Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A novel aurone RNA CAG binder inhibits the huntingtin RNA–protein interaction†
Giovanna
Ballarin‡
abc,
Maddalena
Biasiotto‡
abc,
Annika
Reisbitzer
b,
Marlen
Hegels
c,
Michael
Bolte
 d,
Sybille
Krauß
d,
Sybille
Krauß
 b and
Daria V.
Berdnikova
b and
Daria V.
Berdnikova
 *c
*c
aUniversity of Padova, School of Pharmaceutical Sciences, via Marzolo 5, 35131 Padova, Italy
bInstitut für Biologie, Universität Siegen, Adolf-Reichwein-Str. 2, 57076 Siegen, Germany
cOrganische Chemie II, Universität Siegen, Adolf-Reichwein-Str. 2, 57076 Siegen, Germany. E-mail: berdnikova@chemie-bio.uni-siegen.de
dInstitut für Anorganische Chemie, J.-W.-Goethe-Universität, Max-von-Laue-Str. 7, 60438 Frankfurt-am-Main, Germany
First published on 17th July 2024
Abstract
Huntington's disease (HD) is a devastating, incurable condition whose pathophysiological mechanism relies on mutant RNA CAG repeat expansions. Aberrant recruitment of RNA-binding proteins by mutant CAG hairpins contributes to the progress of neurodegeneration. In this work, we identified a novel binder based on an aurone scaffold that reduces the level of binding of HTT mRNA to the MID1 protein in vitro. The obtained results introduce aurones as a novel platform for the design of functional ligands for disease-related RNA sequences.
Introduction
Short tandem repeats along with other repetitive sequences comprise a substantial fraction of the human genome.1 Being polymorphic and susceptible to mutations, short tandem repeats can elongate yielding repeat expansions, which become toxic after crossing a certain length threshold.2–5 These repeat expansions, scattered throughout the human genome, can lead to more than 40 severe disorders, the majority of which affect the nervous system and are currently incurable.6,7 Most often, repeat expansion disorders are caused by the expansion of CXG trinucleotide sequences. Among them are Huntington's disease (HD), myotonic dystrophy type 1 (DM1), a range of spinocerebellar ataxias, Fuchs corneal dystrophy and others.6,7One of the well-known types of CAG repeat expansion disorders is Huntington's disease (HD), which leads to the progressive degeneration of brain nerve cells.8 The pathophysiological mechanism of HD is based on the expanded CAG repeat formed within exon 1 of the huntingtin (HTT) mRNA. Since the mutant repeat is located within a coding region, it becomes translated into a toxic polyglutamine-containing HTT protein that causes neurodegeneration and other consequences.9 The CAG repeats additionally contribute to the development of HD through another mechanism. The CAG repeat expansions significantly alter the RNA structure because the trinucleotide expansions fold into aberrant hairpins, which never form in a normal RNA. The aberrant hairpins can recruit RNA-binding proteins by providing additional binding sites that are not characteristic of a healthy RNA. Particularly, in the case of HD, proteins involved in translation induction and splice factors can become bound by this mechanism resulting in the loss of regular functions of these proteins and their potentially abnormal behavior.10–14
Understanding of biomolecular mechanisms underlying the development of the repeat expansion disorders paves a way towards potential therapeutical strategies for diagnostics and treatment. One of the strategies relies on selective interactions of the CAG repeat expansions with small organic molecules, which prevents the formation of toxic RNA–protein complexes. Although a range of small organic compounds that target CXG repeat expansion RNAs have been designed,15–17 there is still an urgent need in the development of RNA binders that selectively interact with the HD-associated CAG RNAs and block their aberrant biological functions. Along these lines, we became interested in aurones18–20 – a family of natural and synthetic flavonoids – as potential scaffolds for the design of binders for the CAG RNA. Depending on the substitution pattern, aurone derivatives demonstrate various biological activities, in general, upon selective interactions with proteins (enzymes).18–20 Some aurones possess antioxidant activity21 and antibacterial properties.22 At the same time, interactions of aurones with nucleic acids have been scarcely studied, so far.18 There are reports on the DNA-scission activity of some aurone derivatives23 and fluorescent DNA staining with aurones.24 However, to the best of our knowledge, the RNA-binding properties of aurones have not been described, so far.
