 Open Access Article
Open Access ArticlePushing the limits of mechanoredox RAFT polymerization methods
Jason D. Kaff†
 a,
Mercie N. Hodges†
a,
Mercie N. Hodges† a,
Abdul Moeez
a,
Abdul Moeez b,
Sarah M. Zeitler
b,
Sarah M. Zeitler a,
Lilo D. Pozzo
a,
Lilo D. Pozzo bc and
Matthew R. Golder
bc and
Matthew R. Golder *ab
*ab
aDepartment of Chemistry, University of Washington, Seattle, WA 98195, USA. E-mail: goldermr@uw.edu
bMolecular Engineering & Science Institute, University of Washington, Seattle, WA 98195, USA
cDepartment of Chemical Engineering, University of Washington, Seattle, WA 98195, USA
First published on 12th February 2026
Abstract
Integral aspects of what is considered green chemistry include minimizing solvent use and utilizing energy-efficient methods to synthesize target materials. For polymer synthesis in particular, accessing copolymer sequences derived from immiscible feedstocks and masses in the ultra-high molecular weight regime (>1 MDa) often require specialized methods, extensive optimization, and may consume large amounts of energy. In this work, we report on the synthesis of diverse polyacrylates inspired by the principles of green chemistry using streamlined ball mill grinding methodology. Mechanoredox reversible addition–fragmentation chain-transfer (MR-RAFT) polymerizations are used herein to synthesize multiblock copolymers from immiscible monomers and overcome viscosity restraints to reach ultra-high molecular weights. The ability to access these traditionally challenging-to-synthesize polymers using a (nearly) solvent-free method will enable the discovery of novel materials with minimal environmental impact.
Introduction
As polymers continue to be necessary for constructing high-demand commodity materials, the issue of polymer sustainability has become multifaceted.1,2 Polymers are renowned for their lightweight, durable qualities and low cost of production.3,4 However, they are also castigated for their persistence in the environment, shedding microplastics, and requiring high amounts of energy and solvent to synthesize.1,5–7 Addressing these issues requires a similarly multifaceted approach that considers consumption quantity,8 repurposing polymer waste,9–11 creating degradable or otherwise circular materials,12,13 and finding greener synthetic pathways for existing polymers.14–16 This work strives for the latter goal by generating diverse polyacrylates via mechanochemical polymerizations.Polymerizations are traditionally driven by light, heat, or electricity; however, recent efforts have focused on analogous processes driven by mechanochemistry. While work from Staudinger from nearly 100 years ago,17 as well as more contemporary findings, demonstrate the utility of polymer mechanochemistry for destructive processes,18,19 recent work has highlighted force-mediated reactions as viable means to synthesize myriad polymers.20,21 Indeed, ball mill grinding enables shorter reaction times, lowers energy and solvent consumption, and facilitates the synthesis of materials from immiscible building blocks.16,22,23 Inspired by seminal works from Esser-Kahn,24 Matyjaszewski,25 Bielawski,26 as well as more recent publications from Pang27 and Stenzel,28 previous work from our group explored the utility of ball mill grinding with piezoelectric nanoparticles to drive redox chemistry (i.e., mechanoredox catalysis) for free radical polymerization (FRP) and reversible addition–fragmentation chain transfer (RAFT) polymerization reactions (Fig. 1A).29,30 In this prior work, diblock and random copolymer formulations were synthesized from immiscible building blocks, but additional chain extension was not pursued. Multiblock copolymers have the potential to self-assemble into unique topologies with applications in drug delivery, optoelectronics, and as polyelectrolytes; ball mill mechanochemistry offers experimental simplicity to access these formulations.31–38 Recent work from Kim exemplified this paradigm through the synthesis of “mechano-exclusive” macromolecules using ROMP and free-radical polymerization (Fig. 1A),37 but multiblock structures remained unrealized.
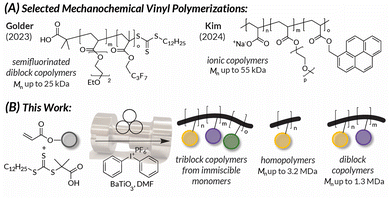 | ||
| Fig. 1 (A) Previous work showcasing mechanochemically synthesized copolymers derived from immiscible vinylic monomers.29,37 (B) This work features the ability MR-RAFT to access triblock copolymers from immiscible monomers and UHMW homopolymers and diblock copolymers. | ||
We were additionally intrigued by the relatively low dispersity (Đ < 1.6) and high molar masses (Mn up to 940 kDa) afforded by our prior mechanoredox FRP work despite the high bulk viscosities realized during the reactions.30 Ultra-high molecular weight (UHMW) polymers (Mn > 106 Da) are industrially relevant and highly desirable materials due to their extensive chain entanglement which endows increased tensile strength, environmental resistance, and adhesive properties.39–44 However, the drastic increase in viscosity that accompanies high chain growth makes the synthesis of UHMW polymers exceptionally challenging, necessitating a large excess of solvent, extended reaction times, or complex and energy-intensive conditions.42–47
In this work, we expand the utility of mechanoredox RAFT (MR-RAFT) by exploring the limits of triblock copolymer synthesis, including examples comprised of mutually immiscible monomers, and accessing UHMW polymers up to 3.2 MDa in a single milling vessel (Fig. 1B). These feats are enabled in part by liquid-assisted grinding (LAG), where a nominal amount of solvent (6% v/w, relative to the mass of reactants) is added to the milling jar to act as a mild lubricant and facilitate productive contact between reagents.48–50 Although its precise role is variable and currently not well understood, we suggest that in our cases the use of a LAG solvent (DMF) may (1) improve reagent compatibility to support the combination of immiscible monomers, (2) marginally temper the viscosity of growing high molar mass weight chains in localized “solution-like” regions, and (3) act as a plasticizer to depress the polymer's Tg and enhance collisional forces from the milling media.48,51,52 Independent of the mechanism, the reduced amount of solvent and energy required to perform MR-RAFT polymerizations make it a sustainable method that we demonstrate herein to have great promise for obtaining novel and synthetically challenging materials.
