Open Access Article
Open Access ArticleBiogenic silver nanoparticles' antibacterial activity and cytotoxicity on human hepatocarcinoma cells (Huh-7)†
Thyerre Santana da Costaa,
Mariana Rodrigues da Silvab,
Júlio César Jerônimo Barbosaac,
Uedson Da Silva Das Nevesa,
Marcelo Bispo de Jesus b and
Ljubica Tasic
b and
Ljubica Tasic *a
*a
aInstitute of Chemistry, Biological Chemistry Laboratory, Universidade Estadual de Campinas, UNICAMP, Campinas, SP 13083-970, Brazil. E-mail: ljubica@unicamp.br
bDepartment of Biochemistry and Tissue Biology, Institute of Biology, Universidade Estadual de Campinas, UNICAMP, Campinas, SP 13083-862, Brazil
cDepartment of Organic Chemistry, Institute of Chemistry, Universidade Estadual de Campinas, UNICAMP, Campinas, SP 13083-970, Brazil
First published on 10th January 2024
Abstract
Exploring diverse synthetic pathways for nanomaterial synthesis has emerged as a promising direction. For example, silver nanoparticles (AgNPs) are synthesized using different approaches yielding nanomaterials with distinct morphological, physical and biological properties. Hence, the present study reports the biogenic synthesis of silver nanoparticles using the aqueous secretome of the fungus Fusarium oxysporum f. sp. cubense (AgNP@Fo) and orange peel extract (AgNP@OR). The physical and morphological properties of synthesized nanoparticles were similar, with AgNP@Fo measuring 56.43 ± 19.18 nm and AgNP@OR measuring 39.97 ± 19.72 nm in size. The zeta potentials for the nanoparticles were low, −26.8 ± 7.55 and −26.2 ± 2.87 mV for AgNP@Fo and AgNP@OR, respectively, demonstrating a similar negative charge. The spherical morphologies of both nanoparticles were evidenced by Scanning Transmission Electron Microscopy (STEM) and Atomic Force Microscopy (AFM). However, despite their similar physical and morphological properties, AgNPs demonstrated different bioactivities. We evaluated and compared the antimicrobial efficacy of these nanoparticles against a range of bacteria, such as Staphylococcus aureus, Enterococcus faecalis, Pseudomonas aeruginosa, and Escherichia coli. The AgNP@Fo showed Minimum Inhibitory Concentration (MIC) values ranging from 0.84 to 1.68 μg mL−1 and were around ten times more potent compared to AgNP@OR. The anticancer activities of both nanoparticles were investigated using human hepatocarcinoma cells (Huh-7), where AgNP@Fo exhibited around 20 times higher cytotoxicity than AgNP@OR with an IC50 value of 0.545 μmol L−1. Anticancer effects were demonstrated by the MTT, confirmed by the calcein-AM assay and fluorescence imaging. This study establishes solid groundwork for future exploration of molecular interactions of nanoparticles synthesized through distinct biosynthetic routes, particularly within bacterial and cancerous cell environments.
1. Introduction
Currently, with wide recognition for their antimicrobial potential, silver nanoparticles (AgNPs) are being applied in the treatment of ulcers, infections, burns, wounds, and for solving other topical problems.1–3 In addition to their antimicrobial properties, AgNPs have potential anticarcinogenic effects.4 They have been found to exhibit inhibitory effects on the growth and proliferation of various cancer cells, making them promising candidates for cancer therapy.5 The unique physicochemical properties of AgNPs, such as their size and high surface area, allow for efficient interactions with cancer cells, leading to cytotoxic effects and apoptosis induction.6,7 These findings highlight the multifunctional nature of AgNPs, making them a fascinating area of research for both antimicrobial and anticancer applications.AgNP synthesis can be carried out through two methods, either “top-down” or “bottom-up.” The top-down approaches involve the reduction of the starting material size, usually achieved through various physical techniques. The bottom-up methods are performed using reducing and capping agents, which can be either chemical or natural (green).8,9 The prevalent approach for synthesizing AgNPs involves chemical reduction, achieved by reducing a silver precursor with substances like sodium borohydride and N,N-dimethylformamide.10,11 However, the use of green methods in AgNP synthesis has witnessed a significant upsurge due to their inherent advantages, such as accessibility, cost-effectiveness, energy efficiency, environmental friendliness, and low toxicity. These environmentally friendly methods involve microorganisms, plants, and irradiation-assisted techniques.12–14 These agents not only facilitate the synthesis process but also provide biomolecule-induced surface modifications that can potentially enhance the nanoparticles' interactions with biological systems.15 Furthermore, despite these notable advantages, the arena of discrepancies in the biological activity of silver nanoparticles fabricated through different biosynthetic routes remains an underexplored domain.16
This study aimed to compare silver nanoparticles biosynthesized by two methods: the aqueous secretome of the fungus Fusarium oxysporum f. sp. cubense and the method using orange peel extract. The physicochemical properties and the biological potential of AgNPs were determined, specifically focusing on their antimicrobial and anticancer activities. A deeper understanding of the efficacy of these nanoparticles as therapeutic agents was sought, potentially leading to the development of novel strategies for combating microbial infections and treating cancer.
Both biosynthesized silver nanoparticles were evaluated for their antimicrobial potential using Gram-positive bacteria (Staphylococcus aureus and Enterococcus faecalis) and Gram-negative bacteria (Pseudomonas aeruginosa and Escherichia coli). Gram-positive and Gram-negative microorganisms have structural differences in their cell walls, which can affect their susceptibility to antimicrobial agents.17 Staphylococcus aureus, Enterococcus faecalis, Pseudomonas aeruginosa and Escherichia coli are microorganisms commonly found in human infections and have significant public health implications.18 These microorganisms are associated with various infections, including skin infections, urinary tract infections, respiratory infections, and hospital-acquired infections.18
Additionally, both biosynthesized silver nanoparticles underwent evaluation for their potential as antitumoral agents, employing the human hepatocarcinoma cell line Huh-7 as the test model. Given that hepatocellular carcinoma (HCC) represents the foremost manifestation of liver cancer and ranks fifth among the most prevalent cancers worldwide, its impact is profound.19 Each year, around 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individuals worldwide confront an HCC diagnosis.20,21 Against this backdrop, the primary objective of this investigation is to ascertain the impact on the viability of Huh-7 cells after their exposure to varying concentrations of AgNPs. Through these assessments, we seek to shed light on the potential utility of these nanoparticles in combating HCC, in addition to opening possibilities for new studies with cancer cell models.
000 individuals worldwide confront an HCC diagnosis.20,21 Against this backdrop, the primary objective of this investigation is to ascertain the impact on the viability of Huh-7 cells after their exposure to varying concentrations of AgNPs. Through these assessments, we seek to shed light on the potential utility of these nanoparticles in combating HCC, in addition to opening possibilities for new studies with cancer cell models.
