 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Assessing inorganic nanoparticle toxicity through omics approaches†
Yanchen
Li
a,
Christopher
Vulpe
 b,
Twan
Lammers
b,
Twan
Lammers
 a and
Roger M.
Pallares
a and
Roger M.
Pallares
 *a
*a
aInstitute for Experimental Molecular Imaging, RWTH Aachen University Hospital, Aachen 52074, Germany. E-mail: rmoltopallar@ukaachen.de
bCenter for Environmental and Human Toxicology, Department of Physiological Sciences, College of Veterinary Medicine, University of Florida, Gainesville, FL 32611, USA
First published on 15th August 2024
Abstract
In the last two decades, the development of nanotechnology has resulted in inorganic nanoparticles playing crucial roles in key industries, ranging from healthcare to energy technologies. For instance, gold and silver nanoparticles are widely used in rapid COVID-19 and flu tests, titania and zinc oxide nanoparticles are commonly found in cosmetic products, and superparamagnetic iron oxide nanoparticles have been clinically exploited as contrast agents and anti-anemia medicines. As a result, human exposure to nanomaterials is continuously increasing, raising concerns about their potential adverse health effects. Historically, the study of nanoparticle toxicity has largely relied on macroscopic observations obtained in different in vitro and in vivo models, resulting in readouts such as median lethal dose, biodistribution profile, and/or histopathological assessment. In recent years, omics methodologies, including transcriptomics, epigenomics, proteomics, metabolomics, and lipidomics, are increasingly used to characterize the biological interactions of nanomaterials, providing a better and broader understanding of their impact and mechanisms of toxicity. These approaches have been able to identify important genes and gene products that mediate toxicological effects, as well as endogenous functions and pathways dysregulated by nanoparticles. Omics methods improve our understanding of nanoparticle biology, and unravel mechanistic insights into nanomedicine-based therapies. This review aims to provide a deeper understanding and new perspectives of omics approaches to characterize the toxicity and biological interactions of inorganic nanoparticles, and improve the safety of nanoparticle applications.
1. Introduction
In the last few decades, the rise of nanotechnology has sparked important transformations across various fields, including medicine, materials science, energy, and environmental sciences.1,2 Nanomaterials, with their unique physicochemical properties and large surface-to-volume ratios, exhibit significantly different biological activities and endogenous behaviors compared to bulk materials. These differences have led to the widespread application of nanomedicine in (pre)clinical settings, such as drug delivery, imaging, and diagnostics.3–5 While this extensive use of nanotechnology has benefited multiple fields, it has also increased human exposure to nanoparticles, raising concerns about potential adverse health effects.Epidemiological data have demonstrated the harmful consequences of airborne ultrafine particles on human health,6,7 a finding further corroborated by animal exposure studies.8 The toxicity of metal and metal oxide nanoparticles is particularly relevant to human health due to their extended residence time and minimal degradation and excretion upon internal exposure.9 Studies indicate that specific physicochemical properties of nanomaterials, such as particle size, shape, surface charge, chemical composition, and colloidal stability, play a crucial role in defining their nano-bio interactions and potential toxicity.10–12 Moreover, due to their unique characteristics and interactions within biological systems, nanoparticles can circulate in the bloodstream,13 and get deposited in multiple organs, including the liver, lungs, spleen, and kidneys, causing non-localized toxicity.14
Conventional in vitro testing predominantly employs cell-based assays with phenotypic readout parameters, focusing on membrane integrity, apoptosis, cell morphology, and proliferation.15 While these methods can provide an overview of toxicity, such as the determination of half-maximal inhibitory concentrations (IC50), they do not provide mechanistic insights into the effects of the nanoparticles or the responses of the organism to the exposures.14
As toxicological assessments advance, the application of omics technologies, such as genomics, transcriptomics, proteomics, and metabolomics (including lipidomics), have emerged as new approaches to gain mechanistic insights into complex interaction mechanisms within biological systems (Fig. 1).16–23 Research using these technologies has identified key targets and biomarkers associated with nanomaterial toxicity, such as those related to oxidative stress and inflammatory gene activation.24–26 Furthermore, in addition to providing mechanistic information, omics approaches are also less sensitive to nanoparticle interferences during the screening readout, which is a common issue with traditional toxicological in vitro assays due to the inherent characteristics of nanoparticles, which can display strong color, fluorescence, chemical activity, and light scattering.27
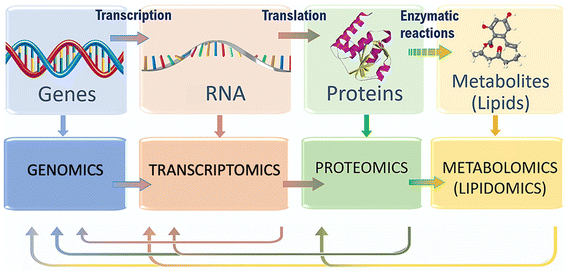 | ||
| Fig. 1 Flow of molecular information within biological systems, and the different omics techniques that can characterize it. Adapted with permission of ref. 20. Copyright Elsevier 2020. | ||
Taken together, understanding the potential toxicity of nanomaterials, and especially their impact on key organs, requires a comprehensive analysis of their biological activities and interactions. The use of advanced toxicity testing techniques, particularly omics-based approaches, facilitates a more comprehensive assessment of nanomaterial safety. This review article summarizes the various approaches, methods, advantages, and limitations of using omics approaches to assess the toxicity of key inorganic nanoparticles, to elucidate their toxicity mechanisms (or lack thereof) and to ensure their safe application in different fields.
2. Omics techniques
In toxicology, omics are used to interpret the response of an organism to an external substance by pooling biomolecules (e.g. DNA, RNA, proteins, and lipids). These profiling techniques are then used to elucidate the molecular mechanisms that result in toxicity. In this section, we provide an overview of the different omics approaches used in the field of toxicology, highlighting the information that is more relevant to the study of nanoparticle toxicity (Fig. S1†). We refer the reader to more specific literature for a more in-depth description of the distinct omics techniques available.282.1. Genomics and transcriptomics
Genomics and transcriptomics are disciplines that seek to understand the composition, structure, and function of genetic material, providing insights into gene expression patterns and regulatory mechanisms.29,30 The distinction lies in genomics focusing on the comprehensive study of an organism's genome, while transcriptomics specifically examines sets of RNA transcribed from the genome.14,29Transcriptomics, focused on genome-wide RNA levels, has evolved considerably with the development of approaches such as DNA microarrays, reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR), and next-generation sequencing (NGS) based RNA sequencing (RNA-seq).31,32 Transcriptomics can unveil the complex responses of organisms to environmental exposures by profiling gene expression.33 Moreover, transcriptomics also enables the examination of short non-coding RNAs, which have emerged as potential biomarkers or therapeutic targets due to their dysregulation in diseases.34–36 Genomics employs genotype arrays,37 exome sequencing,38 and NGS of genomic DNA, among others,39 to discern genetic factors linked to diseases, treatment responses, and future patient prognoses.40,41
Taken together, genomic and transcriptomic profiling can assess the impact of nanomaterial exposures on organisms by reveling changes in the genome and gene expression patterns, providing a comprehensive evaluation of adverse effects and molecular mechanisms associated with nanoparticle exposure and toxicity.
2.2. Epigenomics
Epigenomics investigates reversible modifications of the DNA or DNA-associated proteins, known as epigenetic modifications.42 These alterations, including DNA methylation and histone acetylation, serve as regulators of gene transcription (and, consequently, cellular fate), and are governed by a complex interplay of genetic and environmental factors.43 Furthermore, there is increasing evidence that suggests that the harmful effects of environmental stressors (including those with nanoparticle form), such as smoking, air pollution, and endocrine disruptors, are mediated via epigenetic mechanisms,44 even in prenatal exposures.45Epigenetic modifications can be identified with multiple technologies, including DNA methylation and histone modification assays, as well as DNA microarrays.46 As researchers delve into the intersection of epigenomics and nanotoxicology, they are able to understand to what extent nanoparticles (and other nanomaterials) can induced epigenetic modifications, contributing to a more nuanced understanding of the toxicological impact of nanomaterials at the epigenomic level.
2.3. Proteomics
Proteomics focuses on the comprehensive study of proteins within biological systems, including protein abundance, modifications, and interactions,47 which can be later used to discern the cytotoxic effects of a toxicant or toxin or the response of organisms to chemicals.48,49 Proteomics is primarily explored with mass spectrometry (MS) techniques, which can enable high-throughput analysis of thousands of proteins in complex mixtures.50 For screening of samples with a smaller number of proteins, antibody microarrays can also be used.51 Other proteomics methods include phage display52 and yeast two-hybrid assays53 which focus on identifying protein interactions. Understanding post-translational modifications is also a key focus of proteomics since they have key roles in signaling, enzyme control, turnover, and cellular structure.54 For instance, ongoing efforts target genome-level post-translational modification analyses, which exploit techniques such as two-dimensional electrophoresis,55 fluorescence/stable isotope labeling,56 and various MS approaches.57Proteomics techniques are also commonly used to characterize the impact of biological environments on the nanoparticles, such as the formation of protein corona. This is a shell made by a mixture of environmental proteins that get adsorbed on nanoparticle surfaces when these are exposed to biological media, changing their physicochemical properties and biological impact.58 Proteomics studies have identified that nanoparticle features, such as material composition, particle size, and surface charge, strongly affect the protein corona formation and composition.59–62 These findings are relevant since some studies found that nanoparticles with protein corona have generally less cytotoxicity compared to those without corona.63–65 In the context of nanoparticle toxicology, proteomics stands as a key tool, providing crucial insights into cellular responses and contributing to advances in our understanding of the intricate interplay between nanoparticles and biological systems.
2.4. Metabolomics
Metabolomics focuses on unraveling phenotypic changes induced by chemicals or toxicants through the measurement of alterations in carbohydrate, lipid, and amino acid patterns, among others. Unlike organism-specific transcriptomics and proteomics, metabolomics lacks a fixed code and encompasses the entire metabolome, analyzed either extracellularly (footprint) or intracellularly (fingerprint).66 The workflow involves the separation, enrichment, and quantification of metabolites with nuclear magnetic resonance (NMR) or MS approaches. On the one hand, NMR is known for its reliability in detecting various metabolites,67,68 however, it has limited sensitivity and, usually, it can only identify fewer than 100 metabolites.69,70 On the other hand, MS-based techniques display higher sensitivity and can quantify hundreds to thousands of metabolites. MS is commonly combined with either gas chromatography (GC) or liquid chromatography (LC) for metabolite separation before identification and quantification.71–73 Nevertheless, MS techniques can be costly, require complex sample preparation (e.g. tissue extraction), and provide average reproducibility.Due to the wide variety of potential analytes, metabolomics analyses face several challenges, such as being able to cover broad concentration ranges during the screenings. Furthermore, because metabolites can display rather different chemical structures, their analysis may require the use of different detection techniques.66 Despite these challenges, the field can provide valuable insights into metabolite functions, being particularly well-suited for toxicological studies.