Results and discussion
To synthesize the polymers presented herein (Fig. 1B), tetragonal barium titanate (tet-BaTiO3, 15 wt%) piezoelectric nanoparticles, diphenyl iodonium hexafluorophosphate (radical initiator), and the chain transfer agent (CTA) 2-(dodecylthiocarbonothioylthio)-2-methylpropionic acid (DDMAT) were first added to a 1.5 or 5 mL stainless steel milling jar with one 5 or 8 mm stainless steel milling ball, respectively (Table S1). Under nitrogen, anhydrous DMF (6% v/w) was added for liquid-assisted grinding (LAG) followed by an acrylate monomer that immediately prior was filtered through basic alumina to remove its inhibitor. The milling jar was sealed under an inert atmosphere, transferred to the ball mill and shaken at 30 Hz for ≥3 hours. Subsequent blocks were added under nitrogen in a similar fashion; detailed experimental procedures can be found in the SI. These materials were characterized by gel permeation chromatography with multi-angle light scattering (GPC-MALS), and the monomer conversion was determined by 1H NMR spectroscopy.Utilizing MR-RAFT, we initially synthesized triblock copolymers with ABA sequences, targeting blocks lengths with degrees of polymerization (DPs) from 100 to 500. Our initial efforts demonstrate the modular growth of MR-RAFT polymers that we attribute to the high chain-end fidelity and high monomer conversion (as measured by 1H NMR spectroscopy) previously observed in our earlier MR-RAFT work.29 For instance, in an emblematic ABA copolymer of tert-butyl acrylate (tBA) and methyl acrylate (MA), referred to as poly(tBA-b-MA-b-tBA), the GPC traces (Fig. 2) following the first block (B1) show concomitant decreases in retention time, indicating modular attachment of blocks 2 and 3 (B2 and B3). High chain-end fidelity is supported by these consecutive monomodal GPC traces.
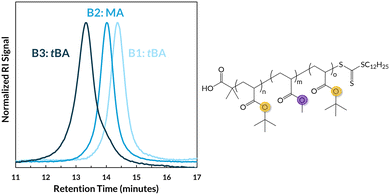 | ||
| Fig. 2 A representative GPC trace of an ABA triblock copolymer, poly(tBA-b-MA-b-tBA), and its structure (Table 1, entry 1). | ||
In our workflow, the Mn of B1 was first determined by GPC-MALS (e.g., 21 kDa, Table 1, entry 1). Then, we elected to use 1H NMR spectroscopy as a practical method to characterize the subsequent chain-extended polymers. For example, with an ABA triblock copolymer, we first normalized diagnostic 1H NMR signals from monomer B (B2) to the known total of monomer A protons (B1) as initially assessed by GPC-MALS. Then, after the addition of the final block (B3), the total mass fraction of monomer A can be referenced back to mass fraction of monomer B; subtracting the known amount of monomer A from B1 (determined by GPC-MALS) can ultimately yield the Mn of B3 (see eqn (S1)–(S4)). This convenient analytical approach allowed us to determine incremental molar masses without needing an accurate dn/dc value for each chain-extended sample. While we recognize this strategy may introduce error into molar mass measurements, the potential for solution-state self-assembly given the diversity of monomers utilized dissuaded us from pursuing conventional GPC column calibration techniques. In this specific example, B2 is shown to have an Mn of 11 kDa and B3 an Mn of 23 kDa (Table 1, entry 1), giving the final triblock copolymer a final molar mass of 55 kDa.
| Entry | A/B/A monomer sequence | Conversion (%) | Mn, MALS B1 (kDa) [DP] | Mn, NMR B2 (kDa) [DP] | Mn, NMR B3 (kDa) [DP] | Đ |
|---|---|---|---|---|---|---|
| [Target DPs] | (B1/B2/B3) | (B1/B2/B3) | ||||
| 1 | tBA/MA/tBA | >95/>95 />95 | 21 [162] | 11 [125] | 21 [167] | <1.1/<1.1/<1.1 |
| [200/200/200] | ||||||
| 2 | 2-MEA/HFBA/2-MEA | 88/>95/95 | 15 [120] | 51 [199] | 17 [127] | <1.1/<1.1/1.2 |
| [100/200/100] | ||||||
| 3 | tBA/HFBA/tBA | >95/>95/>95 | 10 [80] | 22 [88] | 8.2 [64] | <1.1/<1.1/1.3 |
| [100/100/100] |
Differential scanning calorimetry (DSC) further confirmed the presence of two distinct monomer blocks in the final ABA triblock copolymer; two glass transitions were observed, with Tg values of 14 °C and 38 °C corresponding to the MA and tBA blocks, respectively (Fig. S31). Diffusion-ordered spectroscopy (DOSY) was performed on select materials (Fig. S32–S34), likewise confirming the successful addition of new blocks by the presence of a single macromolecular species in each case. Table 1, entry 2 highlights the unique combination of two immiscible monomers, 2-methoxyethyl acrylate (2-MEA) and 2,2,3,3,4,4,4-heptafluorobutyl acrylate (HFBA), into an ABA triblock copolymer. Importantly, high monomer conversions (>86%, Fig. S2–S4) are observed for B1 and B2 in these examples such that chain extension essentially occurs with only the newly added monomer to make a uniform block, rather than random addition of new monomer with unreacted B1 or B2 monomer. Moreover, the trace amount of solvent used enables essentially complete mass recovery from the milling vessels with full conversion.
