Terpene synthases and pathways in animals: enzymology and structural evolution in the biosynthesis of volatile infochemicals†
Dorothea
Tholl
 *a,
Zarley
Rebholz
a,
Alexandre V.
Morozov
*a,
Zarley
Rebholz
a,
Alexandre V.
Morozov
 b and
Paul E.
O'Maille
c
b and
Paul E.
O'Maille
c
aDepartment of Biological Sciences, Virginia Tech, Blacksburg, VA 24060, USA. E-mail: tholl@vt.edu; Tel: +1-540-231-4567
bDepartment of Physics and Astronomy and Center for Quantitative Biology, Rutgers University, Piscataway, NJ 08854, USA
cSRI International, San Diego, CA 92131, USA
First published on 7th March 2023
Abstract
Covering: up to the beginning of 2023
Many animals release volatile or semi-volatile terpenes as semiochemicals in intra- and inter-specific interactions. Terpenes are important constituents of pheromones and serve as chemical defenses to ward off predators. Despite the occurrence of terpene specialized metabolites from soft corals to mammals, the biosynthetic origin of these compounds has largely remained obscure. An increasing number of animal genome and transcriptome resources is facilitating the identification of enzymes and pathways that allow animals to produce terpenes independent of their food sources or microbial endosymbionts. Substantial evidence has emerged for the presence of terpene biosynthetic pathways such as in the formation of the iridoid sex pheromone nepetalactone in aphids. In addition, terpene synthase (TPS) enzymes have been discovered that are evolutionary unrelated to canonical plant and microbial TPSs and instead resemble precursor enzymes called isoprenyl diphosphate synthases (IDSs) in central terpene metabolism. Structural modifications of substrate binding motifs in canonical IDS proteins presumably facilitated the transition to TPS function at an early state in insect evolution. Other arthropods such as mites appear to have adopted their TPS genes from microbial sources via horizontal gene transfer. A similar scenario likely occurred in soft corals, where TPS families with closer resemblance to microbial TPSs have been discovered recently. Together, these findings will spur the identification of similar or still unknown enzymes in terpene biosynthesis in other lineages of animals. They will also help develop biotechnological applications for animal derived terpenes of pharmaceutical value or advance sustainable agricultural practices in pest management.
1 Introduction
All organisms use small molecules for communication within their own species or in interactions with other species.1 Depending on their physicochemical properties, these specialized metabolites facilitate efficient interactions at short and long distance both in water and on land. Natural products of the large class of terpenes or terpenoids (>80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 structures known; https://dnp.chemnetbase.com/;2) represent the most structurally diverse group of molecules used in chemical interactions. Due to their high vapor pressure and volatility, low molecular weight terpenes are ideal “infochemicals” for mediating airborne messages. While volatile terpenes have been studied largely in plants and microbes,3–5 they have received comparatively little attention in animals. This is surprising given the occurrence of volatile terpenes in invertebrates, especially arthropods, but also in different lineages of vertebrates. Even fewer information is available on the enzymatic formation of volatile terpenes in animals, which is partly due to the general notion that specialized metabolites in animals are predominantly derived from other organisms such as host plants or microbial endosymbionts.6,7 Therefore, pathways and reactions involved in de novo biosynthesis of terpene infochemicals in animals are not well understood.
000 structures known; https://dnp.chemnetbase.com/;2) represent the most structurally diverse group of molecules used in chemical interactions. Due to their high vapor pressure and volatility, low molecular weight terpenes are ideal “infochemicals” for mediating airborne messages. While volatile terpenes have been studied largely in plants and microbes,3–5 they have received comparatively little attention in animals. This is surprising given the occurrence of volatile terpenes in invertebrates, especially arthropods, but also in different lineages of vertebrates. Even fewer information is available on the enzymatic formation of volatile terpenes in animals, which is partly due to the general notion that specialized metabolites in animals are predominantly derived from other organisms such as host plants or microbial endosymbionts.6,7 Therefore, pathways and reactions involved in de novo biosynthesis of terpene infochemicals in animals are not well understood.
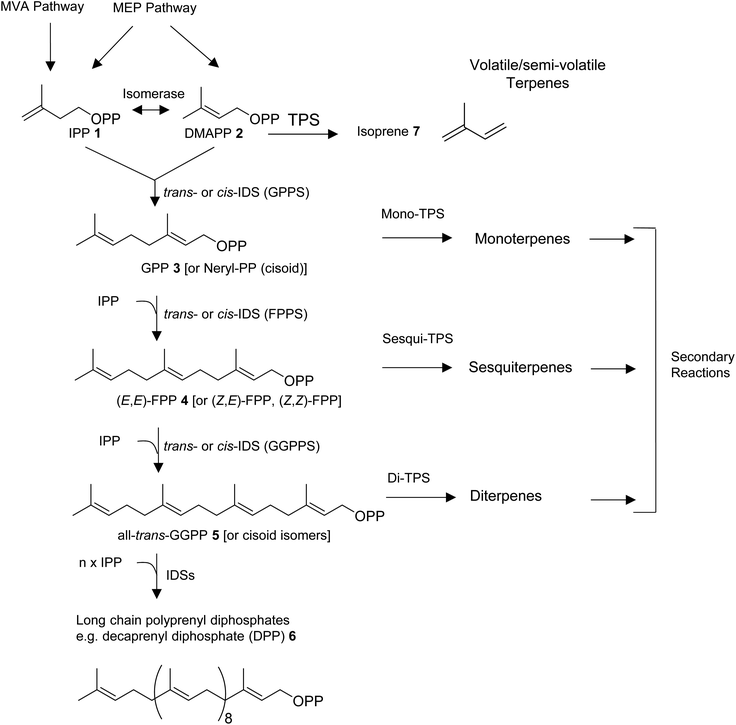
All organisms are capable of making the 5-carbon precursors of terpenes, isopentenyl diphosphate (IPP, 1) and its allylic isomer dimethylallyl diphosphate (DMAPP, 2) by conserved core enzymatic pathways. Plants produce IPP and DMAPP via the plastidial methylerythritol phosphate (MEP) and the non-plastidial mevalonic acid (MVA) pathway.8 Cyanobacteria, parasitic protozoa, and most eubacteria share the MEP pathway with plants, while animals, fungi, and most other eukaryotes rely solely on the MVA pathway in providing the C5-diphosphate precursors for terpene primary and specialized metabolism (Fig. 1).9–11 Isoprenyl diphosphate synthase (IDS) enzymes consecutively combine DMAPP with IPP units in a (1′–4) head-to-tail condensation reaction to synthesize 10-carbon geranyl diphosphate (GPP, 3), 15-carbon farnesyl diphosphate (FPP, 4), 20-carbon geranylgeranyl diphosphate (GGPP, 5), as well as longer prenyl diphosphates required for the formation of non-volatile 30-carbon triterpenes, 40-carbon tetraterpenes, and longer polyprenyl terpenes (e.g. the 50-carbon tail of ubiquinone-10 derived from decaprenyl diphosphate, DPP, 6).8,12,13 Condensation reactions can occur in cis- or trans configuration and are catalyzed by structurally unrelated cis- and trans-IDS enzymes.14 Enzymatic reactions catalyzed by terpene synthases (TPS) then convert DMAPP, GPP, FPP, and GGPP to acyclic or cyclic hemiterpenes (e.g. isoprene, 7), monoterpenes, sesquiterpenes, and diterpenes, respectively (Fig. 1).
Depending on secondary modifications such as hydroxylations, acylations, methylations, glycosylations, and others, the produced terpenes remain volatile or semi-volatile or are converted into non-volatile derivatives. Canonical class I and II enzymes of the TPS superfamily have been studied extensively in plants and microbes.15–17 However, there is growing evidence for the role of various non-canonical TPS enzymes in terpene biosynthesis as recently described by Rudolf and Chang,14 raising questions about the evolution of “unconventional TPSs” in animals. In addition, microbial-type TPSs may have been integrated in animal genomes through horizontal gene transfer as it was found previously for TPS enzymes in lower land plants.18 In this review, we will first provide an overview of the diversity of volatile and semi-volatile terpenes in the animal kingdom. We will then describe recent findings of pathways and TPSs involved in the de novo formation of terpenes in animals, with a particular focus on non-canonical IDS-type enzymes in insects that have adopted TPS activities. Moreover, we will discuss the structural modifications that are likely involved in the transition of these enzymes from IDS to TPS function and compare them with structural features of microbial-type TPSs discovered in octocorals and mites.
2 Volatile terpenes in the animal kingdom
2.1 Invertebrates
Despite the profound structural diversity of terpenes found in marine invertebrates, the pathways of their formation remain obscure. In most cases, it is believed that mollusks sequester these metabolites from their algal diet prior to further biotransformation. For instance, species within the opisthobranch family Aplysiidae (sea hares) are known to derive sesquiterpenes from precursors sequestered from their algal prey.19 Similar dietary sequestrations likely occur for sesquiterpene and diterpene skeletons in nudibranch-sponge predator–prey interactions (e.g. Shen et al.25) and even between specific mollusks and their soft coral prey.19 On the other hand, marine invertebrates are generally believed to accumulate terpenes from microbial sources.26 In particular, it has been suggested that symbiotic dinoflagellates may be the sources of diterpenes found in octocorals.27 While many of the sequestered terpenes may indeed be derived from food sources, it remains unclear how the host further modifies individual skeletons. In other cases, the ability of marine gastropods to synthesize terpene skeletons de novo might have simply been overlooked. A striking example of such capability was recently brought to light with the identification of terpene synthases in coral and sponge genomes (see Section 5). Despite the significant diversity of terpenes in marine mollusks, no accumulation of terpenes has, to the best of our knowledge, been reported in panpulmonate gastropods inhabiting terrestrial or freshwater environments. Moreover, examples of terpene compounds occurring within panpulmonate species are scarce compared to examples in other gastropods and appear restricted to marine genera.28 This astonishing difference clearly indicates dissimilarities in the chemical ecology of food webs and chemically mediated defenses of mollusks in marine and terrestrial ecosystems.
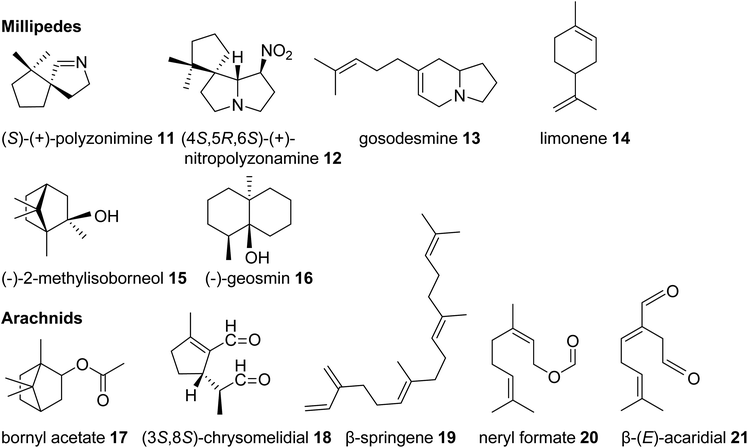 | ||
| Fig. 3 Examples of volatile terpene semiochemicals and derivatives in millipedes and arachnids. No stereochemical configuration is shown in the case the stereochemistry was not determined. | ||
Among the arachnids, the occurrence of volatile terpenes has been reported in the order Opiliones (harvestmen) and different groups of mites. Harvestmen of the species Sclerobunus robustus carry bornyl esters (e.g. bornyl acetate, 17, stereoisomer not reported, Fig. 3) and small amounts of other monoterpenes in their defense secretions.35 Oribatid mites (beetle or moss mites) are known to release the iridoid monoterpene chrysomelidial (18) and the diterpene β-springene (19) from exocrine oil glands.36 Moreover, dust mites (Acariformes, Epidermoptidae) emit the monoterpene ester neryl formate (20) as an aggregation pheromone, and the monoterpene β-acaridial (21) has been found in other acarid mites as sex, aggregation, and alarm pheromone.37,38 It can also be assumed that trombidid mites such as chiggers produce terpenes for chemical interactions based on the surprising finding of TPS genes in the mites' genomes, which presumably have been acquired from microbial sources via horizontal gene transfer (see 3.2).39
One of the most basal insect orders where terpene defenses have been reported are the Phasmatodea or stick insects. Monoterpene iridoids such as actinidine (22) and nepetalactone (cis,trans and/or trans,cis) (23) are disseminated by these insects in defense secretions to deter predators (Fig. 4).45,46 Nepetalactone and other monoterpene iridoids also occur as defenses in rove beetles and as sex pheromones in aphids.42,47 Positioned in the same larger taxonomic clade as stick insects, the Blattodea including cockroaches and termites are well known for releasing terpenes as pheromones and defense metabolites. Roaches of the genus Periplaneta, notably the American cockroach Periplaneta americana, use cyclic sesquiterpenoids called periplanones (e.g. periplanone B, 24) as female-specific aggregation/sex pheromones.48–51 In families of advanced termites (Termitidae and Rhinotermitidae) soldiers release blends of mono-, sesqui-, and/or diterpenes, including the cembrene A- (25) derived multi-cyclic secotrinervitane- (26), trivernitane- (27), and kempene-type (28) diterpenes, as part of their frontal gland defense secretions.52–54 Other constituents of these secretions such as (+)-α-pinene (29) and (E,E)-α-farnesene (30) are thought to act as alarm pheromones55–57 or function as primer pheromones involved in the developmental differentiation of members of the soldier caste.58 Additional functions of terpenes in higher termites include roles as queen sex pheromones [(3R,6E)-nerolidol] (31) and trail pheromones (e.g. cembrene A, 25) in Prorhinotermes simplex.44,59
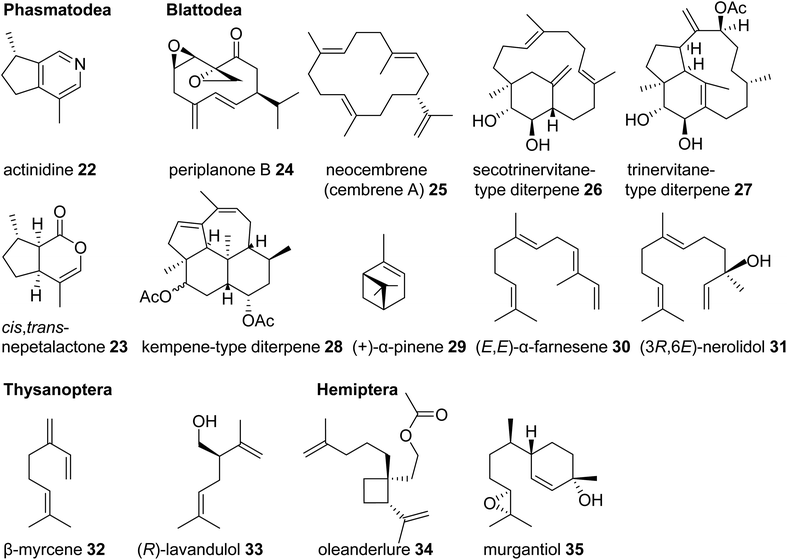 | ||
| Fig. 4 Examples of volatile terpene semiochemicals in stick insects (Phasmatodea), roaches and termites (Blattodea), thrips (Thysanoptera), and hemipteran (true bug) insects. | ||
Another large and diversified group of insects, which has been documented for its dissemination of terpene semiochemicals, comprises the Hemiptera (also referred to a true bugs) and their sister lineage, thrips (Thysanoptera). Many species of thrips use monoterpenes or their derivatives as pheromone or defensive components. For example, the gall-forming thrips, Thlibothrips isunoki, produces a defensive secretion/alarm pheromone containing β-myrcene (32) (Fig. 4) upon disturbance of the host gall, and the monoterpene β-acaridial (21) has been identified in secretions from other gall-forming thrips.60,61 Notably, male aggregation pheromones of flower thrips such as Frankliniella occidentalis are composed of the irregular monoterpene (R)-lavandulol (33) and its ester derivatives,62,63 raising questions about whether these compounds are biosynthesized by irregular TPSs similarly to those identified in plants.64 Interestingly, derivatives of lavandulol and irregular cyclopropane and cyclobutane monoterpenes and sesquiterpenes are also used by hemipteran scale insects and mealy bugs as sex pheromones (e.g. oleanderlure, 34, from the oleander scale, Aspidiotus nerii).65–67 In addition to these compounds, a large number of acyclic and cyclic regular monoterpenes and sesquiterpenes serve as sex or aggregation pheromones and defense compounds in several other hemipteran lineages with stink bugs and related species being the most prominent group (e.g. murgantiol, 35, from the harlequin bug Murgantia histrionica).68 These terpenes will be discussed in more detail below in the context of recent findings of their de novo biosynthesis (see 3.3).