2.5. Lipidomics
Lipidomics is a specialized branch of metabolomics that encompasses membrane-lipidomics and mediator-lipidomics.20 The former involves the characterization of membrane lipid constituents and their structural aspects, while the latter concentrates on the quantification of low-abundance bioactive lipid species. This dynamic field extensively examines individual lipids and lipid-derived species, delving into their structure, function, and interactions with cellular components such as proteins, other lipids, and metabolites.74 Research in lipidomics spans diverse biochemical domains, exploring health and disease conditions across various organs, biological tissues, individual cells, and biofluids.75Ultrahigh-performance liquid chromatography–mass spectrometry (UHPLC-MS)76 is a pivotal tool in lipidomics research, facilitating the rapid and precise lipid profiling in diverse biological samples, which is essential for uncovering disease-related alterations, identifying biomarkers, and studying the impact of nanomaterials on lipid composition and toxicity.8,77
3. Toxicity of noble metal nanoparticles
Noble metal nanoparticles are among the most widely used nanomaterials, with applications in multiple fields ranging from healthcare to catalysis.78–81 Noble metal nanoparticles hold localized surface plasmon resonances (i.e. collective oscillation of their conduction electrons), which yield unique optoelectronic properties.82,83 These plasmon resonances can be tuned by the size and shape of the nanocrystals, and result in strong extinction coefficients, near-field enhancements, and the generation of hot electrons and local heat upon light irradiation. The strong optical properties are commonly used in biosensing (e.g. rapid COVID-19 and flu tests),84 the near-field enhancements are applied in enhanced spectroscopy,85 the hot-electron generation are exploited in catalysis and semiconductor doping,86–88 and the local heat generation is clinically being investigated in photothermal ablation of prostate cancer,89 and is preclinically used for photoacoustic imaging.90,91 Furthermore, some metal noble nanoparticles are commonly used as antibacterial agents.92–94 As a result, human exposure to these nanoparticles is increasing over time and needs to be thoroughly understood.3.1. Toxicity of silver nanoparticles (AgNPs)
Due to their unique physicochemical features, which result in strong optical properties and antibacterial effects (among others), AgNPs are commonly used in a wide range of commercial products, ranging from rapid antigen tests (e.g. COVID and flu tests)95,96 to antibacterial materials.97 The growing use of AgNPs in multiple industries has increased human exposure to these nanoparticles. Hence, a deep understanding of their biological impact is necessary to minimize potential adverse health effects that may originate from their use. It is worth pointing out that ionic and colloidal silver is used as antibacterial agent, especially in burned patients, and silver overload (argyria) is generally considered non-(severely) toxic.98 Hence, a key question in nanotoxicity that needs to be solved is whether AgNPs behave differently.Taken together, these studies with different in vitro skin models indicate that exposures to AgNPs seem to disrupt the cytoskeleton and metabolism (including ATP production), and induce apoptosis through the dysregulation of key biological pathways, such as MAPK signaling pathway.
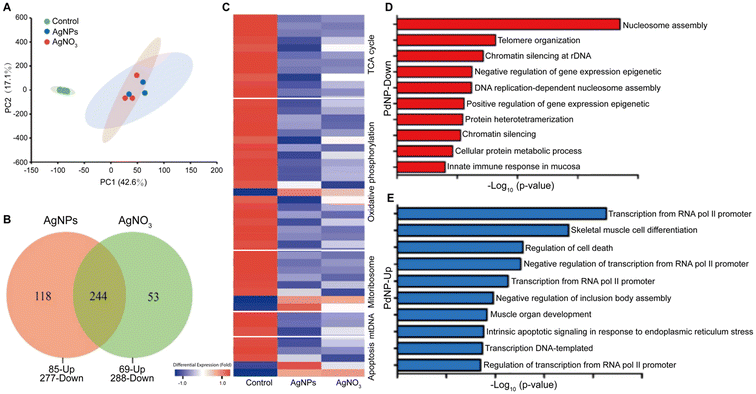 | ||
| Fig. 2 Toxicological effects of AgNPs and PdNPs. Comparison between the toxicological effects of AgNPs and silver nitrate (AgNO3) in gills of common carp, including (A) principal component analysis, (B) Venn diagrams plot, and (C) hierarchical clustering of different dysregulated proteins. The hierarchical clustering also includes the functional labels associated with the dysregulated proteins. Adapted with permission of ref. 110. Copyright Elsevier 2022. Functional groups associated with (D) down- and (E) up-regulated genes by PdNP treatments in human ovarian cancer cells. Adapted with permission of ref. 141. Copyright MDPI (Basel, Switzerland) 2019. | ||
Another study employed a shotgun tandem MS approach to assess the impact of 92 nm AgNPs (coating not reported) on human hepatoma (HepG2) cells.77 Their main conclusion was that treatments with AgNPs resulted in substantial alterations in a wide variety of phosphatidylcholine species. This observation suggested that the toxicity induced by nanoparticles primarily arose from inflammatory processes. It is essential to note, however, that these studies were conducted in liver cancer cells and the toxicogenomic information acquired in these studies may not be entirely applicable to a healthy liver. To date, toxicogenomic studies that use primary cells are extremely rare and most in vitro toxicity studies rely on pathological cells.
Beyond its role in food digestion and nutrient extraction, the digestive system also acts as a host environment to a wide range of microorganisms, including bacteria, archaea, fungi, and viruses, which have a broad impact in many health-related functions, such as resistance to pathogens, metabolism of dietary and pharmaceutical compounds, and control of the immune system, among others.113,114 Therefore, the ingestion of nanoparticles has the potential to also disrupt the normal functioning of the gut microbiota. In this context, a recent transcriptomics study, which involved RNA-Seq and RT-PCR, assessed the in vivo effects of AgNPs on the transcriptome and gut microbiota.115 Notably, the AgNP treatments induced immunotoxicological changes at both the molecular and microbiome levels, which were associated with stress and colon inflammation.
3.2. Toxicity of gold nanoparticles (AuNPs)
AuNPs are nanomaterials with unique optoelectronic properties, which originate from localized surface plasmon resonances.118 These properties depend on the size and morphology of the nanoparticles,119–122 and result in strong extinction coefficients, intense near-field enhancements, and large photothermal conversion effects.123 As a result, AuNPs are commonly used in rapid antigen tests for COVID-19 and flu diagnosis and are being (pre)clinically explored in multiple biosensing assays78 and therapeutic/theranostic formulations.89,124,125 Furthermore, the light-excitation of the localized surface plasmon resonances results in the generation of hot electrons, which can be exploited for catalysis and doping of semiconductors.126 The ever-growing use of AuNPs requires a deep understanding of their biological impact to ensure their safety.Although their toxicological profile has not been as thoroughly studied through omics approaches as in the case of AgNPs, there are a couple of toxicomics studies focused on AuNPs that are worth mentioning. One study compared the toxicological profile of AuNPs and AgNPs on human dermal fibroblasts using RNA-Seq and RT-PCR.127 Notably, at 200 μM, AuNPs were not cytotoxic, despite dysregulating the expression of mRNAs associated with cell cycle, ATP synthesis inhibition, cytoskeleton, and apoptosis. In contrast, AgNPs did cause significant cytotoxicity due to the generation of larger amounts of reactive oxygen species in comparison to AuNPs.
As previously mentioned, several gold nanoformulations are currently being explored in clinical studies.89 A therapeutic approach that relies on AuNPs is photothermal therapy, where solid tumors and atherosclerosis plaques are ablated via the high photothermal conversion efficiency of AuNPs, which concentrate the thermal ablation effect only on the surroundings of the irradiated nanoparticles, providing good therapeutic selectivity. While many preclinical and a few clinical studies have explored the therapeutic benefits of AuNP-based photothermal therapy, only a few have focused on the underlying biological mechanisms. A noteworthy exception is a study where high-resolution LC-MS was used to characterize the impact of photothermal therapy with gold nanorods (AuNRs) on lung cancer cells (A549).128 AuNRs are nanocrystals that absorb light in the near-infrared,129–131 where light penetrates deeper into the tissue.132 The metabolomic data indicated that AuNR-based photothermal treatment exerted toxicity by dysregulating metabolites that contained 18-hydroxyoleate, beta-alanopine and cis-9,10-epoxystearic acid, and phosphorylcholine.128 Several key metabolic pathways were also affected by the photothermal therapy, including those associated with cutin, suberine, and wax biosynthesis, pyruvate and glutamic acid synthesis, and choline metabolism. Taken together, this approach provides a deeper understanding of the mechanisms behind treatment-related cytotoxicity, complementing the conventional focus on macroscopic nanoparticle toxicity.
3.3. Toxicity of palladium nanoparticles (PdNPs)
Despite its high material costs, palladium is commonly used as a catalytic material. When the size of bulk palladium is reduced to the nanoscale, its catalytic activity is enhanced because of the higher surface-to-volume ratio of the nanoparticles. Hence, PdNPs hold significant importance in organic synthesis (e.g. catalyzing carbon–carbon bond forming reactions, such as Suzuki or Heck couplings),133 in the degradation of harmful substances in environmental applications,134 and in conducting materials and printed electronic devices.135 Furthermore, PdNPs may play an important role in hydrogen storage and generation,136 which is vital for the development of hydrogen energy technology. Additionally, PdNPs display strong photothermal capabilities being preclinically used in photoacoustic imaging137–139 and photothermal therapy.140In the field of nanotoxicology, various studies have explored the effects of PdNPs using omics approaches. For instance, one study employed transcriptomics (via RNA-Seq) to analyze the impact of PdNPs on human ovarian cancer cells (SKOV3) (Fig. 2D).141 The study reported that PdNPs primarily downregulated genes associated with nucleosome assembly, telomere organization, and DNA chromatin silencing. Among these processes, nucleosome assembly was the most significantly affected. Furthermore, RNA-seq analysis also demonstrated that the PdNP treatments triggered epigenetic changes that were associated with cell apoptosis. Another transcriptomics study investigated the accumulation of Pd-doped polystyrene nanoparticles in earthworms and its toxic effects.142 The study revealed that both positively and negatively surface-charged nanoparticles could accumulate and move within earthworms, leading to growth inhibition and pathological abnormalities. Positively charged nanoparticles exhibited higher cytotoxicity, likely due to their propensity to form larger aggregations at lower pH, facilitating cellular uptake and apoptosis induction. Gene expression analysis highlighted a more significant impact of positively charged nanoparticles compared to negatively charged ones in both low-dose (0.3 mg kg−1) and high-dose (3 mg kg−1) treatments. Additionally, a proteomics study using western blot techniques identified that PdNPs inhibit cell growth in various cell lines (e.g. Rat-1 rat embryo fibroblasts, CaCo-2 colon adenocarcinoma cells, and HaCaT keratinocyte cells), induce DNA damage, and impact the expression of key cell cycle regulatory proteins, all without triggering apoptosis or oxidative stress.143 These studies collectively contribute to a better understanding of the biological impact (in different biological systems) of PdNPs, which are numerous and diverse.
3.4. Toxicity of tungsten nanoparticles (WNPs)
Compared to their gold and silver counterparts, WNPs have relatively fewer uses. However, as a result of recent advances in industrial technologies combined with tungsten's high plasticity, high melting point, and electron emission ability, WNPs and their derivatives have found niche applications in electronics,144 and coatings and films,145 as well as in the nuclear industry.146 Regarding their toxicity, a transcriptomics study assessed the biological impact of tungsten boride nanoparticles (WBNPs) in HPAEpiC cells (human lung alveolar epithelial cells).147 The study concluded that WBNPs can modulate cytokine/chemokine metabolism, affect angiogenesis, and impede migration/invasion of cancer cells by activating specific genes, such as DACH1, COL3A1, and FOS.3.5. Toxicity of platinum nanoparticles (PtNPs)
In recent years, PtNPs have been increasingly explored in the chemical industry and automotive sector due to their catalytic features.148–152 Additionally, PtNPs have found some applications in the (preclinical) biomedical field, serving as diagnostic mediators, components of medical implants, drug delivery vehicles, and agents for photothermal therapy.153,154Regarding their toxicity, a transcriptomics study evaluated the biological impact of anisotropic lycopene-derived PtNPs on human acute monocytic leukemia (THP-1) cells.155 The study found that PtNPs (in a concentration range between 0 and 12 mM) induced dose-dependent cytotoxicity, oxidative stress, and apoptosis. The underlying mechanisms of toxicity were primarily associated with changes in gene expression related to protein misfolding, mitochondrial function, protein synthesis, inflammatory response, and transcriptional regulation.