2. Experimental section
2.1 Biosynthesis of silver nanoparticles
The first step was to grow a culture of the fungus Fusarium oxysporum in Petri dishes using a culture medium of 0.5% yeast extract, 2% malt extract, and 2% agar. The inoculated medium was left for 7 days at 28 °C for fungus growth. After 7 days, a 500 mL liquid culture medium was made, consisting of 0.5% yeast extract and 2% malt extract. Approximately 1 cm2 of the solid medium with the fungus was removed and transferred to the sterile liquid medium. The 500 mL culture medium was then incubated in a Shaker under 150 rpm agitation at 28 °C for 6 days. The biomass obtained was filtered using filter paper and washed with deionized water. Approximately 10 g of biomass was weighed and put into 100 mL of deionized water. This material was stirred at 150 rpm at 28 °C for 3 days. Next, the mixture might have taken on a pinkish color. The solution with biomass was then filtered using a Büchner funnel and filter paper (Qualy filter paper 9.0 cm diameter, 80 gm−2 grammage, 205 μmol L−1 thickness, 14 μmol L−1 most pores), and the fungal filtrate (FF) was used for the synthesis of AgNP@Fo. Finally, AgNO3 1 mmol L−1 solution was added, and the reaction mixture was kept at 28 °C under protection from light. The formation of AgNP@Fo was investigated by ultraviolet-visible (UV-Vis) spectroscopy in a range of 200 to 800 nm.
2.2 Characterization of the nanoparticles
The sizes of the nanoparticles influence their bioaccumulation and toxicity, and it is of broad importance to perform the characterization of the obtained nanomaterials. The particle diameters, polydispersity indexes, as well as zeta potential measurements were determined by Dynamic Light Scattering (DLS) in a Zetasizer nano-series equipment (Malvern Instruments). The surface plasmon resonance of the nanoparticles was studied using the UV-Vis spectroscopy method with an Agilent HP 8453 spectrophotometer in the wavelength range of 200 to 1000 nm. Furthermore, the morphology and size of silver nanoparticles were evaluated using Scanning Transmission Electron Microscopy (STEM) and Atomic Force Microscopy (AFM) techniques. For DLS, STEM and AFM analyses, the nanoparticles were diluted in MilliQ water 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v) and filtered using a 0.22 μm syringe filter. DLS data was acquired using the Zetasizer software, while microscopy analyses were performed using ImageJ.
10 (v/v) and filtered using a 0.22 μm syringe filter. DLS data was acquired using the Zetasizer software, while microscopy analyses were performed using ImageJ.
2.3 Quantification of silver(I) in colloids of silver nanoparticles
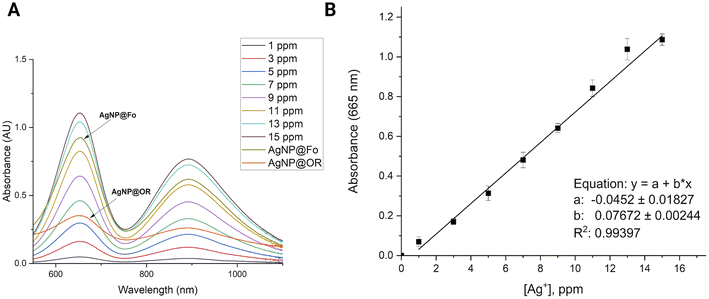
AgNPs were quantified by adapting the method described by Gonzalez and collaborators.22 Initially, standard solutions of Ag+ with varying concentrations ranging from 1 to 15 ppm were prepared. A volume of 1.2 mL of Ag+ standard solutions or AgNP colloids was mixed with 200 μL of a NaAc/AcH buffer solution (pH = 4) and 100 μL of a 3,3′,5,5′-tetramethylbenzidine (TMB) solution (10 mmol L−1 in ethanol). When introducing TMB and a buffer solution (NaAc/AcH) to a transparent aqueous solution containing Ag+, a chemical transformation occurs, resulting in the formation of the oxidized state of TMB (oxTMB) and Ag(0). These solutions were allowed to rest at 5 °C for 24 h. After the oxidation reaction of TMB by Ag+ ions, the solutions were analyzed using a UV-Vis spectrophotometer (typical peaks of oxTMB at ∼655 nm) (Fig. 1).2.4 Antimicrobial activity of silver nanoparticles AgNP@Fo
The bacteria were obtained from the Tropical Culture Collection of the André Tosello Foundation (Campinas, São Paulo, Brazil), and used to determine the AgNP Minimum Inhibitory Concentrations (MIC), which refer to the lowest concentrations of the AgNPs that limit visible bacterial growth.The 96-well microplate dilution technique was used to verify the antimicrobial activity of AgNPs. Each 96-well plate consists of 8 vertical rows (A to H) and 12 horizontal rows (1 to 12). The first vertical row was considered as the positive control (100 μL inoculum + 100 μL sterile culture medium) and the second vertical row as the negative control (100 μL sterile culture medium + 100 μL AgNP colloids). The AgNP stock colloids were diluted in a sterile culture medium, keeping the final volume at 200 μL, with the posterior addition of 50 μL of the inoculum suspension. The culture medium used was Mueller Hinton (MH), a widely used culture medium for Antimicrobial Susceptibility Testing (AST). The plates were incubated at 35 °C for 24 h, and then the reading was performed by visual verification of turbidity. All steps were repeated 3 times for each microorganism separately.
2.5 Nuclear magnetic resonance spectroscopy analyses
The method for acquiring NMR data from intracellular metabolic extracts of the bacteria with the lowest MIC value was adapted from the article by Stanisic and colleagues.23Acquisition of all 1D (1H) and 2D Nuclear Magnetic Resonance – NMR spectra was done on a Bruker Avance III 600 MHz spectrometer, utilizing a TBI – Triple Resonance Broadband Inverse probe at 25 °C. For the NMR analysis and subsequent metabolomics, 2 distinct groups were studied. In all groups, the bacteria was grown in 10 mL of BHI media. For the control group, bacterial cultures were grown for 24 h at 200 rpm and 32 °C. To the culture media of the group treated with AgNP@Fo, 5 mL of bacterial culture was added, after 16 h of shaking. The final concentration of AgNP@Fo must correspond to half the value of the minimum inhibitory concentration (MIC) that will be evaluated. Then, the treated cultures were grown for another 8 h, reaching turbidity at 600 nm of 0.8. By adding methanol, in the ratio 1/1 (v/v) to the bacterial cultures, the growth of bacteria was quenched. Then, the suspensions were centrifuged for 10 min at 7000×g at 4 °C, and the cells were suspended in 30 mL of ice-cold PBS (20 mmol L−1, pH 7.2, 0 °C); this step was repeated twice. The cells were lysed by ultrasonication for 10 s (Ultrasonique). The obtained biological material was centrifuged at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 2 min, then the supernatant was kept, cell residues were washed, and the supernatants were combined and lyophilized. Samples for NMR were prepared by dissolving 15 mg of the biological material into 600 mL of deuterium oxide and spectra were acquired.