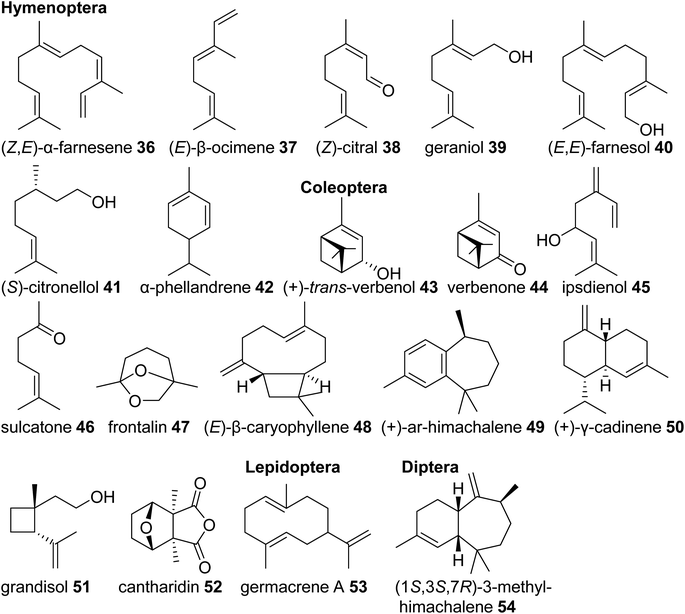
Eusocial species of the Hymenoptera rely on complex communication and defense systems that are mediated by chemicals released from exocrine/secretory glands. The released compounds largely facilitate social organization and colony defense,69 often through the emission of terpenes. For example, the red imported fire ant, Solenopsis invicta, uses isomers of the linear sesquiterpene (Z,E)-α-farnesene (36) in its worker trail pheromone.70 Similar emissions of monoterpenes such as (E)-β-ocimene (37) occur in species of army ants,71 and blends of linear monoterpenes and sesquiterpenes including (Z)-citral (neral) (38), geraniol (39), and farnesol (40) compose the trail pheromone blend secreted from the Nasonov gland of the honey bee Apis mellifera (Fig. 5).72 Other examples include (S)-citronellol (41), a pheromone of male Bombus bumblebees to attract virgin queens,73 and the monoterpenes (+)-limonene [(+)-14)] and α-phellandrene (42) that serve as alarm pheromones and solvents for toxic alkaloids in the poison glands of Myrmicaria ant species.74
Monoterpenes and sesquiterpenes further occur as aggregation pheromone constituents and defense compounds in Coleoptera. Monoterpene alcohols, ketones and acetals from bark beetles of the genera Ips and Dendroctonus (Scolytidae) are among the best studied examples of aggregation pheromones. Investigation of their biosynthetic origins revealed that they are either derived from host tree-specific monoterpenes [cis-/trans-verbenol (43) and verbenone (44) from α-pinene (29)] or formed de novo from endogenous terpene precursors made by the MVA pathway [e.g. ipsdienol (45), sulcatone (46), frontalin (47)] (Fig. 5).43 Elucidation of the ipsdienol biosynthetic pathway, which now has been completed,43 let to the identification of the first endogenous IDS-type TPS enzymes in insects (see 3.3). In other Coleoptera, non-oxygenated sesquiterpenes represent predominant components of their pheromone blends. The invasive, cosmopolitan Asian lady beetle Harmonia axyridis releases a female-specific sex/aggregation pheromone primarily consisting of (E)-β-caryophyllene (48) and isomers.75 Flea beetles of Phyllotreta and Aphthona genera also release sesquiterpenes in the form of himachalene and cadinene isomers as male-specific aggregation pheromones (e.g.49, 50).76 Characteristic terpene defense compounds found in beetles are the iridoid monoterpene chrysomelidial (18) secreted by larvae of leaf beetles, the irregular monoterpene grandisol (51), an aggregation pheromone of the cotton boll weevil, Anthonomus grandis,77 and the highly toxic sesquiterpene cantharidin (52), which is produced by male blister beetles (Meloidae) and transferred to females and offspring through nuptial gifts.78
Lastly, terpene semiochemicals also occur in butterflies (Lepidoptera) and flies (Diptera). In particular, larvae of swallowtail butterflies in the genus Papilio use blends of linear and cyclic mono- and sesquiterpenes such as germacrene A (53) as defense compounds (Fig. 5). Late-instar larvae emit these volatiles from osmeteria, which are fork like defense organs that are everted upon threat.79,80 In another case, the monoterpene (E)-β-ocimene (37) is used by the males of Heliconus butterflies as an anti-aphrodisiac pheromone transferred to females during copulation to repel conspecific males.81 The compound was recently found to be made by an IDS-type TPS enzyme as discussed in 3.3.2. Examples of terpenes found in dipteran sex pheromones include monoterpene and sesquiterpene blends from fruit flies (Ceratitis capitata,82Anastrepha suspensa83) and homosesquiterpene or diterpene constituents (e.g. 3-methyl-himachalene, 54) from the Brazilian sand fly Lutzomyia longipalpis, which is a vector of trypanosome parasites responsible for leishmaniasis disease in humans.84
Taken together, volatile and semi-volatile terpenes of substantial structural diversity are released from species of at least nine insect orders (Fig. 6) ranging from simple acyclic monoterpenes in bumble and honey bees to multi-cyclic diterpenes in termites. The appearance of iridoids and other terpene pheromones and defenses in basal lineages of stick insects and the Blattodea raises questions about how early terpene specialized metabolism emerged in insect evolution. Biosynthetic studies of volatile terpenes in invertebrates (see Section 3) will provide answers to these questions and may also help understand the formation of these infochemicals in vertebrates.
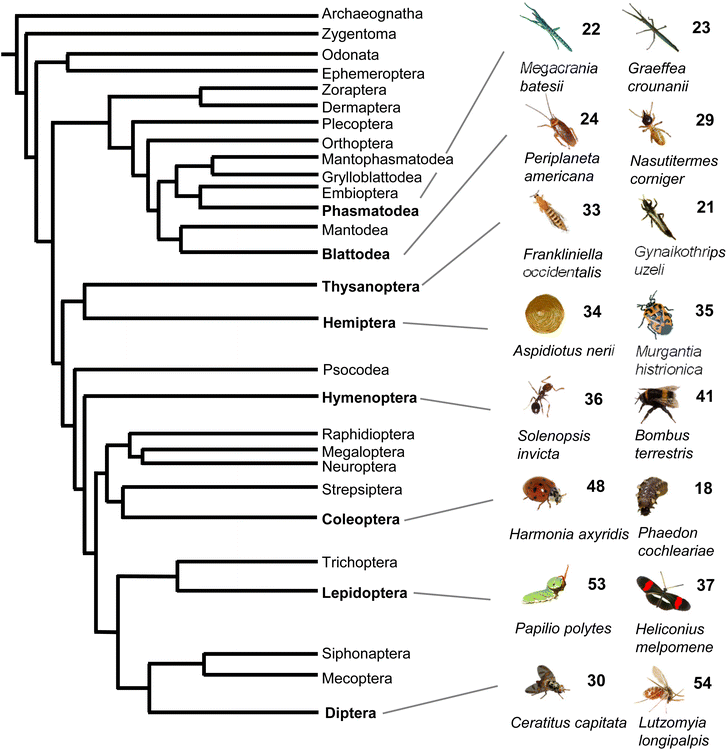 | ||
| Fig. 6 Phylogeny of insect orders and occurrence of volatile terpenes in select insect species representing these orders. Phylogeny of insects adapted from Misof et al.167 with modifications. | ||
2.2 Vertebrates
In comparison to the diverse nature of terpenes found in invertebrate animals, the occurrence of terpene semiochemicals in vertebrates appears rather limited. While chemical communication and scent marking are integral components of vertebrate communication, the reduced abundance of terpenes is not entirely surprising given the expansion of other mechanisms of communication including acoustic, visual, and tactile signals. However, there may be other reasons for the limited use of terpenes in higher animals such as possible adverse physiological effects of specialized terpene compounds. Other causes may include competition and confusion with terpene signals that evolved earlier in plants and lower animals or simply to escape scent-guided predation by other animals. However, there is currently no clear evidence for any of these assumptions.In summary, this survey of specialized terpenes in animals shows that several lineages of animals, especially invertebrates, have integrated terpene compounds in their semiochemical repertoire for intra- and interspecific interactions (Fig. 8). Despite the diversity of terpenes in different animal species, there is also overlap in the constituents of chemical blends. For example, the acyclic diterpene β-springene occurs in mites as well as the spingbock. Whether these metabolites are produced by similar enzymes and if terpene infochemicals are more broadly synthesized de novo in the animal kingdom is in many cases not well understood, with discoveries of genes and enzymes just beginning to emerge.
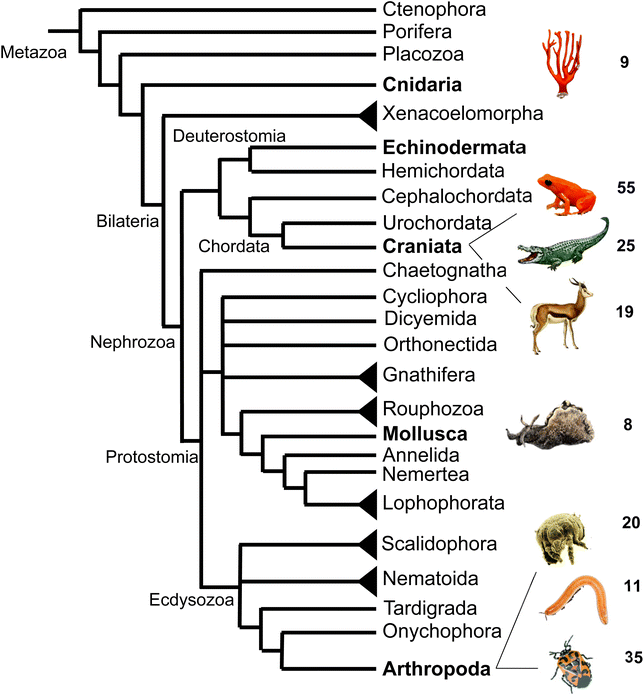 | ||
| Fig. 8 Animal phylogeny and occurrence of volatile terpenes (or volatile precursors in the case of corals) in select species representing different phyla or clades. Species shown from top to bottom are: octocoral species; mantellid frog species, Mantella aurantiaca; alligatorid species; springbok, Antidorcas marsupialis; sea hare, Aplysia brasiliana; dust mite species; polyzoniidan millipede species; pentatomid species, Murgantia histrionica (harlequin bug). Phylogeny adapted from Giribet168 with modifications. | ||
3 Terpene biosynthetic pathways and enzymes in insect pheromone/defense biosynthesis
Since the majority of terpene specialized metabolites has been identified in plants and microbes, terpene biosynthetic enzymes have largely been elucidated in these organisms, and comparatively little attention has been given to the discovery of equivalent enzymatic steps in animals. The fact that animals can sequester specialized metabolites from their food sources or microbial symbionts has complicated the search for de novo biosynthetic pathways. However, an increasing number of high-quality transcriptomes and genomes, which allow detailed genomic and phylogenetic comparisons, facilitate the discovery of terpene biosynthetic genes in animals. Here we review recent findings of enzymes involved in terpene de novo biosynthesis in insects and arachnids.3.1 Biosynthesis of iridoids
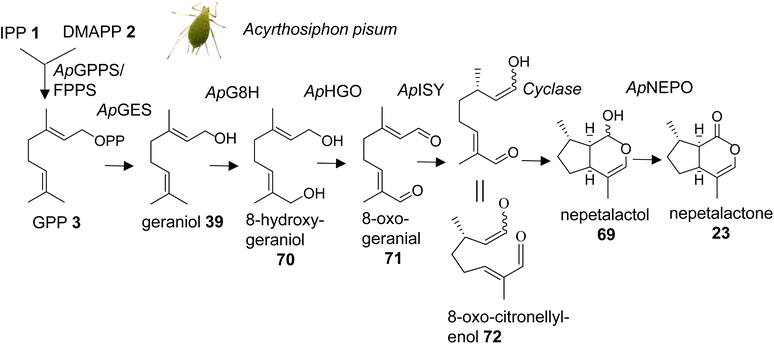
Methylcyclopentanoid monoterpenes or iridoids act as defensive compounds and sex pheromones in a number of different insects. Several of the same compounds also occur in plants where they are involved in defensive activities.104 For instance, nepetalactone (23) is best known as the characteristic iridoid compound of catnip (Nepeta cataria) and functions as an insect repellent, but it also serves as a sex pheromone in aphids.42,105 Gene cluster analysis in Nepeta led to the identification of the nepetalactone biosynthetic pathway in this species.106 An equivalent pathway has been elucidated for the formation of cis–trans-nepetalactol (69), which is a precursor in the biosynthesis of pharmacologically important monoterpene indole alkaloids in Madagascar periwinkle (Catharanthus roseus).107,108 The identification of these pathways in plants raised the question of the existence of similarly evolved pathways in insects. Several enzymatic steps were initially characterized in the formation of iridoid-related dialdehydes such as chrysomelidial (18), in larvae of the chrysomelid leaf beetle Phaedon cochleariae.109–113 Most of these steps have been verified by the just completed identification of the entire biosynthetic pathway of the nepetalactone sex pheromone in the pea aphid Acyrthosiphon pisum (Fig. 9).114 Sexual females of A. pisum exclusively secrete (1R,4aS,7S,7aR)-cis–trans-nepetalactol (69) and (4aS,7S,7aR)-cis–trans-nepetalactone (23) from glands of their hind legs. The elegant study by Köllner et al. determined pathway specific gene candidates by differential gene expression analysis of the hind legs and the non-pheromone producing front legs of sexual females as well as the hind legs of asexual females and males.114 Functional characterization of target gene candidates (Table 1) established a 7-step pathway (Fig. 9), which starts with the formation of GPP (3) by a GPP synthase homolog of the P. cochleariae bifunctional GPP/FPP synthase enzyme that shares the same Co2+/Mg2+ metal dependency. This initial step is followed by the conversion of GPP to geraniol (39) catalyzed by a dolichyldiphosphatase-type homologue (ApGES), and a subsequent hydroxylation to 8-hydroxygeraniol (70) by a cytochrome P450 enzyme of clan 3 (ApG8H). The alcohol is then converted in a two-step oxidation by an NADP-dependent short-chain dehydrogenase (ApHGO) to form 8-oxo-geranial (71). The resulting aldehyde is subsequently reduced to 8-oxo-citronellyl enol (72) and cyclized to cis–trans-nepetalactol by a membrane-bound reductase (ApISY) related to polyprenol type reductases. The final oxidation to cis–trans-nepetalactone is catalyzed by a GMC-type oxidase (ApNEPO).114| Species | Gene/protein | Accession number | Product | Reference |
|---|---|---|---|---|
| a Sesquitrps, sesquiterpenes; * indicates that no accession numbers were listed based on cDNA amplification and gene sequences were synthesized. Nucleotide sequences are listed in the respective publications. | ||||
| Acyrthosiphon | GES | * | 39 | 114 |
| pisum | G8H | ON862918 | 70 | |
| HGO | * | 71 | ||
| ISY | ON862920 | 72, 69 | ||
| NEPO | * | 23 | ||
| Ips pini | GPP | AY953508 | 3, 32 | 118 and 119 |
| Myrcene s | ||||
| Phyllotreta | TPS1 | KT959248 | 74, 75, 76 | 120 |
| striolata | TPS2 | KT959251 | Unidentified sesquitrps | |
| TPS3 | KT959254 | 31 (main product) | ||
| TPS4 | KT959257 | 31 (main product) | ||
| Halyomorpha | TPS1 | MG917093 | 80 | 68 |
| halys | TPS2 | MG870388 | Sesquitrp | |
| Murgantia | TPS1 | MG662378 | 80 | 123 |
| histrionica | Sesquitrps | |||
| Nezara viridula | TPS1 | MG748543 | 82 | 68 and 135 |
| TPS2 | ON934605 | 80 | ||
| Heliconius | Ocimene s | * | 37 | 146 |
| melpomene | HMEL037108g1 | * | 31, 63 | |
| Empoasca onukii | TPS | MH383159 | 39 | 150 |
| Briareum | TC-1 | * | 25 | 162 |
| asbestinum | TC-2 | * | 95 | |
| Capnella | TC-1 | * | 92 | 162 |
| imbricata | ||||
| Dendronephthya | TC-1 | * | 90 | 162 |
| gigantea | TC-2 | * | 25 | |
| Eleutherobia | TC-1 | * | Unidentified sesquitrps | 162 |
| rubra | TC-2 | * | 25 | |
| Erythropodium | TPS1 | OK081311 | 95 | 163 |
| caribaeorum | TPS6 | OK081312 | 93 | |
| Heliopora | TC-1 | * | Unidentified sesquitrps | 162 |
| coerulea | TC-2 | * | 90 | |
| TC-3 | * | 94 | ||
| Paramuricea | TC-1 | * | 94 | 162 |
| biscaya | ||||
| Renilla muelleri | TC-1 | * | 93 | 162 |
| Tubipora musica | TC-1 | * | 25 | 162 |
| TC-2 | * | 91 | ||
| Xenia sp. | TC-1 | * | 96 | 162 |
Interestingly, with the exception of the GPP synthase, which is likely localized in mitochondria, several enzymes of the pathway are presumably associated with the ER membrane (ApGES, ApG8H, ApISY) or targeted to the ER lumen (ApNEPO). This membrane-specific association suggests that the proteins could be organized in the form of a metabolon. Metabolons that modulate metabolic flux and facilitate efficient channeling of intermediates have been documented for a number of secondary metabolic pathways in plants,115 whereas still little is known about such protein complexes in insects.116 A comparison of the aphid-specific enzymes with those identified in leaf beetles indicates that several of the pathway genes evolved independently in these insect lineages. Moreover, a comparison with the plant-specific formation of nepetalactone in Catharanthus and Nepeta reveals that plants and insects employ the same enzymatic steps but use unrelated enzymes. For instance, in plants, TPS enzymes catalyze the formation of geraniol,106,107 whereas this step is mediated by a phosphatase in aphids. In addition, aphids use a membrane bound polyprenol reductase-like protein for the formation of nepetalactol; by contrast, plants employ members of the short-chain dehydrogenase/reductase (SDR) family to catalyze this step.106,108 The finding of these independent iridoid biosynthetic pathways represents a powerful example of convergent evolution in the metabolism of volatile terpene semiochemicals in plants and animals.