4. Toxicity of metal oxide nanoparticles
Metal oxide nanoparticles are a heterogeneous group of nanocrystals that are widely produced and used because of their unique electronic, magnetic, and/or chemical properties.156 Some of the most relevant metal oxide nanoparticles include wide bandgap semiconducting materials (TiO2, CeO2, ZnO) and magnetic crystals (Fe2O3 and Fe3O4). These unique characteristics can be controlled by nanocrystal engineering and endow a wide range of applications, such as clinical imaging, gas sensing, and catalysis.157–1604.1. Toxicity of titania nanoparticles (TiO2NPs)
TiO2NPs exhibit strong ultraviolet (UV) absorption capabilities and resilience to radiation damage.161–163 As a consequence, they are prevalently employed as UV-blocking agents in sunscreen formulations as well as thickening agents in cosmetics.164 Furthermore, TiO2NPs, especially in the anatase form, display photocatalytic activity under UV irradiation. Consequently, titania colloids have become important in key industrial processes, such as photocatalysis, solar energy harnessing, and air and water purification.165–168 The versatility and widespread use of TiO2NPs, particularly in skin cosmetics, require a deep understanding of their toxicity and biological impact.The strong biological impact of titania nanocrystals in the lungs of mice was later also observed at the epigenetic level.173 TiO2NP exposures caused changes in DNA methylation particularly in the promoter regions of genes associated with inflammation and fibrosis, suggesting that abnormal DNA methylation has the potential to be used as biomarkers for pulmonary disease development. Notably, the same study observed that younger mice (5-week-old) displayed greater sensitivity to TiO2NP exposure than older mice (10-week-old) did, as observed by increased methylation, altered gene promoters, and up-regulation in cancer pathways, ultimately resulting in increased pulmonary inflammation and fibrosis. Consistently, a subsequent study identified that exposures to TiO2NPs, even at concentrations well below sublethal levels (0.5 μg mL−1), led to significant changes in DNA methylation,174 indicating that even minimal exposure to these nanoparticles can induce adverse epigenetic effects.
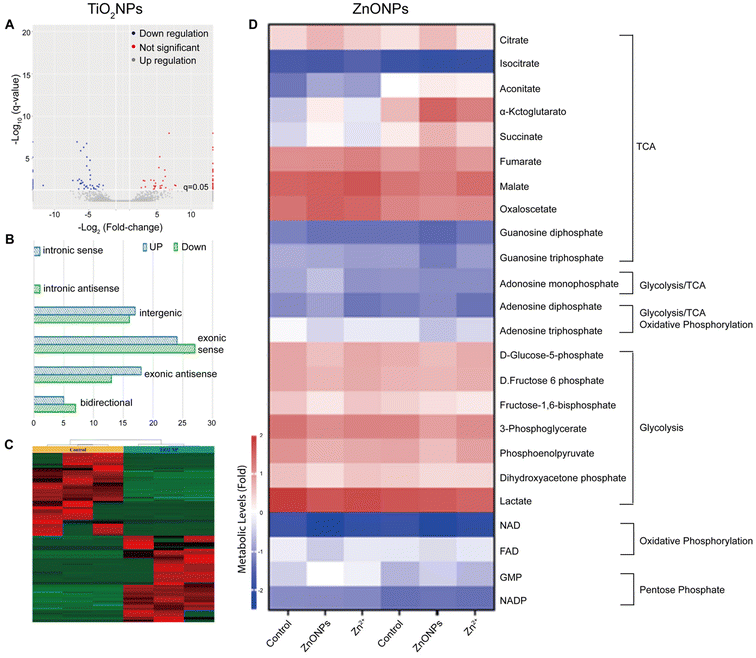 | ||
| Fig. 3 Toxicological profile of TiO2NPs based on lncRNA expression alterations in HepG2. (A) Volcano plot highlighting up-regulated and down-regulated lcnRNAs, (B) histogram with classification of dysregulated lcnRNAs, and (C) heat map and cluster analysis of dysregulated lcnRNAs after exposure to TiO2NPs at concentrations of 0 and 100 μg mL−1. Adapted with permission of ref. 177. Copyright MDPI (Basel, Switzerland) 2022. (D) Metabolomics analysis of zebrafish liver following exposures of 0 or 500 μg L−1 ZnONPs or equivalent dose of zinc salt ions. Adapted with permission of ref. 186. Copyright Elsevier 2022. | ||
4.2. Toxicity of zinc oxide nanoparticles (ZnONPs)
In the last two decades, ZnONPs have become widely used in multiple industries because of their excellent optoelectronic properties, stability, and perceived low toxicity.178 For example, ZnONPs are commonly employed in the manufacturing of sunscreens and cosmetics, as they can effectively block UV radiation.164,179 Additionally, ZnONPs are utilized in the development of antibacterial agents180 and as key components in electronic devices and sensors.181 Due to the widespread applications of ZnONPs, some recent studies have focused on characterizing their toxicological profile through omics approaches, aiming to gain a better understanding of their biological effects and potential uses.In a metabolomics study, male and female zebrafish were exposed to various concentrations of ZnONPs (Fig. 3D).186 Their livers were then subjected to metabolome analysis and energy metabolism assessment. The findings indicated that sub-chronic ZnONP exposures reduced the tricarboxylic acid cycle rate and oxidative phosphorylation, leading to ATP loss and energetic stress in both genders. Females compensated by increasing the fatty acid and amino acid metabolisms, while males solely adjusted their amino acid metabolism. Interestingly, this study highlighted several gender-dependent (in zebrafish) adaptative metabolic responses to mitigate disruptions caused by ZnONP exposures.
4.3. Toxicity of superparamagnetic iron oxide nanoparticles (SPIONs)
SPIONs are iron oxide colloids that display unique magnetic properties which combined with their biocompatibility have resulted in their use in different biomedical applications.189–191 For instance, SPIONs display large magnetic moments that alter the transverse relaxation times (T2) of water protons, decreasing their signal intensity in magnetic resonance imaging (MRI). Hence, in 1996, a SPION-based formulation (ferumoxide) was clinically approved by the US Food and Drugs Administration as an MRI contrast agent for the detection of liver lesions.107 Since then, multiple formulations have been developed for the imaging of other diseases, such as atheroma. Even though many of those diagnostic applications were later discontinued in favor of gadolinium-based contrast agents, SPIONs are still being clinically used for specific applications, including iron replacement therapy in anemic patients, imaging of tumor-associated macrophages, and identification of sentinel lymph nodes.107 Furthermore, a clinical trial has recently started to evaluate SPION-based magnetic hyperthermia to treat advanced pancreatic cancer patients. Beyond therapeutics and diagnostics, the magnetic properties of SPIONs are commonly used for magnetic-based separations, such as in water purification192 and pollutant removal.193Although SPIONs are generally perceived as biocompatible, there have been concerns regarding their biological impact (e.g. DNA damage, oxidative stress, and epigenetic alterations) under specific conditions and formulations.194 Thus, to better understand their endogenous behavior, a recent study explored the epigenetic effects of SPIONs coated with citric acid in human submandibular gland cells.195 Through a combination of colorimetric assays and RT-PCR, the authors concluded that exposures to citric acid-coated SPIONs induced significant alterations in DNA methylation and histone acetylation, which, notably, did not impact cell viability.
Although the adsorption of lipids and proteins onto the surface of nanoparticles has long been known, it was not until the last decade that its impact on nanoparticle behavior in vivo started to be understood.196 In a comprehensive study, researchers exploited proteomic and lipidomic analyses, including LC-MS and a multiple reaction monitoring profiling method, to investigate the formation of protein and lipid corona on 20 nm SPIONs and to explore how this biocorona affects cellular interactions and toxicity.197 To achieve this, the nanoparticles were exposed to isolated bronchoalveolar lavage fluids from both healthy and metabolic syndrome mouse models. The SPIONs exposed to metabolic syndrome samples were uptaken by macrophages to a greater extent than the SPIONs exposed to healthy fluids. Notably, different protein and lipid profiles were identified on the surface of the nanoparticles exposed to the distinct fluids through proteomic and lipidomic analyses. This observation suggested that metabolic syndrome conditions promote protein and lipid corona composition changes in bronchoalveolar lavage fluid, ultimately impacting nanoparticle interactions, which may contribute to the development of acute pulmonary inflammatory responses.
4.4. Toxicity of ceria nanoparticles (CeO2NPs)
CeO2NPs are prized for their rich redox behavior, as cerium can easily shift between +3 and +4 oxidation states, and can act as oxidation switches.198 Thus, CeO2NPs are being exploited to counteract oxidative stress by scavenging reactive oxygen species,199 to serve as core components in biosensors for biomolecule detection,200 to purify wastewater and exhaust gases,201 to act as catalysts in various reactions,202,203 and to function as electrolyte materials in fuel cells.204Because CeO2NPs (and other metal oxide nanostructures such as cobalt oxide nanoparticles, CoONPs) are used in automotive exhaust catalysts, there are concerns regarding their release in air and potential inhalation. A toxicogenomics study explored the biological effects of both CeO2NPs and CoONPs in bronchial and alveolar epithelial cells, two of the main lung cell types that interact with airborne particulates.205 The study focused on understanding inflammatory responses since these cells can produce pro-inflammatory mediators. Notably, although CeO2NPs disrupted the expression of a larger number of genes, CoONPs had a more pronounced impact on immune system-related processes.
The onset of immune responses by inhaled CeO2NPs was further demonstrated in vivo in male Sprague-Dawley rats through lipidomics and transcriptomics.8 The aerosolized 40 nm CeO2NPs triggered inflammation and immune responses, not only in the lungs but in the liver and kidneys, with significant changes in inflammation markers and gene expression, as confirmed by transcriptomics. Moreover, lipidomics profiling of lung tissue further revealed changes to minor lipid species, implying specific rather than general cellular effects. Notably, these inflammatory effects triggered by the CeO2NPs were greater than those observed in toxicomics studies using micron-sized ceria aerosols, corroborating the larger biological reactivity of materials at the nanoscale.
4.5. Toxicity of other metal oxide nanoparticles
Beyond the cases discussed above, several studies have examined the toxicity of other metal oxide nanoparticles. For instance, a study explored the toxicological profile of catalytic vanadium oxide nanoparticles (V2O5NPs) through high-resolution metabolomics in human airway epithelial cells (BEAS-2B).206 Even at non-cytotoxic doses, V2O5NPs induced dose-dependent responses in several major metabolic pathways, including the metabolism of amino sugars, amino acids, fatty acids, carnitine, and nucleotides, some of which have been previously linked to human lung diseases.207In a separate study, the toxicity of low doses (0.5 μg Ni mL−1) of nickel oxide nanoparticles (NiONPs) and nickel nanoparticles (NiNPs), which are used in electronics and catalysis,208 were screened through RNA-Seq in human lung cells (BEAS-2B) after prolonged exposures (six weeks). Both NiONPs and NiNPs caused DNA strand breaks, which were consistent with exposures to nickel chloride salts.
