Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploring open cheminformatics approaches for categorizing per- and polyfluoroalkyl substances (PFASs)†
Bo
Sha‡
 a,
Emma L.
Schymanski‡
a,
Emma L.
Schymanski‡
 *b,
Christoph
Ruttkies
*b,
Christoph
Ruttkies
 c,
Ian T.
Cousins
c,
Ian T.
Cousins
 a and
Zhanyun
Wang
a and
Zhanyun
Wang
 *d
*d
aDepartment of Environmental Science and Analytical Chemistry (ACES), Stockholm University, SE-10691, Stockholm, Sweden
bLuxembourg Centre for Systems Biomedicine, University of Luxembourg, 6 Avenue du Swing, L-4367 Belvaux, Luxembourg. E-mail: emma.schymanski@uni.lu
cDepartment Biochemistry of Plant Interactions, Leibniz Institute of Plant Biochemistry, Weinberg 3, 06120 Halle, Germany
dChair of Ecological Systems Design, Institute of Environmental Engineering, ETH Zürich, 8093 Zürich, Switzerland. E-mail: zhanyun.wang@ifu.baug.ethz.ch
First published on 24th September 2019
Abstract
Per- and polyfluoroalkyl substances (PFASs) are a large and diverse class of chemicals of great interest due to their wide commercial applicability, as well as increasing public concern regarding their adverse impacts. A common terminology for PFASs was recommended in 2011, including broad categorization and detailed naming for many PFASs with rather simple molecular structures. Recent advancements in chemical analysis have enabled identification of a wide variety of PFASs that are not covered by this common terminology. The resulting inconsistency in categorizing and naming of PFASs is preventing efficient assimilation of reported information. This article explores how a combination of expert knowledge and cheminformatics approaches could help address this challenge in a systematic manner. First, the “splitPFAS” approach was developed to systematically subdivide PFASs (for eventual categorization) following a CnF2n+1–X–R pattern into their various parts, with a particular focus on 4 PFAS categories where X is CO, SO2, CH2 and CH2CH2. Then, the open, ontology-based “ClassyFire” approach was tested for potential applicability to categorizing and naming PFASs using five scenarios of original and simplified structures based on the “splitPFAS” output. This workflow was applied to a set of 770 PFASs from the latest OECD PFAS list. While splitPFAS categorized PFASs as intended, the ClassyFire results were mixed. These results reveal that open cheminformatics approaches have the potential to assist in categorizing PFASs in a consistent manner, while much development is needed for future systematic naming of PFASs. The “splitPFAS” tool and related code are publicly available, and include options to extend this proof-of-concept to encompass further PFASs in the future.
Environmental significancePer- and polyfluoroalkyl substances (PFASs) are attracting increasing attention from scientists, regulators and the public. High resolution mass spectrometry has enabled the discovery of many new/overlooked and often only partially characterised PFASs in different environments, yet inconsistent reporting prevents the effective exchange of this vital information. Since identification and categorization of PFASs is an essential first step in determining whether these will have problematic properties, this work explores the potential of open cheminformatics approaches for the systematic categorization of PFASs, using select PFAS categories in the recent OECD list. The structure-based cheminformatics tool provided is implemented flexibly, interpreting structures quickly and has the potential to help scientists, regulators and other interested parties categorize, and thus assess, PFASs. |
1. Introduction
Per- and polyfluoroalkyl substances (PFASs), as currently defined under the OECD/UNEP Global PFC Group, are organic chemicals containing at least one perfluorinated carbon moiety,1i.e., –CF2–. PFASs may exhibit a number of desirable chemical properties, such as high resistance to heat and chemical reactions, as well as hydrophobicity and oleophobicity, in comparison to their non-fluorinated analogues.2 Therefore, since the 1940s, large numbers of PFASs with diverse functional groups and properties have been developed and used widely in numerous industrial and consumer applications.2–7 Since the late 1990s, there has been mounting scientific evidence of the human health risks of many PFASs,8 mirrored with mounting concern in policymakers and the general public. In particular, perfluorooctanesulfonic acid (PFOS), its salts and perfluorooctane sulfonyl fluoride, and perfluorooctanoic acid (PFOA), its salts and PFOA-related compounds, were listed under the UN Stockholm Convention on Persistent Organic Pollutant in 2009 and 2019 for a global phase-out, respectively, and perfluorohexanesulfonic acid (PFHxS), its salts and PFHxS-related compounds are being evaluated for listing under the Stockholm Convention.To date, most studies on the occurrence and effects of PFASs have focused on a limited set of PFASs, namely perfluoroalkyl acids (PFAAs), and several PFAA precursors derived from perfluoroalkane sulfonyl fluorides (i.e., PASF-based compounds) as well as perfluoroalkyl iodides (i.e., n:2 fluorotelomer-based compounds, n:2 FTs),8 see Fig. 1. For the latter, the most commonly studied compounds are those with relatively simple molecular structures, e.g. perfluoroalkane sulfonamides/-amidoethanols (FASAs/FASEs) and fluorotelomer alcohols/sulfonic acids (FTOHs/FTSAs).8Fig. 1 provides an overview of these major PFAS groups and either generic composition information, or specific examples. Additionally, several lists with specific and generic structures are already available online (see ref. 9–14).
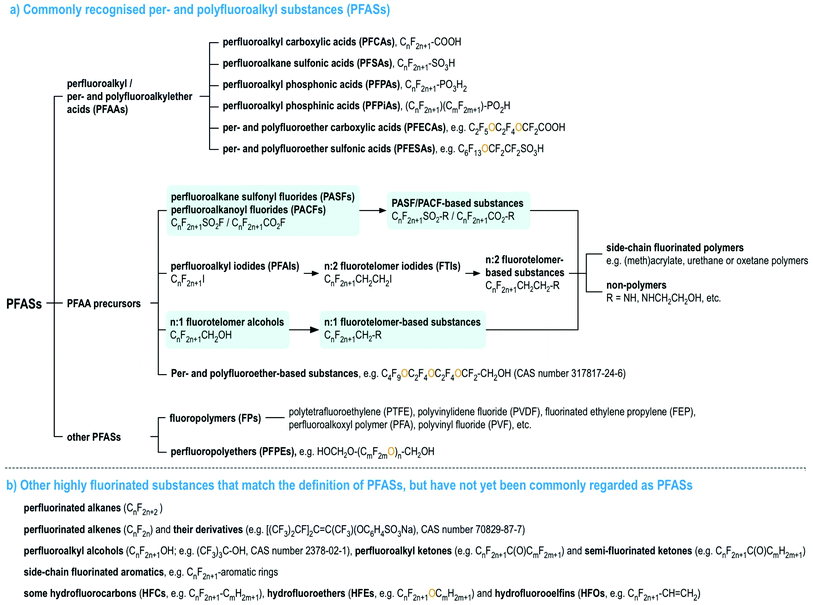 | ||
| Fig. 1 An overview of PFASs (adopted from the OECD report1 with the addition of perfluoroalkanoyl fluorides (PACFs), n:1 fluorotelomer alcohols and their derivatives, highlighted in light blue). Interactive lists with structures and/or generic representations, are available online.9,10 | ||
The main research focus on PFAAs and PFAA precursors with simple molecular structures is due to two main reasons: (1) analytically, they are relatively easier to measure than other PFASs with more complex molecular structures; and (2) analytical standards are generally commercially available. It has been challenging to expand beyond this domain as the chemical composition (let alone analytical reference standards) of most remaining commercial products are not known in the public domain. However, with the increasing accessibility of high resolution mass spectrometry and advancement of non-target screening techniques, as well as increasing exchange of chemical information between authorities and scientists, these factors are becoming less of a barrier for identifying overlooked and unknown PFASs, which can include unreacted reactant residuals and degradation intermediates present in products and in the environment. This has been repeatedly observed in the many recent “non-target” studies on the PFAS-containing aqueous fire-fighting foams and their contaminated sites,15–17 as well as recent reports outlining the extent of PFASs (and other chemicals) in higher order food chain animals such as polar bears18 and near manufacturing plants.19–22 The number of “non-target” studies on PFASs has greatly increased in the past several years and has been reviewed recently.15
Due to the diverse and often complex molecular structures of different PFASs, it may often be challenging to categorize newly identified PFASs in a consistent and coherent manner, particularly for non-technical experts and those who are not familiar with PFASs. The >4700 Chemical Abstracts Service Registry Numbers (CAS_RN) identified in the OECD PFAS list were manually assigned by the same person to certain structure categories. However, such manual categorization efforts cannot be easily reproduced by others due to the high level of expertise required, possible different interpretations of structural traits, and the potential for human errors including oversights and typing errors.