3.2 Horizontal gene transfer of terpene synthases in mites
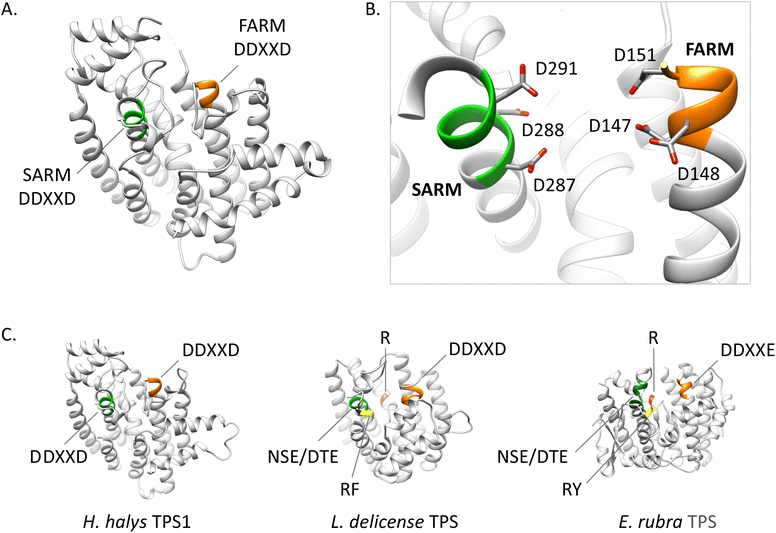
A surprising finding of TPS gene families has recently been made in the genomes of trombidid mites. Larvae in the superfamilies of the Trombiculoidea (chiggers) and Trombidioidea (velvet mites) feed as ectoparasites on vertebrates or other arthropods, respectively.39 Genomes of the chigger Leptotrombidium deliense and the velvet mite Dinothrombium tinctorium were found to contain a family of 39 putative sesqui-TPS genes and a related family of 21 TPS genes, respectively. An additional group of 17 TPS genes was identified in L. delicense. These genes and their encoded proteins are most closely related to fungal and bacterial TPSs, albeit with sequence identity of less than 30%. The absence of homologs in other arthropods or metazoans suggests that the TPS genes are the result of ancient lateral gene transfers from soil-derived bacteria and fungi. This horizontal gene transfer is similar to that of carotenoid biosynthetic genes, which are responsible for the orange coloration in both types of mites.39 The biochemical function of the detected TPS genes and the role of their putative terpene products as pheromones or defense compounds is currently unknown. It remains to be determined why such large gene families have been maintained in the mite genomes and to what extent functionally active genes may be correlated with the release of terpene blends.3.3 IDS-like terpene synthases in insects
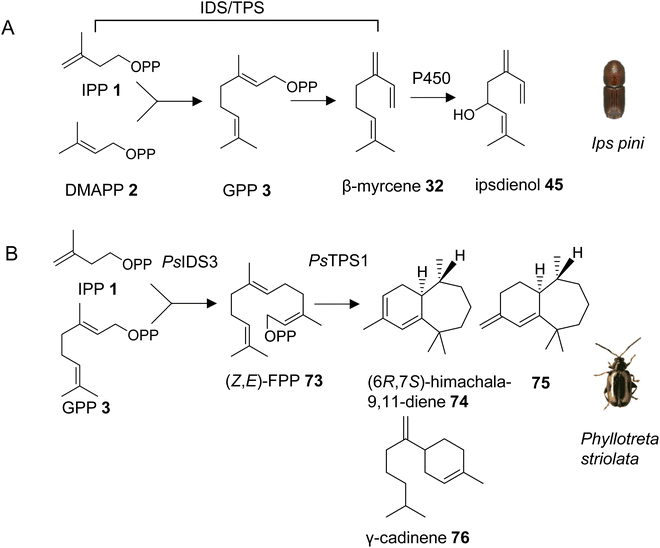
With the exception of the recently discovered TPS genes in mite genomes, insects have generally been thought to lack homologs of canonical microbial and plant TPSs and thus the ability to synthesize specialized terpenes via endogenous pathways. However, early isotope-labeling experiments questioned this notion. For instance, experiments with 14C-labeled precursors in the bark beetle Ips pini provided evidence that the monoterpene aggregation pheromone ipsdienol (45) is synthesized de novo via the MVA pathway.117 A combination of biochemical and transcriptome analyses further determined a coordinated regulation of terpene biosynthetic genes with tissue specificity in the midgut and elevated expression in I. pini males, and upon treatment with juvenile hormone III.43 This approach led to the identification of a bifunctional IDS/TPS enzyme, which makes GPP (3) from IPP (1) and DMAPP (2) and subsequently converts GPP to the ipsdienol precursor myrcene (32) (Fig. 10A, Table 1).118,119 It should be noted that all subsequent steps from myrcene to ipsdienol have also been elucidated.43 The GPP/myrcene synthase was found to be structurally related to canonical IDS enzymes118 and carries two aspartate-rich motifs (DDIMD, NDFKD). These motifs, typically called first and second aspartate rich motifs (FARM and SARM), are characteristic of IDS proteins. The I. pini synthase might be targeted to peroxisomes based on computational predictions of its transit peptide. Despite its similarity to canonical IDS proteins, the I. pini enzyme shares 20% or lower amino acid sequence identity with the I. pini FPP synthase and insect GGPP synthases such as Drosophila melanogaster GGPP synthase, which indicates an early divergence of this protein from canonical insect IDS enzymes.3.3.1.1 Phyllotreta striolata. Almost ten years after the discovery of an IDS-type TPS gene in I. pini, a family of similar genes was identified in a leaf beetle, the striped flea beetle Phyllotreta striolata (Chrysomelidae).120 Four out of five IDS-type genes were functionally characterized as sesquiterpene synthases. Among those, the recombinant protein of the male-expressed PsTPS1 gene was found to convert (Z,E)-FPP (73) to (6R,7S)-himachala-9,11-diene (74), a major constituent of the P. striolata aggregation pheromone, together with five other sesquiterpenes including 75 and γ-cadinene (76) (Fig. 10B, Table 1). Interestingly, PsTPS1 requires a (Z,E)-FPP isomer as a substrate, which is made by an unusual, cis-double bond forming IDS (PsIDS3) from GPP and IPP (Fig. 10B).120 The other functionally active TPS enzymes converted (E,E)-FPP to (E)-nerolidol (31) (PsTPS4) and mixtures of sesquiterpenes (PsTPS2 and 3). These enzymatic products were not detected in vivo since the corresponding genes are expressed at low levels in males and females. PsTPS1 and the P. striolata (E,E)-FPP synthase (PsIDS1) carry putative mitochondrial targeting sequences, indicating a subcellular compartmentalization in the formation of the volatile sesquiterpenes. The study also provided insight into the evolution of the P. striolata TPS genes. Gene structures of PsTPS1, PsIDS1 and PsIDS3 comprised four exons and three introns. The positions of these introns and the intron phases were conserved in canonical IDS genes from other Coleoptera, Lepidoptera and Diptera, which indicated an emergence of IDS-type TPS genes from an IDS progenitor, presumably a FPP synthase.120 In addition, PsIDS1, PsIDS3, and homologs from other insects were found to be under strong purifying selection, indicating a selective removal of deleterious variants to preserve IDS function in core metabolism. In contrast, the IDS-type TPS genes are under relaxed constraints, which is in agreement with the neofunctionalization and diversification of these genes in the evolution of pheromones and chemical communication.
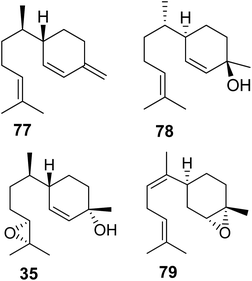
3.3.1.2 Pentatomids. Tholl and collaborators have investigated the presence of IDS-type TPS genes in stink bugs (Pentatomidae), a diverse family in the order of pierce-sucking hemipteran insects, which comprises herbivorous and carnivorous species.121 Due to their ability to easily adapt to different environmental conditions, several species of stink bugs have become important pests with economic impact on agricultural crops in the Neotropics and worldwide.41 Mature males of at least ten genera of pentatomids emit sesquiterpenes with a bisabolane-type skeleton as sex or aggregation pheromones such as (6S,7R)-β-sesquiphellandrene (77) in the red banded stink bug, Piezodorus guildinii, cis-zingiberenol [(3R,6R,7S)-1,10-bisaboladien-3-ol] (78) in the rice stink bug, Oebalus poecilus, 10,11-epoxy-1-bisabolen-3-ol (35) in the harlequin bug, M. histrionica and the brown marmorated stink bug, Halyomorpha halys, and trans/cis-(Z)-α-bisabolene epoxide (79) in the Southern green stink bug, Nezara viridula (Fig. 11).41,122 The compounds are released as mixtures of distinct stereoisomeric composition, sometimes in combination with fatty acid derivatives.41 The structural relationships of the terpene constituents of stink bug pheromones suggests that they could be synthesized de novo by evolutionary related pathways instead of being made from sequestered precursors. This notion is supported by the fact that stink bug specialists and generalists feed on different host plants, many of which do not synthesize bisabolane type sesquiterpenes or make them only in limited amounts.