It is worth noting the potential effects of variable headspace volume in the milling jars upon each addition of liquid monomer, as well as between different trials which may be run at slightly different scales. Although no oxygen is present inside the milling jar, the volume occupied by the reagents may impact the magnitude of the forces applied based on the free volume the milling media is able to travel through. A recent report from Batteas and Felts quantified the collisional forces applied by a vibratory ball mill based on, among other variables, the milling jar's fill ratio; they observed a gradual decrease in both the forces applied and reaction conversion as the fill ratio was increased.53 We propose similar phenomena would be observed with MR-RAFT in our 1.5 and 5 mL milling jars to effect conversion. A specific example highlighting the impact of fill ratio is briefly described in our UHMW studies (vide infra).
We next carried out similar MR-RAFT syntheses targeting ABC triblock copolymers (Fig. 3) to explore additional structural complexity. While Kim demonstrated the use of mechanochemical radical polymerizations to access random copolymers from chemically disparate monomers,33 the use of controlled radical techniques with three separate monomers remained unrealized prior to this work. We again utilized GPC-MALS to measure Mn of B1, establishing an internal DP standard for subsequent analytical measurements using 1H NMR spectroscopy (vide supra). The utility of MR-RAFT was showcased by the inclusion of a fluorinated comonomer at different positions in the ABC sequence where we indeed retain control with each chain extension as evidenced by monomodal GPC traces (Fig. S18 and S19). Again, high conversion was observed for each block with the exception of 2-MEA in poly(tBA-b-HFBA-b-2-MEA) (Table 2, entry 1, B3) which exhibited only 37% conversion; lower conversion may be responsible for a shorter B3 than expected (i.e., DP = 23 with target DP = 50). Nonetheless, this issue does not affect the blocky nature of the material; its DOSY spectrum (Fig. S33) shows a single macromolecular species with resonances corresponding to all three monomers with similar diffusion coefficients.
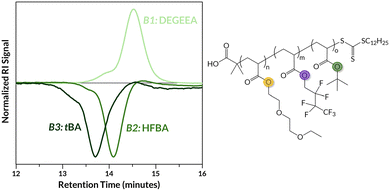 | ||
| Fig. 3 A representative GPC trace of an ABC triblock copolymer, poly(DEGEEA-b-HFBA-b-tBA), and its structure (Table 2, entry 3). | ||
| Entry | A/B/C monomer sequence | Conversion (%) | Mn, MALS B1 (kDa) [DP] | Mn, NMR B2 (kDa) [DP] | Mn, NMR B3 (kDa) [DP] | Đ |
|---|---|---|---|---|---|---|
| [Target DPs] | (B1/B2/B3) | (B1/B2/B3) | ||||
| 1 | tBA/HFBA/2-MEA | >95/>95/37 | 13 [104] | 15 [60] | 3.1 [23] | <1.1/<1.1/<1.1 |
| [100/50/50] | ||||||
| 2 | tBA/DEGEEA/HFBA | >95/>95/87 | 23 [179] | 23 [121] | 28 [107] | <1.1/<1.1/1.2 |
| [200/100/100] | ||||||
| 3 | DEGEEA/HFBA/tBA | >95/86 />95 | 19 [100] | 24 [94] | 17 [133] | <1.1/<1.1/<1.1 |
| [100/100/100] |
While accessing semi-fluorinated acrylate copolymers often requires a fluorinated reaction component (e.g., solvent, catalyst, CTA),54–59 recent work from Chen demonstrates that judicious tuning of the CTA in photo-induced RAFT affords excellent control over molar mass, dispersity, and sequence without these exogenous fluorinated moieties.60 Likewise, MR-RAFT does not require additional fluorinated components to attain semi-fluorinated diblock copolymers. Even when three mutually immiscible monomers are utilized, exhaustive solvent optimization is not required; a small volume of DMF as a LAG solvent is sufficient to effect controlled polymer growth.61
To further emphasize the difference between MR-RAFT and traditional solution-state analogs, we attempted to synthesize poly(tBA-b-DEGEEA-b-HFBA) (Table 2, entry 2) through conventional thermal RAFT in DMF (see SI for Experimental details). Although near complete monomer conversion was observed after 24 h for each block (Fig. S8), no clear shifts in retention time were present in the corresponding GPC traces (Fig. S20), which also exhibited broad, ill-defined shoulders, indicating the presence of multiple macromolecular species. Importantly, the shoulder observed in the final ABC GPC trace exhibits a broad negative RI signal which is typical of fluorinated polymers in chloroform, suggesting some amount of HFBA homopolymerization. Therefore, the vastly different GPC traces measured here suggest that classical solvothermal RAFT is incapable of accessing block copolymers such as those in Table 2 from immiscible monomer feedstocks, as previously noted by Hawker56 and Chen,60 further demonstrating the potential of MR-RAFT to facilitate material discovery.