A comprehensive study combining genomics, transcriptomics, and metabolomics investigated the impact of gadolinium oxide nanoparticles (GdONPs) on 4T1 breast cancer cells.209 The research demonstrated that GdONPs induced apoptosis in 4T1 cells by significantly increasing the production of reactive oxygen species, leading to enhanced DNA damage within the cells. These results were consistent with a study that followed a similar approach and explored the effects of manganese oxide nanoparticles (Mn3O4NPs) at a concentration of 100 μg mL−1 on rat type II alveolar epithelial cells.210 The authors proved that Mn3O4NPs also induced an increase in reactive oxygen species, leading to significant oxidative stress, which triggered the apoptosis of the rat type II alveolar epithelial cells. Other heavy metal oxide nanoparticles, such as molybdenum dioxide nanoparticles (MoO2NPs), have also shown to induce significant toxicity, such as in the liver, through oxidative stress and inflammation, leading to notable changes in the expression of inflammation markers and genes.211
5. Toxicity of other inorganic nanoparticles
5.1. Toxicity of quantum dots
Quantum dots are semiconductor nanocrystals that display shape- and size-dependent optical and electronic properties, which are in between those of bulk semiconductors and discrete atoms.212 For instance, as the size of quantum dots increases (from 2 to 6 nm), their emissions red-shift to longer wavelengths. Quantum dots are widely used in optoelectronic devices, such as light-emitting diode (QLED) screens and lasers.213 Furthermore, they have also been explored as optical probes in preclinical imaging.214 It is worth mentioning that the Noble Prize in Chemistry 2023 was awarded to Professors Ekimov, Brus, and Bawendi for the discovery and synthesis of quantum dots.215–217A proteomic study explored the toxicity of CdTe quantum dots in human monocytes THP-1 and compared it to the toxicity of AuNPs and copper oxide nanoparticles (CuONPs).218 Despite showing similar overall toxicity, CdTe induced the strongest proteomic responses by down-regulating topoisomerases, enzymes that catalyze changes in the topological state of DNA. The authors hypothesized that CdTe may affect the alkylation of DNA or affect chromatin-related proteins, which would be consistent with the cytotoxic mechanisms associated with Cd2+.
5.2. Toxicity of silica nanoparticles (SiO2NPs)
Silicon is one of the most abundant elements on Earth's crust, and nanoparticles made of its oxide have found multiple industrial applications, ranging from fining agents for beverages to cosmetics.219 SiO2NPs can be either crystalline or amorphous. While their molecular composition is the same, both crystalline and amorphous structures can display different properties. For example, some crystalline SiO2NPs possess porous with diameters ranging from 2 to 100 nm that can be filled with different therapeutic and diagnostic agents.215–217 Hence, numerous nanomedicines based on (meso)porous SiO2NPs are currently being explored in clinical trials for sentinel lymph node mapping and oral drug delivery, among others.220 The increasing use of SiO2NPs in biomedical applications and their widespread presence in the food and cosmetic industry requires a thorough characterization of their toxicity.In the last years, there has been an increasing concern about the impact of nanoparticles on male fertility.221,222 In this context, a genomic study explored the toxicity of SiO2NPs in murine spermatocyte cells.223 SiO2NP treatment dysregulated 15 significant miRNAs, which were associated with DNA replication and fatty acid metabolism, identifying the origin of SiO2NPs in spermatocyte cells.
In a separate study, researchers adopted a multifaceted approach by combining proteomics and metabolomics to screen the metabolic disturbances induced by SiO2NPs in liver cells (Fig. 4).224 Through proteomics analysis, they unveiled the aberrant expression of key proteins, including RPL3, HSP90AA1, SOD, PGK1, GOT1, and PNP, which were associated with protein synthesis, folding, oxidative stress, and overall metabolism. Complementing these findings, metabolomics analysis provided further evidence of disrupted metabolic pathways, encompassing glucose metabolism, amino acid metabolism, and nucleotide metabolism. These results highlighted the strong impact of SiO2NPs on cellular metabolic processes.
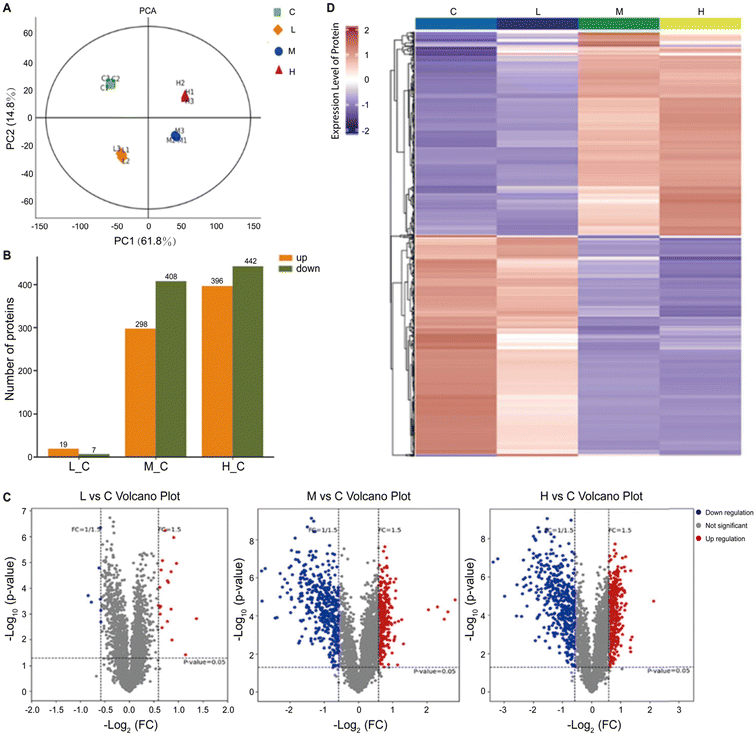 | ||
| Fig. 4 Proteomic analysis of mouse spermatocyte cells exposed to SiO2NPs. In the figure, the control group and the low (12.5 μg mL−1), middle (25 μg mL−1), and high (50 μg mL−1) doses of SiO2NPs are abbreviated as C, L, M, and H, respectively. (A) PCA score of the SiO2NP-treated group and control group. (B) Several upregulated and downregulated differentially expressed proteins in different groups. (C) Volcano plot of proteins identified after exposure to SiO2NPs. (D) Hierarchical clustering heatmap of differentially expressed proteins. Adapted with permission of ref. 224. Copyright Elsevier 2022. | ||
6. Lessons learned and recommendations
6.1. Lack of universal toxicity rules
It is challenging to identify general rules that define the toxicity of nanoparticles, since they come in all sorts of shapes and forms, and can behave differently. For instance, nanoparticles can display sizes ranging from a few nanometers, which result in rapid nanoparticle excretion through renal pathways,225 to several hundreds of nanometers, which yield nanoparticles with large accumulation in fenestrated organs, such as the liver and spleen.108,109 A wide variety of core materials are used to synthesize nanoparticles, which include noble metals, metal oxides, and semiconductors, to name a few (Table S1†). Some of these materials can be degraded or excreted by the body,110 while others cannot and tend to accumulate in tissues for prolonged periods of time.108,109 Moreover, some of these materials can display inherent cytotoxicity, e.g. via reactive oxygen species production,226 or promote acute immune responses.227 Notably, some materials may also possess different properties depending on whether they are found in nanoparticulate form or ionic form, further complicating their toxicity assessment.109 Nanoparticles may have coating or ligand shell on their surface, which can be engineered to improve their colloidal stability or to recognize specific targets.228 The shell characteristics, such as size and charge, strongly affect the fate and toxicity of the nanoparticles both in vitro and in vivo.229,230 For example, nanoparticles with strong positive charge tend to display higher cellular uptake in vitro,231 but also nanoparticles with strong surface charges tend to adsorb large amounts of plasma components on their surface, which can be recognized by phagocytes, yielding nanoparticles with short circulation times in the bloodstream.2326.2. The role of protein corona
In the last decade, the concept of protein (and lipid) corona has emerged in the field of nanoscience. When nanoparticles are exposed to biological fluids, different endogenous biomolecules can adsorb on their surface forming a biomolecular corona.233,234 The composition of the corona strongly affects the behavior and fate of the nanoparticles.235,236 Interestingly, the material core and ligand shell of the nanoparticles can facilitate or (even) prevent the formation of the corona, opening avenues to improve and optimize cell internalization, and biological effects of nanoparticles and nanomedicines.237,2386.3. The importance of the model
Another limitation of many toxicological studies (not only omics) is the use of immortal cancer cell lines.239 These cells are easy to handle since they can continuously grow and divide in laboratory settings. Nevertheless, these cells have unique genetic alterations that are not found in normal healthy cells. Hence, they might display characteristics and functions that are not relevant to any cell type in vivo. Moreover, after multiple passages, the characteristics of immortal cell lines can change, becoming even more distinct from those of healthy cells. Thus, it is important to validate that the characteristics of the cell model are relevant to the aim of the study. Furthermore, in vitro cell models in general are severely limited by their simplicity, which completely ignores the absorption, distribution, metabolism and excretion of the particles.240,241 For instance, to interact with hepatocytes, nanoparticles need to be absorbed, reach the blood circulation, avoid phagocyte uptake, extravasate through the endothelial wall and finally reach the hepatic cells. Some nanoparticles are unlikely to reach the final destination, and the cell culture assays would yield artificial results, while other nanoparticles are likely to be transformed through the process.6.4. The importance of reporting the nanoparticle characteristics
Taken together, the large variability of nanoparticles challenges their toxicological assessment. Conclusions drawn from a toxicomics study using a specific set of nanoparticles may only be valid for that specific set of colloids. Hence, when publishing toxicological studies, it is very important to properly characterize and report the features of the nanoparticles. Unfortunately, this is not always the case, and key characteristics, such as size or shell composition, are not commonly reported in the literature.7. Conclusion
The prevalent use of inorganic nanoparticles in many human-driven activities requires a thorough assessment of nanoparticle biological impact and safety. Omics approaches, including transcriptomics, epigenetics, proteomics, metabolomics, and lipidomics, are improving our understanding of the molecular effects of nanoparticles, uncovering biological functions disrupted by the nanomaterials, and highlighting genes, proteins and other biomarkers associated with their toxicity. In addition, omics techniques can be used to identify endogenous agents that participate in minimizing the harmful effects of the nanoparticles or to better understand the therapeutic mechanisms associated with nanomedicines. So far, a limited number of studies have explored the use of omics to characterize the toxicity of inorganic nanoparticles, however, we expect that these numbers will increase in the future, as omics studies can support the development of nanoparticles with lower biological impact and yield safer industrial and clinical applications.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the Federal Ministry of Education and Research (BMBF) and the Ministry of Culture and Science of the German State of North Rhine-Westphalia (MKW) under the Excellence Strategy of the Federal Government and the Länder through the RWTH Junior Principal Investigator (JPI) fellowship scheme.References
- X. He, H. Deng and H. Hwang, J. Food Drug Anal., 2019, 27, 1–21 CrossRef CAS PubMed.
- S. Bayda, M. Adeel, T. Tuccinardi, M. Cordani and F. Rizzolio, Molecules, 2019, 25, 112 CrossRef PubMed.
- C. Huang, K. G. Neoh, L. Wang, E. Kang and B. Shuter, Contrast Media Mol. Imaging, 2011, 6, 298–307 CrossRef CAS PubMed.
- R. Cheng, F. Feng, F. Meng, C. Deng, J. Feijen and Z. Zhong, J. Controlled Release, 2011, 152, 2–12 CrossRef CAS PubMed.
- J. Pan, D. Wan and J. Gong, Chem. Commun., 2011, 47, 3442–3444 RSC.
- J. E. Thompson, J. Occup. Environ. Med., 2018, 60, 392–423 CrossRef CAS PubMed.
- F. J. Kelly and J. C. Fussell, Philos. Trans. R. Soc., A, 2020, 378, 20190322 CrossRef CAS PubMed.
- C. Guo, S. Robertson, R. J. M. Weber, A. Buckley, J. Warren, A. Hodgson, J. Z. Rappoport, K. Ignatyev, K. Meldrum, I. Römer, S. Macchiarulo, J. K. Chipman, T. Marczylo, M. O. Leonard, T. W. Gant, M. R. Viant and R. Smith, Nanotoxicology, 2019, 13, 733–750 CrossRef CAS PubMed.
- A. V. Singh, P. Laux, A. Luch, C. Sudrik, S. Wiehr, A.-M. Wild, G. Santomauro, J. Bill and M. Sitti, Toxicol. Mech. Methods, 2019, 29, 378–387 CrossRef CAS PubMed.
- X. Li, W. Liu, L. Sun, K. E. Aifantis, B. Yu, Y. Fan, Q. Feng, F. Cui and F. Watari, J. Biomed. Mater. Res., Part A, 2015, 103, 2499–2507 CrossRef CAS PubMed.