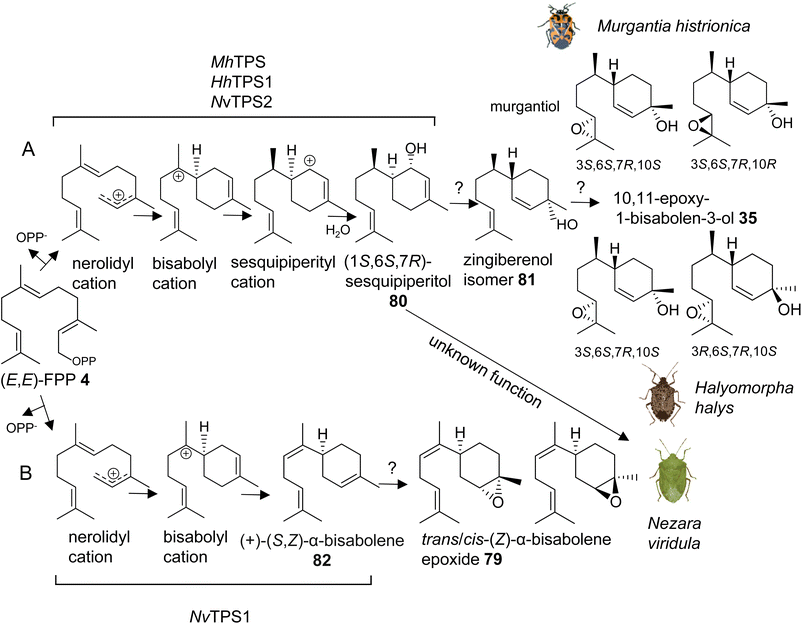
The first investigation of IDS-like TPS genes was performed in the harlequin bug, Murgantia histrionica (tribe Strachiini), which is a crucifer specialist native to Central America and invasive in the southeast of the United States. Mature males of M. histrionica emit an aggregation pheromone, which is composed of the (3S,6S,7R,10S)- and (3S,6S,7R,10R)-stereoisomers of 10,11-epoxy-1-bisabolen-3-ol (35) named murgantiol (Fig. 12A).123,124 The pheromone attracts both males and females as well as nymphs. Sex- and development-specific transcriptome analyses led to the identification of a canonical (E,E)-FPP synthase (MhFPPS) and an IDS-type TPS (MhTPS), which converts (E,E)-FPP (4) to (1S,6S,7R)-1,10-bisaboladien-1-ol, called sesquipiperitol (80), as an intermediate in the pathway leading to murgantiol (Fig. 12A and Table 1).123,125 Sesquipiperitol is also produced in plant species of the Asteraceae, Zingiberaceae, and Cupressaceae families (e.g. Sy and Brown126). MhTPS presumably catalyzes a carbocation mediated reaction typical of a type I TPS enzyme.8 In this reaction, a nerolidyl carbocation is first formed by a metal ion-catalyzed cleavage of the carbon–oxygen bond to release the pyrophosphate moiety of FPP. Next, a bisabolyl cation is generated by a 1,6 ring closure followed by a hydride shift to form a sesquipiperityl cation and subsequent quenching of the carbocation with water (Fig. 12A). Analysis of MhFPPS and MhTPS transcript abundances showed an equal expression of MhFPPS at nymphal and adult stages. By contrast, MhTPS is most highly expressed in mature males and exhibits highest transcript levels in epithelial cells associated with the cuticle of the ventral abdominal sternites, from which the pheromone is likely released.123
Interestingly, the pheromone constituents of the invasive brown marmorated stink bug Halyomorpha halys (Stål) (tribe Cappaeini), which is native to Asia, share the same skeleton as murgantiol but with a different stereoisomeric composition of (3S,6S,7R,10S)-10,11-epoxy-1-bisabolen-3-ol and (3R,6S,7R,10S)-10,11-epoxy-1-bisabolen-3-ol (35) being emitted in a 3.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture (Fig. 12A).127 To investigate whether the pheromones of M. histrionica and H. halys, which are native to different geographical regions, are produced by closely related enzymes or may be the result of convergent evolution, IDS-like genes were mined in H. halys genome and transcriptome resources.128,129 A family of seven IDS-like genes was discovered, of which two were characterized as functionally active sesqui-TPSs (HhTPS1, HhTPS2) (Table 1) and a third was identified as a canonical (E,E)-FPP synthase (HhFPPS).68HhTPS1 was found to be a putative ortholog of MhTPS1, which shares more than 80% amino acid sequence with the M. histrionica enzyme and converts (E,E)-FPP to the same (1S,6S,7R)-sesquipiperitol intermediate (80) as in the M. histrionica pheromone biosynthetic pathway. Similar to M. histrionica, HhTPS1 is most highly expressed in mature males, in agreement with the male-specific release of the pheromone. However, its tissue-specific expression is highest in the fat body, suggesting a different localization of the pheromone-specific enzymes in H. halys and M. histrionica. HhTPS2, which encodes a multi-sesquiterpene synthase, shows comparatively low expression in males and females, and the function of this gene remains unclear.68 The downstream enzymatic steps from sesquipiperitol to 10,11-epoxy-1-bisabolen-3-ol are not yet identified but presumably include a conversion to a zingiberenol isomer (81) and an epoxidation at C10/C11 by a cytochrome P450 monooxygenase enzyme (Fig. 12A). Zingiberenol and sesquipiperitol have also been identified as pheromone constituents of other stink bugs such as the rice stink bugs Oebalus poecilus and Mormidea v-luteum, and the rice stalk stink bug Tibraca limbativentris (tribe Carpocorini), all of which are severe pests in South America.130–132 This finding and the identification of two closely related sesquipiperitol synthases in M. histrionica and H. halys suggest that the enzymatic steps in terpene pheromone formation in these species have been evolutionary conserved independent of their tribe and geographic origin.
1 mixture (Fig. 12A).127 To investigate whether the pheromones of M. histrionica and H. halys, which are native to different geographical regions, are produced by closely related enzymes or may be the result of convergent evolution, IDS-like genes were mined in H. halys genome and transcriptome resources.128,129 A family of seven IDS-like genes was discovered, of which two were characterized as functionally active sesqui-TPSs (HhTPS1, HhTPS2) (Table 1) and a third was identified as a canonical (E,E)-FPP synthase (HhFPPS).68HhTPS1 was found to be a putative ortholog of MhTPS1, which shares more than 80% amino acid sequence with the M. histrionica enzyme and converts (E,E)-FPP to the same (1S,6S,7R)-sesquipiperitol intermediate (80) as in the M. histrionica pheromone biosynthetic pathway. Similar to M. histrionica, HhTPS1 is most highly expressed in mature males, in agreement with the male-specific release of the pheromone. However, its tissue-specific expression is highest in the fat body, suggesting a different localization of the pheromone-specific enzymes in H. halys and M. histrionica. HhTPS2, which encodes a multi-sesquiterpene synthase, shows comparatively low expression in males and females, and the function of this gene remains unclear.68 The downstream enzymatic steps from sesquipiperitol to 10,11-epoxy-1-bisabolen-3-ol are not yet identified but presumably include a conversion to a zingiberenol isomer (81) and an epoxidation at C10/C11 by a cytochrome P450 monooxygenase enzyme (Fig. 12A). Zingiberenol and sesquipiperitol have also been identified as pheromone constituents of other stink bugs such as the rice stink bugs Oebalus poecilus and Mormidea v-luteum, and the rice stalk stink bug Tibraca limbativentris (tribe Carpocorini), all of which are severe pests in South America.130–132 This finding and the identification of two closely related sesquipiperitol synthases in M. histrionica and H. halys suggest that the enzymatic steps in terpene pheromone formation in these species have been evolutionary conserved independent of their tribe and geographic origin.
Another pheromone biosynthetic pathway in stink bugs leads to the formation of trans-/cis-(Z)-α-bisabolene epoxide (79) (Fig. 12B). The epoxide isomers are released by the males of the southern green stink bug Nezara viridula, which is a globally invasive pest with the origin in East Africa.133,134 The same isomers are emitted by the neotropical green stink bug Chinavia impicticornis, a close relative in the same tribe (Nezarini) as N. viridula. Comparative transcriptome analyses of N. viridula mature males and females discovered an N. viridula IDS-type TPS enzyme (NvTPS1), which catalyzes the conversion of (E,E)-FPP to (+)-(S,Z)-α-bisabolene (82) as the likely precursor of the sesquiterpene pheromone (Fig. 12B, Table 1).135 The biosynthetic pathway is presumably localized in glandular cells at the ventral abdomen of mature males, from which the pheromone is emitted.136 Unexpectedly, a functionally active sesquipiperitol synthase gene (NvTPS2) was identified in N. viridula genome and transcriptome resources. The gene encodes a protein with approximately 80% amino acid sequence identity to the corresponding enzymes of M. histrionica and H. halys.68 This finding is surprising since N. viridula does not release sesquipiperitol or any other compound of the murgantiol pheromone complex nor does it store non-volatile derivatives of sesquipiperitol. The conserved status of the sesquipiperitol synthase independent of its role in pheromone biosynthesis suggests that the enzyme had additional or other functions in the common progenitor of these pentatomid species.
3.3.1.3 Genomic organization and evolution of stink bug and hemipteran IDS-like genes. The availability of quality genomes of H. halys and N. viridula (128, https://www.ncbi.nlm.nih.gov/bioproject/PRJEB47893/) has allowed a more detailed investigation of the architecture and genomic position of the IDS-type TPS genes in these species. The six IDS-like genes of H. halys were found to be organized in two separate clusters, each of which most likely emerged by gene duplication (Fig. 13A and B).68 The canonical HhFPPS gene shares only low sequence identity with the IDS-like genes and is positioned independently of the IDS-like clusters, which suggests that this gene is derived from a more ancient duplication event. Genes of the N. viridula IDS-like family are organized in a similar fashion, although in the form of a single three-gene cluster (NvTPS1 and two uncharacterized IDS genes), with NvTPS2 and NvFPPS being positioned separately on two other chromosomes (Fig. 13A).68
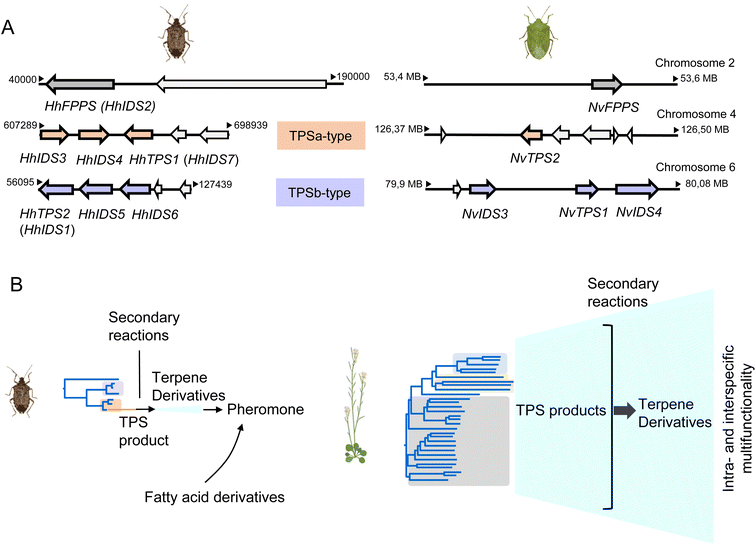 | ||
| Fig. 13 Pentatomid IDS-type gene family organization and size. (A) Genomic organization of IDS-type gene families in Halyomorpha halys (Hh) and Nezara viridula (Nv). Two different clusters represent IDS-type genes of the two pentatomid TPS-a and TPS-b clades (modified from Rebholz et al.68); (B) schematic comparison of TPS gene family sizes in Pentatomids and plants (Arabidopsis thaliana) in relation to volatile terpene product diversity and function. | ||
A closer analysis of the architecture of the H. halys canonical FPPS and IDS-like genes revealed an identical composition of seven introns and eight exons and identical positions of nearly all intron phases. The intron phases are conserved in FPPS genes of other hemipteran and blattodean insects, which provides evidence for a shared ancestral exon-intron structure of IDS genes in these lineages.68 Interestingly, the coleopteran FPPS genes and the IDS-type TPS genes of P. striolata have a reduced number of three introns.120 This difference in gene architecture between representatives of the Hemiptera and Coleoptera supports the hypothesis of an independent emergence of TPS genes from FPPS progenitors in these orders.
To gain a more in-depth understanding of the evolution of IDS-like genes in Pentatomids and Hemiptera that are known to release volatile terpenes as pheromones or defense secretions, Rebholz et al. mined hemipteran IDS-like genes from NCBI nucleotide and transcriptome assembly databases using H. halys FPPS and IDS-like protein sequences as search queries.68 The search resulted in the identification of nearly 300 unique sequences, with 80% classified as canonical type FPPSs and the remaining 20% classified as IDS (FPPS)-like proteins with the potential to function as TPSs (Fig. 14). Phylogenetic analysis of the IDS-like proteins indicated a paralogous division of the pentatomid sequences in two clades, named TPS-a and TPS-b clades, in agreement with the position of the corresponding genes in two different clusters (Fig. 13 and 14). The clades most likely evolved from a common ancestor of the Pentatomidae or possibly the Pentatomoidea superfamily approximately more than 100 million years ago. In agreement with the neofunctionalization of IDS-like genes, and in contrast to the conserved clade of canonical FPPS genes, the pentatomid TPS clades evolved under positive selective pressure.68 However, genes within both TPS clades have undergone limited inter- and intraspecific diversification following clade-specific divergence, which is evidenced by the conservation of sesquipiperitol synthases in different species (Fig. 12). Thus, Pentatomids seemingly have maintained small-size gene families that generate a limited number of terpene intermediates. Limited steps of derivatizations and combinations with other metabolites such as fatty acid derivatives are sufficient to generate species-specific pheromone blends.41 A cross-kingdom comparison shows that pentatomid TPS gene families are notably smaller than those of flowering plants,15,137–139 despite similar evolutionary time spans of more than 100 million years (Fig. 13B). The diversification of plant TPSs into several subfamilies is associated with the synthesis of complex terpene mixtures, which are believed to have multiple functions in attraction and defense and presumably target a larger number of organisms than the smaller compound mixtures released by insects.8 Therefore, it can be assumed that the diversification of TPS genes in Pentatomids and other insects is directed by more specific chemical interactions and perhaps other constraints.
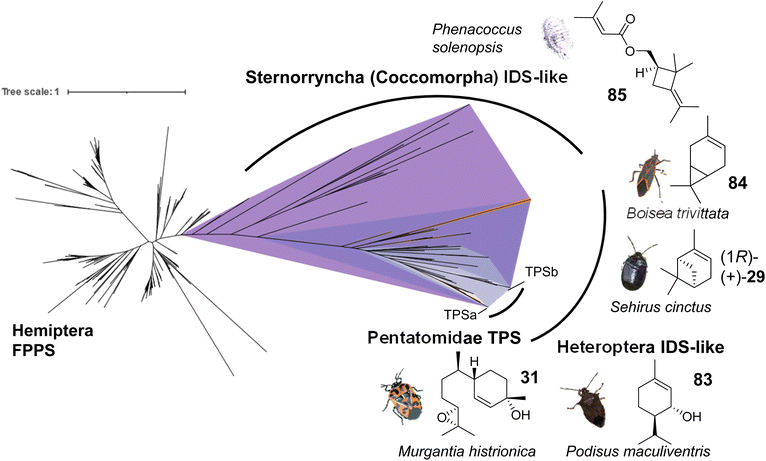 | ||
| Fig. 14 Evolution of putative IDS-like TPSs in hemipteran insects. The phylogram shows canonical FPPS and IDS (FPPS)-like proteins mined from hemipteran genomes and transcriptomes. Branch lengths represent the number of amino acid substitutions per site. A monophyletic clade (purple) of IDS-like sequences clustering with characterized IDS-type TPSs was found to be distributed across species within the terpene-emitting hemipteran suborders Heteroptera and Sternorrhyncha. Select terpene-emitting species are displayed alongside representative terpene compounds. Figure modified from Rebholz et al.68 | ||
Several currently uncharacterized IDS-like genes, which may be associated with the formation of volatile terpenes, have been identified in other species of Pentatomids and the two main suborders of the Hemiptera, the Heteroptera and the Sternorrhyncha. For instance, IDS-like genes with close similarity to sesquipiperitol synthases were found to be expressed in the predatory spined soldier bug, Podisus maculiventris, which releases monoterpene alcohol pheromones including trans-piperitol (83), the C10 analog of sesquipiperitol (Fig. 14).140 Several other families in the heteropteran infraorders Pentatomomorpha and Cimicomorpha are known to secrete monoterpenes for defense or attraction: Acanthosomatidae (shield bugs), Cimicidae (bed bugs), Cycnidae (burrowing bugs), Miridae (plant bugs), Lygaeidae (seed bugs), Pyrrhocoridae (red bugs), Tingidae (lace bugs) and others.68 In conjunction with these findings, IDS-like gene transcripts have been identified in the burrower bug Sehirus cinctus and the boxelder bug Boisea trivittata, which are known to release monoterpenes such as α-pinene (29) and 3-carene (84), respectively (Fig. 14).141–143 In the hemipteran suborder Sternorrhyncha, IDS-like transcripts are present in scale insects (infraorder Coccomorpha) including the lac insect Kerria lacca, which excretes cyclic sesquiterpene acids as lac components.144 Another group of insects in which IDS-like transcripts have been identified are mealybugs (infraorder Coccomorpha) such as the cotton mealybug, Phenacoccus solenopsis, which releases a methylbutenoate ester of the cyclobutane monoterpene R-maconelliol as a sex pheromone (85) (Fig. 14).65 Related irregular monoterpenes (e.g. lavandulol, 33) are also made by plant TPSs that catalyze an irregular coupling between isoprenoid units.145 On the other hand, IDS-like genes are absent in aphids despite the presence of β-farnesene as a well-known alarm pheromone in this hemipteran group.42 Moreover, no IDS-like genes were identified in the suborders Auchenorrhyncha (including cicadas and plant hoppers) and Coleorrhyncha (including moss bugs) in agreement with the lack of terpenes in these orders.68 Overall, the large-scale phylogeny of IDS-like sequences in the Hemiptera supports an ancient emergence of these genes from canonical FPP synthases, possibly in a shared progenitor more than 350 million years ago (Fig. 14), and suggests that volatile terpenes are synthesized de novo in several hemipteran lineages. Further functional characterization of IDS-like genes will provide more insight into the extent of terpene biosynthetic evolution in hemipteran insects.
