction starts from CpdI, which is a triradical system with π*xz1 π*yz1 a2u1 configuration and close-lying doublet and quartet spin states. Our CpdI structure and electronic configuration matches previous studies that also showed it to have close-lying spin-states leading to multi-state reactivity patterns.13,14 The lowest energy (with zero-point correction, ZPE, included) barrier for the first hydrogen atom abstraction only costs ΔE‡ + ZPE = 2.1 (doublet) and 2.6 (quartet) kcal mol−1 in energy from phenol ring A, while the barrier from phenol ring B is ΔE‡ + ZPE = 4.9 kcal mol−1. As such, phenolic hydrogen atom abstraction from ring A will be preferred over hydrogen atom abstraction from phenol group B, probably as a result of the ortho-chloro substitution that better stabilizes the phenolic radical.
Note, all of these barriers are extremely low in energy and much lower than aliphatic C–H atom abstraction reactions by CpdI that typically have barriers above ΔE‡ + ZPE > 10 kcal mol−1.14b For example, hydrogen atom abstraction barriers of 12.4 and 12.5 kcal mol−1 were reported elsewhere for toluene and ethylbenzene activation with a CpdI model.14 Therefore, the O–H bond strength in the phenolic groups of glycopeptides is weak and will be preferentially abstracted even in the presence of aliphatic C–H bonds.
We also calculated the alternative reaction mechanism, whereby CpdI abstracts a hydrogen atom from the phenol group of ring B first followed by a hydrogen atom abstraction from ring A, which is highlighted as the green landscape in Fig. 1. As can be seen from Fig. 1, the energies for hydrogen atom abstraction through pathway B are slightly higher in energy than that for pathway A, and arrive at the radical intermediate 2IM2′, which has the two spins on ring A and B ferromagnetically coupled; hence aromatic cross-linking may be difficult from this intermediate, vide supra.
In addition, a direct pathway from IM1 to IM3 was explored by attack of the phenoxy radical on the ortho C–H position of ring B. Thus, during the aromatic hydroxylation by Cpd I, an electrophilic addition takes place of the iron-oxo onto one of the carbon atoms of the arene. This step is then either followed by ring-closure to form an epoxide intermediate or by proton transfer to form phenolate.15 In our case, however, the resulting geometry scan for electrophilic attack on aromatic ring B by the phenoxyl radical of ring A had a maximum energy of well over 30 kcal mol−1; hence, this pathway cannot compete with the lower energy pathway viaIM2.
Optimized geometries of the reactant structures and the TS1HA optimized transition states are given in Fig. 3. In the reactants the Fe–O interaction is short (1.646 Å in 2Re and 1.641 Å in 4Re), which elongates to about 1.73 Å in the hydrogen atom abstraction transition states. Similar distances were reported for analogous P450 model complexes.13,14,16 Structurally, the transition states (2,4TS1HA,A) are relatively central, with the transferring hydrogen atom roughly midway between the donor and acceptor oxygen atoms: distances of FeO–H of 1.157 (1.162) Å and H–OPh of 1.271 (1.264) Å are obtained for 2TS1HA,A (4TS1HA,A). Also, the angle for the O–H–O group is almost linear with value of 171° as expected of a hydrogen atom abstraction transition state.13,16 The transition states are accompanied by a large imaginary frequency for an O–H–O stretch vibration of i1451 (i1455) cm−1. Interestingly, the 2TS1HA,B transition states are very similar to the pathway A barriers with FeO–H of 1.204 Å and H–OPh of 1.211 Å. The imaginary frequencies for the pathway B transition states are much larger (i1817 cm−1). Hydrogen atom abstraction barriers often feature a large imaginary frequency, which implies that the curvature of the potential energy surface around the hydrogen atom abstraction transition state is steep and narrow.14,17 It is expected that replacing the transferring hydrogen atoms by deuterium atoms will lead to a significant kinetic isotope effect for the reaction, although it may be difficult to experimentally measure these rate constants based on the low free energies of activation predicted by our model.
