Drug delivery by a self-assembled DNA tetrahedron for overcoming drug resistance in breast cancer cells†
Kyoung-Ran
Kim
ab,
Da-Rae
Kim
a,
Taemin
Lee
c,
Ji Young
Yhee
a,
Byeong-Su
Kim
c,
Ick Chan
Kwon
a and
Dae-Ro
Ahn
*ad
aCenter for Theragnosis, Biomedical Research Institute, Korea Institute of Science and Technology, Hwarangno 14-gil 5, Seongbuk-gu, Seoul 136-791, Republic of Korea. E-mail: drahn@kist.re.kr; Fax: +82 2 958 5909; Tel: +82 2 958 6645
bDepartment of Chemistry, College of Science, Yonsei University, Korea
cInterdisciplinary School of Green Energy and KIER-UNIST Advanced Center for Energy, Ulsan National Institute of Science and Engineering (UNIST), Ulsan 689-798, Republic of Korea
dKIST campus, University of Science (UST-KIST), Hwarangno 14-gil 5, Seongbuk-gu, Seoul 136-791, Republic of Korea
First published on 21st January 2013
Abstract
A DNA tetrahedron is employed for efficient delivery of doxorubicin into drug-resistant breast cancer cells. The drug delivered with the DNA nanoconstruct is considerably cytotoxic, whereas free doxorubicin is virtually non-cytotoxic for the drug-resistant cells. Thus, the DNA tetrahedron, made of the inherently natural and biocompatible material, can be a good candidate for the drug carrier to overcome MDR in cancer cells.
With repeated exposure to chemotherapeutic agents, cancer cells become resistant to one or more anticancer drugs.1 This multidrug resistance (MDR) is a main hurdle to obtaining successful anticancer activity. Various mechanisms have been suggested to explain MDR, which include altering membrane transport protein to increase drug efflux, enhancing DNA repair, amending cell cycle regulation to block apoptosis, and detoxification.2 Development and discovery of agents that efficiently overcome MDR with reduced toxicity have been the focus of extensive research.3 In particular, nanocarrier-based drug delivery systems for different drug formulations and modifications have been widely investigated to reverse MDR.4 Nanoparticles travel in and out of cells mainly via endocytosis and exocytosis pathways that are independent of P-glycoprotein (P-gp), a membrane-bound active drug efflux pump, responsible for one of the most important mechanisms involved in many MDR cells.5 Thus, the drug delivery system based on nanoparticles holds promising potential to overcome MDR via highly efficient cellular uptake of the drug-loaded nanoparticles and subsequent rapid release of a large amount of the anticancer drug to induce cytotoxicity. Although many inorganic and organic nanocarriers have been previously employed to enhance the intracellular drug accumulation in MDR cells,6 investigations of new materials are still required to realize sufficient clinical outcomes in cancer therapy.
Recently, DNA nanoconstructs have been reported and proposed as potential candidates for molecular delivery carriers in diagnostics and therapeutics.7 Among them, the DNA tetrahedron has been considered one of the most practical DNA nanoconstructs since it can be self-assembled simply from four DNA strands and prepared in a high yield.8 The recent study by Turberfield et al. demonstrating cellular uptake of the DNA tetrahedron into mammalian cells has indeed provided a great opportunity for the DNA tetrahedron to play important roles in biomedical applications.9 Moreover, DNA can make a physical conjugate with doxorubicin (DOX), a widely used anticancer drug because of the inherent intercalation property of DOX. There have been studies utilizing the DNA–DOX conjugate for effective delivery of DOX into cancer cells.10 To the best of our knowledge, however, there has been no report on evaluation of the DNA tetrahedron as a drug carrier to reverse drug resistance.
In this context, we here prepare a DNA tetrahedron by following the previously reported procedure9 and attempt to use it as a drug carrier for overcoming drug resistance (Fig. 1). On the basis of the DNA-intercalation property of the doxorubicin, we prepare a physical conjugate of the DNA tetrahedron with DOX and demonstrate that the DNA nanostructure can be used as an effective drug carrier to show significant cytotoxicity for MDR cells. Although DNA origami has been employed as a carrier for circumvention of drug resistance very recently,11 the DNA tetrahedron is a far smaller structure capable of achieving the same effect, and thereby more cost-effective and more viable in in vivo drug delivery.