Furthermore, the current development of PFAS terminologies lags behind the rapid development and application of “non-target” screening techniques, particularly for PFASs without a given CAS_RN. As such, the authors of individual studies have often created their own naming conventions (including acronyms) for newly identified PFASs. This leads to the generation of a lot of parallel and often non-intuitive acronyms, potentially prohibiting effective communication among scientists themselves and with other stakeholders, creating barriers for synthesizing knowledge. For instance, “1,1,2,2-tetrahydroperfluorodecanol”, “2-(perfluorooctyl)ethanol”, “8:2 FTOH”, “8:2 fluorotelomer alcohol”, and “PFA 8” are a few of >36 synonyms registered for one single structure (CAS_RN 678-39-7 (ref. 16)). This is not an issue for PFAS studies alone, but is exacerbated for these substances due to high public and scientific interest, as well as the increasing advancement and application of “non-target” studies.15
Some non-target studies17,18 are now using the information included in publically available suspect lists, via e.g. the NORMAN Suspect List Exchange10 and the CompTox Chemicals Dashboard19,20 in their identification efforts. In addition, several groups are investing efforts into naming and categorisation of PFASs. For instance, the US EPA are experimenting with the incorporation of expert knowledge and cheminformatics approaches developed in house,21 recently offering some perspectives on how to name certain groups of PFASs, while Barzen-Hansen et al.22 used a simplified, manual IUPAC-based naming system for the PFASs that they identified in their non-target screening, detailed in the ESI of that publication (pages S6–S7; Table S3 pages S15–S21).†
Recently, an open access approach, ClassyFire,23 was developed to categorize chemicals systematically into a formal chemical ontology. ClassyFire uses chemical structures and structural features to automatically assign chemicals to a predefined taxonomy consisting of up to 11 levels (termed kingdom, superclass, class, subclass, etc.). ClassyFire has been used to annotate over 77 million compounds,23 and the results can be looked up with InChIKeys (the hashed version of the full International Chemical Identifier, InChI)24. Only a few very well-known PFASs were in the dataset used to train ClassyFire, primarily those entries that are in DrugBank25 or T3DB.26 However, new calculations can be performed using structural information provided as Simplified Molecular Input Line Entry System (SMILES),27 InChIs or even the International Union of Pure and Applied Chemistry (IUPAC) name. Results and calculations are available via a freely accessible web server28 at http://classyfire.wishartlab.com.
This background motivates the current study to investigate possible additional automated, open approaches that combine background (expert) knowledge, existing PFAS naming conventions, and cheminformatics to systematically categorize PFASs, particularly in a non-target screening context. In brief, this study consists of two main components: (1) development and testing of a structure manipulation tool, splitPFAS, using simple SMILES27 and the related SMiles ARbitrary Target Specification (SMARTS)29 annotations (explained below) to identify PFASs based on pre-defined structural traits; and (2) investigation of the potential to use the combination of splitPFAS and the ontology-based ClassyFire.23 More specifically, this study focuses on four groups of PFAA precursors: PACF- and PASF- as well as n:1 and n:2 fluorotelomer-based compounds (see Fig. 1) as test subjects (using discrete structures present in the recent OECD PFAS list1). This is because a common terminology for some PFASs in these four groups has been recommended in Buck et al.4 and thus can be used as a reference point to validate the approach. While there are also many other groups of PFASs of interest, e.g. perfluoroether-based substances,1 these were not considered as part of this study, as no commonly used basic rules exist for characterizing, categorizing and naming these structures yet. As there is an ongoing international effort under the leadership of the OECD/UNEP Global PFC Group to establish some harmonized basic rules for these groups of PFASs,30 it is the intention that the approach presented here can be expanded to cover these cases, once this additional information is available in the near future.
2. Methods
This work consisted of three major cheminformatics steps (see Fig. 2), described in detail below. First, the “splitPFAS” method was developed and used to identify whether a given PFAS was within the four PFAS categories of interest in this study. Second, the structures of the PFASs matching these patterns were manipulated according to defined rules and scenarios. The resulting modified structures were used as input for ClassyFire in the third step. The ClassyFire results were then compared with the common terminology recommended by Buck et al.,4 discussed in the Results section.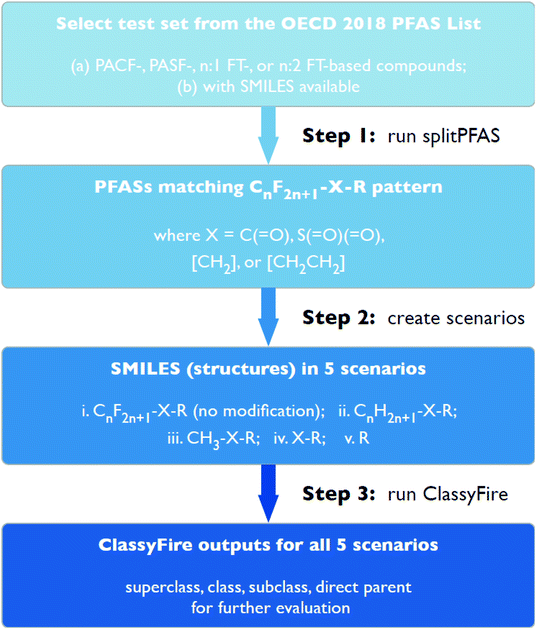 | ||
| Fig. 2 Step-by-step workflow for selecting, splitting, modifying and categorizing PFASs in the current work. | ||
2.1 Groups of PFASs of interest
In this study, the following groups of PFASs from the OECD list (structure categories 101 to 109, 201 to 209, and 401 to 410) were used as a test data set, including:(a) perfluoroalkanoyl (PACF)-based compounds (or PACF derivatives);
(b) perfluoroalkane sulfonyl (PASF)-based compounds (or PASF derivatives); and
(c) n:1* and n:2 fluorotelomer (FT)-based compounds (n:1/n:2 FTs).
*As known commercial n:1 fluorotelomer-based compounds are not derived from the telomerization process, but rather from the reduction of perfluoroalkyl carboxylic acids,3 they are not, strictly speaking, fluorotelomers. Despite this, they are termed “n:1 FT-based compounds” here for readability, since the pattern of the perfluorocarbon:hydrocarbon chain is the same (i.e., n:1 vs. n:2).
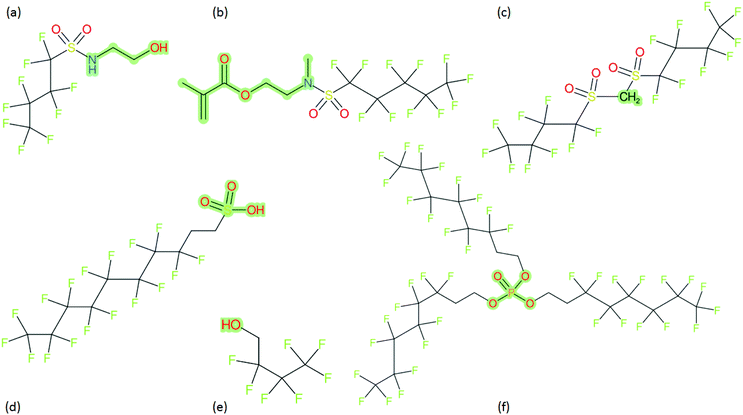
These groups display systematic patterns. The PACF derivatives can be represented with the generic formula CnF2n+1–CO–R, the PASF derivatives as CnF2n+1–SO2–R, and the n:1/n:2 FTs as CnF2n+1–CH2–R/CnF2n+1–CH2CH2–R. Some example PASFs (top row, (a)–(c)) and FTs (bottom row, (d)–(f)) are given in Fig. 3 below, with the “R” group highlighted in green. The corresponding names, CAS_RN, and SMILES (Simplified Molecular Input Line Entry System) code30 of the R group, shown in blue as “RSMILES”, are given in the caption.