Herein, we report a novel aurone derivative that selectively binds to the HD-associated CAG repeat expansion RNA and inhibits the RNA–protein interaction in an RNA pull-down assay in vitro.
Results and discussion
Synthesis
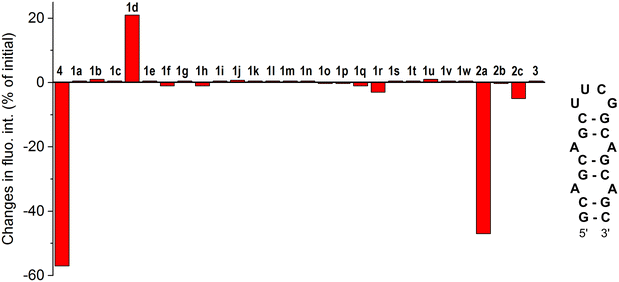
To initially assess the RNA binding potential of aurone ligands and identify the possible hits, a library of twenty-six aurone derivatives 1a–1w and 2a–2c bearing different substituents and one aza-aurone (hemiindigo) derivative 3 (Chart 1) were screened against a short RNA oligonucleotide 5′-GCAGCAGCUUCGGCAGCAGC-3′ comprising two CAG repeats (Fig. 1).25 Known compounds 1a–1e, 1g–1k, 1m–1o, 1r–1u, 1w, 2a–2c and 3 were synthesized in our lab earlier and characterized by comparison with the literature data for melting point values and NMR spectroscopy.21,22,24,26–34 Novel aurone derivatives 1f, 1l, 1p, 1q, and 1v were obtained in this work for the first time and fully characterized by 1D and 2D NMR spectroscopy, mass spectrometry and elemental analysis (ESI†). For compounds 1f and 1w, single-crystal X-ray analysis data were provided for the first time (ESI,† CCDC deposition numbers: 2335536 and 2335537).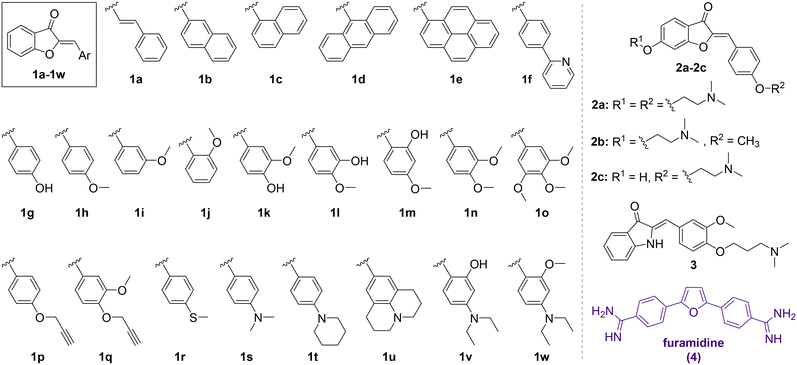 | ||
| Chart 1 Chemical structures of aurone derivatives 1a–1w and 2a–2c and aza-aurone (hemiindigo) 3 used for the screening and the structure of the known CAG RNA binder furamidine (4). | ||
Well-plate screening
The screening of 1a–1w, 2a–2c and 3 against the CAG RNA oligonucleotide was performed using a well plate and the fluorescence output of each ligand was measured without RNA and in the presence of one equivalent of the RNA oligonucleotide at three different excitation wavelengths (λex = 350, 400 and 450 nm). The known CAG RNA binder furamidine (4)35 was included in the screening as a reference compound. The results of the screening are provided in Fig. 1. As can be seen, most of the compounds demonstrated negligible changes in the fluorescence response upon the addition of RNA, indicating an absent or a very weak interaction with the nucleic acid. Notably, the previously reported HIV RNA binder 333 did not associate with CAG RNA. Only for three derivatives out of the twenty-eight screened ones, the changes in fluorescence in the presence of RNA were pronounced. Thus, as expected, the known CAG RNA binder furamidine (4) showed significant fluorescence quenching (57% of the initial intensity). The fluorescence of aurone 2a bearing two alkylamino chains was also remarkably quenched (47% of the initial intensity) upon the addition of the CAG RNA oligonucleotide. Notably, aurones 2b and 2c having only a single alkylamino chain showed small changes in the fluorescence intensity pointing out a crucial binding role of the second alkylamino pendant in 2a. The aurone derivative 1d comprising an anthracenyl moiety demonstrated a moderate fluorescence light-up effect (21% of the initial intensity) in the presence of the CAG RNA oligonucleotide. Due to significant changes in the fluorescence output in the presence of the CAG RNA oligonucleotide, aurone derivatives 1d and 2a were selected for further analysis.Selectivity studies
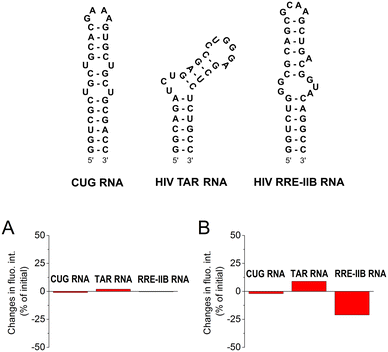