To explore potential self-assembly of the ABC triblock copolymers an emblematic sample, poly(DEGEEA-b-HFBA-b-tBA) (Fig. 3 and Table 2, entry 3), was analyzed using small-angle X-ray scattering (SAXS) (Fig. 4). A sharp principal peak at q = 0.011 Å−1 (d = 2π/q = 57 nm) was observed, along with 7 higher order integer reflections (2q, 3q, 4q, 5q, 7q, 8q, 9q) indicative of a lamellar morphology (Table S3). Interestingly, the second reflection has a higher intensity than that of the first, which has been observed in other ABC triblock copolymers.62 One possible mechanism behind this behavior is the influence of form factor minimums coinciding with structure factor maximums.63 The graded composition between the blocks may also play a role, resulting in electron densities that deviate from step function harmonics.64
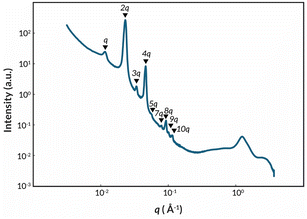 | ||
| Fig. 4 SAXS data of a representative ABC triblock copolymer, poly(DEGEEA-b-HFBA-b-tBA) (Table 2, entry 3), showing a lamellar architecture. d = 57 nm. | ||
Having demonstrated the ability to overcome miscibility restraints using MR-RAFT, we then strove to push the limits of obtainable molar masses towards ultra-high molecular weight (UHMW) polymers. Balancing the rates of termination and propagation, such that the former is much lower, while maintaining chain end fidelity is crucial for obtaining UHMW materials. The high bulk viscosity of such large polymers can limit chain end diffusion, induce phase separation, and give rise to the Trommsdorff effect through uncontrolled propagation.42,45,65,66 Early examples from Sahoo,67 Matyjaszewski,68 and Rzayev69 employed methods based on atom-transfer radical polymerization (ATRP) to access UHMW materials, although scaling solution-state ATRP processes is challenging. While capable of exceeding 9 MDa, a more recent method developed by Matyjaszewski and Maksym that enhances photo-ATRP with high pressure is quite energy-intensive (e.g., 530 nm light and 225 MPa of pressure maintained over 48 h).70 Sumerlin has recently reported on photoiniferter mini-emulsion RAFT polymerizations to overcome viscosity limitations, whereby viscous polymerization events are isolated in aqueous droplets dispersed within a bulk cyclohexane solution, enabling access to UHMW polymers in excess of 1.9 MDa.43,44 Similar strategies allow access to UHMW homopolymers and block copolymers in excess of 5.0 MDa through light-mediated reversible-deactivation radical polymerizations (RDRP), taking advantage of solvophobic water droplets or self-assembled polymer nanoparticles, among other strategies, to maintain low viscosity solutions.43,44,47,71–74 However, in these heterogeneous reaction mixtures, the need for specific surfactants, water-soluble monomers, and/or specialized reactors for maximal light penetration may hinder the scalability and compatibility of such systems.
With these precedents in mind, we surmised that ball mill grinding at high frequencies (i.e., 30 Hz) would sufficiently mix the reaction components and ensure reversible termination remains operative as viscosity increases, thereby mitigating the Trommsdorff effect. To test our hypothesis, we implemented MR-RAFT with significantly increased [M0]/[CTA] (≥18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) for various acrylate monomers and observed moderate to high monomer conversion (43–95%) by 1H NMR spectroscopy (Table 3, entries 1–5, and Fig. S9–S13) over 3–6 h of milling time; these are notably shorter reactions than many complementary methods of synthesizing UHMW polymers.40,42,68,70,73,75,76 The variability in conversion may be attributable to the monomer-dependent change in viscosity with increasing molar mass, creating substantially different force environments that are challenging to predict. GPC traces of these homopolymers reveal retention times approaching the exclusion limit of our columns (Fig. 5), with Mw, MALS = 1.1–3.2 MDa. We elect to report Mw for our UHMW polymers since, unlike Mn, it does not rely on effective column separation.77 Likewise, we cannot reliably determine dispersity for these samples, nor the true experimental DP which is dependent on Mn. In targeting such high molar masses, we also observed limited solubility of certain polymers in our CHCl3 GPC eluent, including poly(DEGEEA), poly(HFBA), poly(2-MEA), and poly(2-hydroxyethyl acrylate), although we suppose MR-RAFT may still be capable of bringing these polymers to the UHMW regime.
000) for various acrylate monomers and observed moderate to high monomer conversion (43–95%) by 1H NMR spectroscopy (Table 3, entries 1–5, and Fig. S9–S13) over 3–6 h of milling time; these are notably shorter reactions than many complementary methods of synthesizing UHMW polymers.40,42,68,70,73,75,76 The variability in conversion may be attributable to the monomer-dependent change in viscosity with increasing molar mass, creating substantially different force environments that are challenging to predict. GPC traces of these homopolymers reveal retention times approaching the exclusion limit of our columns (Fig. 5), with Mw, MALS = 1.1–3.2 MDa. We elect to report Mw for our UHMW polymers since, unlike Mn, it does not rely on effective column separation.77 Likewise, we cannot reliably determine dispersity for these samples, nor the true experimental DP which is dependent on Mn. In targeting such high molar masses, we also observed limited solubility of certain polymers in our CHCl3 GPC eluent, including poly(DEGEEA), poly(HFBA), poly(2-MEA), and poly(2-hydroxyethyl acrylate), although we suppose MR-RAFT may still be capable of bringing these polymers to the UHMW regime.