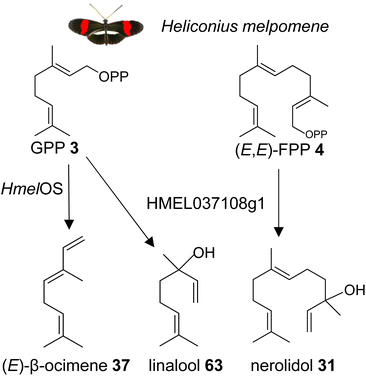 | ||
| Fig. 15 Biosynthesis of the male anti-aphrodisiac (E)-β-ocimene (37) and terpene alcohols by GGPPS-type TPSs in the butterfly Heliconius melpomene (Hmel). Os, ocimene synthase. | ||
It is possible that other insects which use β-ocimene as a pheromone, such as bumble bees and honey bees, have GGPPS-like or perhaps FPPS-like proteins that make β-ocimene. It is curious to note that the H. melpomene β-ocimene synthase is unable to convert GGPP to a diterpene product, which suggests possible constraints in accommodating GGPP as a substrate. This scenario might be different in other insect lineages. For instance, a family of GGPPS-like genes was found to be expressed in soldiers of the nasute termite Nasutitermes takasagoensis.149 The defensive secretions of these termites contain a mixture of diterpenes and monoterpenes, which may be produced by the GGPPS-like enzymes. Based on these findings, it can be assumed that GGPPS-type genes with TPS function have emerged independently multiple times throughout insect evolution. This is supported by the identification of a GGPPS-like TPS in the green tea leafhopper Empoasca onukii, which converts GPP into geraniol (39).150 The authors suggest that geraniol synthase activity is also present in other lepidopteran and coleopteran species.
4 Structural and mechanistic evolution of insect IDS-like TPSs
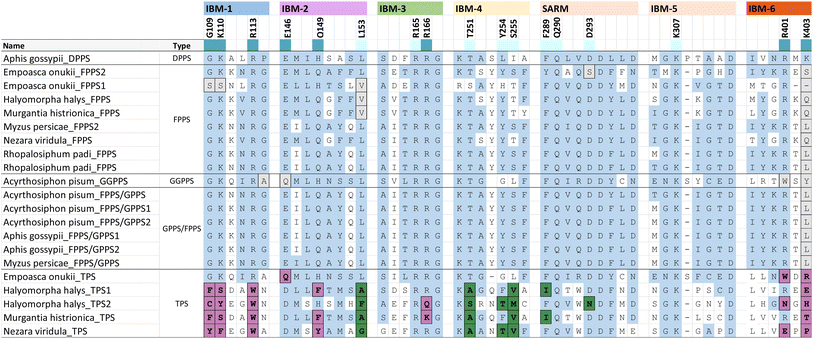
Phylogenetic evidence for the emergence of IDS-like TPS enzymes from IDS progenitors in insects raises the question of which mutations and structural modifications facilitated this evolutionary transition. While extensive experimental proof is still missing, O'Maille and co-workers have developed a structural and mechanistic model for the change in catalytic function from IDS to TPS proteins.151 IDS and TPS enzymes generally share a common alpha-helical protein domain (α-domain) fold,152,153 suggesting an ancient common origin. Whereas TPS protein sequences in microbes and plants no longer have a close evolutionary relationship with IDS proteins of these organisms, the recruitment of IDS to TPS proteins seems to have occurred more recently in insects.IDS proteins carry two conserved aspartate rich motifs (DDxxD) called FARM and SARM, which are positioned on the opposite sides of the active site. These motifs facilitate the cleavage of the diphosphate moiety of the allylic substrate via coordination of Mg2+ ions. Insect IDS-like TPS proteins possess the same motifs but show more frequent substitutions of the first and third aspartate of the SARM (Table 2 and Fig. 16). In addition, aromatic amino acid residues in positions 4 and 5 upstream of the FARM of bonafide FPPS proteins are substituted by non-aromatic residues in IDS-like sesqui-TPSs.120,123,135 Molecular docking of (E,E)-FPP in the active-site cavity of a M. histrionica TPS homology model showed that these residue changes appear to be critical for the positioning of the FPP prenyl side chain into the cavity to facilitate subsequent cyclization.123 Substitutions of the non-aromatic residues in MhTPS with aromatic amino acids led to the loss of TPS activity, confirming this assumption.123 The reciprocal substitutions in the MhFPPS protein did not abolish IDS activity but caused the formation of GGPP instead of FPP due to the ability of the enzyme to accommodate an extended prenyl chain. The loss of aromatic residues upstream of the FARM is typical for long-chain trans-IDS enzymes (≥C20), including insect GGPPSs.146,154 By contrast, the bifunctional GPPS/TPS enzyme from I. pini maintains aromatic amino acids in this position because a presumably smaller-size cavity of this protein is sufficient to accommodate the short chain GPP product/substrate.118,119
| a Extract of IBM motifs from a structure-based sequence alignment of characterized hemipteran IDS and TPS proteins. Residue positions that are ≤ 5 Å away from IPP and highly conserved are colored according to their interaction with the diphosphate moiety (dark blue) or the isoprenyl tail (light blue), respectively. TPS residue substitutions of the diphosphate binding residues and prenyl tail binding residues are marked in purple and green, respectively. Substitutions that deviate from the IBM regular expressions in other insects and animals are shaded in grey. Unshaded residues (white) correspond to variable residues and positions outside of the consensus IPP binding residues. |
|---|
|
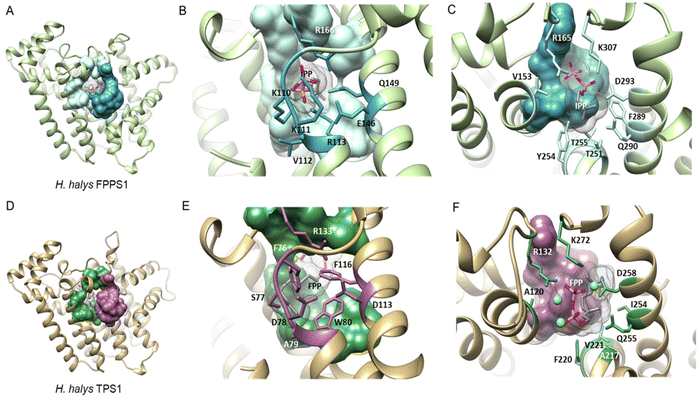 | ||
| Fig. 16 Structural analysis of IPP binding residues in H. halys FPPS and a homology model of the IDS-type H. halys TPS1 enzyme. (A) Structural model of HhFPPS with the IPP binding pocket rendered as a colored surface; (B) HhFPPS residues binding the diphosphate moiety of IPP are marked in dark blue and labeled; (C) HhFPPS residues binding the prenyl tail of IPP are marked in light blue and labeled; (D) structural model of HhTPS1 with a modified IPP binding pocket rendered as a colored surface; (E) HhTPS1 residue substitutions of the diphosphate binding residues in (B) are marked in purple and labeled. Aromatic substitutions in this region favor interactions with the isoprenyl tail of the docked FPP substrate; (F) HhTPS1 residue substitutions of the prenyl tail binding residues in (C) are marked in green and labeled. Substitutions alter the substrate binding region to accommodate a larger isoprenyl diphosphate substrate. Figure adapted from Rebholz et al.151 | ||
While residues upstream of the FARM seem to be critical for a proper positioning of the substrate in insect TPSs, Rebholz et al. hypothesize that the transition from IDS to TPS catalytic function largely depends on a change in the binding or position of the IPP substrate relative to DMAPP.151 To test this hypothesis, a set of 20 IPP-binding residues positioned ≤ 5 Å away from IPP were identified in the crystal structure of a Homo sapiens FPPS in complex with IPP by using the RING web server in combination with residue network interaction analysis in Cytoscape and structural analysis in Chimera.155–157 The identified amino acids comprise basic residues that bind to the diphosphate moiety of IPP, a ring of residues encircling the isoprenyl tail, and residues that interact with both of these moieties. The residues are organized into six IPP binding motifs (IBMs) and were found to be conserved across IDS sequences from diverse organisms including animals, plants and fungi (Table 2 and Fig. 16).
The residues that bind IPP orient the substrate and its prenyl tail in a way that allows condensation with the nascent carbocation formed from DMAPP. Modifications of these critical residues in IDS-type TPS proteins lead to alterations of the electrostatic nature of the IPP binding pocket (Fig. 17). These changes may misalign IPP and DMAPP and disrupt their condensation, which likely allows competing TPS reactions of allylic substrates to occur. In agreement with this assumption, 90% of the IBM residues were found to be modified in characterized insect TPS enzymes. For example, in the first IBM of hemipteran TPSs, the basic diphosphate binding residues are substituted with large aromatic residues (Table 2 and Fig. 16), one of which is also conserved in its equivalent position in plant TPSs. Furthermore, several substitutions of residues interacting with the isoprenyl tail of IPP occur in the fourth IBM motif (Table 2 and Fig. 16). Multiple residue substitutions are also present in the TPSs of Coleoptera and Lepidoptera; however, the substitution patterns are unique among the different taxonomic lineages, supporting independent events of TPS evolution.151
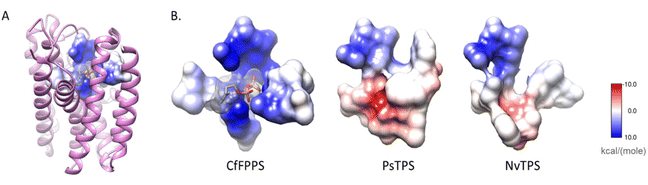 | ||
| Fig. 17 Substitutions in the pyrophosphate-binding region is a common feature of insect TPS enzymes. (A) The structure of a canonical FPPS from the eastern spruce budworm Choristoneura fumiferana (CfFPPS) (PDBid 6b04; light purple ribbons) is shown with IBM-1, 2, 3, and 6 depicted as an atomic surface. The electrostatic character of CfFPPS IBM motifs was mapped onto the surface using UCSF Chimera (blue = basic; red = acidic; grey = neutral/aliphatic); (B) IBM-1, 2, 3, and 6 of CfFPPS and selected insect TPS models are depicted as atomic surfaces for comparison. Figure adapted from Rebholz et al.151 | ||
Lancaster et al. tested whether a re-introduction of the motif KKxR in the IBM1 of MhTPS (Table 2) through replacement of the corresponding SDAW sequence could convert the TPS enzyme into an IDS.123 While no IDS activity was gained, the protein lost TPS activity, indicating an essential role of the SDAW residues in TPS function. A reciprocal substitution in the MhFPPS also caused a loss of IDS activity, which further supports the critical role of the KKxR residues in IPP binding. To fully identify the residues that control the transition between IDS and TPS, O'Maille and co-workers currently perform combinatorial mutations paired with the identification of epistatic residue networks. A similar strategy was applied by Salmon et al. to determine residue substitutions in the transition from linear to cyclic TPSs in plants.158 To probe evolutionary pathways of terpene cyclization, the authors of this study focused on the amino acid substitutions between a β-farnesene (86) synthase (BFS) from Artemisia annua and an amorphadiene synthase (ADS), which produces the bicyclic sesquiterpene amorpha-4,11-diene (87), a precursor of artemisinin (88) (Fig. 18). Structure-based combinatorial protein engineering (SCOPE)159 was employed to construct two libraries of soluble and biochemically active mutant enzymes with ADS substitutions within 6 Angstroms of the BFS active site. Biochemical characterization resulted in the identification of multiple enzyme variants with the ability to generate cyclic terpene products. Chief among these products was the cyclic sesquiterpene alcohol, alpha-bisabolol (89). Quantitative determination of product-specific kinetic rates (kcat,i)s for over 100 unique enzymes allowed to train quantitative models for Michaelis–Menten enzymatic free energies. These models were used to construct a family of biophysical fitness landscapes describing enzyme evolution.160 It was found that most mutations leading to the formation of alpha-bisabolol tended to have adverse effects on the overall magnitudes of product-specific reaction rates except for a previously identified critical gateway mutation (Y402L) that also unlocks the cyclization reaction.158 Overall, this microevolutionary exploration of sequence space allowed the identification of key residues in terpene cyclization.