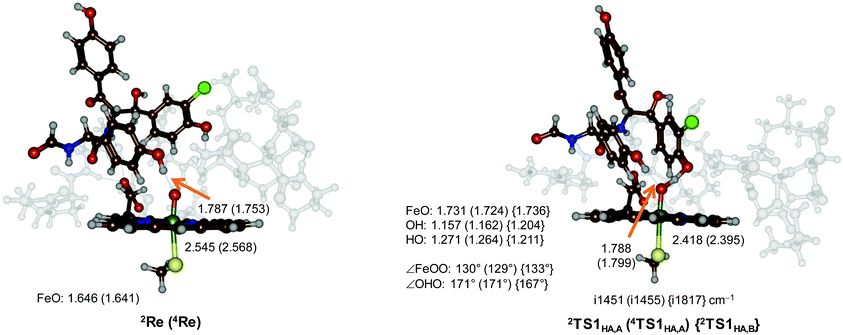 | ||
| Fig. 3 UB3LYP/BS1 optimized geometries of 4,2Re, 4,2TS1HA,A and 2TS1HA,B with bond lengths in angstroms, bond angles in degrees and the imaginary frequency in the transition state in cm−1. | ||
After the hydrogen atom abstraction transition state the systems relax to a radical intermediate (2,4IM1A) with an exothermicity of ΔE + ZPE = −4.9 (−4.9) kcal mol−1 with respect to the reactant complex in the doublet (quartet) spin state for pathway A (Fig. 2). The structures are characterized as iron(III)-hydroxo complexes coupled to a heme and substrate radical, i.e. with electronic configuration π*xz2 π*yz1 a2u1 πSub1. Indeed both radical intermediates have a spin of about 1 on the substrate, on the FeO and on the heme/Cys groups. We tried to swap molecular orbitals to form the alternative electromer as iron(IV)-hydroxo with closed-shell heme; however, during the SCF convergence the original electronic configuration was obtained, and hence the iron(IV) configuration is much higher in energy.
Normally, in substrate hydroxylation by P450 Cpd I, the hydrogen atom abstraction is followed by OH rebound to form alcohol product complexes.18 However, phenolic hydrogen atom abstraction is a one-electron process and no OH rebound is possible. In order to return the catalytic cycle to the resting state, therefore, another one-electron process is needed. Hence, we studied the pathway of the iron-hydroxo species (Compound II, Cpd II) to abstract a second hydrogen atom from another phenol residue. Thus, in the next stage of the catalytic cycle a hydrogen atom abstraction from the other phenolate group of the substrate (ring B) takes place to form iron(III)–water–heme and a substrate with two phenoxyl radicals (IM2). Barriers with respect to IM1A of ΔE‡ + ZPE = 1.6 kcal mol−1 (quartet) and ΔE‡ + ZPE = 2.3 kcal mol−1 (doublet) are found (Fig. 2). Consequently, the two sequential hydrogen atom abstraction steps from the two Tyr residues of the substrate will happen in quick succession with very small rate constants.
The biradical intermediates show radical character on both aromatic rings of the substrate, whereby in 2IM2 an electronic configuration of π*xz2 π*yz1 a2u2 πA1 πB1 was obtained with the two spins on ring A and B antiferromagnetically coupled (one up-spin and one down-spin). From the biradical intermediates IM2 the pathway for aromatic cross-linking was studied in the doublet spin state. By contrast, in the quartet spin state the two spins on the substrate are ferromagnetically coupled (both unpaired electrons in πA and πB are up-spin). As such, in the doublet spin state the two radicals can pair up and form a new C–O bond homolytically, thereby forming a cross-link between the two aromatic residues. On the other hand, in 4IM2 the two aromatic residue unpaired electron spins are ferromagnetically coupled and hence homolytic bond formation will be a spin-forbidden process, see Scheme 3.
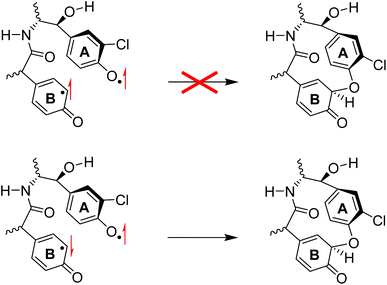 | ||
| Scheme 3 Valence bond description of the aromatic cross-linking steps in ferromagnetic biradical (top) and antiferromagnetic biradical species. | ||
The spin densities on the substrate in 2IM2 give a value of −0.23 on the phenoxyl oxygen of ring A, while the ortho-carbon atom of ring B has a positive spin of 0.31. As such the spin densities indicate a possible homolytic bond formation between the two atoms to generate the cross-linked product. Interestingly, the cyclization step from 2IM2 to 2IM3 is ΔE‡ + ZPE = 20.7 kcal mol−1 endothermic with respect to 2IM2, probably due to the breaking of the aromaticity in one of the rings. Therefore, the aromatic cross-linking step is rate-determining in the reaction mechanism.