To assess the selectivity of interactions of aurones 1d and 2a with CAG RNA, additional well-plate screening was performed using alternative RNA substrates, namely the CUG RNA motif associated with myotonic dystrophy type 1 (DM1) and the regulatory elements of human immunodeficiency virus type 1 (HIV-1) genome RNA – the transactivation response element (TAR) and the stem IIB of the Rev response element (RRE-IIB) (Fig. 2).36,37 The HIV-1 TAR and RRE-IIB RNAs were chosen for this study because their structure allows to test several possible binding modes between the small ligands and RNA, including the stem intercalation, bulge binding and loop binding. In principle, stem intercalation can also take place in the case of the used CUG oligomer comprising an elongated double-helix region (Fig. 2). Like in the case of the CAG RNA oligonucleotide, the fluorescence of aurones 1d and 2a was recorded on the well-plate without RNA and in the presence of 1 equivalent of the corresponding oligonucleotides (Fig. 2). It was found that the addition of these RNA oligonucleotides produced almost no effect on the fluorescence of compound 2a indicating an absent or very weak interaction. For derivative 1d, almost no interaction was detected with the CUG RNA. However, the changes in the fluorescence of 1d in the presence of the TAR RNA (light-up) and RRE-IIB RNA (quenching) were pronounced and, therefore, indicative of binding. Therefore, within the tested RNA sequences, aurone 2a showed clear selectivity towards the CAG trinucleotide repeat motif. At the same time, compound 1d was much less selective towards various RNA sequences (noticeable interactions with HIV TAR and RRE-IIB RNA), although it obviously showed preference for the CAG RNA in comparison to the CUG RNA. Notably, in the case of compound 2a, two alkylamino substituents played a crucial role in the development of the RNA-binding properties and selectivity towards the CAG RNA. For comparison, the core scaffold of derivative 2a, namely 4-methoxyaurone (compound 1h, Fig. 1), did not interact with RNA. Moreover, the related compounds 2b and 2c bearing a single alkylamino substituent either at the coumaranone fragment (2b) or at the phenyl ring (2c) did not show a pronounced interaction with the CAG RNA (Fig. 1). A careful comparison can also be made with the aza-aurone derivative 3 bearing only one alkylamino tail: the presence of a single alkylamino substituent did not provide the affinity towards the CAG RNA (Fig. 1).Determination of the binding constant with the CAG RNA oligonucleotide
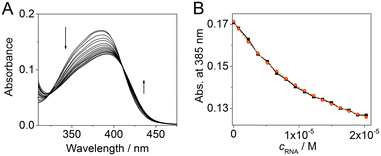
To quantify the interaction of aurone derivatives with the CAG RNA oligonucleotide, spectrophotometric titration was performed (Fig. 3A). Thus, upon the addition of RNA, the absorption spectrum of ligand 2a showed a hypochromic effect along with a moderate red shift of the absorption maximum. During the titration, a clear isosbestic point at 407 nm was formed indicating a single dominating binding mode of 2a as well as homogeneous folding of the RNA motif providing preferentially a single type of the binding pocket. The analysis of the obtained binding isotherm (Fig. 3B) allowed to estimate the stoichiometry and the binding constant of the 2a–RNA complex. Thus, the preferential formation of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ligand–RNA oligonucleotide complexes was observed (for details, see the ESI†). The association constant value is K = (1.4 ± 0.1) × 105 M−1. For derivative 1d, the determination of the binding constant by spectrophotometric titration was not possible due to the aggregation of the compound upon the addition of RNA.