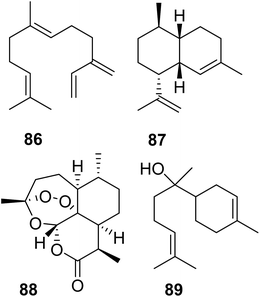 | ||
| Fig. 18 Acyclic and cyclic sesquiterpenes (86–88) in Artemisia annua and example of a cyclic sesquiterpene product (89) generated by an Artemisia β-farnesene synthase mutant variant. | ||
To gain a broader understanding of the emergence of TPS proteins among all insects, Rebholz et al. predicted the presence of putative IDS-like TPSs in several insect lineages beyond the pentatomids.151 To this end, UniProt sequences for polyprenyl synthetases (PFAM id PF00348) in the taxonomic class Insecta were screened for IDS and IDS-like TPS sequences based on distinct residue substitutions in the IBMs. The search identified more than 300 canonical IDS sequences and more than 125 putative TPSs, of which approximately 65% were found to be derived from FPPSs, nearly 23% from GGPPSs, with the rest derived from decaprenyl diphosphate (6) synthase (DPPS) like sequences (Fig. 19). Predicted TPS sequences were present in six insect orders: Blattodea, Hemiptera, Hymenoptera, Coleoptera, Lepidoptera, and Diptera.151
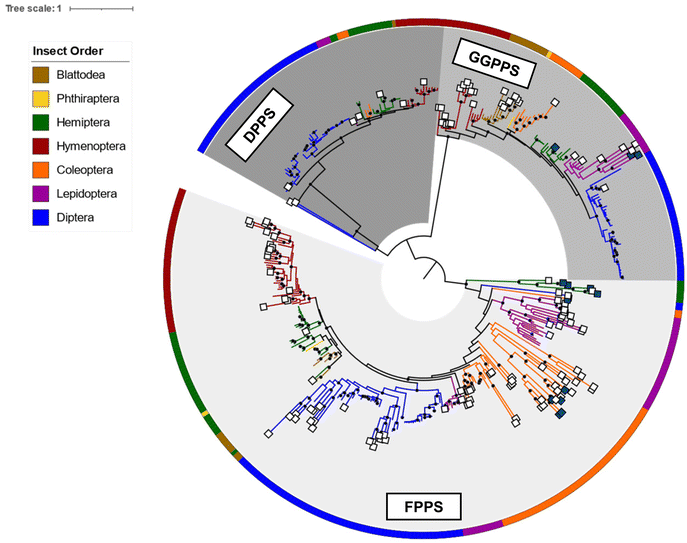 | ||
| Fig. 19 Phylogram of insect IDS and characterized and predicted IDS-like TPS proteins adapted from Rebholz et al.151 Branch lengths are proportional to amino acid substitutions per site. Previously characterized and predicted IDS-like TPS sequences are labeled with dark blue and white squares at branch tips, respectively. Insect orders, from which protein sequences originated, are indicated by colors of the circular perimeter and branches. Results suggest a recurring parallel emergence of TPS activity in IDS-like enzymes within and between insect lineages and IDS enzyme subfamilies. | ||
The analysis revealed a family of FPPS-like TPS proteins specific to the lepidopteran genus Papilio (Swallowtail butterflies), which may be responsible for the formation of linear mono- and sesquiterpenes that are released as defense compounds by Papilio larvae. Within the Lepidoptera, additional GGPPS-like sequences were found in the genome of the monarch butterfly Danaus plexippus plexippus, which emits terpene derivatives from male hairpencils.161 These sequences are monophyletic to the characterized mono-TPSs in H. melpomene. Moreover, GGPPS-like TPSs were predicted within blattodean termites such as Nasutitermes takasagoensis. It is likely that genes of these expanded GGPPS-like families are involved in the production of the described termite terpene metabolites (see 2.1.3). Overall, the findings by Rebholz et al.151 support the notion that insect TPSs originated from IDS genes and that this transition likely occurred via gene duplication and divergence through mutational drift and/or selection. Surprisingly, the study further indicates an independent emergence of TPS function not only within insect orders but also among the IDS subfamilies (FPPS, GGPPS, DPPS) of single species within the same order. This suggests that parallel functionalization of IDS enzymes for volatile and specialized terpene biosynthesis is widespread in insects.
5 Terpene synthases in corals and sponges
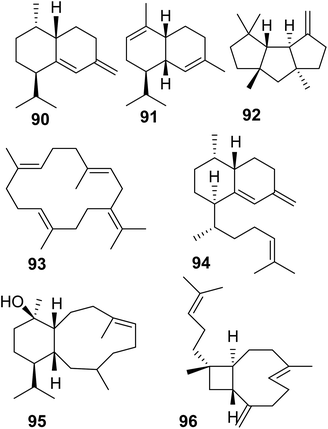
Soft corals (octocorals) are known for their accumulation of diterpenes; however, the biosynthetic origin remained unknown until recently, when a new family of terpene cyclases was discovered in these organisms. Burkhardt et al.162 and Scesa et al.163 identified more than 15 TPSs from genomes and transcriptomes of several octocoral genera. Most of these enzymes were characterized as diterpene cyclases and a few others were found to function as sesquiterpene cyclases, which make the sesquiterpene hydrocarbons nepthene (90), α-muuroladiene (91), and capnellene (92) (Fig. 20 and Table 1). The enzymatic diterpene products such as cembrene A and C (25, 93), elisabethatriene (94), klysimplexin R (95), and xeniaphyllene (96) (Fig. 20 and Table 1) carry the scaffolds for large groups of coral-specific diterpenes and thus represent important semi-volatile precursors in generating the tremendous chemical diversity of diterpene-mediated defenses in corals.Crystal structure analysis of a cembrene A synthase from the coral Eleutherobia rubra revealed that the protein carries the α-helical fold typical of class I TPSs but has three additional helices (Fig. 21).162 The analysis further showed the presence of conserved substrate-binding motifs and residues including the conserved aspartate rich motif, the NSE/DTE motif, and the previously identified arginine “pyrophosphate sensor” involved in carbocation formation (Fig. 21).164 In contrast to plant TPSs, the coral protein carries an RY motif that is conserved in microbial TPSs (e.g. Li et al.165). Its overall closer structural resemblance with microbial TPSs led the authors to speculate that an ancestral gene of the monophyletic coral TPS family might have been acquired by a common progenitor from microbial sources via horizontal gene transfer.
The evolution of these proteins predates the emergence of land plants. Interestingly, the coral TPS genes were found to cluster with P450, acyltransferase, and dehydrogenase genes among others, which presumably encode enzymes involved in secondary reactions of the TPS products.163 These are the first biosynthetic gene clusters found in animals raising questions about their evolution and the potential presence of such clusters in other animal genome. In addition to the finding of terpene cyclases in corals, another family of type I TPS genes has just been discovered in marine sponges of the order Bubarida.169 The corresponding enzymes were shown to make intermediates in the biosynthesis of isonitrile sesquiterpenes but do not install the isonitrile group themselves. In contrast to coral TPSs, the sponge TPS genes do not apppear to be organized in gene clusters. This is the first finding of TPSs that originate from sponge animal hosts independent from their microbial symbionts.169
6 Conclusions and outlook
In the past five to six years, substantial progress has been made in the identification of TPS genes in animals and in our understanding of how volatile terpenes and their derivatives are synthesized de novo in these organisms. The findings of IDS-type TPSs in insect genomes and the recent discovery of TPSs in soft corals and sponges may indicate that endogenous terpene biosynthetic pathways could be more common in animals than previously thought. The ways in which TPS gene functions have been recruited, whether through evolutionary transition from IDS precursors or by ancestral horizontal gene transfer from microbes (Fig. 21), appear to be as versatile as the diverse nature of terpene metabolites and their functions in chemical communication and defense. The finding that the formation of terpenes in corals is associated with assemblies of biosynthetic gene clusters similar to those found in microbes and plants may lead to the discovery of clusters in other animal genomes, especially when longer pathways have been established. To what extent these clustered genes are co-regulated in tissues or single cells in response to social or environmental signals will be of interest for future investigations. Another key question is whether terpene pheromones released from vertebrates such as amphibians and reptiles are the products of endogenous TPS enzymes or perhaps derived from microbial symbionts. Mining of high-quality genomes and gland-specific transcriptomes of these organisms should facilitate the elucidation of the biosynthetic origin of volatile terpene compounds. Finally, the discovery of terpene biosynthetic pathways in animals opens new possibilities for the biotechnological production of volatile terpenes such as species-specific pheromones in the development of alternative pest management strategies. For example, recent advances have been made in the metabolic engineering of fatty acid-derived moth sex pheromones in oilseed crops.166 Similarly, the discovery of terpene biosynthetic gene clusters found in corals will undoubtedly accelerate efforts in the production of bioactive and pharmaceutically valuable compounds.7 Author contributions
ZR participated in the conceptualization of the article, designed figures, wrote parts of the manuscript, and edited the manuscript. PEO designed figures and edited the manuscript. AVM wrote parts of the manuscript and edited the manuscript. DT conceptualized the article, designed figures, and wrote the manuscript. All authors read and approved the final manuscript.8 Conflicts of interest
There are no conflicts to declare.9 Acknowledgements
This work was supported by the National Science Foundation (MCB1920914, MCB1920922, MCB1920925) to AVM, PEO, and DT, respectively, and the US Department of Agriculture National Institute of Food and Agriculture (2016-67013-24759) to DT. Credit to authors and licensing of the images used in modified form in Fig. 6, 8, 9 and 12–14 are listed in ESI Table 1.†10 References
- R. R. Junker, J. Kuppler, L. Amo, J. D. Blande, R. M. Borges, N. M. van Dam, M. Dicke, S. Dötterl, B. K. Ehlers, F. Etl, J. Gershenzon, R. Glinwood, R. Gols, A. T. Groot, M. Heil, M. Hoffmeister, J. K. Holopainen, S. Jarau, L. John, A. Kessler, J. T. Knudsen, C. Kost, A. A. C. Larue-Kontic, S. D. Leonhardt, D. Lucas-Barbosa, C. J. Majetic, F. Menzel, A. L. Parachnowitsch, R. S. Pasquet, E. H. Poelman, R. A. Raguso, J. Ruther, F. P. Schiestl, T. Schmitt, D. Tholl, S. B. Unsicker, N. Verhulst, M. E. Visser, B. T. Weldegergis and T. G. Köllner, New Phytol., 2018, 220, 739–749 CrossRef PubMed.
- D. W. Christianson, Chem. Rev., 2017, 117, 11570–11648 CrossRef CAS PubMed.
- M. C. Lemfack, B. O. Gohlke, S. M. T. Toguem, S. Preissner, B. Piechulla and R. Preissner, Nucleic Acids Res., 2018, 46, D1261–D1265 CrossRef CAS PubMed.
- J. T. Knudsen and J. Gershenzon, in Biology of plant volatiles, ed. E. Pichersky and N. Dudareva, CRC Press, Boca Raton, 2nd edn, 2020, ch. 4, pp. 57–78 Search PubMed.
- E. Pichersky and N. Dudareva, Biology of plant volatiles, CRC Press, Boca Raton, 2nd edn, 2020 Search PubMed.
- M. Morita and E. W. Schmidt, Nat. Prod. Rep., 2018, 35, 357–378 RSC.
- T. Reichstein, J. von Euw, J. A. Parsons and M. Rothschild, Science, 1968, 161, 861–866 CrossRef CAS PubMed.
- D. Tholl, in Biotechnology of isoprenoids, ed. J. Schrader and J. Bohlmann, Springer International Publishing, 2015, vol. 148, pp. 63–106 Search PubMed.
- H. K. Lichtenthaler, in Chloroplast: Basics and Applications, ed. C. A. Rebeiz, C. Benning, H. J. Bohnert, H. Daniell, J. K. Hoober, H. K. Lichtenthaler, A. R. Portis and B. C. Tripathy, 2010, vol. 31, pp. 95–118 Search PubMed.
- M. Rohmer, Nat. Prod. Rep., 1999, 16, 565–574 RSC.
- H. M. Miziorko, Arch. Biochem. Biophys., 2011, 505, 131–143 CrossRef CAS PubMed.
- H. Tao, L. Lauterbach, G. Bian, R. Chen, A. Hou, T. Mori, S. Cheng, B. Hu, L. Lu, X. Mu, M. Li, N. Adachi, M. Kawasaki, T. Moriya, T. Senda, X. Wang, Z. Deng, I. Abe, J. S. Dickschat and T. Liu, Nature, 2022, 606, 414–419 CrossRef CAS PubMed.
- R. Saiki, A. Nagata, T. Kainou, H. Matsuda and M. Kawamukai, FEBS J., 2005, 272, 5606–5622 CrossRef CAS PubMed.
- J. D. Rudolf and C. Y. Chang, Nat. Prod. Rep., 2020, 37, 425–463 RSC.
- F. Chen, D. Tholl, J. Bohlmann and E. Pichersky, Plant J., 2011, 66, 212–229 CrossRef CAS PubMed.
- M. B. Quin, C. M. Flynn and C. Schmidt-Dannert, Nat. Prod. Rep., 2014, 31, 1449–1473 RSC.
- J. S. Dickschat, Nat. Prod. Rep., 2016, 33, 87–110 RSC.
- Q. Jia, G. Li, T. G. Köllner, J. Fu, X. Chen, W. Xiong, B. J. Crandall-Stotler, J. L. Bowman, D. J. Weston, Y. Zhang, L. Chen, Y. Xie, F.-W. Li, C. J. Rothfels, A. Larsson, S. W. Graham, D. W. Stevenson, G. K.-S. Wong, J. Gershenzon and F. Chen, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 12328–12333 CrossRef CAS PubMed.
- Z. H. Chen, Y. W. Guo and X. W. Li, Nat. Prod. Rep., 2022 10.1039/d2np00021k.
- R. B. Pereira, P. B. Andrade and P. Valentão, Mar. Drugs, 2016, 14, 39 CrossRef PubMed.
- C. Lyu, T. Chen, B. Qiang, N. Liu, H. Wang, L. Zhang and Z. Liu, Nucleic Acids Res., 2021, 49, D509–d515 CrossRef CAS PubMed.
- W. Fenical, R. K. Okuda, M. M. Bandurraga, P. Culver and R. S. Jacobs, Science, 1981, 212, 1512–1514 CrossRef CAS PubMed.
- H. Ben, Y. Sawall, A. Al-Sofyani and M. Wahl, Aquat. Biol., 2015, 23, 129–137 CrossRef.
- G. Giordano, M. Carbone, M. L. Ciavatta, E. Silvano, M. Gavagnin, M. J. Garson, K. L. Cheney, I. W. Mudianta, G. F. Russo, G. Villani, L. Magliozzi, G. Polese, C. Zidorn, A. Cutignano, A. Fontana, M. T. Ghiselin and E. Mollo, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 3451–3456 CrossRef CAS PubMed.
- S.-M. Shen, Z.-Y. Zhang, L.-G. Yao, J.-R. Wang, Y.-W. Guo and X.-W. Li, Chin. J. Chem., 2022, 40, 235–246 CrossRef.
- E. P. McCauley, I. C. Piña, A. D. Thompson, K. Bashir, M. Weinberg, S. L. Kurz and P. Crews, J. Antibiot., 2020, 73, 504–525 CrossRef CAS PubMed.
- L. D. Mydlarz, R. S. Jacobs, J. Boehnlein and R. G. Kerr, Chem. Biol., 2003, 10, 1051–1056 CrossRef CAS PubMed.
- C. Avila, Mar. Drugs, 2020, 18, 162 CrossRef CAS PubMed.
- W. A. Shear, Biochem. Syst. Ecol., 2015, 61, 78–117 CrossRef CAS.
- K. Mori and Y. Takagi, Tetrahedron Lett., 2000, 41, 6623–6625 CrossRef CAS.
- J. Smolanoff, A. F. Kluge, J. Meinwald, A. McPhail, R. W. Miller, K. Hicks and T. Eisner, Science, 1975, 188, 734–736 CrossRef CAS PubMed.
- M. F. Hassler, D. P. Harrison, T. H. Jones, C. H. Richart and R. A. Saporito, J. Nat. Prod., 2020, 83, 2764–2768 CrossRef CAS PubMed.
- Y. Kuwahara, Y. Ichiki, M. Morita, T. Tanabe and Y. Asano, J. Chem. Ecol., 2015, 41, 15–21 CrossRef CAS PubMed.
- F. Jüttner and S. B. Watson, Appl. Environ. Microbiol., 2007, 73, 4395–4406 CrossRef PubMed.
- O. Ekpa, J. W. Wheeler, J. C. Cokendolpher and R. M. Duffield, Tetrahedron Lett., 1984, 25, 1315–1318 CrossRef CAS.
- G. Raspotnig, V. Leutgeb, G. Krisper and H. J. Leis, Exp. Appl. Acarol., 2011, 54, 211–224 CrossRef CAS PubMed.
