Open Access Article
Open Access ArticleTranylcypromine specificity for monoamine oxidase is limited by promiscuous protein labelling and lysosomal trapping†
Jonas
Drechsel
 a,
Christina
Kyrousi
b,
Silvia
Cappello
b and
Stephan A.
Sieber
a,
Christina
Kyrousi
b,
Silvia
Cappello
b and
Stephan A.
Sieber
 *a
*a
aDepartment of Chemistry, Technical University of Munich, Lichtenbergstraße 4, 85748 Garching, Germany. E-mail: stephan.sieber@tum.de
bMax Planck Institute of Psychiatry, Kraepelinstraße 2, 80804 Munich, Germany
First published on 12th August 2020
Abstract
Monoamine oxidases MAOA and MAOB catalyze important cellular functions such as the deamination of neurotransmitters. Correspondingly, MAO inhibitors are used for the treatment of severe neuropsychiatric disorders such as depression. A commonly prescribed drug against refractory depression is tranylcypromine, however, the side effects are poorly understood. In order to decipher putative off-targets, we synthesized two tranylcypromine probes equipped with either an alkyne moiety or an alkyne-diazirine minimal photocrosslinker for in situ proteome profiling. Surprisingly, LC–MS/MS analysis revealed low enrichment of MAOA and relatively promiscuous labeling of proteins. Photoprobe labeling paired with fluorescent imaging studies revealed lysosomal trapping which could be largely reverted by the addition of lysosomotropic drugs.
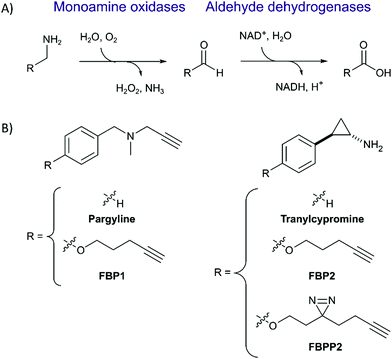
Monoamine oxidases A and B (MAOA and MAOB) catalyze the deamination of neurotransmitters such as norephinephrine, serotonin and dopamine to the corresponding aldehydes which are further processed by e.g. aldehyde dehydrogenases (ALDHs) into the corresponding acids (Fig. 1A). The degradation of these molecules is important for neurotransmission and brain functions such as emotional behaviours.1,2 Thus, inhibition of MAO enzymes has become a viable strategy to treat depression and other nervous system or neurological disorders such as Parkinson's disease.3–6 MAO inhibitors (MAOI) were first introduced in the 1950s but due to side effects and safety concerns are only administered after treatment with other drugs fails. Some commonly reported side effects caused by MAOIs such as tranylcypromine include gastrointestinal disorder, headaches and fatigue.7 Adverse effects are also experienced when MAOIs are administered in combination with primary amine-containing drugs.8 The clearance of amine-containing molecules such as chloroquine relies on MAO deamination; consequently, co-administration with tranylcypromine must be avoided.9 Despite these known side effects, few proteomic studies on the target selectivity of MAOIs have been conducted. Probes based on the pargyline, rasagiline and selegiline scaffolds were investigated by gel-based proteomics as well as whole cell imaging, which confirmed binding to predominantly MAOA and MAOB.10–15 These molecules work by reacting with the covalently bound cofactor FAD upon radical activation.16 Despite these detailed mechanistic insights, a gel-free LC–MS/MS study for off-target profiling of major drugs such as tranylcypromine and pargyline is hitherto unreported. Herein, we synthesize tranylcypromine probes to elucidate the off-target profiles as well as their cellular localization in comparison to pargyline (Fig. 1B). The results indicate that unlike pargyline, tranylcypromine hits MAOA to a lesser degree in comparison to several off-targets including ALDHs. By incorporation of a photolabile reactive group, we additionally show major lysosomal trapping of tranylcypromine, which can be reduced by pre-treatment with lysosomotropic compounds like chloroquine.