- C. Andraos and M. Gulumian, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2021, 13, e1680 CAS.
- S. Borandeh, V. Alimardani, S. S. Abolmaali and J. Seppälä, Chem. Res. Toxicol., 2021, 34, 1386–1402 Search PubMed.
- Nanotoxicology in Safety Assessment of Nanomaterials, ed. H. Louro and M. J. Silva, Springer International Publishing, Cham, 2022, vol. 1357 Search PubMed.
- E. Fröhlich, J. Nanobiotechnol., 2017, 15, 84 CrossRef PubMed.
- A. Aranda, L. Sequedo, L. Tolosa, G. Quintas, E. Burello, J. V. Castell and L. Gombau, Toxicol. In Vitro, 2013, 27, 954–963 CrossRef CAS PubMed.
- A. V. Singh, V. Chandrasekar, N. Paudel, P. Laux, A. Luch, D. Gemmati, V. Tisato, K. S. Prabhu, S. Uddin and S. P. Dakua, Biomed. Pharmacother., 2023, 163, 114784 CrossRef CAS PubMed.
- R. M. Pallares, Y. Li and R. J. Abergel, TrAC, Trends Anal. Chem., 2023, 167, 117251 CrossRef CAS.
- S.-E. Nam, D.-Y. Bae, J.-S. Ki, C.-Y. Ahn and J.-S. Rhee, Mol. Cell. Toxicol., 2023, 19, 3–11 CrossRef CAS.
- R. M. Pallares, D. Faulkner, D. D. An, S. Hébert, A. Loguinov, M. Proctor, J. A. Villalobos, K. A. Bjornstad, C. J. Rosen, C. Vulpe and R. J. Abergel, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2025952118 CrossRef CAS PubMed.
- M. D. Luque De Castro and R. Quiles-Zafra, Talanta, 2020, 219, 121197 CrossRef CAS PubMed.
- R. M. Pallares, D. D. An, S. Hébert, D. Faulkner, A. Loguinov, M. Proctor, J. A. Villalobos, K. A. Bjornstad, C. J. Rosen, C. Vulpe and R. J. Abergel, Mol. Omics, 2022, 18, 237–248 RSC.
- R. M. Pallares, D. D. An, S. Hébert, A. Loguinov, M. Proctor, J. A. Villalobos, K. A. Bjornstad, C. J. Rosen, C. D. Vulpe and R. J. Abergel, ACS Omega, 2022, 7, 34412–34419 CrossRef CAS PubMed.
- R. M. Pallares, D. D. An, S. Hebert, A. Loguinov, M. Proctor, J. A. Villalobos, K. A. Bjornstad, C. J. Rosen, C. Vulpe and R. J. Abergel, Metallomics, 2023, 15, mfad039 CrossRef PubMed.
- T. Xia, M. Kovochich, J. Brant, M. Hotze, J. Sempf, T. Oberley, C. Sioutas, J. I. Yeh, M. R. Wiesner and A. E. Nel, Nano Lett., 2006, 6, 1794–1807 CrossRef CAS PubMed.
- S. J. Kang, B. M. Kim, Y. J. Lee and H. W. Chung, Environ. Mol. Mutagen., 2008, 49, 399–405 CrossRef CAS PubMed.
- S. M. Hussain, K. L. Hess, J. M. Gearhart, K. T. Geiss and J. J. Schlager, Toxicol. In Vitro, 2005, 19, 975–983 CrossRef CAS PubMed.
- E. Fröhlich, Curr. Drug Metab., 2013, 14, 976–988 CrossRef PubMed.
- A. L. Kersey, T.-U. Nguyen, B. Nayak, I. Singh and A. K. Gaharwar, Mater. Today, 2023, 64, 98–120 CrossRef.
- Molecular Profiling: Methods and Protocols, ed. V. Espina and L. A. Liotta, Humana Press, Totowa, NJ, 2012, vol. 823 Search PubMed.
- Y. Hasin, M. Seldin and A. Lusis, Genome Biol., 2017, 18, 83 CrossRef PubMed.
- P.-Y. Wu, R. Chandramohan, J. H. Phan, W. T. Mahle, J. W. Gaynor, K. O. Maher and M. D. Wang, Circ.: Cardiovasc. Genet., 2014, 7, 701–710 Search PubMed.
- R. Hrdlickova, M. Toloue and B. Tian, WIREs RNA, 2017, 8, e1364 CrossRef PubMed.
- R. M. Pallares, E. Jarvis, D. D. An, C. H. Wu, P. Y. Chang and R. J. Abergel, Environ. Adv., 2022, 8, 100245 CrossRef.
- Y. Chen, C. Li, C. Tan and X. Liu, J. Med. Genet., 2016, 53, 359–365 CrossRef CAS PubMed.
- S. P. Barrett and J. Salzman, Development, 2016, 143, 1838–1847 CrossRef CAS PubMed.
- M. W. Feinberg and K. J. Moore, Circ. Res., 2016, 118, 703–720 CrossRef CAS PubMed.
- B. F. Voight, H. M. Kang, J. Ding, C. D. Palmer, C. Sidore, P. S. Chines, N. P. Burtt, C. Fuchsberger, Y. Li, J. Erdmann, T. M. Frayling, I. M. Heid, A. U. Jackson, T. Johnson, T. O. Kilpeläinen, C. M. Lindgren, A. P. Morris, I. Prokopenko, J. C. Randall, R. Saxena, N. Soranzo, E. K. Speliotes, T. M. Teslovich, E. Wheeler, J. Maguire, M. Parkin, S. Potter, N. W. Rayner, N. Robertson, K. Stirrups, W. Winckler, S. Sanna, A. Mulas, R. Nagaraja, F. Cucca, I. Barroso, P. Deloukas, R. J. F. Loos, S. Kathiresan, P. B. Munroe, C. Newton-Cheh, A. Pfeufer, N. J. Samani, H. Schunkert, J. N. Hirschhorn, D. Altshuler, M. I. McCarthy, G. R. Abecasis and M. Boehnke, PLoS Genet., 2012, 8, e1002793 CrossRef CAS PubMed.
- S. B. Ng, E. H. Turner, P. D. Robertson, S. D. Flygare, A. W. Bigham, C. Lee, T. Shaffer, M. Wong, A. Bhattacharjee, E. E. Eichler, M. Bamshad, D. A. Nickerson and J. Shendure, Nature, 2009, 461, 272–276 CrossRef CAS PubMed.
- D. C. Koboldt, K. M. Steinberg, D. E. Larson, R. K. Wilson and E. R. Mardis, Cell, 2013, 155, 27–38 CrossRef CAS PubMed.
- J. He and J. Gai, in Plant Genotyping, ed. Y. Shavrukov, Springer US, New York, NY, 2023, vol. 2638, pp. 123–146 Search PubMed.
- V. Tam, N. Patel, M. Turcotte, Y. Bossé, G. Paré and D. Meyre, Nat. Rev. Genet., 2019, 20, 467–484 CrossRef CAS PubMed.
- K. C. Wang and H. Y. Chang, Circ. Res., 2018, 122, 1191–1199 CrossRef CAS PubMed.
- A. Piunti and A. Shilatifard, Science, 2016, 352, aad9780 CrossRef PubMed.
- R. Barouki, E. Melén, Z. Herceg, J. Beckers, J. Chen, M. Karagas, A. Puga, Y. Xia, L. Chadwick, W. Yan, K. Audouze, R. Slama, J. Heindel, P. Grandjean, T. Kawamoto and K. Nohara, Environ. Int., 2018, 114, 77–86 CrossRef CAS PubMed.
- M. H. Rabotnick, J. Ehlinger, A. Haidari and J. M. Goodrich, Mol. Cell. Endocrinol., 2023, 578, 112046 CrossRef CAS PubMed.
- Roadmap Epigenomics Consortium, A. Kundaje, W. Meuleman, J. Ernst, M. Bilenky, A. Yen, A. Heravi-Moussavi, P. Kheradpour, Z. Zhang, J. Wang, M. J. Ziller, V. Amin, J. W. Whitaker, M. D. Schultz, L. D. Ward, A. Sarkar, G. Quon, R. S. Sandstrom, M. L. Eaton, Y.-C. Wu, A. R. Pfenning, X. Wang, M. Claussnitzer, Y. Liu, C. Coarfa, R. A. Harris, N. Shoresh, C. B. Epstein, E. Gjoneska, D. Leung, W. Xie, R. D. Hawkins, R. Lister, C. Hong, P. Gascard, A. J. Mungall, R. Moore, E. Chuah, A. Tam, T. K. Canfield, R. S. Hansen, R. Kaul, P. J. Sabo, M. S. Bansal, A. Carles, J. R. Dixon, K.-H. Farh, S. Feizi, R. Karlic, A.-R. Kim, A. Kulkarni, D. Li, R. Lowdon, G. Elliott, T. R. Mercer, S. J. Neph, V. Onuchic, P. Polak, N. Rajagopal, P. Ray, R. C. Sallari, K. T. Siebenthall, N. A. Sinnott-Armstrong, M. Stevens, R. E. Thurman, J. Wu, B. Zhang, X. Zhou, A. E. Beaudet, L. A. Boyer, P. L. De Jager, P. J. Farnham, S. J. Fisher, D. Haussler, S. J. M. Jones, W. Li, M. A. Marra, M. T. McManus, S. Sunyaev, J. A. Thomson, T. D. Tlsty, L.-H. Tsai, W. Wang, R. A. Waterland, M. Q. Zhang, L. H. Chadwick, B. E. Bernstein, J. F. Costello, J. R. Ecker, M. Hirst, A. Meissner, A. Milosavljevic, B. Ren, J. A. Stamatoyannopoulos, T. Wang and M. Kellis, Nature, 2015, 518, 317–330 CrossRef PubMed.
- Z. Zhang, S. Wu, D. L. Stenoien and L. Paša-Tolić, Annu. Rev. Anal. Chem., 2014, 7, 427–454 CrossRef CAS PubMed.
- T. Rabilloud and P. Lescuyer, Proteomics, 2015, 15, 1051–1074 CrossRef CAS PubMed.
- T. Qiao and X. Wang, Cell Biol. Toxicol., 2019, 35, 289–291 CrossRef PubMed.
- R. J. Rotello and T. D. Veenstra, Curr. Protein Pept. Sci., 2021, 22, 121–133 CrossRef CAS PubMed.
- H. Zhu, M. Bilgin and M. Snyder, Annu. Rev. Biochem., 2003, 72, 783–812 CrossRef CAS PubMed.
- W. Li and N. B. Caberoy, Appl. Microbiol. Biotechnol., 2010, 85, 909–919 CrossRef CAS PubMed.
- A. Paiano, A. Margiotta, M. De Luca and C. Bucci, Curr. Protoc. Protein Sci., 2019, 95, e70 CrossRef PubMed.
- D. Tsikas, Amino Acids, 2021, 53, 485–487 CrossRef CAS PubMed.
- Z. W. Lai, Y. Yan, F. Caruso and E. C. Nice, ACS Nano, 2012, 6, 10438–10448 CrossRef CAS PubMed.
- Y. Zhang, B. R. Fonslow, B. Shan, M.-C. Baek and J. R. Yates, Chem. Rev., 2013, 113, 2343–2394 CrossRef CAS PubMed.
- T. Chen, J. Zhao, J. Ma and Y. Zhu, Genomics, Proteomics Bioinf., 2015, 13, 36–39 CrossRef PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- R. Bilardo, F. Traldi, A. Vdovchenko and M. Resmini, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 14, e1788 CAS.
- S. J. Park, Int. J. Nanomed., 2020, 15, 5783–5802 CrossRef CAS PubMed.
- V. Pareek, A. Bhargava, V. Bhanot, R. Gupta, N. Jain and J. Panwar, J. Nanosci. Nanotechnol., 2018, 18, 6653–6670 CrossRef CAS PubMed.