- A. C. Skelton, M. M. Cameron, J. A. Pickett and M. A. Birkett, J. Med. Entomol., 2010, 47, 798–804 CrossRef CAS PubMed.
- N. Shimizu, N. Mori and Y. Kuwahara, Biosci., Biotechnol., Biochem., 2003, 67, 1732–1736 CrossRef CAS PubMed.
- X. Dong, K. Chaisiri, D. Xia, S. D. Armstrong, Y. Fang, M. J. Donnelly, T. Kadowaki, J. W. McGarry, A. C. Darby and B. L. Makepeace, Gigascience, 2018, 7, giy127 Search PubMed.
- M. Müller and G. Buchbauer, Flavour Fragrance J., 2011, 26, 357–377 CrossRef.
- D. C. Weber, A. Khrimian, M. C. Blassioli-Moraes and J. G. Millar, in Invasive stink bugs and related species (Pentatomoidea): Biology, higher systematics, semiochemistry, and management, ed. J. E. McPherson, CRC Press, Boca Raton, Florida, 2018, pp. 677–725 Search PubMed.
- J. A. Pickett, R. K. Allemann and M. A. Birkett, Nat. Prod. Rep., 2013, 30, 1277–1283 RSC.
- C. I. Keeling, C. Tittiger, M. MacLean and G. J. Blomquist, in Insect pheromone biochemistry and molecular biology, ed. G. J. Blomquist and R. G. Vogt, Academic Press, San Diego, 2021, pp. 123–154 Search PubMed.
- J. Havlíčková, K. Dolejšová, M. Tichý, V. Vrkoslav, B. Kalinová, P. Kyjaková and R. Hanus, Z. Naturforsch. C, 2019, 74, 251–264 CrossRef PubMed.
- T. A. K. Prescott, J. Bramham, O. Zompro and S. K. Maciver, Biochem. Syst. Ecol., 2009, 37, 759–760 CrossRef CAS.
- R. M. Smith, J. J. Brophy, G. W. K. Cavill and N. W. Davies, J. Chem. Ecol., 1979, 5, 727–735 CrossRef CAS.
- D. B. Weibel, N. J. Oldham, B. Feld, G. Glombitza, K. Dettner and W. Boland, Insect Biochem. Mol. Biol., 2001, 31, 583–591 CrossRef CAS PubMed.
- C. J. Persoons, P. E. J. Verwiel, F. J. Ritter, E. Talman, P. J. F. Nooijen and W. J. Nooijen, Tetrahedron Lett., 1976, 24, 2055–2058 CrossRef.
- E. Talman, P. E. J. Verwiel, F. J. Ritter and C. J. Persoons, Tetrahedron Lett., 1978, 17, 227–235 CAS.
- H. Takegawa and S. Takahashi, Appl. Entomol. Zool., 1989, 24, 435–440 CrossRef CAS.
- S. Takahashi, K. Watanabe, S. Saito and Y. Nomura, Appl. Entomol. Zool., 1995, 30, 357–360 CrossRef CAS.
- T. Hirukawa, T. Shudo and T. Kato, J. Chem. Soc., Perkin Trans. 1, 1993, 2, 217–225 RSC.
- J. C. Braekman, D. Daloze, A. Dupont, J. M. Pasteels and R. Ottinger, Bull. Soc. Chim. Belg., 1984, 93, 291–298 CrossRef CAS.
- G. D. Prestwich, B. A. Solheim, J. Clardy, F. G. Pilkiewicz, I. Miura, S. P. Tanis and K. Nakanishi, J. Am. Chem. Soc., 1977, 99, 8082–8083 CrossRef CAS.
- J. Sobotnik, A. Jirosova and R. Hanus, J. Insect Physiol., 2010, 56, 1012–1021 CrossRef CAS PubMed.
- G. D. Prestwich, Annu. Rev. Entomol., 1984, 29, 201–232 CrossRef CAS.
- C. Everaerts, O. Bonnard, J. M. Pasteels, Y. Roisin and W. A. König, Experientia, 1990, 46, 227–230 CrossRef CAS.
- M. R. Tarver, E. A. Schmelz, J. R. Rocca and M. E. Scharf, J. Chem. Ecol., 2009, 35, 256–264 CrossRef CAS PubMed.
- D. Sillam-Dusses, B. Kalinova, P. Jiros, A. Brezinova, J. Cvacka, R. Hanus, J. Sobotnik, C. Bordereau and I. Valterova, J. Insect Physiol., 2009, 55, 751–757 CrossRef CAS PubMed.
- K. Haga, T. Suzuki, S. Kodama and Y. Kuwahara, Appl. Entomol. Zool., 1990, 25, 138–139 CrossRef CAS.
- T. Suzuki, K. Haga, W. S. Leal, S. Kodama and Y. Kuwahara, Appl. Entomol. Zool., 1989, 24, 222–228 CrossRef CAS.
- J. G. C. Hamilton, D. R. Hall and W. D. J. Kirk, J. Chem. Ecol., 2005, 31, 1369–1379 CrossRef CAS PubMed.
- S. Niassy, A. Tamiru, J. G. C. Hamilton, W. D. J. Kirk, R. Mumm, C. Sims, W. J. de Kogel, S. Ekesi, N. K. Maniania, K. Bandi, F. Mitchell and S. Subramanian, J. Chem. Ecol., 2019, 45, 348–355 CrossRef CAS PubMed.
- Z. A. Demissie, L. A. E. Erland, M. R. Rheault and S. S. Mahmoud, J. Biol. Chem., 2013, 288, 6333–6341 CrossRef CAS PubMed.
- J. Tabata and R. T. Ichiki, J. Chem. Ecol., 2016, 42, 1193–1200 CrossRef CAS PubMed.
- J. Einhorn, A. Guerrero, P. H. Ducrot, F. D. Boyer, M. Gieselmann and W. Roelofs, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 9867–9872 CrossRef CAS PubMed.
- A. M. El-Sayed, C. R. Unelius, A. Twidle, V. Mitchell, L.-A. Manning, L. Cole, D. M. Suckling, M. F. Flores, T. Zaviezo and J. Bergmann, Tetrahedron Lett., 2010, 51, 1075–1078 CrossRef CAS.
- Z. Rebholz, J. Lancaster, H. Larose, A. Khrimian, K. Luck, M. E. Sparks, K. L. Gendreau, L. Shewade, T. G. Köllner, D. C. Weber, D. E. Gundersen-Rindal, P. O'Maille, A. V. Morozov and D. Tholl, Insect Biochem. Mol. Biol., 2023, 152, 103879 CrossRef CAS PubMed.
- M. Inwood and P. Morgan, Myrmecol. News, 2008, 11, 79–90 Search PubMed.
- R. K. Vander Meer, F. Alvarez and C. S. Lofgren, J. Chem. Ecol., 1988, 14, 825–838 CrossRef CAS PubMed.
- S. J. Keegans, J. Billen, E. D. Morgan and O. A. Gokcen, J. Chem. Ecol., 1993, 19, 2705–2719 CrossRef CAS PubMed.
- J. A. Pickett, I. H. Williams, M. C. Smith and A. P. Martin, J. Chem. Ecol., 1981, 7, 543–554 CrossRef CAS PubMed.
- R. Kubo and M. Ono, Entomol. Sci., 2010, 13, 167–173 CrossRef.
- M. Kaib and H. Dittebrand, Chemoecology, 1990, 1, 3–11 CrossRef.
- B. Fassotte, C. Fischer, D. Durieux, G. Lognay, E. Haubruge, F. Francis and F. J. Verheggen, PLoS One, 2014, 9, e115011 CrossRef PubMed.
- R. J. Bartelt, A. A. Cossé, B. W. Zilkowski, D. Weisleder and F. A. Momany, J. Chem. Ecol., 2001, 27, 2397–2423 CrossRef CAS PubMed.
- N. Mitlin and P. A. Hedin, J. Insect Physiol., 1974, 20, 1825–1831 CrossRef CAS PubMed.
- J. E. Carrel, M. H. McCairel, A. J. Slagle, J. P. Doom, J. Brill and J. P. McCormick, Experientia, 1993, 49, 171–174 CrossRef CAS PubMed.
- K. Honda, J. Chem. Ecol., 1981, 7, 1089–1113 CrossRef CAS PubMed.
- H. Omura, K. Honda and P. Feeny, J. Chem. Ecol., 2006, 32, 1999–2012 CrossRef CAS PubMed.
- S. Schulz, C. Estrada, S. Yildizhan, M. Boppré and L. E. Gilbert, J. Chem. Ecol., 2008, 34, 82–93 CrossRef CAS PubMed.
- R. Baker, R. H. Herbert and G. G. Grant, J. Chem. Soc., Chem. Commun., 1985, 12, 824–825 RSC.
- P. E. A. Teal, J. A. Meredith and Y. Gomez-Simuta, Arch. Insect Biochem. Physiol., 1999, 42, 225–232 CrossRef CAS PubMed.
- C. N. Spiegel, P. Jeanbourquin, P. M. Guerin, A. M. Hooper, S. Claude, R. Tabacchi, S. Sano and K. Mori, J. Insect Physiol., 2005, 51, 1366–1375 CrossRef CAS PubMed.
- M. Menke, K. Melnik, P. S. Peram, I. Starnberger, W. Hödl, M. Vences and S. Schulz, Eur. J. Org. Chem., 2018, 2018, 2651–2656 CrossRef CAS PubMed.
- M. Menke, P. S. Peram, I. Starnberger, W. Hödl, G. F. Jongsma, D. C. Blackburn, M. O. Rödel, M. Vences and S. Schulz, Beilstein J. Org. Chem., 2016, 12, 2731–2738 CrossRef CAS PubMed.
- I. Starnberger, D. Poth, P. S. Peram, S. Schulz, M. Vences, J. Knudsen, M. F. Barej, M. O. Rodel, M. Walzl and W. Hodl, Biol. J. Linn. Soc., 2013, 110, 828–838 CrossRef PubMed.
- C. Nowack, P. S. Peram, S. Wenzel, A. Rakotoarison, F. Glaw, D. Poth, S. Schulz and M. Vences, J. Zool., 2017, 303, 72–81 CrossRef.
- S. Yildizhan, J. van Loon, A. Sramkova, M. Ayasse, C. Arsene, C. ten Broeke and S. Schulz, ChemBioChem, 2009, 10, 1666–1677 CrossRef CAS PubMed.
- T. Chuman, J. Sivinski, R. R. Heath, C. O. Calkins, J. H. Tumlinson, M. A. Battiste, R. L. Wydra, L. Strekowski and J. L. Nation, Tetrahedron Lett., 1988, 29, 6561–6563 CrossRef CAS.
- D. Vanderwel, B. Johnston and A. C. Oehlschlager, Insect Biochem. Mol. Biol., 1992, 22, 875–883 CrossRef CAS.
- B. P. C. Smith, Y. Hayasaka, M. J. Tyler and B. D. Williams, Aust. J. Zool., 2004, 52, 521–530 CrossRef CAS.
- D. L. Mattern, W. D. Scott, C. A. McDaniel, P. J. Weldon and D. E. Graves, J. Nat. Prod., 1997, 60, 828–831 CrossRef CAS PubMed.
- J. W. Avery, A. Shafagati, A. Turner, J. W. Wheeler and P. J. Weldon, Biochem. Syst. Ecol., 1993, 21, 533–534 CrossRef CAS.
- S. Schulz, K. Krückert and P. J. Weldon, J. Nat. Prod., 2003, 66, 34–38 CrossRef CAS PubMed.
- S. García-Rubio, A. B. Attygalle, P. J. Weldon and J. Meinwald, J. Chem. Ecol., 2002, 28, 769–781 CrossRef PubMed.
- B. V. Burger, M. Leroux, H. S. C. Spies, V. Truter and R. C. Bigalke, Z. Naturforsch., C: Biosci., 1981, 36, 340–343 CrossRef CAS.
- J. W. Wheeler, L. E. Rasmussen, F. Ayorinde, I. O. Buss and G. L. Smuts, J. Chem. Ecol., 1982, 8, 821–835 CrossRef CAS PubMed.
- T. E. Goodwin, F. D. Brown, R. W. Counts, N. C. Dowdy, P. L. Fraley, R. A. Hughes, D. Z. Liu, C. D. Mashburn, J. D. Rankin, R. S. Roberson, K. D. Wooley, E. L. Rasmussen, S. W. Riddle, H. S. Riddle and S. Schulz, J. Nat. Prod., 2002, 65, 1319–1322 CrossRef CAS PubMed.
- C. G. Faulkes, J. S. Elmore, D. A. Baines, B. Fenton, N. B. Simmons and E. L. Clare, PeerJ, 2019, 7, e7734 CrossRef PubMed.
- E. J. Elwell, D. Walker and S. Vaglio, Animals, 2021, 11, 2091 CrossRef PubMed.
- M. Salamon and N. W. Davies, J. Chem. Ecol., 1998, 24, 1659–1676 CrossRef CAS.
- D. J. King, R. M. Gleadow and I. E. Woodrow, New Phytol., 2006, 172, 440–451 CrossRef CAS PubMed.
- C. S. Mathela and N. Joshi, Nat. Prod. Commun., 2008, 3, 945–950 CrossRef CAS.
- M. A. Birkett, A. Hassanali, S. Hoglund, J. Pettersson and J. A. Pickett, Phytochemistry, 2011, 72, 109–114 CrossRef CAS PubMed.
- B. R. Lichman, G. T. Godden, J. P. Hamilton, L. Palmer, M. O. Kamileen, D. Zhao, B. Vaillancourt, J. C. Wood, M. Sun, T. J. Kinser, L. K. Henry, C. Rodriguez-Lopez, N. Dudareva, D. E. Soltis, P. S. Soltis, C. R. Buell and S. E. O'Connor, Sci. Adv., 2020, 6, eaba0721 CrossRef CAS PubMed.
- K. Miettinen, L. M. Dong, N. Navrot, T. Schneider, V. Burlat, J. Pollier, L. Woittiez, S. van der Krol, R. Lugan, T. Ilc, R. Verpoorte, K. M. Oksman-Caldentey, E. Martinoia, H. Bouwmeester, A. Goossens, J. Memelink and D. Werck-Reichhart, Nat. Commun., 2014, 5, 4175 CrossRef CAS.
- F. Geu-Flores, N. H. Sherden, V. Courdavault, V. Burlat, W. S. Glenn, C. Wu, E. Nims, Y. Cui and S. E. O'Connor, Nature, 2012, 492, 138–142 CrossRef CAS PubMed.
- R. R. Bodemann, P. Rahfeld, M. Stock, M. Kunert, N. Wielsch, M. Groth, S. Frick, W. Boland and A. Burse, Proc. R. Soc. B, 2012, 279, 4126–4134 CrossRef CAS PubMed.
- S. Frick, R. Nagel, A. Schmidt, R. R. Bodemann, P. Rahfeld, G. Pauls, W. Brandt, J. Gershenzon, W. Boland and A. Burse, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 56–61 CrossRef PubMed.
- P. Rahfeld, W. Haeger, R. Kirsch, G. Pauls, T. Becker, E. Schulze, N. Wielsch, D. Wang, M. Groth, W. Brandt, W. Boland and A. Burse, Insect Biochem. Mol. Biol., 2015, 58, 28–38 CrossRef CAS PubMed.