2.2 SMILES and SMARTS-based manipulations with splitPFAS
The systematic patterns, visible from the structures shown in Fig. 3 and the generic formulas given above, render these substances suitable for basic cheminformatics manipulations based on SMARTS,29 an extension of SMILES able to specify substructures via e.g. wildcard atoms and logical operators (see Reference for further details). In fact, the green highlighting in Fig. 3 is performed using SMARTS functionality in the chemical drawing software used here, CDK Depict.31,32 These systematic patterns mean that it would be possible to split the molecule into two parts, the perfluoroalkyl part (CnF2n+1) and the R group, using a SMARTS-based recognition of the alpha carbon on the PFAS chain and the “dividing group” (which we will term “X” in this manuscript). In other words, using the test subjects CnF2n+1–CO–R, CnF2n+1–SO2–R, CnF2n+1–CH2–R, and CnF2n+1–CH2CH2–R as examples, all substances satisfy the pattern CnF2n+1–X–R where X is CO, SO2, CH2 or CH2CH2. Using this information, it is possible to come up with some simple SMARTS codes to catch these cases:PACF derivatives: FC(F)([C,F])C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/b_char_e001.gif) O)[!$(C(F)(F)); !$(F)] X = C( O)[!$(C(F)(F)); !$(F)] X = C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
PASF derivatives: FC(F)([C,F])S(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/b_char_e001.gif) O)( O)(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/b_char_e001.gif) O)[!$(C(F)(F)); !$(F)] X = S( O)[!$(C(F)(F)); !$(F)] X = S(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)( O)(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
| n:2 FTs: FC(F)([C,F])[CH2][CH2][!$(C(F)(F)); !$(F)] X = [CH2][CH2] |
| n:1 FTs: FC(F)([C,F])[CH2][!$(C(F)(F)); !$(F)] X = [CH2] |
As SMARTS can be inherently tricky for users not intimately acquainted with SMILES, let alone SMARTS notation, “splitPFAS”, a program written in Java using the Chemistry Development Kit (CDK)32 was created to implement this SMARTS-based pattern search with a simple input file that requires only the SMILES/SMARTS of the dividing group “X”, along with several options controlling the output. The SMARTS codes above can be interpreted as follows: (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) refers to a double bonded oxygen, [CH2] specifies a carbon with exactly 2 hydrogens attached. FC(F)([C,F]) specifies a CF2 attached to either another F or C, i.e., this detects the “alpha” carbon of the perfluorinated chain, while [!$(C(F)(F)); !$(F)] means that X (the SMARTS code in bold above) is not adjacent to a CF2 group or an F and thus identifies the R part of CnF2n+1–X–R.
O) refers to a double bonded oxygen, [CH2] specifies a carbon with exactly 2 hydrogens attached. FC(F)([C,F]) specifies a CF2 attached to either another F or C, i.e., this detects the “alpha” carbon of the perfluorinated chain, while [!$(C(F)(F)); !$(F)] means that X (the SMARTS code in bold above) is not adjacent to a CF2 group or an F and thus identifies the R part of CnF2n+1–X–R.
The SMARTS detecting the PFAS alpha carbon (both parts of the non-bolded SMARTS code above) can be adjusted by advanced users via the optional input “pacs” (PFAS alpha carbon SMARTS). The “splitPFAS” approach was integrated into the “MetFragTools” suite (current version 2.4.5 (ref. 33)), with source code and documentation available on GitHub.34 Accompanying R scripts and functions are documented and available for use via the RChemMass package in GitHub35 and as part of the ESI,† along with user instructions on how to use splitPFAS. The SMARTS implemented by default in the current version were designed to handle the case studies in the proof-of-concept approach described here, i.e., focusing on saturated, linear isomers of the perfluoroalkyl part (CnF2n+1). Other forms of the perfluoroalkyl part (e.g., unsaturated and/or branched or cyclic isomers) can be captured (e.g., in future studies) by adjusting the SMARTS with the “pacs” option described above.
The order of the SMARTS in the splitPFAS input file (example available online)36 is important, as it determines the processing order of the list of PFASs. For instance, the order used here is:
C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
S(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)( O)(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
| [CH2][CH2] |
| [CH2] |
2.3 Calculation with ClassyFire using different scenarios
As mentioned in the introduction, ClassyFire uses chemical structures and structural features to automatically categorize chemicals into a specially designed ontology. Pre-calculated results, as well as results from new calculations can be accessed via a freely accessible web server at http://classyfire.wishartlab.com. In this study, the web server was accessed using InChIKeys (to retrieve pre-calculated results) and SMILES (for new calculations) from the OECD PFAS list via R. The script is available in the ESI.The ClassyFire workflow contains four steps: (1) preprocessing of the chemical entity; (2) feature extraction; (3) rule-based category assignment and category reduction; and (4) selection of the direct parent.32 Briefly, the categorization starts with the calculation of the physico-chemical (e.g. mass and pKa) and structural properties (e.g. number of aromatic or aliphatic rings) of the query compound. Then, a list of structural features is generated based on a combination of property calculations and superstructure search, which is performed on a built-in library of over 9000 manually designed SMARTS patterns and Markush structures.23 Each feature in the list is then assigned to a category in the taxonomy according to a manually compiled dictionary, which contains the weighting and category of each feature. After that, a non-redundant list of chemical categories is constructed and the category of the largest structural feature that describes the compound is selected as the direct parent. However, when the largest structural feature is less informative in describing the compound, the category of the most descriptive feature is defined as the direct parent. Such cases are handled by a manually compiled set of exceptions in ClassyFire. In ClassyFire, the taxonomy categories are defined by unambiguous, computable structural rules, and are named using a consensus-based nomenclature. In this study, four outputs from ClassyFire (superclass, class, subclass and direct parent) were evaluated for their potential to be used in systematic categorization and naming of PFASs by comparing with the common terminology recommended by Buck et al.4
To explore how different structures may influence the ClassyFire results, especially as ClassyFire was not developed with PFASs in mind, the PFASs of interest (i.e., PACF derivatives, PASF derivatives, and n:1/n:2 FTs) were manipulated using splitPFAS into five scenarios. To start, the SMILES of the structure CnF2n+1–X–R, was split into the fluorinated (CnF2n+1), dividing group (X), and non-fluorinated functional group (R) parts using splitPFAS. These were then used in various combinations, with each scenario documented below in terms of the pattern CnF2n+1–X–R. The SMILES codes of the structures resulting from the following scenarios were then taken as inputs for ClassyFire. The scenarios were:
(i) CnF2n+1–X–R The structure was not modified;
(ii) CnH2n+1–X–R The structure was converted into a non-fluorinated analogue (i.e., replacing F with H in the PFAS part);
(iii) H3C–X–R The fluorinated part was discarded and a methyl added to X, which was re-combined with R to form H3C–X–R and thus compensated for the missing PFAS chain;
(iv) X–R As in scenario (iii), but only the SMILES of X–R;
(v) R As in scenario (iv), but only the SMILES of R.