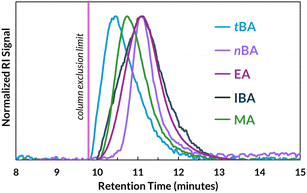 | ||
| Fig. 5 GPC traces of UHMW acrylate homopolymers (Table 3, entries 1–5). IBA = isobornyl acrylate. | ||
| Entry | Monomer | [M0]/[CTA] | Milling Time (h) | Conversion (%) | Mn, theo (MDa) | Mw, MALS B1 (MDa) | Mn, NMR B2 (MDa) |
|---|---|---|---|---|---|---|---|
| [est. DP]a | [DP] | ||||||
| a Experimental DP is properly calculated from Mn, but since we report Mw due to its independence from column separation, we can only approximate DP based on Mw.b For the UHMW diblock copolymers, Mn is reported for the first block since it is far enough removed from the exclusion limit of our columns to afford good separation. Therefore, the DPs reported for entries 6 and 7 are the true experimental DPs. | |||||||
| 1 | tBA | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
3 | 95 | 2.3 | 2.0 | — |
[16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000] 000] |
|||||||
| 2 | nBA | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
6 | 43 | 2.6 | 1.2 | — |
[9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100] 100] |
|||||||
| 3 | EA | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
6 | 77 | 2.0 | 1.1 | — |
[11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000] 000] |
|||||||
| 4 | IBA | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
6 | 54 | 4.2 | 3.2 | — |
[12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000] 000] |
|||||||
| 5 | MA | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
6 | 68 | 3.0 | 1.4 | — |
[16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000] 000] |
|||||||
| 6 | tBA/2-MEA | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000/5 000/5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
3/6 | 85/92 | 1.9/0.65 | 0.63b | 0.55 |
[4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900] 900] |
[4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200] 200] |
||||||
| 7 | tBA/EA | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000/10 000/10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
3/6 | >95/87 | 1.3/1.0 | 0.60b | 0.71 |
[4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700] 700] |
[7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100] 100] |
||||||
To probe the effect of fill ratio (vide supra) on the synthesis of these UHMW species, we prepared two 5 mL milling jars with analogous reactions at different scales (see Table S2), targeting a DP of 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for methyl acrylate (MA). Surprisingly, the jar with the higher fill ratio (0.60) yielded a larger poly(MA) (1.4 MDa, Table 3, entry 5) than the jar with the lower fill ratio (0.49, 0.83 MDa). This unusual finding points towards the markedly different and often unpredictable force environments experienced inside the milling jars even with slight changes in the polymerization scale. Further investigation into this phenomenon is ongoing.
000 for methyl acrylate (MA). Surprisingly, the jar with the higher fill ratio (0.60) yielded a larger poly(MA) (1.4 MDa, Table 3, entry 5) than the jar with the lower fill ratio (0.49, 0.83 MDa). This unusual finding points towards the markedly different and often unpredictable force environments experienced inside the milling jars even with slight changes in the polymerization scale. Further investigation into this phenomenon is ongoing.
Importantly, we also demonstrate chain end fidelity is retained at high molar masses; a 0.63 MDa poly(tBA) was further extended through MR-RAFT with a second monomer, 2-methoxyethyl acrylate (2-MEA), to afford an UHMW diblock copolymer with a final Mn of 1.16 MDa (Table 3, entry 6, and Fig. S21). A similar result was obtained with poly(tBA-b-EA) (Table 3, entry 7, and Fig. S22) that surpassed 1.3 MDa.
Ball mill grinding has been used extensively to depolymerize and degrade materials, yet counterintuitively, we are able to access UHMW polymers mechanochemically which should ostensibly invoke chain cleavage.19,52,78 Our findings may be a result of the increasing viscosity that coincides with chain growth, which would dampen the forces applied by the milling media to an extent that mitigates backbone cleavage. Mark–Houwink–Sakurada plots of the UHMW homopolymers reported (Fig. S24–S28) all show slopes consistent with non-branched materials (>0.5), suggesting homolytic backbone cleavage is not occurring.79,80
Conclusion
In conclusion, we have demonstrated the broader applications of MR-RAFT to access novel block copolymer formulations and ultra-high molecular weight materials. The chain extension of diblock macro-CTAs into triblock copolymers with ABA or ABC architectures was successful using a range of acrylate monomers. A notable example combining immiscible monomers, poly(DEGEEA-b-HFBA-b-tBA), self assembles into well-defined lamellae as assessed by small angle X-ray scattering. We also pushed the limits of molar masses obtainable through MR-RAFT, accessing homopolymers up to 3.2 MDa and diblock copolymers over 1.3 MDa. The implementation of liquid-assisted grinding with minimal DMF enabled these complementary processes, which represent a key step towards sustainable polymer synthesis. We envision MR-RAFT facilitating rapid material discovery due to its broad monomer compatibility, essentially complete material recovery, and relatively short reaction times. With mechanochemistry as a growing industrial interest, MR-RAFT shows potential for large-scale, green polymerizations.Author contributions
J. D. K., M. N. H., S. M. Z., and M. R. G. conceived of the ideas. M. N. H. and J. D. K. conducted synthetic experiments and analyzed polymers in solution. A. M. and L. D. P. conducted SAXS experiments, processed the data, and analyzed the results. J. D. K., M. N. H. and M. R. G. wrote the manuscript. All authors discussed and edited the manuscript. J. D. K. designed and drew the Table of Contents image.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article (experimental procedures, NMR spectra, GPC traces, sample calculations) have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5py01061f.Acknowledgements
This work was supported by generous start-up funds from the University of Washington. M. N. H. acknowledges the University of Washington Clean Energy Institute for a graduate research fellowship. The authors also acknowledge the use of facilities and instrumentation supported by the NSF through the MRI program (DMR-211-6265) and the UW Molecular Engineering Materials Center (MEM-C), an NSF MRSEC (DMR-2308979). This material is based in part upon work supported by the state of Washington through the University of Washington Clean Energy Institute. NMR spectroscopy resources are supported under NIH S10 OD030224-01A1. The authors would also like to thank Nicholas P. Serck for his assistance in collecting and processing the DOSY spectra.References
- R. Geyer, J. R. Jambeck and K. L. Law, Sci. Adv., 2017, 3, 25–29 CrossRef PubMed.