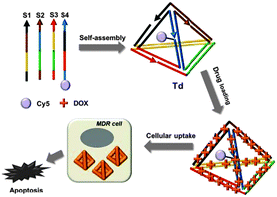 | ||
| Fig. 1 Schematic diagram of the drug-loaded DNA tetrahedron and its cytotoxic effect for drug-resistant cells. | ||
Our DNA tetrahedron (Td) was assembled by using the sequences adopted from Turberfield's DNA tetrahedron.9 A fluorescence dye either Cy5 (Cy5-S4) or fluorescein (FAM-S4) was labelled at one DNA strand in the DNA tetrahedron (ESI,† Table S1). Construction of the self-assembled DNA tetrahedron was verified in 6% non-denaturing polyacrylamide gel electrophoresis (PAGE) (Fig. 2a). The hydrodynamic size of Td was 9.08 (±0.67) nm as determined by dynamic light scattering (DLS) (Fig. 2b). The nanoconstruct was also characterized by the atomic force microscopy (AFM) image showing the tetrahedron in the dried state to be about 2–3 nm in height, which was consistent with the previous results (Fig. 2c).12
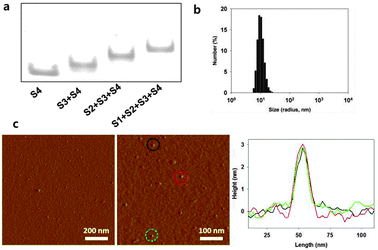 | ||
| Fig. 2 Characterization of DNA tetrahedron. (a) Native PAGE to verify assembly of Td. (b) DLS data of the Td. (c) AFM images of Td. | ||
The DNA tetrahedron itself was efficiently delivered into both the drug-sensitive (MCF-7) and doxorubicin-resistant (MCF-7/ADR) breast cancer cell lines (Fig. 3a). Flow cytometry-based quantitative analysis of the delivery also clearly showed uptake of Td in the two breast cancer cell lines (Fig. 3b). The possibility that the cellular uptake might be driven by the small portion of the polymeric mis-assembled product often observed in the PAGE could be precluded, since the polymeric product extracted from the gel could not be delivered into the cells (ESI,† Fig. S1). To investigate the pathway mediating the cellular uptake of the DNA tetrahedron, MCF-7 cells were incubated with Td at 37 °C and 4 °C. The amount of delivered Td was estimated by the fluorescence intensity of the Cy5 label in each cell lysate after normalization based on the whole protein amount measured using the Bradford assay. Much lower cellular uptake at 4 °C than at 37 °C revealed that Td was delivered via endocytosis. Of the three endocytosis inhibitors applied, amiloride and genistein effectively inhibited the Td uptake, indicating that Td was internalized via macropinocytosis and caveolae-mediated endocytosis pathways (Fig. 3c).13
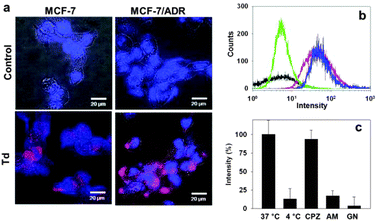 | ||
| Fig. 3 Delivery of Td into breast cancer cells. (a) Fluorescence microscopic images of MCF-7 (left) and MCF-7/ADR (right) cells treated with Cy5-labeled Td (bottom) compared with the control images (top). (b) Flow cytometry analysis for the cellular uptake of Td into MCF-7 (red) or MCF-7/ADR cells (blue). The black and green traces represent the untreated MCF-7 and MCF-7/ADR cells, respectively. (c) Average cellular fluorescence intensity of Td in MCF-7 cell lysates from the cells cultured in the presence of three different endocytosis inhibitors: chlorpromazine (CPZ), amiloride (AM), and genistein (GN). Fluorescence intensity is normalized to the total amount of cellular proteins. | ||
To incorporate DOX into the DNA tetrahedron, DOX was incubated with the Td (DOX@Td), and the unloaded DOX was removed by G25 gel filtration. The intercalated amount of DOX per DNA tetrahedron was determined by comparing the concentration of DNA and the concentration of DOX, which was calculated by using the extinction coefficient of Cy5 at 633 nm (ε = 230![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 M−1 cm−1) and that of DOX at 480 nm (ε = 10
400 M−1 cm−1) and that of DOX at 480 nm (ε = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 410 M−1 cm−1), respectively. As a result, 26 DOX molecules per Td were found to be loaded.