The cross-linking transition state (2TS3Cyc) is shown in Fig. 4. The cross-linking bond is very long (2.024 Å) and the imaginary frequency of i267 cm−1 displays the stretch vibration along this bond. The Fe–O distance is long (2.163 Å) and reflects the weak interaction of a water molecule bound to iron(III). This distance is similar to previous calculations on resting state complexes.19
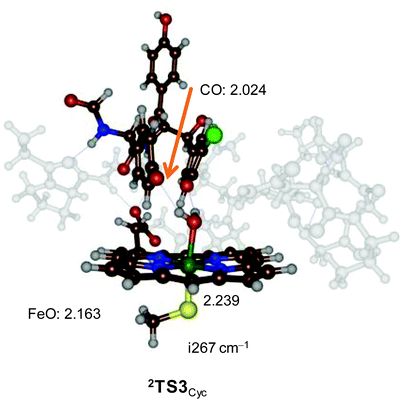 | ||
| Fig. 4 UB3LYP/BS1 optimized geometry of 2TS3Cyc with bond lengths in angstroms and the imaginary frequency in the transition state in cm−1. | ||
After the transition state (2TS3Cyc) the system relaxes to intermediate 2IM3 with a covalent ether bond between the two aromatic rings, but still has the ipso-proton bound to one of the ring carbon atoms of ring B. After 2IM3 a barrierless proton transfer from the ipso-position to the phenoxyl group of ring B leads to the cyclic product 2PCyc, which is lower in energy than 2IM2 by 0.7 kcal mol−1. As the ipso-proton is located close to the phenoxyl oxygen atom, we expect a quick and direct proton transfer, although a water assisted proton transfer may be possible as well.
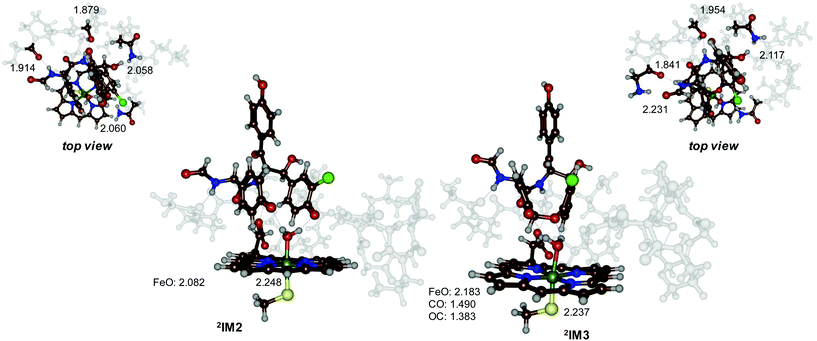
To understand the features of the cross-linking step better, we display in Fig. 5 the optimized geometries of 2IM2 and 2IM3 side-by-side. Both structures have an iron(III)–heme with a water molecule in the distal position bound. The Fe–O and Fe–S distances; therefore, are very similar, namely 2.082 Å and 2.248 Å for 2IM2 and 2.183 Å and 2.237 Å for 2IM3, respectively. A further analysis of the structures of 2IM2 and 2IM3 shows that several hydrogen bonds around the substrate have weakened due to the cross-linking. Thus, in 2IM2 the substrate interacts with the peptide carbonyl group of Pro286 (1.914 Å), the alcohol group of Thr287 (1.879 Å) and the amide group of Gln232 (2.058 Å). In 2IM3 these three interactions elongate to 2.231 Å, 1.954 Å and 2.117 Å, respectively. In addition, the phenol group of ring A forms a hydrogen bond with the side-chain of Asn240. In 2IM2 the hydrogen bond is 2.060 Å, while in 2IM3 it has broken and the two groups are at a distance of 4.132 Å. Furthermore, the interaction of the water ligand to this oxygen atom is elongated from 1.788 to 1.954 Å. Therefore, the local interactions inside the protein may prevent the cyclization to take place within the enzyme and make the overall process thermoneutral in the binding pocket while it may be strongly exothermic in solution.