- F. Pederzoli, G. Tosi, M. A. Vandelli, D. Belletti, F. Forni and B. Ruozi, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9, e1467 Search PubMed.
- W. Wohlleben, M. D. Driessen, S. Raesch, U. F. Schaefer, C. Schulze, B. V. Vacano, A. Vennemann, M. Wiemann, C. A. Ruge, H. Platsch, S. Mues, R. Ossig, J. M. Tomm, J. Schnekenburger, T. A. J. Kuhlbusch, A. Luch, C.-M. Lehr and A. Haase, Nanotoxicology, 2016, 10, 970–980 CrossRef CAS PubMed.
- S. Schöttler, G. Becker, S. Winzen, T. Steinbach, K. Mohr, K. Landfester, V. Mailänder and F. R. Wurm, Nat. Nanotechnol., 2016, 11, 372–377 CrossRef PubMed.
- C. Pisani, J.-C. Gaillard, M. Odorico, J. L. Nyalosaso, C. Charnay, Y. Guari, J. Chopineau, J.-M. Devoisselle, J. Armengaud and O. Prat, Nanoscale, 2017, 9, 1840–1851 RSC.
- K. D. La Luz-Hdez, in Metabolomics, ed. U. Roessner, InTech, 2012 Search PubMed.
- R. T. McKay, in Metabolomics and Its Impact on Health and Diseases, ed. V. Ghini, K. A. Stringer and C. Luchinat, Springer International Publishing, Cham, 2022, vol. 277, pp. 73–116 Search PubMed.
- G. A. Nagana Gowda and D. Raftery, in Cancer Metabolomics, ed. S. Hu, Springer International Publishing, Cham, 2021, vol. 1280, pp. 19–37 Search PubMed.
- J. L. Markley, R. Brüschweiler, A. S. Edison, H. R. Eghbalnia, R. Powers, D. Raftery and D. S. Wishart, Curr. Opin. Biotechnol., 2017, 43, 34–40 CrossRef CAS PubMed.
- A.-H. Emwas, R. Roy, R. T. McKay, L. Tenori, E. Saccenti, G. A. N. Gowda, D. Raftery, F. Alahmari, L. Jaremko, M. Jaremko and D. S. Wishart, Metabolites, 2019, 9, 123 CrossRef CAS PubMed.
- W. B. Dunn, D. I. Broadhurst, H. J. Atherton, R. Goodacre and J. L. Griffin, Chem. Soc. Rev., 2011, 40, 387–426 RSC.
- R. Madsen, T. Lundstedt and J. Trygg, Anal. Chim. Acta, 2010, 659, 23–33 CrossRef CAS PubMed.
- G. J. Patti, O. Yanes and G. Siuzdak, Nat. Rev. Mol. Cell Biol., 2012, 13, 263–269 CrossRef CAS PubMed.
- X. Han and R. W. Gross, J. Lipid Res., 2022, 63, 100164 CrossRef CAS PubMed.
- H. F. Avela and H. Sirén, Clin. Chim. Acta, 2020, 510, 123–141 CrossRef CAS PubMed.
- D. Masuero, D. Škrab, G. Chitarrini, M. Garcia-Aloy, P. Franceschi, P. Sivilotti, G. Guella and U. Vrhovsek, Metabolites, 2021, 11, 827 CrossRef CAS PubMed.
- J. A. Adeyemi, C. A. Sorgi, A. R. T. Machado, A. T. Ogunjimi, L. G. A. Gardinassi, V. Nardini, L. H. Faccioli, L. M. G. Antunes and F. Barbosa, Arch. Toxicol., 2020, 94, 2625–2636 CrossRef CAS PubMed.
- R. M. Pallares, N. T. K. Thanh and X. Su, Nanoscale, 2019, 11, 22152–22171 RSC.
- B. Klębowski, J. Depciuch, M. Parlińska-Wojtan and J. Baran, Int. J. Mol. Sci., 2018, 19, 4031 CrossRef PubMed.
- G. Doria, J. Conde, B. Veigas, L. Giestas, C. Almeida, M. Assunção, J. Rosa and P. V. Baptista, Sensors, 2012, 12, 1657–1687 CrossRef CAS PubMed.
- M. Azharuddin, G. H. Zhu, D. Das, E. Ozgur, L. Uzun, A. P. F. Turner and H. K. Patra, Chem. Commun., 2019, 55, 6964–6996 RSC.
- S. Li, M. Meng Lin, M. S. Toprak, D. K. Kim and M. Muhammed, Nano Rev., 2010, 1, 5214 CrossRef PubMed.
- C. J. Murphy, T. K. Sau, A. M. Gole, C. J. Orendorff, J. Gao, L. Gou, S. E. Hunyadi and T. Li, J. Phys. Chem. B, 2005, 109, 13857–13870 CrossRef CAS PubMed.
- R. M. Pallares, F. Kiessling and T. Lammers, Nanomedicine, 2023, 18, 413–416 CrossRef CAS PubMed.
- J. Langer, D. Jimenez De Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. García De Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Käll, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J.-F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J.-M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L.-L. Tay, K. G. Thomas, Z.-Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzán, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- D. T. Thompson, Nano Today, 2007, 2, 40–43 CrossRef.
- I. Saldan, Y. Semenyuk, I. Marchuk and O. Reshetnyak, J. Mater. Sci., 2015, 50, 2337–2354 CrossRef CAS.
- I. Kriegel, F. Scotognella and L. Manna, Phys. Rep., 2017, 674, 1–52 CrossRef CAS.
- R. Zhang, F. Kiessling, T. Lammers and R. M. Pallares, Drug Delivery Transl. Res., 2023, 13, 378–385 CrossRef PubMed.
- Y. Liu, W. Ma and J. Wang, Curr. Pharm. Des., 2018, 24, 2719–2728 CrossRef CAS PubMed.
- W. Li and X. Chen, Nanomedicine, 2015, 10, 299–320 CrossRef CAS PubMed.
- L. Danai, L. A. Rolband, V. A. Perdomo, E. Skelly, T. Kim and K. A. Afonin, Nanomedicine, 2023, 18, 769–782 CrossRef CAS PubMed.
- M. Alavi, Expert Rev. Anti-Infect. Ther., 2022, 20, 897–906 CrossRef CAS PubMed.
- T. Bruna, F. Maldonado-Bravo, P. Jara and N. Caro, Int. J. Mol. Sci., 2021, 22, 7202 CrossRef CAS PubMed.
- A. R. Ghosh, A. N. Bhooshitha, H. M. Chandan and K. L. Krishna, Indian J. Pharm. Educ. Res., 2021, 55, 319–329 CrossRef CAS.
- S. Park, Y.-S. Ko, S. J. Lee, C. Lee, K. Woo and G. Ko, Environ. Sci. Pollut. Res., 2018, 25, 27021–27030 CrossRef CAS PubMed.
- P. Velmurugan, S.-M. Lee, M. Cho, J.-H. Park, S.-K. Seo, H. Myung, K.-S. Bang and B.-T. Oh, Appl. Microbiol. Biotechnol., 2014, 98, 8179–8189 CrossRef CAS PubMed.
- B. S. Atiyeh, M. Costagliola, S. N. Hayek and S. A. Dibo, Burns, 2007, 33, 139–148 CrossRef PubMed.
- F. L. Filon, M. Mauro, G. Adami, M. Bovenzi and M. Crosera, Regul. Toxicol. Pharmacol., 2015, 72, 310–322 CrossRef PubMed.
- J. Ma, X. Lü and Y. Huang, J. Biomed. Nanotechnol., 2011, 7, 263–275 CrossRef CAS PubMed.
- Y. Huang, X. Lü and J. Ma, J. Biomed. Nanotechnol., 2014, 10, 3304–3317 CrossRef CAS PubMed.
- J. Carrola, V. Bastos, I. Jarak, R. Oliveira-Silva, E. Malheiro, A. L. Daniel-da-Silva, H. Oliveira, C. Santos, A. M. Gil and I. F. Duarte, Nanotoxicology, 2016, 10, 1105–1117 CrossRef CAS PubMed.
- S. Bakand, A. Hayes and F. Dechsakulthorn, Inhalation Toxicol., 2012, 24, 125–135 CrossRef CAS PubMed.
- S. Sonwani, S. Madaan, J. Arora, S. Suryanarayan, D. Rangra, N. Mongia, T. Vats and P. Saxena, Front. Sustainable Cities, 2021, 3, 690444 CrossRef.
- R. Foldbjerg, E. S. Irving, Y. Hayashi, D. S. Sutherland, K. Thorsen, H. Autrup and C. Beer, Toxicol. Sci., 2012, 130, 145–157 CrossRef CAS PubMed.
- J. Jang, S. Park and I.-H. Choi, Biomolecules, 2021, 11, 234 CrossRef CAS PubMed.
- R. M. Pallares, F. M. Mottaghy, V. Schulz, F. Kiessling and T. Lammers, J. Nucl. Med., 2022, 63, 1802–1808 CrossRef CAS PubMed.
- Y.-N. Zhang, W. Poon, A. J. Tavares, I. D. McGilvray and W. C. W. Chan, J. Controlled Release, 2016, 240, 332–348 CrossRef CAS PubMed.
- K. Kawata, M. Osawa and S. Okabe, Environ. Sci. Technol., 2009, 43, 6046–6051 CrossRef CAS PubMed.
- Q.-Q. Xiang, Y.-H. Kang, L.-H. Lian, Z.-Y. Chen, P. Wang, J.-M. Hu and L.-Q. Chen, Aquat. Toxicol., 2022, 252, 106318 CrossRef CAS PubMed.
- S.-F. Thai, C. P. Jones, B. L. Robinette, H. Ren, B. Vallanat, A. A. Fisher and K. T. Kitchin, J. Nanosci. Nanotechnol., 2021, 21, 5414–5428 CrossRef CAS PubMed.
- S. Gioria, P. Urbán, M. Hajduch, P. Barboro, N. Cabaleiro, R. La Spina and H. Chassaigne, Toxicol. In Vitro, 2018, 50, 347–372 CrossRef CAS PubMed.
- K. Williams, J. Milner, M. D. Boudreau, K. Gokulan, C. E. Cerniglia and S. Khare, Nanotoxicology, 2015, 9, 279–289 CrossRef CAS PubMed.
- X.-L. Wang, N. Yu, C. Wang, H.-R. Zhou, C. Wu, L. Yang, S. Wei and A.-J. Miao, ACS Nano, 2022, 16, 19002–19012 CrossRef CAS PubMed.
- S. Wang, X. Kang, H. Alenius, S. H. Wong, P. Karisola and H. El-Nezami, Food Chem. Toxicol., 2022, 169, 113368 CrossRef CAS PubMed.
- B. Cui, L. Ren, Q.-H. Xu, L.-Y. Yin, X.-Y. Zhou and J.-X. Liu, Aquat. Toxicol., 2016, 177, 295–305 CrossRef CAS PubMed.
- C. Lu, Y. Lv, G. Kou, Y. Liu, Y. Liu, Y. Chen, X. Wu, F. Yang, J. Luo and X. Yang, Ecotoxicol. Environ. Saf., 2022, 243, 113993 CrossRef CAS PubMed.
- R. Sardar, A. M. Funston, P. Mulvaney and R. W. Murray, Langmuir, 2009, 25, 13840–13851 CrossRef CAS PubMed.
- R. M. Pallares, X. Su, S. H. Lim and N. T. K. Thanh, J. Mater. Chem. C, 2016, 4, 53–61 RSC.
- R. Zhang, F. Kiessling, T. Lammers and R. M. Pallares, J. Phys. Chem. C, 2022, 126, 18580–18585 CrossRef CAS.
- Colloidal synthesis of plasmonic nanometals, ed. L. M. Liz-Marzán, Jenny Stanford Publishing, Singapore, 2020 Search PubMed.