- S. B. Borrelle, J. Ringma, K. L. Law, C. C. Monnahan, L. Lebreton, A. McGivern, E. Murphy, J. Jambeck, G. H. Leonard, M. A. Hilleary, M. Eriksen, H. P. Possingham, H. De Frond, L. R. Gerber, B. Polidoro, A. Tahir, M. Bernard, N. Mallos, M. Barnes and C. M. Rochman, Science, 2020, 369, 1515–1518 CrossRef CAS PubMed.
- D. Feldman, Des. Monomers Polym., 2008, 11, 1–15 CrossRef CAS.
- D. M. Evans, R. Parsons, P. Jackson, S. Greenwood and A. Ryan, Global Environ. Change, 2020, 65, 102166 CrossRef.
- A. Ragusa, A. Svelato, C. Santacroce, P. Catalano, V. Notarstefano, O. Carnevali, F. Papa, M. C. A. Rongioletti, F. Baiocco, S. Draghi, E. D'Amore, D. Rinaldo, M. Matta and E. Giorgini, Environ. Int., 2021, 146, 106274 CrossRef CAS PubMed.
- C. Fuschi, H. Pu, M. MacDonell, K. Picel, M. Negri and J. Chen, ACS Sustainable Chem. Eng., 2022, 10, 14074–14091 CrossRef CAS.
- H. Brunn, G. Arnold, W. Körner, G. Rippen, K. G. Steinhäuser and I. Valentin, Environ. Sci. Eur., 2023, 35, 20 CrossRef CAS.
- B. Bahramian, A. Fathi and F. Dehghani, Polym. Degrad. Stab., 2016, 133, 174–181 CrossRef CAS.
- O. Guselnikova, O. Semyonov, E. Sviridova, R. Gulyaev, A. Gorbunova, D. Kogolev, A. Trelin, Y. Yamauchi, R. Boukherroub and P. Postnikov, Chem. Soc. Rev., 2023, 52, 4755–4832 RSC.
- A. A. Garforth, S. Ali, J. Hernández-Martínez and A. Akah, Curr. Opin. Solid State Mater. Sci., 2004, 8, 419–425 CrossRef CAS.
- M. Okan, H. M. Aydin and M. Barsbay, J. Chem. Technol. Biotechnol., 2019, 94, 8–21 CrossRef CAS.
- X.-H. Wang, Chin. J. Polym. Sci., 2022, 40, 431–432 CrossRef CAS.
- L. T. J. Korley, T. H. Epps, B. A. Helms and A. J. Ryan, Science, 2021, 373, 66–69 CrossRef CAS PubMed.
- F. Kurul, B. Doruk and S. N. Topkaya, Discover Chem., 2025, 2, 68 CrossRef.
- P. Anastas and N. Eghbali, Chem. Soc. Rev., 2010, 39, 301–312 RSC.
- K. J. Ardila-Fierro and J. G. Hernández, ChemSusChem, 2021, 14, 2145–2162 CrossRef CAS PubMed.
- H. Staudinger and W. Heuer, Ber. Dtsch. Chem. Ges., 1934, 67, 1159–1164 CrossRef.
- J. Zhou, T. Hsu and J. Wang, Angew. Chem., Int. Ed., 2023, 62, e202300768 CrossRef CAS PubMed.
- S. Aydonat, A. H. Hergesell, C. L. Seitzinger, R. Lennarz, G. Chang, C. Sievers, J. Meisner, I. Vollmer and R. Göstl, Polym. J., 2024, 56, 249–268 CrossRef CAS.
- X. Zhang, C. Lu and M. Liang, J. Polym. Res., 2009, 16, 411–419 CrossRef CAS.
- J. Ghosh, S. Ghorai, S. Bhunia, M. Roy and D. De, Polym. Eng. Sci., 2018, 58, 74–85 CrossRef CAS.
- S. Zhang, L. Han, H. Bai, C. Li, X. Wang, Z. Yang, M. Zai, H. Ma and Y. Li, ACS Sustainable Chem. Eng., 2021, 9, 8053–8058 CrossRef CAS.
- V. Canale, M. Kamiński, W. Trybała, M. Abram, K. Marciniec, X. Bantreil, F. Lamaty, J. R. Parkitna and P. Zajdel, ACS Sustainable Chem. Eng., 2023, 11, 16156–16164 CrossRef CAS.
- H. Mohapatra, M. Kleiman and A. P. Esser-Kahn, Nat. Chem., 2017, 9, 135–139 CrossRef CAS.