To gain insight into the details of the reaction mechanism and why the cross-linking is overall endothermic, we calculated the bond dissociation energies (BDE) of various O–H bonds of the substrate (Sub-H) as well as some of the C–O bonds in possible cross-linked products. BDEs were calculated according to eqn (1), where we take the substrate (Sub-H), a hydrogen atom and the substrate with a hydrogen atom removed (Sub˙) individually.20 The difference in energy for the structures in eqn (1) is then taken as the BDE. The calculated BDE values for the various bond strengths are shown in Fig. 6.
| Sub-H → Sub˙ + H˙ + BDESub-H | (1) |
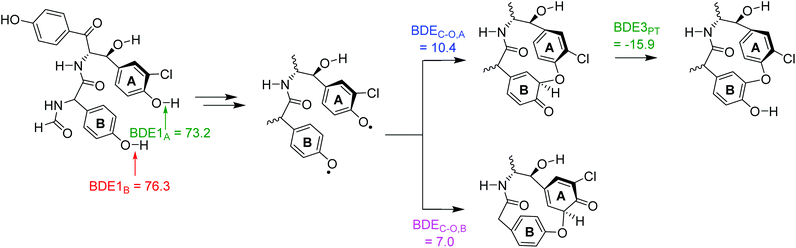 | ||
| Fig. 6 Bond dissociation energies (in kcal mol−1) for the reactant (left) and possible cross-linked products (right). | ||
The phenolic O–H bonds are very weak: the BDE for the phenol group of ring A is BDE1A = 73.2 kcal mol−1, while the one for ring B is BDE1B = 76.3 kcal mol−1. As such the bond strength of the O–H bond in the phenol group of ring A is weaker than the one in ring B, probably due to the ortho-chloride substitution. Based on these BDE values; therefore, we expect preferential hydrogen atom abstraction from the phenol group of ring A over that of ring B, which is indeed observed in our potential energy landscape seen in Fig. 1 above. These BDE values are low in energy for typical C–H/O–H bonds and match the low transition state barriers observed. Moreover, the values confirm that phenolic hydrogen atom abstraction is a low-energy process for P450 CpdI and should be preferred over aliphatic or aromatic hydroxylation reactions. It also explains why in P450 enzymes there usually are no Tyr residues in the substrate-binding pocket closely positioned to the heme as Cpd I will rapidly react with phenolic O–H groups.
Next, we calculated the BDEs for the C–O bonds that are formed during the cross-linking process, whereby we considered a linkage between the phenolate of ring A with a carbon atom of ring B or the phenolate of ring B with a carbon atom of ring A. For these BDECO-A and BDECO-B calculations, we considered an isolated substrate structure and calculated the energy difference between the bound and biradical isomers. The strongest bond is formed between the phenolate of ring A with ring B: BDECO-A is calculated to be 10.4 kcal mol−1, while BDECO-B is only 7.0 kcal mol−1. Consequently, based on the difference in BDE values in the possible products, the most likely reaction path should lead to a bond between the phenolate oxygen of ring A with a carbon atom from ring B. The low value of these bond energies highlights the energetic cost of breaking the aromaticity in the benzene ring to weaken the C–H bond to enable its removal.
Finally, we calculated the proton-transfer energy for the final step in the reaction mechanism that moves the ipso-proton to the ketone group (BDE3PT). The difference in energy between the two isomers, where one structure had the ipso-proton bound to the aromatic ring, while the second was a phenol. The final proton transfer step is exergonic by ΔG = −15.9 kcal mol−1 and, therefore, should proceed fast and give the phenol as most stable structure.
Experimental
All the quantum mechanical calculations were done using density functional theory (DFT) methods which are implemented in Gaussian-09.21 In previous work we extensively tested and benchmarked models and methods for P450 reaction mechanisms and reproduced experimental structures and rate constants well.22 All calculations used the unrestricted B3LYP hybrid density functional method,23 in combination with a basis set containing an LANL2DZ+ECP on iron and 6-31G* on the rest of the atoms (basis set BS1).24 Full geometry optimizations, frequencies and scan calculations were run for all structures using UB3LYP/BS1 in the gas-phase. Subsequent single point calculations with the polarized continuum model (CPCM)25 were performed with a dielectric constant mimicking chlorobenzene, and a triple-ζ quality basis set (basis set BS2) containing LACV3P++ECP on iron and 6-311+G* on the rest of the atoms.Conclusions
In summary, a set of DFT calculations on the cross-linking of aromatic peptide residues by a P450 model CpdI is reported. We show that sequential hydrogen atom abstraction from two adjacent phenol groups of Tyr residues gives rise to a relatively stable biradical species and an iron(III)–water–heme complex. The antiferromagnetically coupled biradical can react further through aromatic ring-closure followed by proton transfer to form the cross-linked product. The work further shows that the rate-determining step in the mechanism is the attack of the phenoxyl radical of ring A on one of the aromatic carbon atoms to form a covalent bond, which disrupts the aromaticity in the arene, which is restored after proton transfer from that position to the ketone group.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The Punjab Education Endowment Funds (PEEF) Pakistan is thanked for a PhD scholarship to HSA.Notes and references
- (a) M. Sono, M. P. Roach, E. D. Coulter and J. H. Dawson, Chem. Rev., 1996, 96, 2841 CrossRef CAS PubMed; (b) Cytochrome P450: Structure, Mechanism and Biochemistry, ed. P. R. Ortiz de Montellano, Kluwer Academic/Plenum Publishers, New York, 3rd edn, 2005 Search PubMed; (c) A. W. Munro, H. M. Girvan and K. J. McLean, Nat. Prod. Rep., 2007, 24, 585 RSC; (d) Handbook of Porphyrin Science, ed. K. M. Kadish, K. M. Smith and R. Guilard, World Scientific Publishing Co., New Jersey, 2010 Search PubMed; (e) T. L. Poulos, Chem. Rev., 2014, 114, 3919 CrossRef CAS PubMed; (f) X. Huang and J. T. Groves, J. Biol. Inorg. Chem., 2017, 22, 185 CrossRef CAS PubMed.