- Z. Xi, R. Zhang, F. Kiessling, T. Lammers and R. M. Pallares, ACS Biomater. Sci. Eng., 2024, 10, 38–50 CrossRef CAS PubMed.
- E. C. Dreaden, A. M. Alkilany, X. Huang, C. J. Murphy and M. A. El-Sayed, Chem. Soc. Rev., 2012, 41, 2740–2779 RSC.
- R. Zhang, S. Thoröe-Boveleth, D. N. Chigrin, F. Kiessling, T. Lammers and R. M. Pallares, J. Nanobiotechnol., 2024, 22, 115 CrossRef CAS PubMed.
- Z. Xi, R. Zhang, S. Thoröe-Boveleth, F. Kiessling, T. Lammers and R. M. Pallares, ACS Appl. Nano Mater., 2023, 6, 17336–17346 CrossRef CAS.
- J. Zhang, G. Chen, D. Guay, M. Chaker and D. Ma, Nanoscale, 2014, 6, 2125–2130 RSC.
- Y. Huang, X. Lü, Y. Qu, Y. Yang and S. Wu, Biomaterials, 2015, 37, 13–24 CrossRef CAS PubMed.
- M. Hu, C. Wen, J. Liu, P. Cai, N. Meng, X. Qin, P. Xu, Z. Li and X.-C. Lin, ACS Appl. Bio Mater., 2023, 6, 1886–1895 CrossRef CAS PubMed.
- B. Nikoobakht and M. A. El-Sayed, Chem. Mater., 2003, 15, 1957–1962 CrossRef CAS.
- H.-H. Chang and C. J. Murphy, Chem. Mater., 2018, 30, 1427–1435 CrossRef CAS PubMed.
- R. M. Pallares, Y. Wang, S. H. Lim, N. T. K. Thanh and X. Su, Nanomedicine, 2016, 11, 2845–2860 CAS.
- S. Stolik, J. A. Delgado, A. Pérez and L. Anasagasti, J. Photochem. Photobiol., B, 2000, 57, 90–93 CrossRef CAS PubMed.
- C. Deraedt and D. Astruc, Acc. Chem. Res., 2014, 47, 494–503 CrossRef CAS PubMed.
- S. Sounderajan, G. Kiran Kumar, S. A. Kumar, A. C. Udas and G. Venkateswaran, Talanta, 2009, 78, 1122–1128 CrossRef CAS PubMed.
- D. Astruc, C. Ornelas and J. Ruiz, Acc. Chem. Res., 2008, 41, 841–856 CrossRef CAS PubMed.
- U. Lange, T. Hirsch, V. M. Mirsky and O. S. Wolfbeis, Electrochim. Acta, 2011, 56, 3707–3712 CrossRef CAS.
- M. Song, N. Liu, L. He, G. Liu, D. Ling, X. Su and X. Sun, Nano Res., 2018, 11, 2796–2808 CrossRef CAS.
- M. Chen, S. Chen, C. He, S. Mo, X. Wang, G. Liu and N. Zheng, Nano Res., 2017, 10, 1234–1248 CrossRef CAS.
- L. Nie, M. Chen, X. Sun, P. Rong, N. Zheng and X. Chen, Nanoscale, 2014, 6, 1271–1276 RSC.
- X. Huang, S. Tang, X. Mu, Y. Dai, G. Chen, Z. Zhou, F. Ruan, Z. Yang and N. Zheng, Nat. Nanotechnol., 2011, 6, 28–32 CrossRef CAS PubMed.
- S. Gurunathan, M. Qasim, C. H. Park, M. Arsalan Iqbal, H. Yoo, J. H. Hwang, S. J. Uhm, H. Song, C. Park, Y. Choi, J.-H. Kim and K. Hong, Nanomaterials, 2019, 9, 787 CrossRef CAS PubMed.
- Y. Zhou, G. He, H. Jiang, K. Pan and W. Liu, Environ. Int., 2023, 174, 107899 CrossRef CAS PubMed.
- I. Iavicoli, M. Farina, L. Fontana, D. Lucchetti, V. Leso, C. Fanali, V. Cufino, A. Boninsegna, K. Leopold, R. Schindl, D. Brucker and A. Sgambato, Toxicol. In Vitro, 2017, 42, 191–199 CrossRef CAS PubMed.
- J. Liang, N. Li, Z. Zhao, L. Ma, X. Wang, S. Li, X. Liu, T. Wang, Y. Du, G. Lu, J. Han, Y. Huang, D. Su and Q. Li, Angew. Chem., Int. Ed., 2019, 58, 15471–15477 CrossRef CAS PubMed.
- W. Si, Y. Yin, D. Cao, Y. Hu, X. Kang, J. Liu, X. Wang, Y. Xu, R. Yang and Q. Gu, Constr. Build. Mater., 2022, 340, 127700 CrossRef CAS.
- H. Xu, L.-L. He, Y.-F. Pei, C.-Z. Jiang, W.-Q. Li and X.-H. Xiao, Tungsten, 2021, 3, 20–37 CrossRef.
- H. Türkez, M. E. Arslan, E. Sönmez, A. Tatar, M. Açikyildiz and F. Geyikoğlu, Chem.-Biol. Interact., 2017, 273, 257–265 CrossRef PubMed.
- M. Jeyaraj, S. Gurunathan, M. Qasim, M.-H. Kang and J.-H. Kim, Nanomaterials, 2019, 9, 1719 CrossRef CAS PubMed.
- S. McFarlane, C. P. K. Manchee, J. W. Silverstone, J. Veinot and A. Meldrum, J. Visualized Exp., 2013, 50256 Search PubMed.
- Md. A. R. Khan, M. S. A. Mamun and M. H. Ara, Microchem. J., 2021, 171, 106840 CrossRef CAS.
- D. Pedone, M. Moglianetti, E. De Luca, G. Bardi and P. P. Pompa, Chem. Soc. Rev., 2017, 46, 4951–4975 RSC.
- P. V. Asharani, Y. Lianwu, Z. Gong and S. Valiyaveettil, Nanotoxicology, 2011, 5, 43–54 CrossRef CAS PubMed.
- R. E. Doherty, I. V. Sazanovich, L. K. McKenzie, A. S. Stasheuski, R. Coyle, E. Baggaley, S. Bottomley, J. A. Weinstein and H. E. Bryant, Sci. Rep., 2016, 6, 22668 CrossRef CAS PubMed.
- Z. Wang, L. Chen, C. Huang, Y. Huang and N. Jia, J. Mater. Chem. B, 2017, 5, 3498–3510 RSC.
- S. Gurunathan, M. Jeyaraj, H. La, H. Yoo, Y. Choi, J. T. Do, C. Park, J.-H. Kim and K. Hong, Int. J. Mol. Sci., 2020, 21, 440 CrossRef CAS PubMed.
- M. S. Chavali and M. P. Nikolova, SN Appl. Sci., 2019, 1, 607 CrossRef CAS.
- J. A. Makawana, C. B. Sangani, Y.-F. Yao, Y.-T. Duan, P.-C. Lv and H.-L. Zhu, Mini-Rev. Med. Chem., 2016, 16, 1303–1320 CrossRef CAS PubMed.
- E. Lee, Y. S. Yoon and D.-J. Kim, ACS Sens., 2018, 3, 2045–2060 CrossRef CAS PubMed.
- Y. Ren, W. Xie, Y. Li, J. Ma, J. Li, Y. Liu, Y. Zou and Y. Deng, ACS Cent. Sci., 2021, 7, 1885–1897 CrossRef CAS PubMed.
- M. Rahman, Nanotheranostics, 2023, 7, 424–449 CrossRef PubMed.
- H. Y. Son, B. I. Koo, J. B. Lee, K. R. Kim, W. Kim, J. Jang, M. S. Yoon, J.-W. Cho and Y. S. Nam, ACS Appl. Mater. Interfaces, 2018, 10, 27344–27354 CrossRef CAS PubMed.
- S. Li, W. Yin, Y. Li, J. Sun, M. Zhu, Z. Liu and T. Deng, Nanoscale, 2019, 11, 14912–14920 RSC.
- L. C. Mohr, A. P. Capelezzo, C. R. D. M. Baretta, M. A. P. M. Martins, M. A. Fiori and J. M. M. Mello, Polym. Test., 2019, 77, 105867 CrossRef CAS.
- T. G. Smijs and S. Pavel, Nanotechnol., Sci. Appl., 2011, 4, 95–112 CrossRef CAS PubMed.
- A. Ayati, A. Ahmadpour, F. F. Bamoharram, B. Tanhaei, M. Mänttäri and M. Sillanpää, Chemosphere, 2014, 107, 163–174 CrossRef CAS PubMed.
- S. Villa, V. Caratto, F. Locardi, S. Alberti, M. Sturini, A. Speltini, F. Maraschi, F. Canepa and M. Ferretti, Materials, 2016, 9, 771 CrossRef PubMed.
- C.-Y. Wu, K.-J. Tu, J.-P. Deng, Y.-S. Lo and C.-H. Wu, Materials, 2017, 10, 566 CrossRef PubMed.
- M.-C. Wu, K.-C. Hsiao, Y.-H. Chang and K. Kordás, ACS Appl. Nano Mater., 2019, 2, 1970–1979 CrossRef CAS.
- K. Fujita, M. Horie, H. Kato, S. Endoh, M. Suzuki, A. Nakamura, A. Miyauchi, K. Yamamoto, S. Kinugasa, K. Nishio, Y. Yoshida, H. Iwahashi and J. Nakanishi, Toxicol. Lett., 2009, 191, 109–117 CrossRef CAS PubMed.
- S. Montalvo-Quiros and J. L. Luque-Garcia, Food Chem. Toxicol., 2019, 127, 197–205 CrossRef CAS PubMed.
- M. Husain, A. T. Saber, C. Guo, N. R. Jacobsen, K. A. Jensen, C. L. Yauk, A. Williams, U. Vogel, H. Wallin and S. Halappanavar, Toxicol. Appl. Pharmacol., 2013, 269, 250–262 CrossRef CAS PubMed.
- B. Li, Y. Ze, Q. Sun, T. Zhang, X. Sang, Y. Cui, X. Wang, S. Gui, D. Tan, M. Zhu, X. Zhao, L. Sheng, L. Wang, F. Hong and M. Tang, PLoS One, 2013, 8, e55563 CrossRef CAS PubMed.
- Y. Ma, Y. Guo, H. Ye, K. Huang, Z. Lv and Y. Ke, Ecotoxicol. Environ. Saf., 2019, 176, 1–10 CrossRef CAS PubMed.
- D. D. Deobagkar, N. Patil and W. N. Gade, Int. J. Nanomed., 2016, 11, 4509–4519 CrossRef PubMed.
- A. B. Herman, D. Tsitsipatis and M. Gorospe, Mol. Cell, 2022, 82, 2252–2266 CrossRef CAS PubMed.
- S. Panni, R. C. Lovering, P. Porras and S. Orchard, Biochim. Biophys. Acta, Gene Regul. Mech., 2020, 1863, 194417 CrossRef CAS PubMed.
- J. Shi, Y. Zhang, Y. Ma, Z. Chen and G. Jia, Toxics, 2022, 10, 724 CrossRef CAS PubMed.
- J. Jiang, J. Pi and J. Cai, Bioinorg. Chem. Appl., 2018, 2018, 1–18 CrossRef PubMed.
- K. Girigoswami, M. Viswanathan, R. Murugesan and A. Girigoswami, Mater. Sci. Eng., C, 2015, 56, 501–510 CrossRef CAS PubMed.
- S. V. Gudkov, D. E. Burmistrov, D. A. Serov, M. B. Rebezov, A. A. Semenova and A. B. Lisitsyn, Front. Phys., 2021, 9, 641481 CrossRef.
- P. Fan, D. Zhang, Y. Wu, J. Yu and T. P. Russell, Adv. Funct. Mater., 2020, 30, 2002932 CrossRef CAS.