1 ligand–RNA oligonucleotide complexes was observed (for details, see the ESI†). The association constant value is K = (1.4 ± 0.1) × 105 M−1. For derivative 1d, the determination of the binding constant by spectrophotometric titration was not possible due to the aggregation of the compound upon the addition of RNA.
RNA pull-down experiments
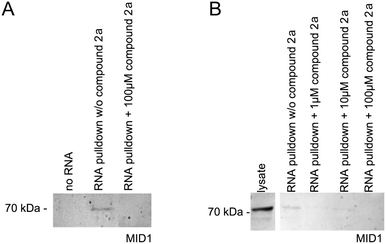
As has been shown previously, the MID1 protein binds its target HTT mRNA at its CAG-repeat in a length-dependent manner.11,12,35 Therefore, we used HTT exon 1 transcripts containing the CAG-repeat region (for details, see the ESI†) to test if compounds 2a and 1d affect the binding between the MID1 protein and its target mRNA by performing RNA–protein pull-down assays. To perform this experiment, biotinylated RNA-oligos were incubated with cell extracts that contained the MID1 protein in the presence or the absence of 2a and 1d. The RNA–protein complexes were then isolated using streptavidin beads, and the RNA-bound proteins were analyzed by western blot detecting MID1 (Fig. 4). As a negative control, an experiment without RNA was performed. As expected, MID1 was detected in the samples without compound 2a (positive control). At the same time, in the presence of 2a, the binding of MID1 to HTT RNA was suppressed (Fig. 4A). The dose-dependence assay (Fig. 4B) showed that aurone 2a provided the inhibition of the RNA–MID1 interactions at all tested doses (final concentrations from 1 μM to 100 μM). The pull-down assay with compound 1d did not reveal an inhibiting effect on the RNA–protein interactions (Fig. S1, ESI†). The possible reason for this is the low selectivity of 1d towards the CAG RNA motif (vide supra).Conclusions
In summary, we have identified novel CAG RNA binder 2a that inhibits the toxic RNA–MID1 protein interaction in vitro in the Huntington's disease model. To the best of our knowledge, this is the first example of an RNA binder based on an aurone scaffold, which, therefore, provides a proof-of-principle for the application of aurone flavonoids as a platform for the design of RNA-targeting ligands.Data availability
The data supporting this article have been included as part of the ESI.† Crystallographic data for 1f and 1w have been deposited at the CCDC under deposition numbers 2335536 and 2335537.Author contributions
D. V. B. and S. K. conceived and designed the experiments. G. B., M. B. (Maddalena Biasiotto), A. R., M. H. and D. V. B. performed the experiments. M. B. (Michael Bolte) carried out the single-crystal X-ray analysis. D. V. B. and S. K. performed the supervision, analysed the data and wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
G. B. and M. B. are thankful to the ERASMUS Exchange Program for the fellowships. D. V. B. thanks the University of Siegen for financial support. We thank Ms. Sandra Uebach and Mr. Sören Steup (University of Siegen, Germany) for technical assistance.Notes and references
- A. Jasinska and W. J. Krzyzosiak, FEBS Lett., 2004, 567, 136 CrossRef CAS PubMed.