000×g for 2 min, then the supernatant was kept, cell residues were washed, and the supernatants were combined and lyophilized. Samples for NMR were prepared by dissolving 15 mg of the biological material into 600 mL of deuterium oxide and spectra were acquired.
2.6 Huh-7 cell culture
Human hepatoma cells (Huh-7, No. JCRB0403) were obtained from the Japanese Collection of Research Bioresources Cell Bank (JCRB, Japan). Huh-7 cells were cultured as a monolayer in a T25 cell culture flask containing 5 mL of Dulbecco's Modified Eagle's Medium Low Glucose (DMEM LG, Life Technologies, Canada) supplemented with 10% Fetal Bovine Serum (FBS, Gibco, South America), and 1% Penicillin/Streptomycin (Pen/Strep) (Gibco, Grand Island – USA). The cells were maintained in a Panasonic incubator at 37 °C, 95% humidity, and 5% CO2. Cells were passed as required for maintenance when confluency reached approximately 80%. In all experiments, cells were cultured for a maximum of 1 month, after which a new vial of Huh-7 cells was thawed and used for 1 month. Cultures were determined to be free of mycoplasma at each thaw using the direct DNA staining method and inspected under fluorescence microscopy. All experiments were performed with cells free of mycoplasma.2.7 MTT assay
After 24 h cell exposure to AgNP@OR or AgNP@Fo, 10 μL of MTT solution (5 mg mL−1) (Invitrogen, Oregon – USA) were added to three wells. In the blank group, 10 μL of non-supplemented LG-DMEM was added. Next, the plate was incubated at 37 °C in an atmosphere of 5% CO2 and protected from light. After 2.5 h, the MTT and blank solutions were replaced by 100 μL of dimethyl sulfoxide – DMSO (Synth, Brazil) to solubilize the formazan crystals. Then, the plate was kept under agitation for 15 min. After this, the absorbance reading (at a wavelength of 570 nm) of each well (treated with MTT and blank) was performed in the Cytation 5 Hybrid Multidetection Reader (BioTek Instruments, Inc., Winooski, VT, USA).24The results were extracted into Excel spreadsheets, where the calculations were performed to obtain cell viability as follows:25
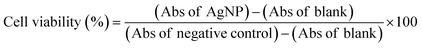 | (1) |
For the viability calculation, at least three independent replicates were performed, each of the replicates contained triplicates of wells (for example, three wells treated with AgNP@OR or AgNP@Fo at each of the concentrations of the curve, three wells treated with negative control, in this case, non-supplemented DMEM medium), three wells were the blank, where there are cells and treatment, but without the addition of MTT. In an independent repeat, the arithmetic mean of the three wells for each AgNP was used to calculate the arithmetic mean of the relative cell viability. And, the final arithmetic mean of cell viability for each AgNP was calculated from the three independent replicates.
Then, the calculated cell viability data and the respective concentrations of AgNP@OR and AgNP@Fo were plotted in GraphPad Prism 8.0.2.24 software to perform IC50 calculation by linear regression log(inhibitor) vs. response – variable slope (four parameters).
The results were extracted into Excel spreadsheets, where the calculations were performed to obtain cell viability as follows:25
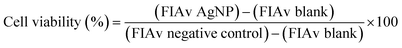 | (2) |
For this calculation were used:
• Average of at least three values of calcein fluorescence intensities (FIAv) referring to the treatment in triplicate with AgNP@OR or AgNP@Fo at each of its concentrations.
• FIAv of blank (mean of three wells without MTT addition);
• FIAv of negative control (cells treated with non-supplemented DMEM LG medium).
Then, the calculated cell viability data and the respective concentrations of AgNP@OR and AgNP@Fo were plotted in GraphPad Prism 8.0.2 software to perform IC50 calculation by linear regression log(inhibitor) vs. response – variable slope (four parameters).
2.8 Imaging assay based on commercial live/dead cell viability using the image of Hoechst, propidium iodide and calcein
In addition to MTT and calcein AM assay, an image-based assay using the dyes – Hoechst, propidium iodide and calcein was performed to evaluate cell viability. Hoechst 33342 (Sigma-Aldrich®) is used to determine the total cell count as it binds DNA from living or non-living cells. Propidium iodide (PI) (ThermoFisher®) also binds DNA, but it is impermeant to healthy cells. Therefore, PI-stained cells are considered non-viable cells. Calcein-AM, when converted to calcein by intracellular esterases indicate viable cells.After 24 h of cell exposure to AgNP@OR or AgNP@Fo solutions, treatments were replaced by 50 μL of a solution containing calcein (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000, v/v), PI (1
2000, v/v), PI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 v/v) and Hoechst (1