- (a) B. Meunier, S. P. de Visser and S. Shaik, Chem. Rev., 2004, 104, 3947 CrossRef CAS PubMed; (b) I. G. Denisov, T. M. Makris, S. G. Sligar and I. Schlichting, Chem. Rev., 2005, 105, 2253 CrossRef CAS PubMed; (c) P. R. Ortiz de Montellano, Chem. Rev., 2010, 110, 932 CrossRef CAS PubMed.
- (a) K. L. Dunbar, D. H. Scharf, A. Litomska and C. Hertweck, Chem. Rev., 2017, 117, 5521 CrossRef CAS PubMed; (b) M. Pickl, S. Kurakin, F. G. Cantú Reinhard, P. Schmid, A. Pöcheim, C. K. Winkler, W. Kroutil, S. P. de Visser and K. Faber, ACS Catal., 2019, 9, 565 CrossRef CAS PubMed.
- (a) G. H. Posner and P. M. O'Neill, Acc. Chem. Res., 2004, 37, 397 CrossRef CAS PubMed; (b) F. P. Guengerich and F. K. Yoshimoto, Chem. Rev., 2018, 118, 6573 CrossRef CAS PubMed.
- (a) R. D. Süssmuth and W. Wohlleben, Appl. Microbiol. Biotechnol., 2004, 63, 344 CrossRef PubMed; (b) C. T. Walsh and Y. Tang, Biochemistry, 2018, 57, 3987 CrossRef PubMed.
- See, e.g. (a) C. Feng, Q. Wei, C. Hu and Y. Zou, Org. Lett., 2019, 21, 3114 CrossRef CAS PubMed; (b) V. Agarwal, J. M. Blanton, S. Podell, A. Taton, M. A. Schorn, J. Busch, Z. Lin, E. W. Schmidt, P. R. Jensen, V. J. Paul, J. S. Biggs, J. W. Golden, E. E. Allen and B. S. Moore, Nat. Chem. Biol., 2017, 13, 537 CrossRef CAS PubMed.
- (a) K. Zerbe, K. Woithe, D. B. Li, F. Vitali, L. Bigler and J. A. Robinson, Angew. Chem., Int. Ed., 2004, 43, 6709 CrossRef CAS PubMed; (b) C. Brieke, M. Peschke, K. Haslinger and M. J. Cryle, Angew. Chem., Int. Ed., 2015, 54, 15715 CrossRef CAS PubMed.
- K. Zerbe, O. Pylypenko, F. Vitali, W. Zhang, S. Rouset, M. Heck, J. W. Vrijbloed, D. Bischoff, B. Bister, R. D. Süssmuth, S. Pelzer, W. Wohlleben, J. A. Robinson and I. Schlichting, J. Biol. Chem., 2002, 277, 47476 CrossRef CAS PubMed.
- (a) M. T. Green, Curr. Opin. Chem. Biol., 2009, 13, 84 CrossRef CAS PubMed; (b) F. Ogliaro, S. Cohen, S. P. de Visser and S. Shaik, J. Am. Chem. Soc., 2000, 122, 12892 CrossRef CAS.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455 CAS.