First, we set out to directly decipher the specificity of tranylcypromine in living cells. For this, we designed two probe scaffolds, to account for both the irreversible and reversible binding partners of the compound (Fig. 1B). In the case of targeting irreversible binders such as flavin-dependent enzymes, we synthesized probe FBP2 which exhibits a terminal alkyne linked via an ether to the benzene ring of tranylcypromine.17 In brief, the synthesis was initiated by amine protection of (trans)-2-(4-hydroxyphenyl)cyclopropanamine followed by Mitsunobu reaction and acidic deprotection (Scheme 1). To complement our analysis with the detection of putative reversible binders, we synthesized probe FBPP2 bearing a diazirine photocrosslinker moiety via an analogous synthetic strategy (Scheme 1). Prior to proteomic studies with FBP2, we determined apparent IC50 values for MAOA and MAOB using an AzBTS/peroxidase-coupled assay and compared them to tranylcypromine (Fig. S1, ESI†). Satisfyingly, the alkyne handle did not strongly affect the inhibition of MAOA (IC50 = 0.5 μM) was largely comparable, whereas a decrease in inhibition of MAOB (IC50 = 2.3 μM) was observed. As a reference, we also prepared a structurally unrelated probe (FBP1) based on the pargyline scaffold as reported previously.10 This probe was already validated to target MAO enzymes via gel-based studies.
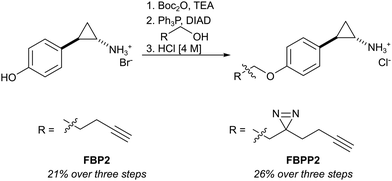 | ||
| Scheme 1 Synthesis of tranylcypromine-derived flavin binding probe (FBP2) and flavin binding photo-probe (FBPP2). | ||
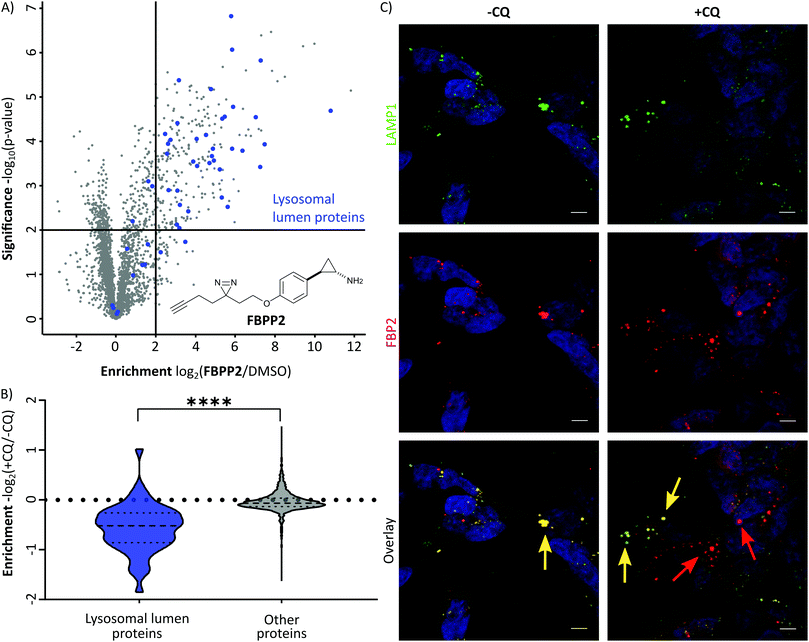
With these probes in hand, we turned our attention to the profiling of tranylcypromine in human cell lines. We selected SH-SY5Y due to its widespread use as a model cell line for studying neuronal function and neurodegenerative disorders like Parkinson's disease, as well as HeLa for a complementary approach.18 First we determined optimal conditions for FBP2 by concentration dependent gel-based labelling experiments (Fig. S2, ESI†). For subsequent target identification and quantification, SH-SY5Y and HeLa cells were treated with either FBP1 or FBP2 at 100 μM for 1 h; DMSO treated samples were included as control. After lysis, proteins were conjugated to biotin azide using click chemistry. Biotin-labelled target proteins were enriched on avidin beads, and further processed by tryptic digest for quantitative LC–MS/MS analysis and label-free quantification (Fig. 2A). In accordance with previous work, pargyline probe FBP1 revealed MAOA as the major target with an 81-fold enrichment over DMSO treated SH-SY5Y cells, followed by only one other significantly enriched protein, ALDH1B1, throughout both cell lines (Fig. S3A and B, ESI†).10 While MAOB was among the top hits in HeLa cells, no enrichment could be observed in SH-SY5Y. In contrast, labelling with the tranylcypromine derived probe FBP2 resulted in a comparatively low 3.6-fold enrichment of MAOA and no MAOB labelling in both cell lines (Fig. 2B and Fig. S3C, ESI†). Strikingly, multiple off-targets were observed, including ALDHs, heme oxygenase 2 and cathepsin L1 as significantly enriched hits (Fig. 2B, Fig. S3C (ESI†) and Table 1). To further validate these results, we performed a competitive analysis by pre-treating the cells with an excess of unmodified tranylcypromine and pargyline followed by probe incubation. Through this, MAOA and several off-targets including ALDHs and cathepsin L1 could be outcompeted by addition of both parent drugs in either cell line (Fig. 2C and Fig. S4, ESI†). Again, FBP2 revealed binding to more proteins than FBP1, confirming its higher