- Z. Wang, Z. Wang, X. Pan, L. Fu, S. Lathwal, M. Olszewski, J. Yan, A. E. Enciso, Z. Wang, H. Xia and K. Matyjaszewski, ACS Macro Lett., 2018, 7, 275–280 CrossRef CAS PubMed.
- H. Y. Cho and C. W. Bielawski, Angew. Chem., Int. Ed., 2020, 59, 13929–13935 CrossRef CAS PubMed.
- M. Zhou, Y. Zhang, G. Shi, Y. He, Z. Cui, X. Zhang, P. Fu, M. Liu, X. Qiao and X. Pang, ACS Macro Lett., 2023, 12, 26–32 CrossRef CAS PubMed.
- M. D. Nothling, J. E. Daniels, Y. Vo, I. Johan and M. H. Stenzel, Angew. Chem., Int. Ed., 2023, 62(20), e202218955 CrossRef CAS PubMed.
- P. Chakma, S. M. Zeitler, F. Baum, J. Yu, W. Shindy, L. D. Pozzo and M. R. Golder, Angew. Chem., Int. Ed., 2023, 62, e202215733 CrossRef CAS PubMed.
- S. M. Zeitler, P. Chakma and M. R. Golder, Chem. Sci., 2022, 13, 4131–4138 RSC.
- O. Okay, Polymer, 1999, 40, 4117–4129 CrossRef CAS.
- C. P. Hsu and L. J. Lee, Polymer, 1993, 34, 4516–4523 CrossRef CAS.
- E. J. Kim, J. J. Shin, G. S. Lee, S. Kim, S. Park, J. Park, Y. Choe, D. Lee, J. Choi, J. Bang, Y. H. Kim, S. Li, S.-M. Hur, J. G. Kim and B. J. Kim, Macromolecules, 2022, 55, 1590–1599 CrossRef CAS.
- W. Zhang, M. Huang, S. al Abdullatif, M. Chen, Y. Shao-Horn and J. A. Johnson, Macromolecules, 2018, 51, 6757–6763 CrossRef CAS.
- K. Ponnusamy, R. P. Babu and R. Dhamodharan, J. Polym. Sci., Part A: Polym. Chem., 2013, 51, 1066–1078 CrossRef CAS.
- G. S. Lee, B. R. Moon, H. Jeong, J. Shin and J. G. Kim, Polym. Chem., 2019, 10, 539–545 RSC.
- G. S. Lee, H. S. Lee, N. Kim, H. G. Shin, Y. H. Hwang, S. J. Lee and J. G. Kim, Macromolecules, 2024, 57, 9408–9418 CrossRef CAS.
- A. Sarkar, R. Sasmal, A. Das, A. Venugopal, S. S. Agasti and S. J. George, Angew. Chem., Int. Ed., 2021, 60, 18209–18216 CrossRef CAS PubMed.
- J. D. Marquez, K. A. Stewart, K. C. Stevens, B. J. Ryder, T. H. Le, N. B. Wei, W. J. Choi, Y. Huang, B. S. Sumerlin and A. M. Evans, J. Am. Chem. Soc., 2025, 147, 29223–29231 CrossRef CAS PubMed.
- C. Hwang, S. Shin, D. Ahn, H. J. Paik, W. Lee and Y. Yu, ACS Appl. Mater. Interfaces, 2023, 15, 58905–58916 CrossRef CAS PubMed.
- Z. An, ACS Macro Lett., 2020, 9, 350–357 CrossRef CAS PubMed.
- S. Zhu, W. Kong, S. Lian, A. Shen, S. P. Armes and Z. An, Nat. Synth., 2025, 4, 15–30 CrossRef CAS.
- C. L. G. Davidson, M. E. Lott, L. Trachsel, A. J. Wong, R. A. Olson, D. I. Pedro, W. G. Sawyer and B. S. Sumerlin, ACS Macro Lett., 2023, 12, 1224–1230 CrossRef PubMed.
- R. A. Olson, M. E. Lott, J. B. Garrison, C. L. G. Davidson, L. Trachsel, D. I. Pedro, W. G. Sawyer and B. S. Sumerlin, Macromolecules, 2022, 55, 8451–8460 CrossRef CAS.
- J. M. Nölle, S. Primpke, K. Müllen, P. Vana and D. Wöll, Polym. Chem., 2016, 7, 4100–4105 RSC.
- N. P. Truong, M. V. Dussert, M. R. Whittaker, J. F. Quinn and T. P. Davis, Polym. Chem., 2015, 6, 3865–3874 RSC.
- L. E. Diodati, A. J. Wong, M. E. Lott, A. G. Carter and B. S. Sumerlin, ACS Appl. Polym. Mater., 2023, 5, 9714–9720 CrossRef CAS.
- M. E. Skala, S. M. Zeitler and M. R. Golder, Chem. Sci., 2024, 15, 10900–10907 RSC.
- I. D’Abbrunzo and D. Hasa, CrystEngComm, 2026 10.1039/D5CE00778J.
- P. Ying, J. Yu and W. Su, Adv. Synth. Catal., 2021, 363, 1246–1271 CrossRef CAS.
- S. A. Salami, M. H. Manyeruke, C. I. Ezekiel, U. N. Ndagano, J. B. Safari, S. O. Amusat and R. W. M. Krause, Res. Chem., 2025, 18, 102675 CAS.