- J. M. Fine, T. Gordon, L. C. Chen, P. Kinney, G. Falcone and W. S. Beckett, J. Occup. Environ. Med., 1997, 39, 722–726 CrossRef CAS PubMed.
- L. C. Rohrs, Arch. Intern. Med., 1957, 100, 44 CrossRef.
- N. Hadrup, F. Rahmani, N. R. Jacobsen, A. T. Saber, P. Jackson, S. Bengtson, A. Williams, H. Wallin, S. Halappanavar and U. Vogel, Nanotoxicology, 2019, 13, 1275–1292 CrossRef CAS PubMed.
- X. Pei, H. Jiang, G. Xu, C. Li, D. Li and S. Tang, Int. J. Mol. Sci., 2022, 23, 6724 CrossRef CAS PubMed.
- X. Wang, S. Chen, Y. Qin, H. Wang, Z. Liang, Y. Zhao, L. Zhou and C. J. Martyniuk, Aquat. Toxicol., 2022, 253, 106333 CrossRef CAS PubMed.
- H. H. Gustafson, D. Holt-Casper, D. W. Grainger and H. Ghandehari, Nano Today, 2015, 10, 487–510 CrossRef CAS PubMed.
- Z. Doumandji, R. Safar, M. Lovera-Leroux, S. Nahle, H. Cassidy, D. Matallanas, B. Rihn, L. Ferrari and O. Joubert, Cell Biol. Toxicol., 2020, 36, 65–82 CrossRef PubMed.
- R. Jin, B. Lin, D. Li and H. Ai, Curr. Opin. Pharmacol., 2014, 18, 18–27 CrossRef CAS PubMed.
- C. Blanco-Andujar, A. Walter, G. Cotin, C. Bordeianu, D. Mertz, D. Felder-Flesch and S. Begin-Colin, Nanomedicine, 2016, 11, 1889–1910 CrossRef CAS PubMed.
- Y. Li, R. Zhang, R. Barmin, E. Rama, M. Schoenen, F. Schrank, V. Schulz, I. Slabu, F. Kiessling, T. Lammers and R. M. Pallares, Nanoscale Adv., 2024 10.1039/D4NA00378K.
- H. Nogueira, S. Toma, A. Silveira Jr. and K. Araki, J. Braz. Chem. Soc., 2020, 31, 11 Search PubMed.
- A. Kumar, G. Sharma, M. Naushad and S. Thakur, Chem. Eng. J., 2015, 280, 175–187 CrossRef CAS.
- N. Singh, G. J. S. Jenkins, R. Asadi and S. H. Doak, Nano Rev., 2010, 1, 5358 CrossRef PubMed.
- R. S. Bonadio, M. Cunha, J. P. F. Longo, R. B. Azevedo and M. J. PoÇas-Fonseca, J. Nanosci. Nanotechnol., 2020, 20, 1454–1462 CrossRef CAS PubMed.
- P. C. Ke, S. Lin, W. J. Parak, T. P. Davis and F. Caruso, ACS Nano, 2017, 11, 11773–11776 CrossRef CAS PubMed.
- L. Xia, S. Alqahtani, C. R. Ferreira, U. K. Aryal, K. Biggs and J. H. Shannahan, Nanomaterials, 2022, 12, 2022 CrossRef CAS PubMed.
- C. Sun, H. Li and L. Chen, Energy Environ. Sci., 2012, 5, 8475 RSC.
- A. Nemmar, S. Al-Salam, S. Beegam, P. Yuvaraju and B. Ali, Biomolecules, 2019, 9, 376 CrossRef CAS PubMed.
- S. De, S. Mohanty and S. K. Nayak, Bioprocess Biosyst. Eng., 2015, 38, 1671–1683 CrossRef CAS PubMed.
- S. Chaudhary, P. Sharma, D. Singh, A. Umar and R. Kumar, ACS Sustainable Chem. Eng., 2017, 5, 6803–6816 CrossRef CAS.
- S. Balaji, B. K. Mandal, L. Vinod Kumar Reddy and D. Sen, Bioengineering, 2020, 7, 26 CrossRef CAS PubMed.
- R. M. Pallares, S. L. Karstens, T. Arino, A. M. Minor and R. J. Abergel, ACS Appl. Nano Mater., 2023, 6, 8141–8145 CrossRef CAS.
- G. Lu, H. Zheng, J. Lv, G. Wang and X. Huang, J. Power Sources, 2020, 480, 229091 CrossRef CAS.
- S. Verstraelen, S. Remy, E. Casals, P. De Boever, H. Witters, A. Gatti, V. Puntes and I. Nelissen, Toxicol. Lett., 2014, 228, 157–169 CrossRef CAS PubMed.
- X. He, Z. R. Jarrell, M. R. Smith, V. T. Ly, Y. Liang, M. Orr, Y.-M. Go and D. P. Jones, Toxicol. Appl. Pharmacol., 2023, 459, 116327 CrossRef CAS PubMed.
- P. R. Suma, R. A. Padmanabhan, S. R. Telukutla, R. Ravindran, A. K. G. Velikkakath, C. D. Dekiwadia, W. Paul, M. Laloraya, S. M. Srinivasula, S. V. Bhosale and R. S. Jayasree, Free Radicals Biol. Med., 2020, 161, 198–211 CrossRef CAS PubMed.
- A. R. Gliga, S. Di Bucchianico, E. Åkerlund and H. L. Karlsson, Nanomaterials, 2020, 10, 649 CrossRef CAS PubMed.
- B. Yu, X. Lu, X. Feng, T. Zhao, J. Li, Y. Lu, F. Ye, X. Liu, X. Zheng, Z. Shen, X. Jin, W. Chen and Q. Li, Int. J. Nanomed., 2023, 18, 7713–7728 CrossRef CAS PubMed.
- R. Frick, B. Müller-Edenborn, A. Schlicker, B. Rothen-Rutishauser, D. O. Raemy, D. Günther, B. Hattendorf, W. Stark and B. Beck-Schimmer, Toxicol. Lett., 2011, 205, 163–172 CrossRef CAS PubMed.
- G. M. Saladino, B. Brodin, R. Kakadiya, M. S. Toprak and H. M. Hertz, Sci. Adv., 2024, 10, eadl2267 CrossRef CAS PubMed.
- S. M. Reimann and M. Manninen, Rev. Mod. Phys., 2002, 74, 1283–1342 CrossRef CAS.
- J. Wu, S. Chen, A. Seeds and H. Liu, J. Phys. D: Appl. Phys., 2015, 48, 363001 CrossRef.
- R. E. Bailey, A. M. Smith and S. Nie, Phys. E, 2004, 25, 1–12 CrossRef CAS.
- R. M. Pallares, P. Agbo, X. Liu, D. D. An, S. S. Gauny, S. E. Zeltmann, A. M. Minor and R. J. Abergel, ACS Appl. Mater. Interfaces, 2020, 12, 40078–40084 CrossRef CAS PubMed.
- S. Jafari, H. Derakhshankhah, L. Alaei, A. Fattahi, B. S. Varnamkhasti and A. A. Saboury, Biomed. Pharmacother., 2019, 109, 1100–1111 CrossRef CAS PubMed.
- S. Porrang, S. Davaran, N. Rahemi, S. Allahyari and E. Mostafavi, Int. J. Nanomed., 2022, 17, 1803–1827 CrossRef PubMed.
- N. K. Tarasova, A. Gallud, A. J. Ytterberg, A. Chernobrovkin, J. R. Aranzaes, D. Astruc, A. Antipov, Y. Fedutik, B. Fadeel and R. A. Zubarev, J. Proteome Res., 2017, 16, 689–697 CrossRef CAS PubMed.
- F. Akhter, A. A. Rao, M. N. Abbasi, S. A. Wahocho, M. A. Mallah, H. Anees-ur-Rehman and Z. A. Chandio, Silicon, 2022, 14, 8295–8310 CrossRef CAS.
- T. I. Janjua, Y. Cao, C. Yu and A. Popat, Nat. Rev. Mater., 2021, 6, 1072–1074 CrossRef CAS PubMed.
- M. Iftikhar, A. Noureen, M. Uzair, F. Jabeen, M. Abdel Daim and T. Cappello, Int. J. Environ. Res. Public Health, 2021, 18, 1758 CrossRef CAS PubMed.
- J.-P. Klein, L. Mery, D. Boudard, C. Ravel, M. Cottier and D. Bitounis, Int. J. Mol. Sci., 2022, 24, 576 CrossRef PubMed.
- G. Zhou, L. Ren, H. Yin, J. Liu, X. Li, J. Wang, Y. Li, Y. Sang, Y. Zhao, X. Zhou and Z. Sun, NanoImpact, 2021, 23, 100348 CrossRef CAS PubMed.
- Y. Zhu, Y. Zhang, Y. Li, C. Guo, Z. Fan, Y. Li, M. Yang, X. Zhou, Z. Sun and J. Wang, J. Hazard. Mater., 2022, 434, 128820 CrossRef CAS PubMed.
- H. S. Choi, W. Liu, P. Misra, E. Tanaka, J. P. Zimmer, B. I. Ipe, M. G. Bawendi and J. V. Frangioni, Nat. Biotechnol., 2007, 25, 1165–1170 CrossRef CAS PubMed.
- Z. Yu, Q. Li, J. Wang, Y. Yu, Y. Wang, Q. Zhou and P. Li, Nanoscale Res. Lett., 2020, 15, 115 CrossRef CAS PubMed.
- R. Mohammapdour and H. Ghandehari, Adv. Drug Delivery Rev., 2022, 180, 114022 CrossRef CAS PubMed.
- K. Bhattacharjee and B. L. V. Prasad, Chem. Soc. Rev., 2023, 52, 2573–2595 RSC.
- E. Fröhlich, Int. J. Nanomed., 2012, 5577 CrossRef PubMed.
- C. He, Y. Hu, L. Yin, C. Tang and C. Yin, Biomaterials, 2010, 31, 3657–3666 CrossRef CAS PubMed.
- J. Mosquera, I. García and L. M. Liz-Marzán, Acc. Chem. Res., 2018, 51, 2305–2313 CrossRef CAS PubMed.
- D. Docter, D. Westmeier, M. Markiewicz, S. Stolte, S. K. Knauer and R. H. Stauber, Chem. Soc. Rev., 2015, 44, 6094–6121 RSC.
- M. Hadjidemetriou and K. Kostarelos, Nat. Nanotechnol., 2017, 12, 288–290 CrossRef CAS PubMed.
- E. Casals and V. F. Puntes, Nanomedicine, 2012, 7, 1917–1930 CrossRef CAS PubMed.
- T. Kopac, Int. J. Biol. Macromol., 2021, 169, 290–301 CrossRef CAS PubMed.
- C. C. Fleischer and C. K. Payne, Acc. Chem. Res., 2014, 47, 2651–2659 CrossRef CAS PubMed.
- G. Caracciolo, S. Palchetti, V. Colapicchioni, L. Digiacomo, D. Pozzi, A. L. Capriotti, G. La Barbera and A. Laganà, Langmuir, 2015, 31, 10764–10773 CrossRef CAS PubMed.
- V. Palmieri and G. Caracciolo, Nanoscale Adv., 2022, 4, 3300–3308 RSC.
- M. Carter and J. Shieh, in Guide to Research Techniques in Neuroscience, Elsevier, 2015, pp. 295–310 Search PubMed.
- L. Yuan, Q. Chen, J. E. Riviere and Z. Lin, J. Drug Delivery Sci. Technol., 2023, 83, 104404 CrossRef CAS PubMed.
- A. Zhang, K. Meng, Y. Liu, Y. Pan, W. Qu, D. Chen and S. Xie, Adv. Colloid Interface Sci., 2020, 284, 102261 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr02328e |
| This journal is © The Royal Society of Chemistry 2024 |