promiscuity throughout the cell lines. With a panel of putative off-targets in hand, we performed initial binding and activity assays to monitor the consequences of tranylcypromine treatment. We obtained recombinant HMOX2 and ALDH2, two representative proteins with importance for cellular functions, and confirmed labelling with the probe (Fig. S5, ESI†). However, we were not able to monitor a reduction of their activity upon compound treatment, suggesting, that at least for these proteins inhibition seems not to be the main consequence of drug binding (data not shown). Nevertheless, it cannot be excluded that alteration of protein pathways by impaired protein–protein interactions contribute to a global cellular dysregulation. This notion was further supported by labelling with the photoprobe FBPP2, which enriched even more proteins (Fig. 3A and Fig. S6A, ESI†). Strikingly, competition with tranylcypromine confirmed binding to proteins, e.g. CTSL and ALDH1B1, detected with FBP2, yet no additional off-targets were confirmed for the drug (Fig. S6B, ESI†). Interestingly though, when analysing the proteins by cellular compartment, strong prevalence of lysosomal targets was found (Fig. 3A and Fig. S6A, ESI†). This emphasizes that, in line with the primary amino functionality, the compound is trapped in acidic organelles and physically interacts with binding partners in this compartment likely due to spatial proximity.19
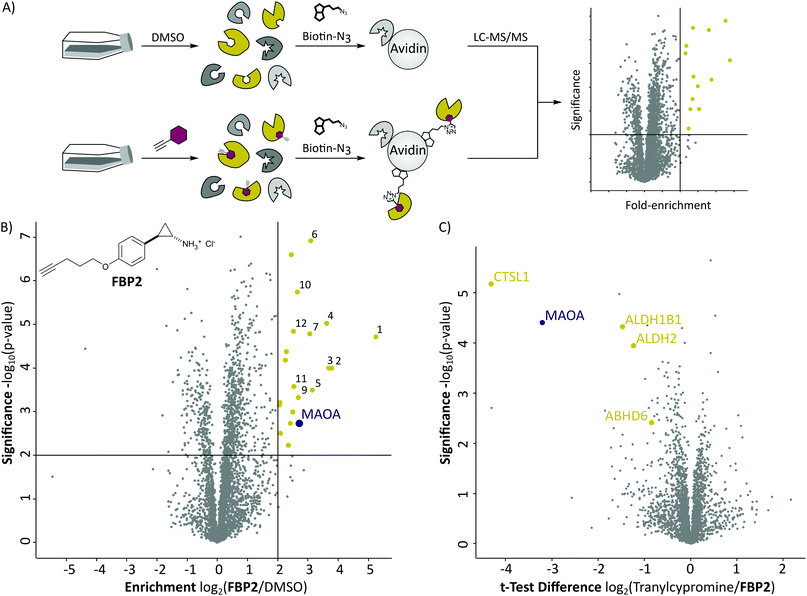 | ||
| Fig. 2 (A) Schematic depiction of a label-free quantitative proteomic experiment using avidin beads for target protein enrichment. (B) Results of target enrichment experiments using FBP2 in the human cancer cell line HeLa with 100 μM probe concentration. The scatter plot shows statistical significance of protein enrichment levels over protein enrichment ratios from probe treated to control cells. Cut offs are at a p-value < 0.01 and 4-fold enrichment (indicated by solid lines). Proteins within these set criteria are highlighted in yellow and described in more detail in the corresponding Table 1. MAOA is highlighted in purple. (C) Result of the corresponding competition experiment using FBP2 in the human cancer cell line HeLa. The scatter plots show statistical significance of protein enrichment levels over protein enrichment ratios from tranylcypromine treated to control cells. | ||
| No. | Gene name | Enrichment | Significance |
|---|---|---|---|
| log2(Probe/DMSO) | −log10(p-value) | ||
| 1 | BST2 | 5.24 | 4.72 |
| 2 | CTSL1 | 3.78 | 4.00 |
| 3 | PNPLA4 | 3.69 | 4.00 |
| 4 | HNRNPDL | 3.63 | 5.03 |
| 5 | PQBP1 | 3.14 | 3.50 |
| 6 | ALDH2 | 3.10 | 6.92 |
| 7 | ABHD6 | 3.06 | 4.79 |
| 8 | MAOA | 2.72 | 2.72 |
| 9 | CHMP1B | 2.68 | 3.32 |
| 10 | ALDH1B1 | 2.64 | 5.74 |
| 11 | C19orf43 | 2.53 | 3.57 |
| 12 | HMOX2 | 2.51 | 4.64 |
As the therapeutic MAO targets are located in mitochondria, lysosomal trapping for tranylcypromine lowers the effective dose at the cellular compartment of interest. To explore the possibility of manipulating this effect, we repeated photo-affinity labelling with cells that were either pre-treated with chloroquine or DMSO. Chloroquine has been long marketed for the treatment of malaria, but regained interest in the COVID-19 crisis for its antiviral effect by increasing lysosomal pH.20 The result of this experiment nicely shows a very significant decrease of FBPP2 binding to proteins annotated to be located in the lysosomal lumen (Fig. 3B). As the mean enrichment of all other proteins remains nearly unaltered, it emphasises the strong interaction of tranylcypromine with lysosomal proteins.