The rationale behind these scenarios is as follows. Scenario (i) formed the base case; ideally this case would yield the desired categorization results, but as ClassyFire was not trained on many PFASs, this was not expected initially in all cases. Scenario (ii) was created to determine whether, instead, ClassyFire could generate sufficiently informative results on the analogous non-fluorinated structure (as alkyl chains are generally far more prevalent than perfluoroalkyl chains). To remove the influence of the perfluorinated carbon chain on the results entirely, scenario (iv) was conceived. This initially generated many errors that could be resolved by adding a methyl group; this became scenario (iii). An additional concern with scenario (iv), which was easier to implement than scenario (iii), was that the replacement of a (perfluoro)alkyl chain with a sole hydrogen (a result of SMILES manipulation) could lead to miscategorization of the functional group (e.g. an ether becomes an alcohol). Since splitPFAS could actually already separate the perfluorinated part and the functional group “X”, finally scenario (v), containing only the R group, was used as the simplest case to assess the potential of ClassyFire for categorization.
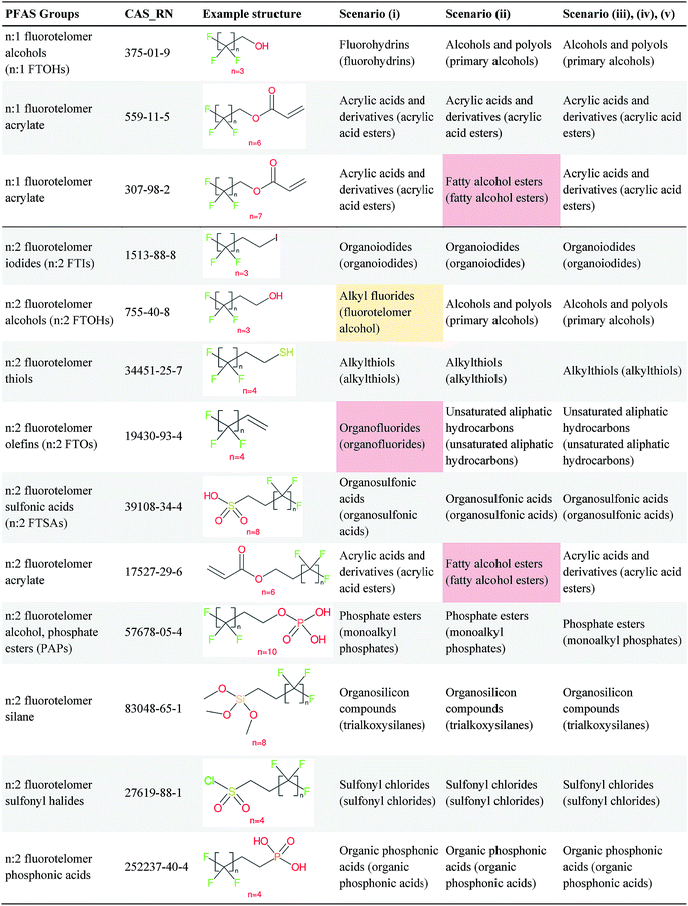
Several examples of scenarios (i) to (iii) are shown in Fig. 4, giving one selected compound for each major case (i.e. PASF, PACF, n:1 FT, n:2 FT). The “X” group is shown in green; thus the X–R and R groups in scenarios (iv) and (v) can be interpreted easily from the column showing scenario (iii). While the splitPFAS method in the Java program can handle structures that result in multiple perfluorinated carbon chains or multiple non-fluorinated parts after splitting (e.g.Fig. 3(c) and (f)), these were not taken into further consideration for ClassyFire at this stage, primarily for simplicity in presenting the results at this proof-of-concept stage, but are discussed further below.
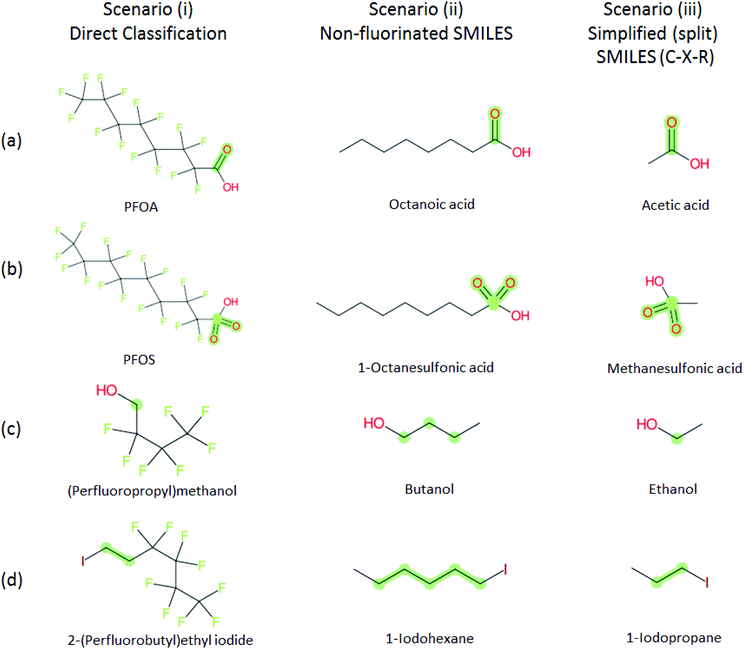 | ||
Fig. 4 Scenarios (i), (ii) and (iii) for X = (a) C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) (b) S( O) (b) S(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)( O)(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) (c) [CH2] (d) [CH2][CH2]. The corresponding compound information is in the ESI.† Green highlights indicate the SMARTS pattern (“X”). O) (c) [CH2] (d) [CH2][CH2]. The corresponding compound information is in the ESI.† Green highlights indicate the SMARTS pattern (“X”). | ||
3. Results
3.1 Results from splitPFAS
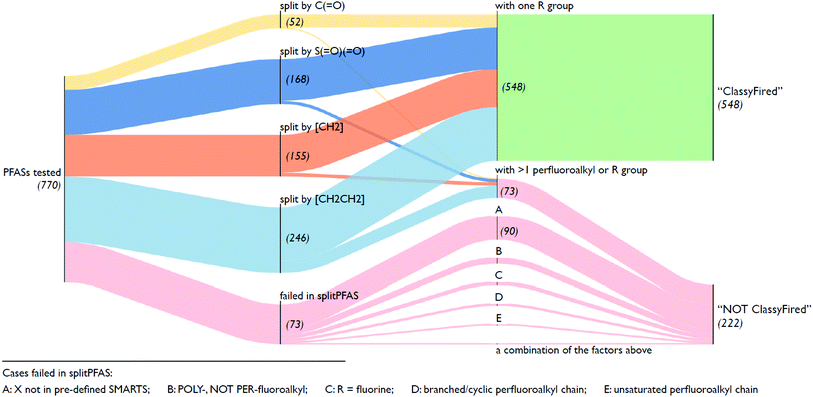
As mentioned in Section 2.1, the splitPFAS tool was used to split the input SMILES from the selected OECD PFASs following the “CnF2n+1–X–R” pattern according to the given SMARTS of the dividing group “X” (listed above). The output of splitPFAS includes the SMILES of “X”, “CnF2n+1–X” and the R group (separated by “|” if more than one), as well as the number of PFAS parts and an error message if the splitting failed. Several examples are given in Table 1; the complete results are in the ESI.† As mentioned above, the order of SMARTS in the file determines the processing order and thus the order of the given SMARTS may have an influence on the results. For instance, if [CH2] is listed before [CH2][CH2] in the SMARTS file, the latter entry would be useless, as all SMILES matching this pattern would also match the preceding SMARTS and thus have already been processed. The results below were processed using the order given in Section 2.2.