410 M−1 cm−1), respectively. As a result, 26 DOX molecules per Td were found to be loaded.
For the drug delivery study, the internalization and the efficiency of DOX@Td and free DOX were initially analysed by fluorescence microscopy (Fig. 4a and b). When treated with free DOX, the MCF-7/ADR cells showed relatively low drug accumulation, as compared to that of the wild-type MCF-7 cell line (Fig. 4a and b, second columns). The drug in the DNA tetrahedron, however, was delivered into the drug-resistant cells as efficiently as into the wild-type cells. According to the microscopic images after nuclear staining, DOX@Td appeared and stayed widely in the cytoplasm of both MCF-7 and MCF-7/ADR cells (Fig. 4a and b, third columns). These results proved that the DNA tetrahedron could be used as an efficient drug carrier and also indicated that the drug delivered into the MCF-7/ADR cells with the DNA construct was not effluxed by the action of P-gp. When analysed by flow cytometry, the accumulation of DOX in the wild-type cells was dependent on the concentration of the treated drug (Fig. 4c and ESI,† Fig. S2). In the MCF-7/ADR cells, however, the drug was not accumulated when the cells were treated with free DOX, which is consistent with the microscopy image. In the case of DOX@Td, the drug was delivered into both cell types in a concentration-dependent manner. Different from free DOX that interacts with P-gp on the cell membrane, the drug delivered by the DNA nanostructure-based carrier may avoid the recognition by the efflux pump by means of protection in an endosome when entering the cell, leading to intracellular drug accumulation, as similarly observed in other nanostructured drug carriers.5
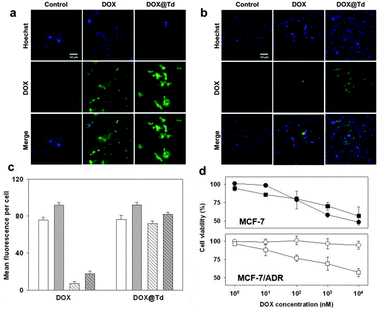 | ||
| Fig. 4 (a) MCF-7 cells treated with 10 nM DOX or DOX@Td. (b) MCF-7/ADR cells treated with 10 nM DOX or DOX@Td. Scale bar: 50 μm. (c) Drug delivery efficiency to the MCF-7 (solid) and MCF-7/ADR cells (striped) at 10 nM (white) and 50 nM DOX (gray) determined by flow cytometry. (d) Cytotoxicity of DOX (circles) and DOX@Td (squares). | ||
In the cell viability test, while no significant cytotoxicity of the DNA tetrahedron itself was observed in both breast cancer cell lines, the drug-loaded DNA tetrahedron showed dose-dependent toxicity (ESI,† Fig. S3). Since biocompatibility of drug carriers is of utmost importance in drug delivery technology, the utilization of the non-cytotoxic DNA tetrahedron as a drug carrier creates a new class of drug delivery systems. Moreover, various chemical modifications can be easily introduced in the DNA construct for fine-tuning of the biophysical properties of the carrier including drug loading efficiency and controlled release.
In terms of the drug concentration-dependent cell viability, about 50% of MCF-7 cells were viable after 24 h treatment with 10 μM free DOX (Fig. 4d, closed circles). Similar viability was observed by treatment with DOX@Td (closed squares) containing the same concentration of the drug. In MCF-7/ADR cells, however, free DOX showed significantly decreased cytotoxicity by resulting in more than 90% viable cells (open circles) possibly because of the drug efflux pump on the cell membrane of the MDR cells. In contrast to free DOX, the same amount of drug delivered with Td (open squares) was still considerably cytotoxic also against the drug-resistant cells, leading to the viability profile of MCF-7/ADR cells similar to that of MCF-7 cells. These results clearly confirmed that DOX in the DNA tetrahedron could reverse drug-resistant cells with enhanced uptake and bypass the efflux process.
To examine the possible mechanism for the release of DOX from the DNA nanoconstruct, the in vitro release of DOX from the DNA tetrahedron was performed in phosphate-buffered saline (PBS) solutions. Since the DNA nanoconstruct was delivered through endocytosis, the release of DOX was monitored at pH 5.0, the endosomal pH as well as at pH 7.4, the physiological condition. For the initial 3 h, less than 10% of DOX was released from the drug carrier (ESI,† Fig. S4a). The release proceeded slowly but continuously for 10 h until the maximum release was achieved. The release was dramatically increased by lowering the pH from 7.4 to 5.0, which suggested that the drug release could be accelerated during endosomal retention of the carriers. This enhanced release at pH 5.0 was due to partial disassembly of the DNA tetrahedron as observed in the PAGE analysis (ESI,† Fig. S4b). This is consistent with the release pattern of DOX intercalated in other types of DNA constructs.14 In addition, degradation of the DNA construct by intracellular nucleases might also influence the drug release (ESI,† Fig. S5).