To visualize these intriguing findings, we used fluorescence imaging of SH-SY5Y cells treated with FBP2. In accordance to our previous results, we saw predominant localization of the probe in the lysosome (as visualized by the specific marker for LAMP1) (Fig. 3C). Most importantly, when pre-treating the cells with lysosomotropic compounds, either chloroquine or tamoxifen, a clear release of the tranylcypromine probe from the lysosomes was detectable (Fig. 3C and Fig. S7, ESI†).
Conclusions
Due to its unique binding mechanism to monoamine oxidases and use in various diseases, tranylcypromine has been subject to extensive scientific research, yet many aspects of this compound, like side effects or drug–drug-interactions remain hitherto unknown. The aim of this study, was to utilize modern LC–MS/MS technology, in order to find reversible and irreversible protein targets of tranylcypromine. With our novel probe, we were able to show that tranylcypromine, in contrast to pargyline, irreversibly binds to many off-targets. Additionally, by using a photoprobe as well as fluorescence imaging, we proved strong lysosomal trapping of the compound that can be reduced by addition of the lysosomotropic compounds chloroquine and tamoxifen. Co-administration of therapeutic doses of chloroquine and tranylcypromine is known to cause severe side effects and was therefore not investigated further. As our results show a release of tranylcypromine from the lysosome, these findings could become a starting point for new studies, looking into the possible use of lysosomotropic drugs to decrease tranylcypromine dosage.Note
MS-data are available via ProteomeXchange with identifier PXD018580.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the European Research Council (ERC) and the European Union's Horizon 2020 research and innovation programme (grant agreement no. 725085, CHEMMINE, ERC consolidator grant). We thank K. Bäuml and M. Wolff for technical assistance and I. Hübner, S. Hacker and S. Ruddell for careful proof reading.References
- R. McDermott, D. Tingley, J. Cowden, G. Frazzetto and D. D. P. Johnson, Monoamine oxidase A gene (MAOA) predicts behavioral aggression following provocation, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(7), 2118–2123 CrossRef CAS PubMed.
- D. Kim, S. H. Baik, S. Kang, S. W. Cho, J. Bae, M.-Y. Cha, M. J. Sailor, I. Mook-Jung and K. H. Ahn, Close Correlation of Monoamine Oxidase Activity with Progress of Alzheimer's Disease in Mice, Observed by in Vivo Two-Photon Imaging, ACS Cent. Sci., 2016, 2(12), 967–975 CrossRef CAS PubMed.
- M. B. H. Youdim, D. Edmondson and K. F. Tipton, The therapeutic potential of monoamine oxidase inhibitors, Nat. Rev. Neurosci., 2006, 7(4), 295–309 CrossRef CAS PubMed.
- P. Riederer and G. Laux, MAO-inhibitors in Parkinson's Disease, Experimental Neurobiology, 2011, 20(1), 1–17 CrossRef PubMed.
- S. K. Al-Nuaimi, E. M. Mackenzie and G. B. Baker, Monoamine oxidase inhibitors and neuroprotection: a review, Am. J. Ther., 2012, 19(6), 436–448 CrossRef PubMed.
- R. Shi, Q. Wu, C. Xin, H. Yu, K.-L. Lim, X. Li, Z. Shi, C.-W. Zhang, L. Qian, L. Li and W. Huang, Structure-Based Specific Detection and Inhibition of Monoamine Oxidases and Their Applications in Central Nervous System Diseases, Chembiochem, 2019, 20(12), 1487–1497 CAS.