Fig. 5 illustrates an overview of the results from splitPFAS. In total, out of the 770 compounds selected from the latest OECD PFAS list (i.e., those with structure code 101–109, 201–209 and 401–410), splitPFAS performed as designed for 621 compounds (52, 168, 155 and 246 were split by “C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)”, “S(
O)”, “S(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)(
O)(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)”, “[CH2]” and “[CH2][CH2]”, respectively). Among them, 548 compounds (50, 156, 142, and 200 for compounds split by “C(
O)”, “[CH2]” and “[CH2][CH2]”, respectively). Among them, 548 compounds (50, 156, 142, and 200 for compounds split by “C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)”, “S(
O)”, “S(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)(
O)(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)”, “[CH2]” and “[CH2][CH2]”, respectively) match the pattern “CnF2n+1–X–R” and were further used as inputs in ClassyFire. The others that were correctly split using splitPFAS (73 compounds) had either two or more “CnF2n+1” or “R” groups and were not used as inputs in ClassyFire, primarily for simplicity at this proof-of-concept phase. As mentioned above, splitPFAS was run with the SMARTS [CH2][CH2] (for n:2 FTs) before [CH2] (for n:1 FTs) to ensure that these cases were treated correctly. The remaining 149 compounds were not correctly split using splitPFAS because their molecular structures were outside the patterns pre-defined in the current version of splitPFAS, including:
O)”, “[CH2]” and “[CH2][CH2]”, respectively) match the pattern “CnF2n+1–X–R” and were further used as inputs in ClassyFire. The others that were correctly split using splitPFAS (73 compounds) had either two or more “CnF2n+1” or “R” groups and were not used as inputs in ClassyFire, primarily for simplicity at this proof-of-concept phase. As mentioned above, splitPFAS was run with the SMARTS [CH2][CH2] (for n:2 FTs) before [CH2] (for n:1 FTs) to ensure that these cases were treated correctly. The remaining 149 compounds were not correctly split using splitPFAS because their molecular structures were outside the patterns pre-defined in the current version of splitPFAS, including:
(1) the perfluoroalkyl chain was branched or cyclic (10 compounds),
(2) the perfluoroalkyl chain was unsaturated (7 compounds),
(3) the fluoroalkyl chain was not perfluorinated (23 compounds),
(4) the R group was a single F atom (15 compounds),
(5) the dividing groups (X) were outside the SMARTS notation used in splitPFAS (90 compounds, see Section 2.2), and
(6) a combination of the factors above (4 compounds).
Details on these cases (and possible extensions to resolve them in future studies) are discussed further in Section 4 below.
In addition, the splitPFAS results were compared with the manually curated structure codes given in the latest OECD PFAS list.1,13 In total, eleven compounds were identified as being mislabeled in this list (one PACF was an n:1 FT, two PASFs were in fact n:2 FTs, one n:2 FTs were PASFs, and eight n:1 FTs were rather perfluoroalkene derivatives). These entries (a list is provided in the ESI†) will be communicated back to the OECD/UNEP Global PFC Group for possible revisions in the next OECD PFAS list. This demonstrates that splitPFAS has the potential to assist in categorizing PFAS automatically and detect human error, thus supporting experts in this work, which is becoming more challenging with the thousands of PFAS structures now being documented.
As this OECD PFAS list was the basis for this investigation, and as CAS_RN and name are the primary identifiers in this list, we refer to specific examples throughout this manuscript using the CAS_RN from this list for clarity and to allow a more compact presentation of the results below.
3.2 Results from ClassyFire
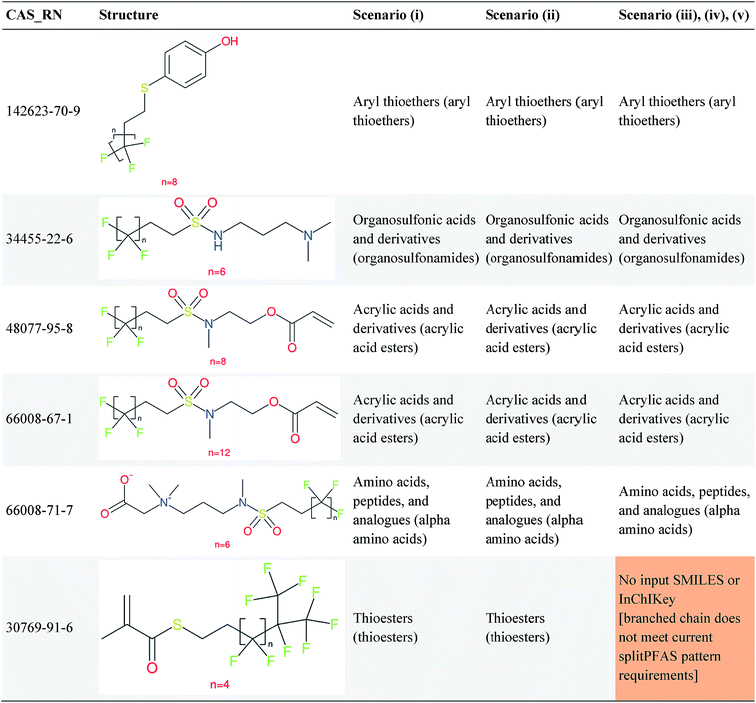
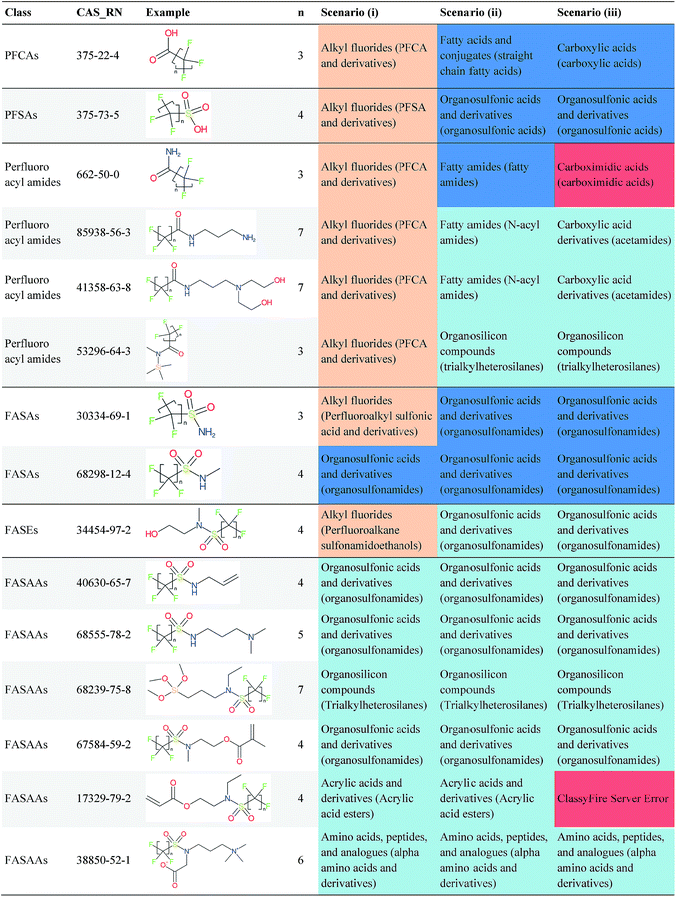
Overall, ClassyFire returned results in the vast majority of cases. Out of the 548 compounds (50 PFACs, 156 PFSCs, 142 n:1 FTs and 200 n:2 FTs), ClassyFire failed to return results in only two cases, both scenario (i) for n:2 FTs. These cases, CAS_RN 26650-09-9 and 26650-10-2 consistently returned server errors in ClassyFire (e.g. query IDs 3540761 and 3541037) and it is likely that ClassyFire cannot process these properly (both are thiocyanic acids). These cases have been reported to the developers.The ClassyFire results for scenario (i) vary considerably across different compounds (see Tables 2–4), with a few exceptions where ClassyFire has been fine tuned to recognize certain PFASs (e.g., see the “direct parent names” of row 5 in Table 2, row 1–7 and 9 in Table 4). This suggests that the current version of ClassyFire alone is not suitable for systematic categorization of PFASs, but does have the potential to be adjusted to do so.