- P. Rahfeld, R. Kirsch, S. Kugel, N. Wielsch, M. Stock, M. Groth, W. Boland and A. Burse, Proc. R. Soc. B, 2014, 281, 20140842 CrossRef PubMed.
- N. Fu, Z. L. Yang, Y. Pauchet, C. Paetz, W. Brandt, W. Boland and A. Burse, Insect Biochem. Mol. Biol., 2019, 113, 103212 CrossRef CAS PubMed.
- T. G. Köllner, A. David, K. Luck, F. Beran, G. Kunert, J. J. Zhou, L. Caputi and S. E. O'Connor, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2211254119 CrossRef PubMed.
- T. Laursen, J. Borch, C. Knudsen, K. Bavishi, F. Torta, H. J. Martens, D. Silvestro, N. S. Hatzakis, M. R. Wenk, T. R. Dafforn, C. E. Olsen, M. S. Motawia, B. Hamberger, B. L. Moller and J. E. Bassard, Science, 2016, 354, 890–893 CrossRef CAS PubMed.
- M. Sugumaran, K. Nellaiappan, C. Amaratunga, S. Cardinale and T. Scott, Arch. Biochem. Biophys., 2000, 378, 393–403 CrossRef CAS PubMed.
- S. J. Seybold, D. R. Quilici, J. A. Tillman, D. Vanderwel, D. L. Wood and G. J. Blomquist, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 8393–8397 CrossRef CAS PubMed.
- A. B. Gilg, J. C. Bearfield, C. Tittiger, W. H. Welch and G. J. Blomquist, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 9760–9765 CrossRef CAS PubMed.
- A. B. Gilg, C. Tittiger and G. J. Blomquist, Naturwissenschaften, 2009, 96, 731–735 CrossRef CAS PubMed.
- F. Beran, P. Rahfeld, K. Luck, R. Nagel, H. Vogel, N. Wielsch, S. Irmisch, S. Ramasamy, J. Gershenzon, D. G. Heckel and T. G. Köllner, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2922–2927 CrossRef CAS PubMed.
- A. Čokl, A. M. Dias, M. C. B. Moraes, M. Borges and R. A. Laumann, J. Insect Behav., 2017, 30, 741–758 CrossRef.
- D. Tholl, in Insect pheromone biochemistry and molecular biology, ed. G. J. Blomquist and R. G. Vogt, Academic Press, San Diego, 2nd edn, 2021, pp. 269–281 Search PubMed.
- J. Lancaster, A. Khrimian, S. Young, B. Lehner, K. Luck, A. Wallingford, S. K. B. Ghosh, P. Zerbe, A. Muchlinski, P. E. Marek, M. E. Sparks, J. G. Tokuhisa, C. Tittiger, T. G. Köllner, D. C. Weber, D. E. Gundersen-Rindal, T. P. Kuhar and D. Tholl, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E8634–E8641 CAS.
- A. Khrimian, S. Shirali, K. E. Vermillion, M. A. Siegler, F. Guzman, K. Chauhan, J. R. Aldrich and D. C. Weber, J. Chem. Ecol., 2014, 40, 1260–1268 CrossRef CAS PubMed.
- M. E. Sparks, J. H. Rhoades, D. R. Nelson, D. Kuhar, J. Lancaster, B. Lehner, D. Tholl, D. C. Weber and D. E. Gundersen-Rindal, Insects, 2017, 8, 55 CrossRef PubMed.
- L. K. Sy and G. D. Brown, Phytochemistry, 1997, 45, 537–544 CrossRef CAS.
- A. Khrimian, A. J. Zhang, D. C. Weber, H. Y. Ho, J. R. Aldrich, K. E. Vermillion, M. A. Siegler, S. Shirali, F. Guzman and T. C. Leskey, J. Nat. Prod., 2014, 77, 1708–1717 CrossRef CAS PubMed.
- M. E. Sparks, R. Bansal, J. B. Benoit, M. B. Blackburn, H. Chao, M. Chen, S. Cheng, C. Childers, H. Dinh, H. V. Doddapaneni, S. Dugan, E. N. Elpidina, D. W. Farrow, M. Friedrich, R. A. Gibbs, B. Hall, Y. Han, R. W. Hardy, C. J. Holmes, D. S. T. Hughes, P. Ioannidis, A. M. Cheatle Jarvela, J. S. Johnston, J. W. Jones, B. A. Kronmiller, F. Kung, S. L. Lee, A. G. Martynov, P. Masterson, F. Maumus, M. Munoz-Torres, S. C. Murali, T. D. Murphy, D. M. Muzny, D. R. Nelson, B. Oppert, K. A. Panfilio, D. P. Paula, L. Pick, M. F. Poelchau, J. Qu, K. Reding, J. H. Rhoades, A. Rhodes, S. Richards, R. Richter, H. M. Robertson, A. J. Rosendale, Z. J. Tu, A. S. Velamuri, R. M. Waterhouse, M. T. Weirauch, J. T. Wells, J. H. Werren, K. C. Worley, E. M. Zdobnov and D. E. Gundersen-Rindal, BMC Genomics, 2020, 21, 227 CrossRef PubMed.
- M. E. Sparks, K. S. Shelby, D. Kuhar and D. E. Gundersen-Rindal, PLoS One, 2014, 9, e111646 CrossRef PubMed.
- M. Borges, M. Birkett, J. R. Aldrich, J. E. Oliver, M. Chiba, Y. Murata, R. A. Laumann, J. A. Barrigossi, J. A. Pickett and M. C. B. Moraes, J. Chem. Ecol., 2006, 32, 2749–2761 CrossRef CAS PubMed.
- M. W. M. de Oliveira, M. Borges, C. K. Z. Andrade, R. A. Laumann, J. A. F. Barrigossi and M. C. Blassioli-Moraes, J. Agric. Food Chem., 2013, 61, 7777–7785 CrossRef CAS PubMed.
- A. A. C. Moliterno, D. J. De Melo and P. H. G. Zarbin, J. Chem. Ecol., 2021, 47, 1–9 CrossRef CAS PubMed.
- J. R. Aldrich, J. E. Oliver, W. R. Lusby, J. P. Kochansky and J. A. Lockwood, J. Exp. Zool., 1987, 244, 171–175 CrossRef CAS.
- V. E. Harris and J. W. Todd, Entomol. Exp. Appl., 1980, 27, 117–126 CrossRef.
- J. Lancaster, B. Lehner, A. Khrimian, A. Muchlinski, K. Luck, T. G. Köllner, D. C. Weber, D. E. Gundersen-Rindal and D. Tholl, J. Chem. Ecol., 2019, 45, 187–197 CrossRef CAS PubMed.
- B. W. Cribb, K. N. Siriwardana and G. H. Walter, J. Morphol., 2006, 267, 831–840 CrossRef PubMed.
- S. Kumar, C. Kempinski, X. Zhuang, A. Norris, S. Mafu, J. C. Zi, S. A. Bell, S. E. Nybo, S. E. Kinison, Z. D. Jiang, S. Goklany, K. B. Linscott, X. L. Chen, Q. D. Jia, S. D. Brown, J. L. Bowman, P. C. Babbitt, R. J. Peters, F. Chen and J. Chappell, Plant Cell, 2016, 28, 2632–2650 CAS.
- G. L. Li, T. G. Köllner, Y. B. Yin, Y. F. Jiang, H. Chen, Y. Xu, J. Gershenzon, E. Pichersky and F. Chen, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14711–14715 CrossRef CAS PubMed.
- A. Muchlinski, X. Chen, J. T. Lovell, T. G. Köllner, K. A. Pelot, P. Zerbe, M. Ruggiero, L. Callaway, S. Laliberte, F. Chen and D. Tholl, Front. Plant Sci., 2019, 10, 1144 CrossRef PubMed.
- J. R. Aldrich, W. R. Lusby and J. P. Kochansky, Experientia, 1986, 42, 583–585 CrossRef CAS.
- J. P. Farine, O. Bonnard, R. Brossut and J. L. Le Quere, J. Chem. Ecol., 1992, 18, 1673–1682 CrossRef CAS PubMed.
- B. S. Krall, B. W. Zilkowski, S. L. Kight, R. J. Bartelt and D. W. Whitman, J. Chem. Ecol., 1997, 23, 1951–1962 CrossRef CAS.
- J. R. Aldrich, S. P. Carroll, J. E. Oliver, W. R. Lusby, A. A. Rudmann and R. M. Waters, Biochem. Syst. Ecol., 1990, 18, 369–376 CrossRef CAS.
- E. D. Morgan, Biosynthesis in insects, RSC Publishing, Cambridge, UK, Advanced Edition edn., 2010 Search PubMed.
- S. B. Rivera, B. D. Swedlund, G. J. King, R. N. Bell, C. E. Hussey, D. M. Shattuck-Eidens, W. M. Wrobel, G. D. Peiser and C. D. Poulter, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 4373–4378 CrossRef CAS PubMed.
- K. Darragh, A. Orteu, D. Black, K. J. R. P. Byers, D. Szczerbowski, I. A. Warren, P. Rastas, A. Pinharanda, J. W. Davey, S. Fernanda Garza, D. Abondano Almeida, R. M. Merrill, W. O. McMillan, S. Schulz and C. D. Jiggins, PLoS Biol., 2021, 19, e3001022 CrossRef CAS PubMed.
- S. Schulz, C. Estrada, S. Yildizhan, M. Boppre and L. E. Gilbert, J. Chem. Ecol., 2008, 34, 82–93 CrossRef CAS PubMed.
- K. Darragh, G. Montejo-Kovacevich, K. M. Kozak, C. R. Morrison, C. M. E. Figueiredo, J. S. Ready, C. Salazar, M. Linares, K. Byers, R. M. Merrill, W. O. McMillan, S. Schulz and C. D. Jiggins, Ecol. Evol., 2020, 10, 3895–3918 CrossRef PubMed.
- M. Hojo, S. Shigenobu, K. Maekawa, T. Miura and G. Tokuda, Insect Biochem. Mol. Biol., 2019, 111, 103177 CrossRef CAS PubMed.
- Y. Zhou, X. Liu and Z. Yang, Biomolecules, 2019, 9, 808 CrossRef CAS PubMed.
- Z. Rebholz, L. Shewade, K. Kaler, H. Larose, F. Schubot, D. Tholl, A. V. Morozov and P. E. O'Maille, Protein Sci., submitted Search PubMed.
- C. M. Starks, K. W. Back, J. Chappell and J. P. Noel, Science, 1997, 277, 1815–1820 CrossRef CAS PubMed.
- C. A. Lesburg, G. Zhai, D. E. Cane and D. W. Christianson, Science, 1997, 277, 1820–1824 CrossRef CAS PubMed.
- F. H. Wallrapp, J.-J. Pan, G. Ramamoorthy, D. E. Almonacid, B. S. Hillerich, R. Seidel, Y. Patskovsky, P. C. Babbitt, S. C. Almo, M. P. Jacobson and C. D. Poulter, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E1196–E1202 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- D. Piovesan, G. Minervini and S. C. Tosatto, Nucleic Acids Res., 2016, 44, W367–W374 CrossRef CAS PubMed.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Genome Res., 2003, 13, 2498–2504 CrossRef CAS PubMed.
- M. Salmon, C. Laurendon, M. Vardakou, J. Cheema, M. Defernez, S. Green, J. A. Faraldos and P. E. O'Maille, Nat. Commun., 2015, 6, 6143 CrossRef CAS PubMed.
- M. Dokarry, C. Laurendon and P. E. O'Maille, in Natural product biosynthesis by microorganisms and plants, part A, ed. D. A. Hopwood, 2012, vol. 515, pp. 21–42 Search PubMed.
- A. Ballal, C. Laurendon, M. Salmon, M. Vardakou, J. Cheema, M. Defernez, P. E. O'Maille and A. V. Morozov, Mol. Biol. Evol., 2020, 37, 1907–1924 CrossRef CAS PubMed.
- J. Meinwald, A. M. Chalmers, T. E. Pliske and T. Eisner, Tetrahedron Lett., 1968, 9, 4893–4896 CrossRef.
- I. Burkhardt, T. De Rond, P. Y.-T. Chen and B. S. Moore, Nat. Chem. Biol., 2022, 18, 664–669 CrossRef CAS PubMed.
- P. D. Scesa, Z. Lin and E. W. Schmidt, Nat. Chem. Biol., 2022, 18, 659–663 CrossRef CAS PubMed.
- P. Baer, P. Rabe, K. Fischer, C. A. Citron, T. A. Klapschinski, M. Groll and J. S. Dickschat, Angew. Chem., Int. Ed., 2014, 53, 7652–7656 CrossRef CAS PubMed.
- R. Li, W. K. Chou, J. A. Himmelberger, K. M. Litwin, G. G. Harris, D. E. Cane and D. W. Christianson, Biochemistry, 2014, 53, 1155–1168 CrossRef CAS PubMed.
- Y. H. Xia, H. L. Wang, B. J. Ding, G. P. Svensson, C. Jarl-Sunesson, E. B. Cahoon, P. Hofvander and C. Löfstedt, J. Chem. Ecol., 2021, 47, 950–967 CrossRef CAS PubMed.
- B. Misof, S. Liu, K. Meusemann, R. S. Peters, A. Donath, C. Mayer, P. B. Frandsen, J. Ware, T. Flouri, R. G. Beutel, O. Niehuis, M. Petersen, F. Izquierdo-Carrasco, T. Wappler, J. Rust, A. J. Aberer, U. Aspöck, H. Aspöck, D. Bartel, A. Blanke, S. Berger, A. Böhm, T. R. Buckley, B. Calcott, J. Chen, F. Friedrich, M. Fukui, M. Fujita, C. Greve, P. Grobe, S. Gu, Y. Huang, L. S. Jermiin, A. Y. Kawahara, L. Krogmann, M. Kubiak, R. Lanfear, H. Letsch, Y. Li, Z. Li, J. Li, H. Lu, R. Machida, Y. Mashimo, P. Kapli, D. D. McKenna, G. Meng, Y. Nakagaki, J. L. Navarrete-Heredia, M. Ott, Y. Ou, G. Pass, L. Podsiadlowski, H. Pohl, B. M. von Reumont, K. Schütte, K. Sekiya, S. Shimizu, A. Slipinski, A. Stamatakis, W. Song, X. Su, N. U. Szucsich, M. Tan, X. Tan, M. Tang, J. Tang, G. Timelthaler, S. Tomizuka, M. Trautwein, X. Tong, T. Uchifune, M. G. Walzl, B. M. Wiegmann, J. Wilbrandt, B. Wipfler, T. K. Wong, Q. Wu, G. Wu, Y. Xie, S. Yang, Q. Yang, D. K. Yeates, K. Yoshizawa, Q. Zhang, R. Zhang, W. Zhang, Y. Zhang, J. Zhao, C. Zhou, L. Zhou, T. Ziesmann, S. Zou, Y. Li, X. Xu, Y. Zhang, H. Yang, J. Wang, J. Wang, K. M. Kjer and X. Zhou, Science, 2014, 346, 763–767 CrossRef CAS PubMed.
- G. Giribet, Org. Diversity Evol., 2016, 16, 419–426 CrossRef.
- K. Wilson, T. de Rond, I. Burkhardt, T. S. Steele, R. J. B. Schäfer, S. Podell, E. E. Allen and B. S. Moore, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2220934120 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2np00076h |
| This journal is © The Royal Society of Chemistry 2023 |