- (a) M. G. Quesne, T. Borowski and S. P. de Visser, Chem. – Eur. J., 2016, 22, 2562 CrossRef CAS PubMed; (b) S. Ghafoor, A. Mansha and S. P. de Visser, J. Am. Chem. Soc., 2019, 141, 20278 CrossRef CAS PubMed.
- J. Rittle and M. T. Green, Science, 2010, 330, 933 CrossRef CAS PubMed.
- (a) S. Shaik, D. Kumar, S. P. de Visser, A. Altun and W. Thiel, Chem. Rev., 2005, 105, 2279 CrossRef CAS PubMed; (b) D. Li, Y. Wang and K. Han, Coord. Chem. Rev., 2012, 256, 1137 CrossRef CAS; (c) M. R. A. Blomberg, T. Borowski, F. Himo, R.-Z. Liao and P. E. M. Siegbahn, Chem. Rev., 2014, 114, 3601 CrossRef CAS PubMed.
- (a) S. P. de Visser, D. Kumar, S. Cohen, R. Shacham and S. Shaik, J. Am. Chem. Soc., 2004, 126, 8362 CrossRef CAS PubMed; (b) S. Shaik, D. Kumar and S. P. de Visser, J. Am. Chem. Soc., 2008, 130, 10128 CrossRef CAS PubMed.
- (a) M. G. Quesne, D. Senthilnathan, D. Singh, D. Kumar, P. Maldivi, A. B. Sorokin and S. P. de Visser, ACS Catal., 2016, 6, 2230 CrossRef CAS; (b) X.-X. Li, V. Postils, W. Sun, A. S. Faponle, M. Solà, Y. Wang, W. Nam and S. P. de Visser, Chem. – Eur. J., 2017, 23, 6406 CrossRef CAS PubMed.
- (a) F. G. Cantú Reinhard, M. A. Sainna, P. Upadhyay, G. A. Balan, D. Kumar, S. Fornarini, M. E. Crestoni and S. P. de Visser, Chem. – Eur. J., 2016, 22, 18608 CrossRef PubMed; (b) C. Colomban, A. H. Tobing, G. Mukherjee, C. V. Sastri, A. B. Sorokin and S. P. de Visser, Chem. – Eur. J., 2019, 25, 14320 CrossRef CAS PubMed.
- P. Barman, F. G. Cantú Reinhard, U. K. Bagha, D. Kumar, C. V. Sastri and S. P. de Visser, Angew. Chem., Int. Ed., 2019, 58, 10639 CrossRef CAS PubMed.
- S. Shaik, S. Cohen, S. P. de Visser, P. K. Sharma, D. Kumar, S. Kozuch, F. Ogliaro and D. Danovich, Eur. J. Inorg. Chem., 2004, 207 Search PubMed.
- (a) H.-P. Hersleth, U. Ryde, P. Rydberg, C. H. Görbitz and K. K. Andersson, J. Inorg. Biochem., 2006, 100, 460 CrossRef CAS PubMed; (b) P. R. Balding, C. S. Porro, K. J. McLean, M. J. Sutcliffe, J.-D. Maréchal, A. W. Munro and S. P. de Visser, J. Phys. Chem. A, 2008, 112, 12911 CrossRef CAS PubMed; (c) K. D. Dubey and S. Shaik, Acc. Chem. Res., 2019, 52, 389 CrossRef PubMed.
- S. P. de Visser, J. Am. Chem. Soc., 2010, 132, 1087 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Gaussian, Inc., Wallingford CT, 2009 Search PubMed.
- (a) S. Kumar, A. S. Faponle, P. Barman, A. K. Vardhaman, C. V. Sastri, D. Kumar and S. P. de Visser, J. Am. Chem. Soc., 2014, 136, 17102 CrossRef CAS PubMed; (b) F. G. Cantú Reinhard, A. S. Faponle and S. P. de Visser, J. Phys. Chem. A, 2016, 120, 9805 CrossRef PubMed.
- (a) A. D. Becke, Phys. Rev. A, 1988, 38, 3098 CrossRef CAS PubMed; (b) C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785 CrossRef CAS PubMed.
- W. R. Wadt and P. J. Hay, J. Chem. Phys., 1985, 82, 284 CrossRef CAS.
- M. Cossi, G. Scalmani, N. Rega and V. Barone, J. Chem. Phys., 2002, 117, 43 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Tables with absolute and relative energies, group spin densities and charges of optimized geometries discussed here as well as Cartesian coordinates. See DOI: 10.1039/d0ob01023e |
| This journal is © The Royal Society of Chemistry 2020 |