Considering the ClassyFire results across PFASs and the respective scenarios, the potential of using ClassyFire as a basis for PFAS naming is elaborated further below in terms of two groups: (1) n:1 and n:2 fluorotelomer-based compounds, and (2) PACF and PASF derivatives.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2– moiety, but not the –N(CH3)CH2CH2– moiety. Therefore, for these cases it seems key pieces of information are missing in the ClassyFire results that would be necessary to name the PFASs correctly. While other parts of the ClassyFire output (other than sub-class name and direct parent name) were also considered, the general pattern described here holds over all output types.
CH2– moiety, but not the –N(CH3)CH2CH2– moiety. Therefore, for these cases it seems key pieces of information are missing in the ClassyFire results that would be necessary to name the PFASs correctly. While other parts of the ClassyFire output (other than sub-class name and direct parent name) were also considered, the general pattern described here holds over all output types.
3.3 Combining splitPFAS and ClassyFire
In summary, splitPFAS worked as designed, and could successfully distinguish different predefined patterns of PFASs and thus be used to categorize and identify PFASs of interest. The cases that were not considered in this manuscript are discussed in more detail below. In contrast, the ClassyFire results were more mixed. Among the five scenarios examined for PFASs, scenario (iii) appears to be most reasonable for future use. For PFASs with simple molecular structures, it seems that ClassyFire results, when combined with splitPFAS results, could potentially be a good basis for systematically naming PFASs, whereas for more complex structures, the ClassyFire results are not yet sufficient for such purpose and more extensive training or development of ClassyFire may be needed for PFASs. In the following section, these results are assessed and discussed in more detail to propose possible strategies and next steps to further improve this concept.4 Discussion
4.1 Overall
The results presented above indicate a few general trends, which will be discussed here with the perspective of scaling this up to future categorization/naming efforts of a greater range of PFASs. In general, splitPFAS is able to identify pre-defined PFAS patterns as designed and thus holds the potential for automated categorisation of PFASs. ClassyFire yielded interpretable results for n:1/n:2 FTs with rather simple functional groups, although the categories were sometimes a little broad, while for the more complex functional groups, the classification seems to correspond with only part of the functional group. ClassyFire also generally yielded interpretable results for the PASF/PACF-based derivatives, but for these cases the direct classification (scenario (i)) was less useful, since the splitPFAS output already takes care of the pattern that required classification. In processing the ClassyFire results, several examples appeared where compound-specific rules seem to be incorporated into ClassyFire, for instance the n:2 fluorotelomer alcohols (e.g. row 5, Table 2) and Table 4, row 1. For the latter, the sub-class name “alkyl fluorides” does not make much sense in the context of the structure, but the direct parent name (perfluoroalkyl carboxylic acid and derivatives) is very specific.The results demonstrate that the combination of expert knowledge and cheminformatics techniques will be needed to improve the characterization, categorization and naming of PFASs – if the patterns can be represented systematically in a cheminformatics format, this expert knowledge and lists of substances can be combined to form a large training set to generate PFAS-specific rules for ClassyFire, which could then be accessible to the community and thus available to research groups performing e.g. non-target screening of PFASs. This sharing of various expertise will be critical to move the field forwards.
A logical next step to build on this work would be to expand the SMARTS definitions for the dividing group “X” to cover other major PFAS groups (i.e., those not considered in this manuscript) and to adjust the PFAS alpha carbon SMARTS, if necessary, to capture some of the (few) specialised cases that fail to split properly. These cases are discussed in more detail in Section 4.2 below. The results above show that output from splitPFAS is, at this stage, already enough to assist categorizing PFASs and in curating lists, and would potentially provide the detailed training set needed to generate a specialised set of rules for a highly customized ClassyFire for PFASs. Future work should investigate whether a resulting specialised ClassyFire-based ontology, based on splitPFAS categorization, could be used for automated naming of PFASs; currently the results do not yet appear to capture the detail of the R groups to produce sufficiently informative names. As splitPFAS is able to divide PFASs into a variety of different scenarios, it will be possible to investigate several different options in future work, once further SMARTS groups are defined. It is interesting to note, especially with respect to potential future efforts, that scenario (iii) was the most promising input into ClassyFire when scenario (i) failed to yield good results. While scenario (iii) was originally prepared by adjusting splitPFAS outputs in an R script (see ESI†), this scenario has been directly incorporated into splitPFAS for future use.
4.2 Extending splitPFAS beyond the original scope
This study focused on structures in the various selected groups on the OECD list (i.e., structure code 101 to 109, 201 to 209 and 401 to 410). Six major cases were identified that did not fit the patterns defined currently in the splitPFAS approach, or the approach taken here in general; here we discuss these in more detail with specific examples. Most cases mentioned in Section 3.1 above are shown in Table 5, with an example structure and an explanation. Since these are best viewed side-by-side, we refer the reader to the table for more information on these cases.For one special case, branched fluorotelomer structures, the SMARTS [CH2][CH] was included in early splitPFAS calculations via the splitPFAS SMARTS input file, to capture these cases and include possible branched and ring FT structures (i.e., where the branching occurs on the FT part, the one or two non-fluorinated carbons). However, this pattern caused incorrect splitting results for some compounds, such as breaking down of ring structures in the “R group” (e.g. CAS_RN 1765-92-0) or yielding more than one “R group” (e.g. CAS_RN 38550-34-4). After removing the [CH2][CH] pattern, those compounds could be correctly split by [CH2]. Therefore, given the complexity of the structure of PFASs, it was decided not to consider this case in this investigation, as they do not strictly follow the n:1 or n:2 FT patterns chosen. It is, however, possible to process them with the existing splitPFAS method. Again, the patterns and the order of the patterns should be carefully selected when using splitPFAS in order to achieve optimal splitting results. For greater clarity, it is likely that subsets of lists should be processed using different SMARTS lists as input for different group of compounds to avoid such conflicts in patterns, i.e., first processing simple cases and then adjusting splitPFAS inputs to account for more complicated cases and run these only on those entries that fail the simple cases. This is discussed further below.
For a further special case, perfluoroalkene derivatives, no example is shown in Table 5. These examples all failed due to a combination of factors, including the presence of an unsaturated perfluoroalkyl chain and the fact that X did not match the functional groups chosen. However, as these cases do exist in the list, future efforts should consider the possibility of unsaturation in the perfluoroalkyl chain, as well as linear and branched perfluoroalkyl chains, and ring structures. The necessary features to do this are already built into the splitPFAS approach.
In light of the results presented here and all cases in this section, the functionality of the original splitPFAS was extended to allow users to adjust the SMARTS used to identify where to “split” the structures, accessible via the option “pacs” (PFAS Alpha Carbon SMARTS).33 Care should be taken when trying new SMARTS for the “pacs” and “X” groups, to avoid incorrect splitting, it is likely that optimal results will be achieved when experts in PFASs and cheminformatics join forces to design optimal SMARTS codes for various PFAS groups.
4.3 Issues caused by tautomeric structures
Cheminformatics approaches also have their limitations, and tautomeric structures are often difficult cases to handle. While it is often easy for a trained chemist to see the equivalence in tautomers due to resonance, this can be very difficult to program into a computer (even the InChI algorithm has several tautomer-related issues). A variety of established cheminformatics toolkits exist; here we have used the CDK, whereas ClassyFire is largely implemented using ChemAxon.23 While these are generally compatible, differences in structural interpretation can occur, especially for large and challenging structures with several tautomeric forms. Furthermore, choosing to work off the efficient SMILES notation (which is semi-human readable, as done here) rather than more information-rich formats like MOL formats can exacerbate this, as each SMILES has to be interpreted by the toolkit into a richer form for manipulation. Two entries in this work where this appears to have happened are highlighted red in Table 4 (rows 3 and 4) and the suspected tautomerization shown in Fig. 6. This structure was depicted as drawn on the left on the 4 major open depiction tools displayed in AMBIT37 (https://apps.ideaconsult.net/ambit2/depict using the SMILES NC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C(F)(F)C(F)(F)C(F)(F)F in the respective field) and is also displayed as such on the CompTox Chemicals Dashboard, which uses ChemAxon for depiction, so it is not clear how the reinterpretation happened in ClassyFire to yield a false classification (carboximidic acid instead of perfluoroacyl amide). While cases such as these will happen with any automated approach, they are relatively rare and could be captured in the future using a consensus tautomer approach; chemical databases like PubChem38 and the CompTox Chemicals Dashboard19 and others are continually improving their handling of tautomers.