1000 v/v) and Hoechst (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 v/v) in FluoroBrite DMEM. The plates were incubated for 30 min at 37 °C in an atmosphere of 5% CO2 and protected from light. Then, fluorescence images of each well were acquired with the 10× objective using the image multi-mode reader of Cytation 5 (Biotek Instruments, USA). The filters used for imaging were: GFP for calcein (excitation/emission = 469/525 nm), PI filter for PI (excitation/emission = 531/647 nm), DAPI filter for Hoechst (excitation/emission = 377/447 nm). All images in the GFP channel had their background subtracted in Image J (process > subtract background), with a rolling ball radius of 1800 pixels. It also reduced the brightness of the images to −38. Phase contrast images were also obtained with the 10× objective, which were also acquired.24–27
1000 v/v) in FluoroBrite DMEM. The plates were incubated for 30 min at 37 °C in an atmosphere of 5% CO2 and protected from light. Then, fluorescence images of each well were acquired with the 10× objective using the image multi-mode reader of Cytation 5 (Biotek Instruments, USA). The filters used for imaging were: GFP for calcein (excitation/emission = 469/525 nm), PI filter for PI (excitation/emission = 531/647 nm), DAPI filter for Hoechst (excitation/emission = 377/447 nm). All images in the GFP channel had their background subtracted in Image J (process > subtract background), with a rolling ball radius of 1800 pixels. It also reduced the brightness of the images to −38. Phase contrast images were also obtained with the 10× objective, which were also acquired.24–27
3. Results and discussions
3.1 Physicochemical characterization of AgNP@Fo and AgNP@OR
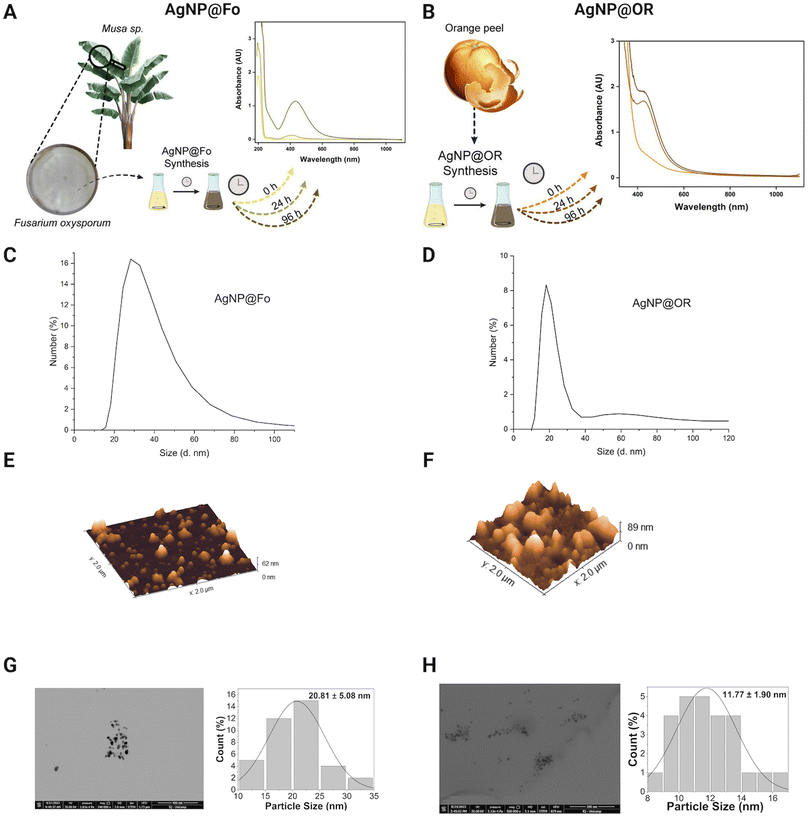
To evaluate the physicochemical characteristics of biosynthesized silver nanoparticles, we determined their hydrodynamic radii, zeta potentials, and polydispersity indexes. By assessing the nanoparticle size using DLS and STEM, we can understand the dimensions of the particles, which play a crucial role in their interactions with biological systems and other materials.28 The zeta potential measurements provide information about the surface charge of the nanoparticles, revealing the presence of functional groups that influence their stability and potential for aggregation.28,29 Additionally, the determination of the polydispersion index provides insights to the uniformity of particle sizes within the solution, indicating the level of homogeneity in the nanoparticle dispersion.29The formation of AgNPs was monitored exploring UV-Vis spectra (Fig. 2A and B). The wavelengths from 350 to 500 nm, characteristic for silver nanoparticles,28 were observed. Some studies indicate that spherical silver nanoparticles exhibit a plasmon absorption band near 400 nm.30,31 AgNP@Fo showed the maximum at 433 nm indicative for the spherical morphology of the nanomaterial. Similarly, in the case of AgNP@OR, a surface plasmon resonance (SPR) was detected around 426 nm. The spherical morphologies of the nanoparticles were confirmed in AFM (Fig. 2E and F) and STEM (Fig. 2G and H).
After 96 h of biosynthesis, AgNP@Fo exhibited a medium nanoparticle size of 56.43 ± 19.18 nm, determined using DLS (Fig. 2C), and 20.81 ± 5.08 nm using STEM (Fig. 2G). The main difference between DLS and STEM lies in the approach used to measure the size of nanoparticles. DLS provides information about the average hydrodynamic size of particles in solution, while STEM offers direct images of the size and shape of individual nanoparticles.26,27
The zeta potentials of these nanoparticles, when diluted in Milli-Q water at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 v/v ratio, was measured and negative zeta potential values were attributed to the presence of carboxylate groups in the biological samples, playing a role in the stabilization of AgNPs.26 The zeta potential of AgNP@Fo was −26.8 ± 7.55 mV. This stabilization led to a polydispersion index (PDI) of 0.163, indicating a relatively homogeneous dispersion of nanoparticles within the colloid. The concentration of AgNP@Fo was calculated to be 0.401 ± 0.099 mmol L−1 using the calibration curve depicted in Fig. 1, closely aligning with the theoretically expected concentration of 0.5 mmol L−1, thereby falling within the anticipated range, as indicated by Fig. S1 and Table S1.†
10 v/v ratio, was measured and negative zeta potential values were attributed to the presence of carboxylate groups in the biological samples, playing a role in the stabilization of AgNPs.26 The zeta potential of AgNP@Fo was −26.8 ± 7.55 mV. This stabilization led to a polydispersion index (PDI) of 0.163, indicating a relatively homogeneous dispersion of nanoparticles within the colloid. The concentration of AgNP@Fo was calculated to be 0.401 ± 0.099 mmol L−1 using the calibration curve depicted in Fig. 1, closely aligning with the theoretically expected concentration of 0.5 mmol L−1, thereby falling within the anticipated range, as indicated by Fig. S1 and Table S1.†
On the other hand, AgNP@OR nanoparticles showed smaller average sizes 39.97 ± 19.72 nm, determined using DLS (Fig. 2D), and 11.77 ± 1.90 nm using STEM (Fig. 2H). The zeta potential for these nanoparticles was −26.2 ± 2.87 mV, demonstrating a similar negative charge as observed for the AgNP@Fo. The sample exhibited a PDI value of 0.414, consistent with previous research suggesting that AgNPs synthesized from fruit and vegetable extracts often display moderate PDIs, from 0.3 to 0.5.29 Additionally, the concentration of AgNP@OR was determined to be 0.465 ± 0.01 mmol L−1 (Fig. S1 and Table S1†).