In conclusion, we have prepared a DNA tetrahedron and observed that the DNA tetrahedron was easily delivered into the drug-resistant breast cancer cells as well as the wild-type cells without additional transfection agents. This advantageous character of the DNA tetrahedron for drug delivery was combined with the DOX binding property of DNA to provide a drug delivery method particularly for multidrug resistant MCF-7/ADR cells. This drug delivery system could significantly inhibit the growth of the MDR cells, because of the highly efficient internalization of DOX by the DNA nanoconstruct and the effective intracellular release of DOX circumventing the efflux pathway of the MDR cells. Since DNA is a purely natural material with low immunogenicity15 and is expected to be eventually degraded into a non-toxic state, the DNA tetrahedron therefore is a potentially good candidate for a drug carrier to be clinically used to overcome drug resistance in cancer cells.
This study was supported by a grant funded by Korea Institute of Science and Technology, a grant of the Basic Science Research Program (2011-0009172) and a grant of the Proteogenomic Research Program through the National Research Foundation of Korea funded by the Ministry of Education, and grants of the Korea Healthcare technology R&D Project, the Ministry of Health & Welfare, Republic of Korea (A090147 and A121191).
Notes and references
- D. B. Longley and P. G. Johnston, J. Pathol., 2005, 205, 275 CrossRef CAS.
- E. Borowski, M. M. Bontemps-Gracz and A. Piwkowska, Acta Biochim. Pol., 2005, 52, 609 CAS.
- G. Szakacs, J. K. Paterson, J. A. Ludwig, C. Booth-Genthe and M. M. Gottesman, Nat. Rev. Drug Discovery, 2006, 5, 219 CrossRef CAS.
- R. R. Patil, S. A. Guhagarkar and P. V. Devarajan, Crit. Rev. Ther. Drug Carrier Syst., 2008, 25, 1 CrossRef CAS.
- H. L. Wong, R. Bendayan, A. M. Rauth, H. Y. Xue, K. Babakhanian and X. Y. Wu, J. Pharmacol. Exp. Ther., 2006, 317, 1372 CrossRef CAS.
- A. Shapira, Y. D. Livney, H. J. Broxterman and Y. G. Assaraf, Drug Resist. Updates, 2011, 14, 150 CrossRef CAS.
- R. Chhabra, J. Sharma, Y. Liu, S. Rinker and H. Yan, Adv. Drug Delivery Rev., 2010, 62, 617 CrossRef CAS.
- R. P. Goodman, I. A. T. Schaap, C. F. Tardin, C. M. Erben, R. M. Berry, C. F. Schmidt and A. J. Turberfield, Science, 2005, 310, 1661 CrossRef CAS.
- A. S. Walsh, H. Yin, C. M. Erben, M. J. A. Wood and A. J. Turberfield, ACS Nano, 2011, 5, 5427 CrossRef CAS.
- V. Bagalkot, O. C. Farokhzad, R. Langer and S. Jon, Angew. Chem., Int. Ed., 2006, 45, 8149 CrossRef CAS.
- Q. Jiang, C. Song, J. Nangreave, X. Liu, L. Lin, D. Qiu, Z.-G. Wang, G. Zou, X. Liang, H. Yan and B. Ding, J. Am. Chem. Soc., 2012, 134, 13396 CrossRef CAS.
- N. Y. Wong, C. Zhang, L. H. Tan and Y. Lu, Small, 2011, 7, 1427 CrossRef CAS.
- Rajpal, A. Mann, R. Khanduri, R. J. Naik and M. Ganguli, J. Controlled Release, 2012, 157, 260 CrossRef CAS.
- M. Chang, C.-S. Yang and D.-M. Huang, ACS Nano, 2011, 5, 6156 CrossRef CAS.
- P. R. Bouchard, R. M. Hutabarat and K. M. Thompson, Annu. Rev. Pharmacol. Toxicol., 2010, 50, 237 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and additional data. See DOI: 10.1039/c3cc38693g |
| This journal is © The Royal Society of Chemistry 2013 |