O)C(F)(F)C(F)(F)C(F)(F)F in the respective field) and is also displayed as such on the CompTox Chemicals Dashboard, which uses ChemAxon for depiction, so it is not clear how the reinterpretation happened in ClassyFire to yield a false classification (carboximidic acid instead of perfluoroacyl amide). While cases such as these will happen with any automated approach, they are relatively rare and could be captured in the future using a consensus tautomer approach; chemical databases like PubChem38 and the CompTox Chemicals Dashboard19 and others are continually improving their handling of tautomers.
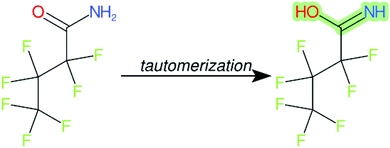 | ||
| Fig. 6 The categorization of the perfluoroacyl amide as a carboximidic acid by ClassyFire (Table 4, rows 3 and 4) is likely due to tautomerization at some point during the ClassyFire workflow. | ||
5. Outlook
In this study, two cheminformatics approaches (i.e., splitPFAS and ClassyFire) were evaluated to explore the potential of using such automated, open tools to enable stakeholders to systematically categorize and name PFASs. In particular, splitPFAS has proven useful to identify specific PFAS patterns and thus can be helpful in systematic categorization of PFASs in general. For example, splitPFAS has successfully identified a number of cases where PFASs were assigned incorrect structure codes/categories in the OECD PFAS list. Therefore, one particular future use of splitPFAS can be to assist stakeholders, particularly those who are not familiar with the complex PFAS class, in curating and processing long lists of PFASs with pre-defined structure categories. Regulators and manufacturers may be able to use splitPFAS to process their own inventories and identify certain PFASs of interest (e.g. PFOA-related compounds under the Stockholm Convention). While splitPFAS holds a promising future, it should also be noticed that the predefined structure categories used here are still limited, as this study focused on proof-of-concept. In the future, splitPFAS should be developed to encompass a wider range of PFASs by defining further major dividing groups X, e.g. X = “P(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)”. Further work should also be done to capture the cases that are not yet perfectly handled, such as (1) branched and cyclic perfluoroalkyl chains, (2) unsaturated perfluoroalkyl chains, (3) polyfluoroalkyl chains (e.g. H- or Cl-CnF2n–R) and (4) perfluoroalkyl ether chains (e.g. CnF2n+1–O–CmF2m+1). While the rules to be used by splitPFAS in some of these areas are yet to be defined, the functionality is built in and ready to be applied and it is likely that extensions to the SMARTS used in splitPFAS could provide useful functionality for several different audiences.
O)”. Further work should also be done to capture the cases that are not yet perfectly handled, such as (1) branched and cyclic perfluoroalkyl chains, (2) unsaturated perfluoroalkyl chains, (3) polyfluoroalkyl chains (e.g. H- or Cl-CnF2n–R) and (4) perfluoroalkyl ether chains (e.g. CnF2n+1–O–CmF2m+1). While the rules to be used by splitPFAS in some of these areas are yet to be defined, the functionality is built in and ready to be applied and it is likely that extensions to the SMARTS used in splitPFAS could provide useful functionality for several different audiences.
In contrast, using ontology-based approaches such as ClassyFire in systematic categorisation and naming of PFASs warrants greater investigation and discussion. The results do not appear sufficiently detailed at this stage to provide enough information for systematic naming. However, a more detailed training set, created using e.g., the splitPFAS approach, may yield sufficient specialized rules in the future to enable this.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors acknowledge fruitful discussions with Steffen Neumann (IPB Halle) and are grateful to the reviewers for their detailed comments. ELS and CR acknowledge funding by the SOLUTIONS project (grant agreement 603437), supported by the EU Seventh Framework Programme. CR was also supported by European Commission H2020 project PhenoMeNal Grant EC654241. ELS is supported by the Luxembourg National Research Fund (FNR) for project A18/BM/12341006. The PhD work of BS is funded by the Swedish Research Council (FORMAS), Grant 2016-00644. ZW gratefully acknowledges funding for his research from the Swiss Federal Office for the Environment (FOEN).References
- OECD, Toward a new comprehensive global database of per- and polyfluoroalkyl substances (PFASs): Summary report on updating the OECD 2007 list of per- and polyfluorinated substances (PFASs), Report ENV/JM/MONO(2018)7, 2018. http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=ENV-JM-MONO(2018)7%26doclanguage=en Search PubMed.
- E. Kissa, Fluorinated surfactants and repellents, Marcel Dekker, New York, 2nd edn, revised and expanded, 2001. ISBN: 978-0-8247-0472-8 Search PubMed.
- R. E. Banks, B. E. Smart and J. C. Tatlow, ed. Organofluorine Chemistry, Springer US, Boston, MA, 1994 Search PubMed.
- R. C. Buck, J. Franklin, U. Berger, J. M. Conder, I. T. Cousins, P. de Voogt, A. A. Jensen, K. Kannan, S. A. Mabury and S. P. van Leeuwen, Perfluoroalkyl and polyfluoroalkyl substances in the environment: Terminology, classification, and origins, Integr. Environ. Assess. Manage., 2011, 7, 513–541 CrossRef CAS PubMed.
- Z. Wang, I. T. Cousins, M. Scheringer, R. C. Buck and K. Hungerbühler, Global emission inventories for C4–C14 perfluoroalkyl carboxylic acid (PFCA) homologues from 1951 to 2030, Part I: production and emissions from quantifiable sources, Environ. Int., 2014, 70, 62–75 CrossRef CAS PubMed.
- Z. Wang, I. T. Cousins, M. Scheringer, R. C. Buck and K. Hungerbühler, Global emission inventories for C4–C14 perfluoroalkyl carboxylic acid (PFCA) homologues from 1951 to 2030, part II: The remaining pieces of the puzzle, Environ. Int., 2014, 69, 166–176 CrossRef CAS PubMed.
- UNEP, Report of the Persistent Organic Pollutants Review Committee on the work of its fourteenth meeting - Risk profile on perfluorohexane sulfonic acid (PFHxS), its salts and PFHxS-related compounds, Report UNEP/POPS/POPRC.14/6/Add.1, UNEP, 2018 Search PubMed.
- Z. Wang, J. C. DeWitt, C. P. Higgins and I. T. Cousins, A Never-Ending Story of Per- and Polyfluoroalkyl Substances (PFASs)?, Environ. Sci. Technol., 2017, 51, 2508–2518 CrossRef CAS PubMed.
- US Environmental Protection Agency, CompTox Chemicals Dashboard: PFAS Lists, https://comptox.epa.gov/dashboard/chemical_lists/?search=PFAS, accessed 17 March 2019 Search PubMed.
- NORMAN Network, NORMAN Suspect List Exchange, https://www.norman-network.com/nds/SLE/, accessed 9 June 2019 Search PubMed.
- X. Trier and D. Lunderberg, S9 | PFASTRIER | PFAS Suspect List: fluorinated substances, Zenodo Dataset, DOI:10.5281/zenodo.2621989, 2015, accessed 7 July 2019.
- S. Fischer, S14 | KEMIPFAS | PFAS Highly Fluorinated Substances List: KEMI, Zenodo Dataset, DOI:10.5281/zenodo.2621525, 2017, accessed 7 July 2019.