- G. I. Peterson, W. Ko, Y. J. Hwang and T. L. Choi, Macromolecules, 2020, 53, 7795–7802 CrossRef CAS.
- E. Nwoye, K. Floyd, J. Batteas and J. Felts, RSC Mechanochem., 2025, 2, 911–922 RSC.
- H. Gong, Y. Zhao, X. Shen, J. Lin and M. Chen, Angew. Chem., Int. Ed., 2018, 57, 333–337 CrossRef CAS PubMed.
- H. Gong, Y. Gu and M. Chen, Synlett, 2018, 1543–1551 CAS.
- E. H. Discekici, A. Anastasaki, R. Kaminker, J. Willenbacher, N. P. Truong, C. Fleischmann, B. Oschmann, D. J. Lunn, J. R. De Alaniz, T. P. Davis, C. M. Bates and C. J. Hawker, J. Am. Chem. Soc., 2017, 139, 5939–5945 CrossRef CAS PubMed.
- N. G. Taylor, S. H. Chung, A. L. Kwansa, R. R. Johnson, A. J. Teator, N. J. B. Milliken, K. M. Koshlap, Y. G. Yingling, Y. Z. Lee and F. A. Leibfarth, Chem. – Eur. J., 2020, 26, 9982–9990 CrossRef CAS PubMed.
- Q. Quan, H. Wen, S. Han, Z. Wang, Z. Shao and M. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 24319–24327 CrossRef CAS PubMed.
- Y. Zhao, M. Ma, X. Lin and M. Chen, Angew. Chem., Int. Ed., 2020, 59, 21470–21474 CrossRef CAS PubMed.
- Q. Quan, H. Gong and M. Chen, Polym. Chem., 2018, 9, 4161–4171 RSC.
- G. A. Bowmaker, Chem. Commun., 2013, 49, 334–348 RSC.
- Y. Tanaka, H. Hasegawa, T. Hashimoto, A. Ribbe, K. Sugiyama, A. Hirao and S. Nakahama, Polym. J., 1999, 31, 989–994 CrossRef CAS.
- S. Mani, R. A. Weiss, S. F. Hahn, C. E. Williams, M. E. Cantino and L. H. Khairallah, Polymer, 1998, 39, 2023–2033 CrossRef CAS.
- T. S. Bailey, C. M. Hardy, T. H. Epps and F. S. Bates, Macromolecules, 2002, 35, 7007–7017 CrossRef CAS.
- T. J. Tulig and M. Tirrell, Macromolecules, 1982, 15, 459–463 CrossRef CAS.
- Y. Suzuki, D. S. Cousins, Y. Shinagawa, R. T. Bell, A. Matsumoto and A. P. Stebner, Polym. J., 2019, 51, 423–431 CrossRef CAS.
- V. Percec, T. Guliashvili, J. S. Ladislaw, A. Wistrand, A. Stjerndahl, M. J. Sienkowska, M. J. Monteiro and S. Sahoo, J. Am. Chem. Soc., 2006, 128, 14156–14165 CrossRef CAS PubMed.
- R. Nicolaÿ, Y. Kwak and K. Matyjaszewski, Angew. Chem., Int. Ed., 2010, 49, 541–544 CrossRef PubMed.
- J. K. D. Mapas, T. Thomay, A. N. Cartwright, J. Ilavsky and J. Rzayev, Macromolecules, 2016, 49, 3733–3738 CrossRef CAS.
- R. Bernat, G. Szczepaniak, K. Kamiński, M. Paluch, K. Matyjaszewski and P. Maksym, Chem. Commun., 2023, 60, 843–846 RSC.
- C. B. Eades, K. C. Stevens, D. E. Cabrera, M. K. Vereb, M. E. Lott, J. I. Bowman and B. S. Sumerlin, Chem. Sci., 2025, 16, 5573–5578 RSC.
- M. E. Lott, L. Trachsel, E. Schué, C. L. G. Davidson, R. A. Olson S, D. I. Pedro, F. Chang, Y. Hong, W. G. Sawyer and B. S. Sumerlin, Macromolecules, 2024, 57, 4007–4015 CrossRef CAS.
- R. N. Carmean, M. B. Sims, C. A. Figg, P. J. Hurst, J. P. Patterson and B. S. Sumerlin, ACS Macro Lett., 2020, 9, 613–618 CrossRef CAS PubMed.
- R. N. Carmean, T. E. Becker, M. B. Sims and B. S. Sumerlin, Chem, 2017, 2, 93–101 CAS.
- Y. Bai, J. He and Y. Zhang, Angew. Chem., Int. Ed., 2018, 57, 17230–17234 CrossRef CAS PubMed.
- C. Zhou, Z. Zhang, W. Li and M. Chen, Angew. Chem., Int. Ed., 2024, 63(2), e202314483 CrossRef CAS PubMed.
- J. B. Matson, A. Q. Steele, M. D. Schulz and J. D. Mase, Polym. Chem., 2024, 15, 127–142 RSC.
- A. Briš, D. Margetić and V. Štrukil, Green Chem., 2025, 27, 14401–14435 RSC.
- N. T. Hoai, A. Sasaki, M. Sasaki, H. Kaga, T. Kakuchi and T. Satoh, Biomacromolecules, 2011, 12, 1891–1899 CrossRef CAS PubMed.
- S. Podzimek and T. Vlcek, J. Appl. Polym. Sci., 2001, 82, 454–460 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work |
| This journal is © The Royal Society of Chemistry 2026 |