- J. Huang, F. Liu, H. Tang, H. Wu, L. Li, R. Wu, J. Zhao, Y. Wu, Z. Liu and J. Chen, Tranylcypromine Causes Neurotoxicity and Represses BHC110/LSD1 in Human-Induced Pluripotent Stem Cell-Derived Cerebral Organoids Model, Front. Neurol., 2017, 8, 626 CrossRef PubMed.
- E. Spina and M. G. Scordo, Clinically significant drug interactions with antidepressants in the elderly, Drugs Aging, 2002, 19(4), 299–320 CrossRef CAS PubMed.
- S. Bleakley, Antidepressant drug interactions: evidence and clinical significance, Prog. Neurol. Psychiatry, 2016, 20(3), 21–27 CrossRef.
- J. M. Krysiak, J. Kreuzer, P. Macheroux, A. Hermetter, S. A. Sieber and R. Breinbauer, Activity-based probes for studying the activity of flavin-dependent oxidases and for the protein target profiling of monoamine oxidase inhibitors, Angew. Chem., Int. Ed., 2012, 51(28), 7035–7040 CrossRef CAS PubMed.
- L. Li, C.-W. Zhang, J. Ge, L. Qian, B.-H. Chai, Q. Zhu, J.-S. Lee, K.-L. Lim and S. Q. Yao, A Small-Molecule Probe for Selective Profiling and Imaging of Monoamine Oxidase B Activities in Models of Parkinson's Disease, Angew. Chem., Int. Ed., 2015, 54(37), 10821–10825 CrossRef CAS PubMed.
- H. Fang, H. Zhang, L. Li, Y. Ni, R. Shi, Z. Li, X. Yang, B. Ma, C. Zhang, Q. Wu, C. Yu, N. Yang, S. Q. Yao and W. Huang, Rational Design of a Two-Photon Fluorogenic Probe for Visualizing Monoamine Oxidase A Activity in Human Glioma Tissues, Angew. Chem., Int. Ed., 2020, 59, 7536 CrossRef CAS PubMed.
- X. Wu, W. Shi, X. Li and H. Ma, A Strategy for Specific Fluorescence Imaging of Monoamine Oxidase A in Living Cells, Angew. Chem., Int. Ed., 2017, 56(48), 15319–15323 CrossRef CAS PubMed.
- X. Wu, L. Li, W. Shi, Q. Gong, X. Li and H. Ma, Sensitive and Selective Ratiometric Fluorescence Probes for Detection of Intracellular Endogenous Monoamine Oxidase A, Anal. Chem., 2016, 88(2), 1440–1446 CrossRef CAS PubMed.
- Z. Yang, W. Li, H. Chen, Q. Mo, J. Li, S. Zhao, C. Hou, J. Qin and G. Su, Inhibitor structure-guided design and synthesis of near-infrared fluorescent probes for monoamine oxidase A (MAO-A) and its application in living cells and in vivo, Chem. Commun., 2019, 55(17), 2477–2480 RSC.
- C. Binda, M. Li, F. Hubalek, N. Restelli, D. E. Edmondson and A. Mattevi, Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(17), 9750–9755 CrossRef CAS PubMed.
- K. L. Kirk, S. Yoshida and G. Haufe, Synthesis and Biochemical Evaluation of Fluorinated Monoamine Oxidase Inhibitors, Fluorine Health, 2008, 661–697 CAS.
- H. Xicoy, B. Wieringa and G. J. M. Martens, The SH-SY5Y cell line in Parkinson's disease research: a systematic review, Mol. Neurodegener., 2017, 12(1), 10 CrossRef PubMed.
- A. M. Zuhl, C. E. Nolan, M. A. Brodney, S. Niessen, K. Atchison, C. Houle, D. A. Karanian, C. Ambroise, J. W. Brulet, E. M. Beck, S. D. Doran, B. T. O'Neill, C. W. Ende, C. Chang, K. F. Geoghegan, G. M. West, J. C. Judkins, X. Hou, D. R. Riddell and D. S. Johnson, Chemoproteomic profiling reveals that cathepsin D off-target activity drives ocular toxicity of β-secretase inhibitors, Nat. Commun., 2016, 7, 13042 CrossRef CAS PubMed.
- S. Lu, T. Sung, N. Lin, R. T. Abraham and B. A. Jessen, Lysosomal adaptation: how cells respond to lysosomotropic compounds, PLoS One, 2017, 12(3), e0173771 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cb00048e |
| This journal is © The Royal Society of Chemistry 2020 |