- S. M. Mirkin, Nature, 2007, 447, 932 CrossRef CAS PubMed.
- G. Liu and M. Leffak, Cell Biosci., 2012, 2, 7 CrossRef CAS PubMed.
- H. Fan and J.-Y. Chu, Genomics, Proteomics Bioinf., 2007, 5, 7 CrossRef CAS PubMed.
- B. Swinnen, W. Robberecht and L. Van Den Bosch, EMBO J., 2019, 39, e101112 CrossRef PubMed.
- H. Paulson, Handb. Clin. Neurol., 2018, 147, 105 Search PubMed.
- A. P. Lieberman, V. G. Shakkottai and R. L. Albin, Annu. Rev. Pathol., 2019, 14, 1 CrossRef CAS PubMed.
- F. O. Walker, Lancet, 2007, 369, 218 CrossRef CAS PubMed.
- C. A. Ross and S. J. Tabrizi, Lancet Neurol., 2011, 10, 83 CrossRef CAS PubMed.
- J. Schilling, M. Broemer, I. Atanassov, Y. Duernberger, I. Vorberg, C. Dieterich, A. Dagane, G. Dittmar, E. Wanker, W. van Roon-Mom, J. Winter and S. Krauß, J. Mol. Biol., 2019, 431, 1869 CrossRef CAS PubMed.
- N. Griesche, J. Schilling, S. Weber, M. Rohm, V. Pesch, F. Matthes, G. Auburger and S. Krauss, Front. Cell. Neurosci., 2016, 10, 226 Search PubMed.
- S. Krauß, N. Griesche, E. Jastrzebska, C. Chen, D. Rutschow, C. Achmüller, S. Dorn, S. M. Boesch, M. Lalowski, E. Wanker, R. Schneider and S. Schweiger, Nat. Commun., 2013, 4, 1511 CrossRef PubMed.
- R. Nalavade, N. Griesche, D. P. Ryan, S. Hildebrand and S. Krauss, Cell Death Dis., 2013, 4, e752 CrossRef CAS PubMed.
- J. Schilling, N. Griesche and S. Krauß, Mechanisms of RNA-Induced Toxicity in Diseases Characterised by CAG Repeat Expansions, eLS, John Wiley & Sons, Ltd, Chichester, 2016 Search PubMed.
- Q. Chen, T. Yamada, K. Miyagawa, A. Murata, M. Shoji and K. Nakatani, Bioorg. Med. Chem., 2024, 98, 117580 CrossRef CAS PubMed.
- S. M. Meyer, C. C. Williams, Y. Akahori, T. Tanaka, H. Aikawa, Y. Tong, J. L. Childs-Disney and M. D. Disney, Chem. Soc. Rev., 2020, 49, 7167 RSC.
- A. K. Verma, E. Khan, S. R. Bhagwat and A. Kumar, Mol. Neurobiol., 2020, 57, 566 CrossRef CAS PubMed.
- A. Alsayari, A. B. Muhsinah, M. Z. Hassan, M. J. Ahsan, J. A. Alshehri and N. Begum, Eur. J. Med. Chem., 2019, 166, 417 CrossRef CAS PubMed.
- G. Sui, T. Li, B. Zhang, R. Wang, H. Hao and W. Zhou, Bioorg. Med. Chem., 2021, 29, 115895 CrossRef CAS PubMed.
- I. Mazziotti, G. Petrarolo and C. La Motta, Molecules, 2022, 27, 2 CrossRef CAS PubMed.