- Z. Wang, S25 | OECDPFAS | List of PFAS from the OECD, Zenodo Dataset, DOI:10.5281/zenodo.2648776, 2018, accessed 7 July 2019.
- Y. Liu, L. D'Agostino, E. Schymanski and J. Martin, S46 | PFASNTREV19 | List of PFAS reported in Non-Target HRMS Studies (Liu et al. 2019), Zenodo Dataset, DOI:10.5281/zenodo.2656744, 2019, accessed 7 July 2019.
- Y. Liu, L. A. D'Agostino, G. Qu, G. Jiang and J. W. Martin, High-Resolution Mass Spectrometry (HRMS) Methods for Nontarget Discovery and Characterization of Poly- and Per-fluoroalkyl Substances (PFASs) in Environmental and Human Samples, TrAC, Trends Anal. Chem. DOI:10.1016/j.trac.2019.02.021.
- US Environmental Protection Agency, Chemistry Dashboard 8:2 Fluorotelomer alcohol, https://comptox.epa.gov/dashboard/DTXSID7029904, accessed 4 June 2019 Search PubMed.
- Y. Wang, N. Yu, X. Zhu, H. Guo, J. Jiang, X. Wang, W. Shi, J. Wu, H. Yu and S. Wei, Suspect and Nontarget Screening of Per- and Polyfluoroalkyl Substances in Wastewater from a Fluorochemical Manufacturing Park, Environ. Sci. Technol., 2018, 52, 11007–11016 CrossRef CAS PubMed.
- N. Yu, H. Guo, J. Yang, L. Jin, X. Wang, W. Shi, X. Zhang, H. Yu and S. Wei, Non-Target and Suspect Screening of Per- and Polyfluoroalkyl Substances in Airborne Particulate Matter in China, Environ. Sci. Technol., 2018, 52, 8205–8214 CrossRef CAS PubMed.
- A. J. Williams, C. M. Grulke, J. Edwards, A. D. McEachran, K. Mansouri, N. C. Baker, G. Patlewicz, I. Shah, J. F. Wambaugh, R. S. Judson and A. M. Richard, The CompTox Chemistry Dashboard: a community data resource for environmental chemistry, J. Cheminf., 2017, 9, 61 Search PubMed.
- US Environmental Protection Agency, CompTox Chemicals Dashboard: Chemical Lists Page, https://comptox.epa.gov/dashboard/chemical_lists, accessed 17 March 2019 Search PubMed.
- G. Patlewicz, A. M. Richard, A. J. Williams, C. M. Grulke, R. Sams, J. Lambert, P. D. Noyes, M. J. DeVito, R. N. Hines, M. Strynar, A. Guiseppi-Elie and R. S. Thomas, A Chemical Category-Based Prioritization Approach for Selecting 75 Per- and Polyfluoroalkyl Substances (PFAS) for Tiered Toxicity and Toxicokinetic Testing, Environ. Health Perspect., 2019, 127, 014501 CrossRef PubMed.
- K. A. Barzen-Hanson, S. C. Roberts, S. Choyke, K. Oetjen, A. McAlees, N. Riddell, R. McCrindle, P. L. Ferguson, C. P. Higgins and J. A. Field, Discovery of 40 Classes of Per- and Polyfluoroalkyl Substances in Historical Aqueous Film-Forming Foams (AFFFs) and AFFF-Impacted Groundwater, Environ. Sci. Technol., 2017, 51, 2047–2057 CrossRef CAS PubMed.
- Y. Djoumbou Feunang, R. Eisner, C. Knox, L. Chepelev, J. Hastings, G. Owen, E. Fahy, C. Steinbeck, S. Subramanian, E. Bolton, R. Greiner and D. S. Wishart, ClassyFire: automated chemical classification with a comprehensive, computable taxonomy, J. Cheminf., 2016, 8, 61 Search PubMed.
- S. Heller, A. McNaught, S. Stein, D. Tchekhovskoi and I. Pletnev, InChI – the worldwide chemical structure identifier standard, J. Cheminf., 2013, 5, 7 CAS.
- D. S. Wishart, Y. D. Feunang, A. C. Guo, E. J. Lo, A. Marcu, J. R. Grant, T. Sajed, D. Johnson, C. Li, Z. Sayeeda, N. Assempour, I. Iynkkaran, Y. Liu, A. Maciejewski, N. Gale, A. Wilson, L. Chin, R. Cummings, D. Le, A. Pon, C. Knox and M. Wilson, DrugBank 5.0: a major update to the DrugBank database for 2018, Nucleic Acids Res., 2018, 46, D1074–D1082 CrossRef CAS PubMed.
- D. Wishart, D. Arndt, A. Pon, T. Sajed, A. C. Guo, Y. Djoumbou, C. Knox, M. Wilson, Y. Liang, J. Grant, Y. Liu, S. A. Goldansaz and S. M. Rappaport, T3DB: the toxic exposome database, Nucleic Acids Res., 2015, 43, D928–D934 CrossRef CAS PubMed.
- Daylight Chemical Information Systems, Inc., SMILES – A Simplified Chemical Language, http://www.daylight.com/dayhtml/doc/theory/theory.smiles.html, accessed 13 April 2019 Search PubMed.
- University of Alberta, ClassyFire Web Server Version 1.0, http://classyfire.wishartlab.com/, accessed 4 June 2019 Search PubMed.
- Daylight Chemical Information Systems, Inc., SMARTS - A Language for Describing Molecular Patterns, http://www.daylight.com/dayhtml/doc/theory/theory.smarts.html, accessed 13 April 2019 Search PubMed.
- SETAC, SETAC PFAS Focused Topic Meeting Abstract Book (Abstract 6), https://pfas.setac.org/wp-content/uploads/2019/08/FINAL-PFAS-abstract-book-v5.pdf, 2019, accessed 20 August 2019 Search PubMed.
- J. Mayfield, CDK Depict Web Interface, http://simolecule.com/cdkdepict/depict.html, 2019, accessed 30 October 2018 Search PubMed.
- E. L. Willighagen, J. W. Mayfield, J. Alvarsson, A. Berg, L. Carlsson, N. Jeliazkova, S. Kuhn, T. Pluskal, M. Rojas-Chertó, O. Spjuth, G. Torrance, C. T. Evelo, R. Guha and C. Steinbeck, The Chemistry Development Kit (CDK) v2.0: atom typing, depiction, molecular formulas, and substructure searching, J. Cheminf., 2017, 9, 33 Search PubMed.
- C. Ruttkies, splitPFAS Download (jar file), https://msbi.ipb-halle.de/%7Ecruttkie/metfrag/MetFrag2.4.5-Tools.jar, accessed 5 July 2019 Search PubMed.
- C. Ruttkies, splitPFAS Source Code (GitHub), https://github.com/ipb-halle/MetFragRelaunched/tree/master/MetFragTools/src/main/java/de/ipbhalle/metfrag/split, accessed 5 July 2019 Search PubMed.
- E. L. Schymanski, RChemMass: splitPFAS code, https://github.com/schymane/RChemMass/blob/master/R/SplitPFAS.R, accessed 13 April 2019 Search PubMed.
- E. L. Schymanski, RChemMass: Example Files, https://github.com/schymane/RChemMass/tree/master/inst/extdata, accessed 13 April 2019 Search PubMed.
- N. Jeliazkova and V. Jeliazkov, AMBIT RESTful web services: an implementation of the OpenTox application programming interface, J. Cheminf., 2011, 3, 18 CAS.
- S. Kim, P. A. Thiessen, E. E. Bolton, J. Chen, G. Fu, A. Gindulyte, L. Han, J. He, S. He, B. A. Shoemaker, J. Wang, B. Yu, J. Zhang and S. H. Bryant, PubChem Substance and Compound databases, Nucleic Acids Res., 2016, 44, D1202–D1213 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9em00321e |
| ‡ Shared first authors. |
| This journal is © The Royal Society of Chemistry 2019 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)