3.2 Antimicrobial activity
AgNP@Fo demonstrated significant antimicrobial activity, with minimum inhibitory concentrations (MICs) ranging from 0.84 to 1.68 μg mL−1, exhibiting lower MIC values for Gram-negative bacteria (Table 1 and Fig. S2†). Nonetheless, AgNP@OR exhibited a broader range of MIC values, ranging from 10.8 to 27.0 μg mL−1. Interestingly, AgNP@OR showed no antimicrobial activity against Staphylococcus aureus within the tested concentration range, as summarized in Table 1 and Fig. S3.† The mechanism by which silver nanoparticles (AgNPs) combat bacteria is not fully comprehended at present. Nevertheless, there exist several theories elucidating the antibacterial effects of silver nanoparticles: (1) production of reactive oxygen species, (2) liberation of Ag+ ions from AgNPs causing protein denaturation by binding with sulfhydryl groups, and (3) attachment of AgNPs to bacteria, resulting in subsequent damage to the bacterial structures.32–36| Microorganism | MIC AgNP@Fo (μg mL−1) | MIC AgNP@OR (μg mL−1) |
|---|---|---|
| Pseudomonas aeruginosa (CCT 1476) (Gram-negative) | 0.84 | 27.0 |
| Staphylococcus aureus (CCT1485) (Gram-positive) | 1.68 | — |
| Escherichia coli (CCT1457) (Gram-negative) | 0.84 | 27.0 |
| Enterococcus faecalis (CCT 1494) (Gram-positive) | 1.68 | 10.8 |
Using fungi as a reducing agent and stabilizer of biogenic silver nanoparticles has become very attractive due to the production of large amounts of proteins, easy handling and low toxicity of the residues.37,38 The antimicrobial activity is believed to result from the tendency of nanoparticles to interact with bacteria. AgNPs accumulate on the surfaces of the bacterial membrane, causing the inhibition of cellular proteins, consequently leading to bacterial cell death.38 According to the obtained MIC values, the antimicrobial activity of AgNP@Fo is quite promising and exhibits a greater antimicrobial potential, which can be up to around 10 times more potent, compared to certain published studies that evaluated AgNPs synthesized under different conditions, making a significant contribution to this field of study.32,37
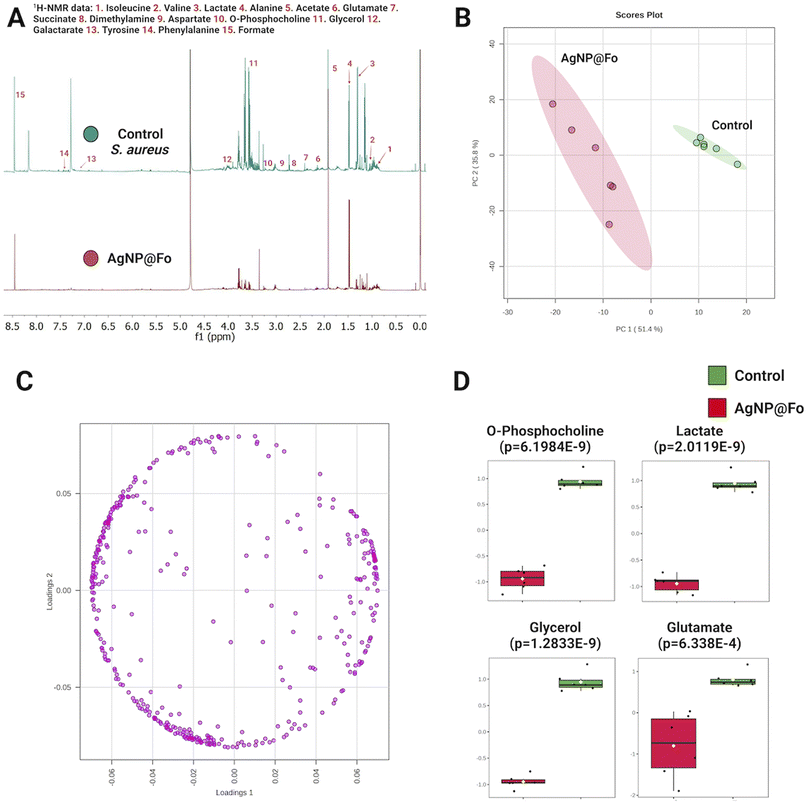
Using the 1H-NMR, we aimed to evaluate the change in the metabolic profile of S. aureus in response to the action of silver nanoparticles (AgNP@Fo) (Fig. 3). These results aim to improve our understanding of the impact of AgNP@Fo on bacteria at a metabolic level and can be reproduced for the other bacteria tested.
The overlapping of metabolic profiles between the control sample (which did not undergo treatment with AgNP@Fo) and the sample treated with nanoparticles is illustrated in Fig. 3A. To confirm the difference between these groups, we employed a chemometrics analysis, represented in the Principal Component Analysis (PCA) plot in Fig. 3B. This analysis confirmed that AgNP@Fo nanoparticles had a significant impact on the metabolic level of bacterial cells. In Fig. 3C, we demonstrate the corresponding loading plot of the PCA, which shows the individual contributions of metabolites to the observed variation in metabolic data. This allows us to identify the metabolites that most influenced the separation between the control and AgNP@Fo-treated groups.
To assess which metabolites were most affected, meaning where there was a more significant variation in relative concentrations, we conducted a t-test analysis (Fig. 3D). In this analysis, we identified a pronounced decrease in the concentrations of metabolites such as phosphocholine, lactate, glycerol, and glutamate. These metabolites play essential roles in crucial metabolic pathways for bacterial survival. For example, phosphocholine39 is fundamental in cell membrane synthesis, lactate is involved in energy production,40 glycerol plays a central role in lipid biosynthesis,41 and glutamate participates in amino acid metabolism and the tricarboxylic acid cycle.42 The significant reduction of these metabolites has important implications for understanding the effects of AgNP@Fo nanoparticles on bacterial metabolism.
The variance in biological potential observed between AgNP@Fo and AgNP@OR might be linked to proteins secreted by the Fusarium oxysporum fungus and those present in orange biomass, potentially serving as both reducing and stabilizing agents. In prior research conducted by our research group, using a distinct strain of Fusarium oxysporum43 and orange biomass44 for silver nanoparticle biosynthesis, the anchoring of these proteins through nitrogen and sulfur-containing groups was noted. Understanding the protein anchoring process is crucial in determining how nanoparticles interact with their environment. This interaction could impact the controlled release of silver ions, thus influencing the antimicrobial activity of the nanoparticles. The way proteins bind to the nanoparticles may not only affect particle stability but also their potential for internalization by microorganisms, a factor critical to their antimicrobial effectiveness.43,44
3.3 Cytotoxicity evaluation of AgNP@OR and AgNP@Fo in Huh-7 cells
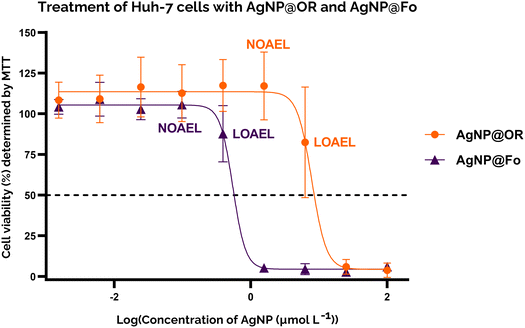
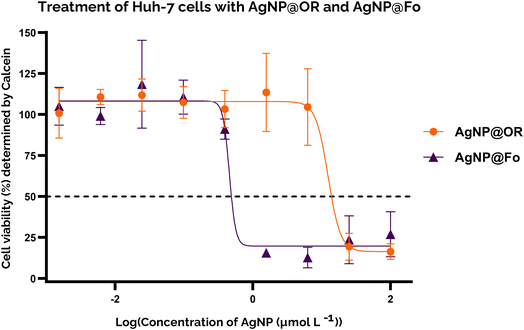
To gain insight into the cytotoxicity profile of AgNP@OR and AgNP@Fo, Huh-7 cells were used and the half-maximal concentration for cell viability (IC50) was determined by MTT and calcein assay. The treatment concentrations of AgNP@OR and AgNP@Fo ranged from 0.0015 to 100 μmol L−1. Other relevant parameters were calculated, such as the No Observable Adverse Effect Level (NOAEL), the highest dose at which the harmful effect is not observed and the Lowest Observable Adverse Effect Level (LOAEL), the lowest dose at which the harmful effect is observed.44,45For the MTT assay, the IC50 values for Huh-7 exposure to AgNP@OR and AgNP@Fo for 24 h were determined using a nine-point nonlinear regression model with GraphPad Prism 8.0.2 software (Fig. 4). For AgNP@OR, the NOAEL was calculated to be 1.56 μmol L−1, the LOAEL 6.25 μmol L−1, and the IC50 value was 7.833 μmol L−1. For AgNP@Fo, the NOAEL was calculated at 0.097 μmol L−1, LOAEL 0.39 μmol L−1, and the IC50 value was 0.545 μmol L−1. Therefore, the cytotoxicity profile determined using the MTT assay suggests that AgNP@Fo has significantly higher cytotoxicity towards Huh-7 cells than AgNP@OR.
For the calcein assay, the cytotoxic profile was determined using a similar approach to the MTT assay; for comparison, the same concentrations of AgNP@OR and AgNP@Fo were tested, and the same parameters were determined (Fig. 5). For AgNP@OR, the NOAEL was determined to be 6.25 μmol L−1, the LOAEL 25 μmol L−1, and the IC50 value was 12.36 μmol L−1. Compared to the values determined by MTT, the NOAEL and LOAEL values were one tested concentration higher, and the IC50 value was similar. For AgNP@Fo, the NOAEL was determined to be 0.097 μmol L−1, the LOAEL 0.39 μmol L−1, and the IC50 value was 0.4544 μmol L−1. Compared to the values determined by MTT, the NOAEL and LOAEL values were the same, and the IC50 value was similar. Therefore, the MTT and calcein results agreed, suggesting that AgNP@Fo is more cytotoxic than AgNP@OR, increasing the robustness of the results.
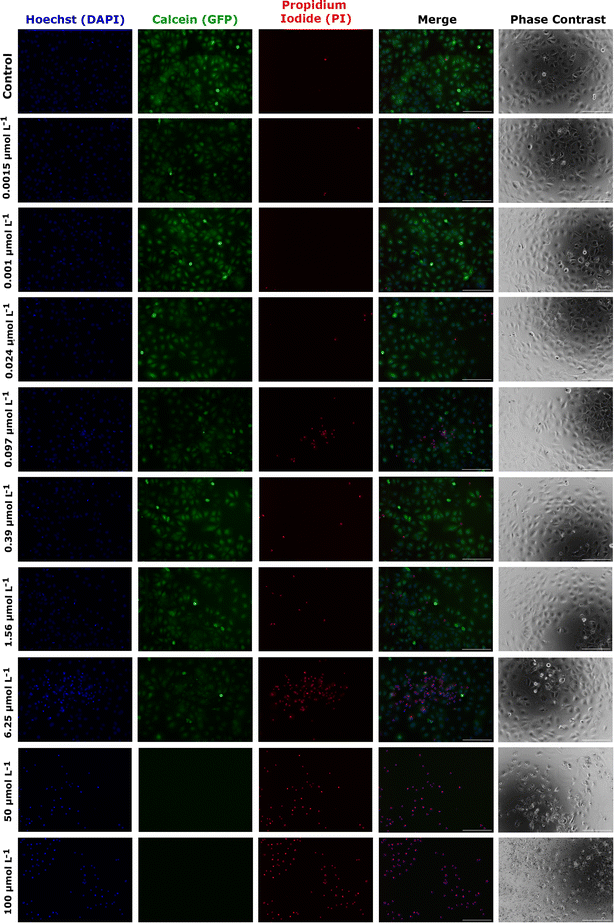
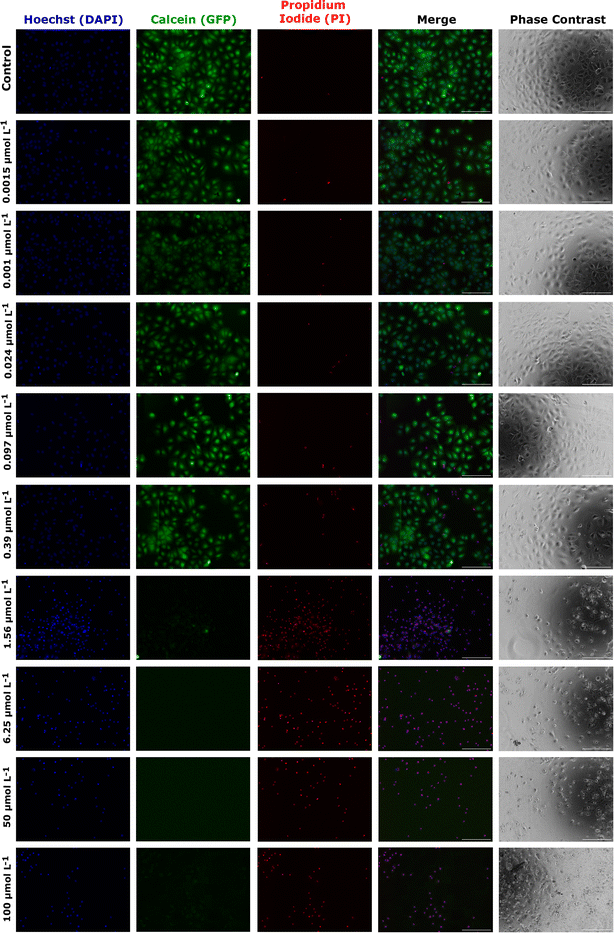
As an alternative and complementary measure of cytotoxic response, the fluorescence and phase contrast images of cells stained with calcein, PI, and Hoechst were also used to monitor the toxic response of Huh-7 cells to AgNP@OR and AgNP@Fo MTT exposure. Fig. 6 shows that from a concentration of 6.25 μmol L−1 AgNP@OR, more Huh-7 cells have a nucleus stained with PI, indicating non-viable cells, i.e., cells that lose the integrity of plasma membrane. From the concentration of 6.25 μmol L−1 AgNP@OR, no cells stained with calcein were observed, i.e., no viable cells. In the phase contrast images, it can be seen that cells treated with AgNP@OR at 6.25 and 100 μmol L−1 have a different morphology than the healthy cells treated with the negative control (culture medium); they are smaller, rounded, and detached. Fig. 7 shows that from a concentration of 0.39 μmol L−1 AgNP@Fo, a higher number of Huh-7 cells have a cell nucleus stained with the PI fluorophore. From a concentration of 1.56 μmol L−1 AgNP@Fo, no cells are stained with calcein, i.e., viable cells. In the phase-contrast images, cells treated with concentrations greater than 1.56 mol L−1 AgNP@Fo show a different morphology than healthy cells treated with the negative control (culture medium), being smaller, rounded, and detached from the bottom.
AgNP@Fo was approximately 20 times more cytotoxic than AgNP@OR, this was supported by the MTT assay, the calcein-AM assay and fluorescence images. One possible explanation for the cytotoxicity profile differences between AgNP@OR and AgNP@Fo is a consequence of the physicochemical properties of nanoparticles. As both nanoparticles are very similar in size and shape (the size of AgNP@Fo is 56.43 ± 19.18 nm and the size of AgNP@OR is 39.97 ± 19.72 nm, both being spherical), the differences in cytotoxicity could be due to surface modifications of these two particles and their interaction with Huh-7 cells. To fully understand the reasons for this discrepancy in anticancer activity between nanoparticles, further in-depth investigations are needed to identify the specific chemical compounds present in each type of nanoparticles, as well as their biological properties and mechanisms of action. Based on these findings, it will be possible to develop more effective and targeted cancer treatments.43–51
In summary, this study aimed to explore the antimicrobial and anticancer potential to improve our understanding of the impact of biogenic silver nanoparticles. In addition, it paves the way for future investigations into the AgNP interactions with cancer cells, thus broadening the horizons for advanced research in this area.
4. Conclusions
In summary, our results support the hypothesis that the biosynthesis of the silver nanoparticles determines their biological role. AgNP@Fo showed impressive antimicrobial activity, with MIC values approximately 10-fold lower than AgNP@OR. Additionally, AgNP@Fo showed almost 20 times higher cytotoxicity than AgNP@OR in Huh-7 cells, with an IC50 value of 0.545 μmol L−1. Elucidating the mechanisms underlying these activities would undoubtedly improve our understanding of the relationship between physicochemical properties and biological applications. This knowledge, along with safety and efficacy in clinical trials, is paramount to improving the rational development of nanomaterial applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the funding by Sao Paulo Research Foundation (FAPESP, Grants #2023/02338-0, #2022/07854-4, #2020/01218-3, #2023/06874-4 and #2020/08615-8) and CNPq (INCTBio) for financial support.References
- C. Selvaraj, S. Sakkiah, W. Tong and H. Hong, Food Chem. Toxicol., 2018, 112, 495–506 CrossRef CAS PubMed.
- H. Liu, H. Zhang, J. Wang and J. Wei, Arabian J. Chem., 2020, 13, 1011–1019 CrossRef CAS.
- R. H. Ahmed and D. E. Mustafa, Int. Nano Lett., 2020, 10, 1–14 CrossRef CAS.
- L. A. B. Ferreira, F. Garcia-Fossa, A. Radaic, N. Durán, W. J. Fávaro and M. B. de Jesus, Eur. J. Pharm. Biopharm., 2020, 151, 162–170 CrossRef CAS PubMed.
- K. Jadhav, S. Deore, D. Dhamecha, R. Hr, S. Jagwani, S. Jalalpure and R. Bohara, ACS Biomater. Sci. Eng., 2018, 4, 892–899 CrossRef CAS PubMed.
- B. Plackal Adimuriyil George, N. Kumar, H. Abrahamse and S. S. Ray, Sci. Rep., 2018, 8, 14368 CrossRef PubMed.
- S. Gurunathan, K. J. Lee, K. Kalishwaralal, S. Sheikpranbabu, R. Vaidyanathan and S. H. Eom, Biomaterials, 2009, 30, 6341–6350 CrossRef CAS PubMed.
- G. Magdy, E. Aboelkassim, S. Elhaleem and F. Belal, Microchem. J., 2024, 196, 109615 CrossRef CAS.
- G. Magdy, E. Aboelkassim, R. A. El-Domany and F. Belal, Sci. Rep., 2022, 12, 21395 CrossRef CAS PubMed.
- C. Quintero-Quiroz, N. Acevedo, J. Zapata-Giraldo, L. E. Botero, J. Quintero, D. Zárate-Trivinõ, J. Saldarriaga and V. Z. Pérez, Biomater. Res., 2019, 23, 27 CrossRef PubMed.
- R. Vishwanath and B. Negi, Curr. Res. Green Sustainable Chem., 2021, 4, 100205 CrossRef CAS.
- R. Majumdar and P. K. Kar, Sci. Rep., 2023, 13, 7415 CrossRef CAS PubMed.
- M. Guilger, T. Pasquoto-Stigliani, N. Bilesky-Jose, R. Grillo, P. C. Abhilash, L. F. Fraceto and R. De Lima, Sci. Rep., 2017, 7, 44421 CrossRef CAS PubMed.
- M. Guilger-Casagrande and R. de Lima, Front. Bioeng. Biotechnol., 2019, 7, 287 CrossRef PubMed.
- N. Rodríguez-Barajas, U. de Jesús Martín-Camacho and A. Pérez-Larios, Curr. Top. Med. Chem., 2022, 22, 2506–2526 CrossRef PubMed.
- V. Mulens-Arias, J. M. Rojas and D. F. Barber, Nanomaterials, 2020, 10, 837 CrossRef CAS PubMed.
- M. A. Pfaller, R. K. Flamm, L. R. Duncan, J. M. Streit, M. Castanheira and H. S. Sader, Diagn. Microbiol. Infect. Dis., 2018, 91, 77–84 CrossRef CAS PubMed.
- R. Soltani, H. Fazeli, R. B. Najafi and A. Jelokhanian, Iran. J. Pharm. Res., 2017, 16, 290–296 CAS.
- C. C. Chen, C. Y. Chen, S. H. Ueng, C. Hsueh, C. T. Yeh, J. Y. Ho, L. F. Chou and T. H. Wang, Cell Death Dis., 2018, 9, 543 CrossRef PubMed.
- M. Kawamoto, T. Yamaji, K. Saito, Y. Shirasago, K. Satomura, T. Endo, M. Fukasawa, K. Hanada and N. Osada, Front. Genet., 2020, 11, 546106 CrossRef CAS PubMed.
- R. Laube, A. H. Sabih, S. I. Strasser, L. Lim, M. Cigolini and K. Liu, J. Gastroenterol. Hepatol., 2021, 36, 618–628 CrossRef PubMed.
- R. A. González-Fuenzalida, Y. Moliner-Martínez, M. González-Béjar, C. Molins-Legua, J. Verdú-Andres, J. Pérez-Prieto and P. Campins-Falcó, Anal. Chem., 2013, 85, 10013–10016 CrossRef PubMed.
- D. Stanisic, N. L. Fregonesi, C. H. N. Barros, J. G. M. Pontes, S. Fulaz, U. J. Menezes, J. L. Nicoleti, T. L. P. Castro, N. Seyffert, V. Azevedo, N. Durán, R. W. Portela and L. Tasic, RSC Adv., 2018, 8, 40778–40786 RSC.
- T. Omanovic Kolaric, T. Kizivat, V. Mihaljevic, M. Zjalic, I. Bilic-Curcic, L. Kuna, R. Smolic, A. Vcev, G. Y. Wu and M. Smolic, Curr. Issues Mol. Biol., 2022, 44, 3465–3480 CrossRef CAS PubMed.
- S. Kamiloglu, G. Sari, T. Ozdal and E. Capanoglu, Food Front., 2020, 1, 332–349 CrossRef.
- M. L. Swift, J. Chem. Inf. Model., 1997, 37, 411–412 CrossRef CAS.
- M. Tenopoulou, T. Kurz, P. T. Doulias, D. Galaris and U. T. Brunk, Biochem. J., 2007, 403, 261–266 CrossRef CAS PubMed.
- M. K. Rasmussen, J. N. Pedersen and R. Marie, Nat. Commun., 2020, 11, 23337 Search PubMed.
- M. Danaei, M. Dehghankhold, S. Ataei, F. Hasanzadeh Davarani, R. Javanmard, A. Dokhani, S. Khorasani and M. R. Mozafari, Pharmaceutics, 2018, 10, 57 CrossRef PubMed.
- P. V. Kamat, J. Phys. Chem. B, 2002, 106, 7729–7744 CrossRef CAS.
- J. J. Mock, M. Barbic, D. R. Smith, D. A. Schultz and S. Schultz, J. Chem. Phys., 2002, 116, 6755–6759 CrossRef CAS.
- C. H. N. Barros, S. Fulaz, D. Stanisic and L. Tasic, Antibiotics, 2018, 7, 69 CrossRef CAS PubMed.
- S. S. dos Santos, R. A. A. de Couto, I. R. da Silva, M. R. M. Aouada, V. R. L. Costantino, L. P. da Costa and G. F. Perotti, J. Braz. Chem. Soc., 2023, 34, 705–712 CAS.
- L. Du, X. Lei, J. Wang, L. Wang, Q. Zhong, X. Fang, P. Li, B. Du, Y. Wang and Z. Liao, Int. Rev. Immunol., 2022, 41, 45–56 CrossRef CAS PubMed.
- G. Gahlawat and A. R. Choudhury, RSC Adv., 2019, 9, 12944–12967 RSC.
- S. K. Kailasa, T. J. Park, J. V. Rohit and J. R. Koduru, Nanopart. Pharmacother., 2019, 15, 2555–2562 Search PubMed.
- R. M. Elamawi, R. E. Al-Harbi and A. A. Hendi, Egyptian Journal of Biological Pest Control, 2018, 28, 28 CrossRef.
- D. Wang, B. Xue, L. Wang, Y. Zhang, L. Liu and Y. Zho, Sci. Rep., 2021, 11, 10356 CrossRef CAS PubMed.
- F. Pizzo, D. Gadaleta and E. Benfenati, Methods Mol. Biol., 2022, 1425, 163–176 CrossRef PubMed.
- Y. Kadowaki, T. Hirano, K. Fujita, T. Kawano, T. Matsunaga, K. Yoshinaga and M. Suzuki, Auris, Nasus, Larynx, 2021, 48, 57–64 CrossRef PubMed.
- K. Vinasco, H. M. Mitchell, N. O. Kaakoush and N. Castaño-Rodríguez, Biochim. Biophys. Acta, Rev. Cancer, 2019, 1872, 188309 CrossRef CAS PubMed.
- M. Zhao, Z. Jiang, H. Cai, Y. Li, Q. Mo, L. Deng, H. Zhong, T. Liu, H. Zhang, J. X. Kang and F. Fenga, mBio, 2020, 11, 190 Search PubMed.
- D. Ballottin, S. Fulaz, F. Cabrini, J. Tsukamoto, N. Durán, O. L. Alves and L. Tasic, Mater. Sci. Eng., C, 2017, 75, 582–589 CrossRef CAS PubMed.
- C. H. N. de Barros, G. C. F. Cruz, W. Mayrink and L. Tasic, Nanotechnol., Sci. Appl., 2018, 11, 1–14 CrossRef CAS PubMed.
- Y. Yong, Y. Zhou, K. Liu, G. Liu, L. Wu and B. Fang, Front. Microbiol., 2021, 12, 759170 CrossRef PubMed.
- T. H. Tseng, C. J. Wang, Y. J. Lee, Y. C. Shao, C. H. Shen, K. C. Lee, S. Y. Tung and H. C. Kuo, Int. J. Mol. Sci., 2022, 23, 5102 CrossRef CAS PubMed.
- L. Tasic, N. Durán, W. J. Fávaro, S. Alborés and T. S. da Costa, J. Braz. Chem. Soc., 2023, 34, 897–905 Search PubMed.
- P. E. Porporato, N. Filigheddu, J. M. B. S. Pedro, G. Kroemer and L. Galluzzi, Cell Res., 2018, 28, 265–280 CrossRef CAS PubMed.
- X. Fu, Y. Shi, T. Qi, S. Qiu, Y. Huang, X. Zhao, Q. Sun and G. Lin, Signal Transduction Targeted Ther., 2020, 5, 262 CrossRef PubMed.
- J. Wang, Y. Li and G. Nie, Nat. Rev. Mater., 2021, 6, 766–783 CrossRef CAS PubMed.
- A. B. B. Oliveira, F. R. De Moraes, N. M. Candido, I. Sampaio, A. S. Paula, A. De Vasconcellos, T. C. Silva, A. H. Miller, P. Rahal, J. G. Nery and M. F. Calmon, J. Proteome Res., 2016, 15, 4337–4348 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra07733k |
| This journal is © The Royal Society of Chemistry 2024 |