- A. Detsi, M. Majdalani, C. A. Kontogiorgis, D. Hadjipavlou-Litina and P. Kefalas, Bioorg. Med. Chem., 2009, 17, 8073 CrossRef CAS PubMed.
- S. Venkateswarlu, G. K. Panchagnula, A. L. Gottumukkala and G. V. Subbaraju, Tetrahedron, 2007, 63, 6909 CrossRef CAS.
- L. Huang, M. E. Wall, M. C. Wani, H. Navarro, T. Santisuk, V. Reutrakul, E. K. Seo, N. R. Farnsworth and A. D. Kinghorn, J. Nat. Prod., 1998, 61, 446 CrossRef CAS PubMed.
- N. Shanker, O. Dilek, K. Mukherjee, D. W. McGee and S. L. Bane, J. Fluoresc., 2011, 21, 2173 CrossRef CAS PubMed.
- S. Peng, P. Guo, X. Lin, Y. An, K. H. Sze, M. H. Y. Lau, Z. S. Chen, Q. Wang, W. Li, J. K.-L. Sun, S. Y. Ma, T.-F. Chan, K.-F. Lau, J. C. K. Ngo, K. M. Kwan, C.-H. Wong, S. L. Lam, S. C. Zimmerman, T. Tuccinardi, Z. Zuo, H. Y. Au-Yeung, H.-M. Chow and H. Y. E. Chan, Proc. Natl. Acad. Sci. U. S. A., 2012, 118, e2022940118 CrossRef PubMed.
- D. V. Berdnikova, S. Steup, M. Bolte and M. Suta, Chem. – Eur. J., 2023, 29, e202300356 CrossRef CAS PubMed.
- D. V. Berdnikova, Chem. – Eur. J., 2024, 30, e202304237 CrossRef CAS PubMed.
- I. Hawkins and S. T. Handy, Tetrahedron, 2013, 69, 9200 CrossRef CAS.
- L. Ma, Y. Sun, D. Cao, H. Chen, Z. Liu and Q. Fang, Spectrochim. Acta, Part A, 2013, 103, 120 CrossRef CAS PubMed.
- R. S. Varma and M. Varma, Tetrahedron Lett., 1992, 33, 5937 CrossRef CAS.
- M. Morimoto, H. Fukumoto, T. Nozoe, A. Hagiwara and K. Komai, J. Agric. Food Chem., 2007, 55, 700 CrossRef CAS PubMed.
- K. Manjulatha, S. Srinivas, N. Mulakayala, D. Rambabu, M. Prabhakar, K. M. Arunasree, M. Alvala, M. V. B. Rao and M. Pal, Bioorg. Med. Chem. Lett., 2012, 22, 6160 CrossRef CAS PubMed.
- (a) D. V. Berdnikova, Chem. Commun., 2019, 55, 8402 RSC; (b) D. V. Berdnikova, Beilstein J. Org. Chem., 2019, 15, 2822 CrossRef CAS PubMed.
- K.-F. Liew, K.-L. Chan and C.-Y. Lee, Eur. J. Med. Chem., 2015, 94, 195 CrossRef CAS PubMed.
- F. Matthes, S. Massari, A. Bochicchio, K. Schorpp, J. Schilling, S. Weber, N. Offermann, J. Desantis, E. Wanker, P. Carloni, K. Hadian, O. Tabarrini, G. Rossetti and S. Krauss, ACS Chem. Neurosci., 2018, 9, 1399 CrossRef CAS PubMed.
- J. Karn, J. Mol. Biol., 1999, 293, 235 CrossRef CAS PubMed.
- S. M. Kingsman and A. J. Kingsman, The regulation of human immunodeficiency virus type-1 gene expression in EJB Reviews, Springer, Berlin, Heidelberg, 1996 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthetic procedures and characterization of novel compounds, 1H and 13C NMR spectra, single-crystal X-ray analysis data, and description of biological experiments. CCDC 2335536 and 2335537. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4md00403e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |