Enhancement of anammox performance in a novel non-woven fabric membrane bioreactor (nMBR)†
Long-Fei Rena,
Shuang Lianga,
Huu Hao Ngob,
Wenshan Guob,
Shou-Qing Ni*a,
Cui Liuc,
Yuan-Kun Zhaoad and
Daisuke Hirae
aShandong Provincial Key Laboratory of Water Pollution Control and Resource Reuse, School of Environmental Science and Engineering, Shandong University, No. 27 Shanda South Road, Jinan 250100, Shandong, PR China. E-mail: sqni@sdu.edu.cn
bCentre for Technology in Water and Wastewater, School of Civil and Environmental Engineering, University of Technology Sydney, Sydney, NSW 2007, Australia
cDepartment of Mathematics and Statistics, Texas Tech University, Broadway and Boston, Lubbock, TX 79409-1042, USA
dSchool of Civil and Environmental Engineering, Georgia Institute of Technology, North Ave NW, Atlanta, GA 30332, USA
eDepartment of Applied Life Science, Faculty of Biotechnology and Life Science, Sojo University, 4-22-1 Ikeda, Kumamoto 860-0082, Japan
First published on 28th September 2015
Abstract
To reduce operating costs and membrane fouling of conventional membrane bioreactors (cMBR), a novel MBR using a non-woven fabric membrane (nMBR) was constructed and the performance of the two MBRs was compared for anaerobic ammonium oxidation (anammox) cultivation. The results showed that the start-up period for the nMBR (44 days) was notably shorter than that for the cMBR (56 days), meanwhile the nMBR achieved a 2-times higher nitrogen removal rate (231.5 mg N per L per d) compared to the cMBR (112.3 mg N per L per d). Illumina MiSeq sequencing showed that Candidatus Kuenenia and Candidatus Jettenia were the main distinguished anammox bacteria. FISH analysis revealed that anammox bacteria predominated in both reactors, especially in the nMBR (58%) corresponding to a qPCR analysis of 1.07 × 109 copies per mL (day 120). N2O emission analysis confirmed the advantage of the nMBR in N2O reduction to reduce the influence of greenhouse gas emission while treating identical nitrogen. These results clearly demonstrated that nMBRs could be a prospective choice for anammox start-up and performance enhancement.
1. Introduction
Anaerobic ammonium oxidation (anammox) is a promising biotechnology to treat ammonium-containing wastewater, especially with low biological degradable carbon.1,2 In this process, anammox bacteria, namely Planctomycetales, convert ammonium to dinitrogen using nitrite as an electron acceptor under strict anaerobic conditions (eqn (1)). Anammox has been implemented in various full-scale applications for treating ammonium-rich wastewater, such as sludge digestion water and industrial wastewater.2,3 As an efficient and economical alternative to traditional nitrification and denitrification processes, the anammox process avoids the addition of external carbon and aeration, resulting in remarkable savings of operation cost associated with higher nitrogen removal rates (NRR).4–6| NH4+ + 1.32NO2− + 0.06HCO3− + 0.13H+ → 1.02N2 + 0.26NO3− + 0.066CH2O0.5N0.15 + 2.03H2O | (1) |
However, the start-up of the anammox process is mainly affected by the availability of anammox biomass due to its slow-growing rate (0.072 per d measured at 32 °C).7,8 Even a quite unimpressive biomass loss via the effluent or another approach may impede the start-up of the anammox process severely because of the deficient biomass. Moreover, the severe sludge washout caused by granule floatation could lead to instability or even system collapses, particularly at high nitrogen loading.9 Thus, an efficient system or operation strategy is required in order to minimize biomass run off with effluent.
Massive efforts towards reducing biomass loss via the choice of suitable reactor configurations have been made by researchers. Conventional membrane bioreactors (cMBRs) using hollow fiber modules have been regarded as suitable reactors to start up the anammox process as they can achieve high biomass retention.10,11 Several studies have proved that submerged MBRs are excellent tools for enriching slow-growing microorganisms,12 especially anammox bacteria.11 In MBRs, membrane modules can not only function as biofilters, but also as biofilm carriers,13 which is beneficial to the formation of biomass aggregates, attached biofilms and granular sludge. With higher biomass density, these sludge forms are more efficient to deal with high load nitrogen pollutants than flocculated sludge.10,14,15
Considering the cost of hollow fiber membranes, MBRs using non-woven fabric (nMBRs) have attracted increasing interest in biofilm formation and biomass retention. Non-woven fabric membranes are porous with abundant small hollow areas, which are efficient in attaching anammox biomass, leading to enhanced anammox reactor performance.13,16,17 With larger pore size compared to hollow fiber membrane, the separation mode of nMBRs is broadly categorized as macro-filtration, while that of cMBRs is categorized as micro-filtration. Therefore, the operating transmembrane pressures (TMPs) of cMBRs (the TMP of micro-filtration <100–200 kPa) are generally higher than those of nMBRs (the TMP of macro-filtration <25 kPa).18–20 Based on the low TMP which is employed for monitoring membrane fouling, the membrane fouling of non-woven fabric including membrane pore blocking, cake formation and biofouling could also be alleviated to some extent.19
Many researchers have explored the feasibility of anammox start-up with submerged anaerobic cMBRs.10,11,14,21 However, there are few studies devoted to using nMBRs to cultivate anammox bacteria. Meng et al.13 employed a nMBR to evaluate the performance of a novel anammox biofilm process using anammox sludge as seed sludge. The results showed that NRR of about 1.6 kg N per m3 per d could be achieved. The non-woven modules as well as the attached biofilm also prevented the loss of free bacteria effectively. Although they analyzed the composition of the bacterial community in the reactor, the definite enumeration of anammox bacteria was not reported. Therefore, further investigation of the components and amount of biocenosis during the anammox start-up period using mixed sludge is needed. In another aspect, Meng et al.13 laid emphasis on investigating the nitrogen removal limit of non-woven modules with seeding mature anammox bacteria, while more attention was paid to explore a program with more potential for municipal and industrial applications using mixed sludge with few anammox bacteria in this research. The concentration and load of nitrogen pollutant also were set to a scale nearing reality.
This study focuses on comparing the anammox performance of two submerged MBRs, namely a cMBR and a nMBR. Both reactors were inoculated with the same type of seed sludge for anammox start-up. Start-up time and nitrogen removal performance were compared between these two reactors. Furthermore, high-throughput sequencing (Illumina MiSeq sequencing), fluorescence in situ hybridization (FISH) and real-time quantitative PCR (qPCR) were employed to investigate the composition and quantities of the bacterial communities in the reactors. The variation of N2O emission was also examined during the entire anammox cultivation process. Since the analysis of the bacterial community in a nMBR has not been explored in previous research, it could provide valuable information for quick start-up of the anammox process and its application.
2. Materials and methods
2.1. Experimental set-up
The configuration of the submerged MBR in this study is depicted in Fig. 1, which shows a double-walled cylindrical column with an inner diameter of 14 cm and a height of 50 cm. The only difference between the two MBRs was the selection of membrane modules (Fig. 1(5), cMBR: hollow fiber membrane and nMBR: non-woven fabric). The hollow fiber membrane module (absolute pore size: 0.1 μm) consisted of 100 tubes (diameter ca. 1 mm, length ca. 300 mm) and the non-woven fabric module (a pore size of 0.1 mm approximately) was composed of two same rectangular modules (24 cm × 10 cm). The filtration area of both membrane modules was around 0.01 m2.The peristaltic influent pump (BT100-2J, LongerPump, China) was used to adjust the influent rate for controlling the hydraulic retention time (HRT). A liquid level sensor was placed at a height of 39 cm in the reactor to maintain an effective volume of 6 L. A level-controlled peristaltic effluent pump (BT300-2J, LongerPump, China) was used for drainage. The TMP of the membrane module was monitored by a vacuum gauge connected with an effluent pump. A thermostatic bath was connected to the double wall of the reactor to maintain the temperature at 33 °C. Both MBR reactors were also covered with tinfoil for light avoidance.
2.2. Operational strategy
The seed sludge was a mixture of 1/4 anammox sludge and 3/4 anaerobic granular sludge. Anammox sludge was taken from a lab-scale UASB reactor which was used for anammox process start-up22 and the anaerobic sludge was obtained from a running UASB reactor. 4 L mixed sludge was divided equally to control the initial mixed liquor suspended solids (MLSS) at 4000 ± 50 mg L−1 in cMBR and nMBR.The composition of synthetic wastewater for anammox bacteria enrichment is shown in Table 1 and the trace elements were composed of EDTA: 20.0 g L−1, FeSO4: 5.0 g L−1, ZnSO4·7H2O: 0.43 g L−1, CoCl2·6H2O: 0.24 g L−1, MnCl2·4H2O: 0.99 g L−1, CuSO4·5H2O: 0.25 g L−1, Na2MoO4·2H2O: 0.22 g L−1, NiCl2·6H2O: 0.19 g L−1, Na2SeO4·10H2O: 0.21 g L−1 and H3BO4: 0.014 g L−1. The ratio of ammonium to nitrite concentration in synthetic wastewater was set to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.20. In order to maintain anaerobic conditions in each reactor, the synthetic wastewater was fed to the reactor after deoxygenation by flushing with argon gas. The pH of the influent varied in the range of 7.5–8.0 without intended control. The injection rate was adjusted to maintain the initial HRT at 48 h (the first 10 days was set to 96 h for anammox bacteria in seed sludge to adapt to the new environment), and then the HRT was shortened by increasing the injection rate when the removal of ammonium and nitrite was effective (>90%) and stable. The membrane module was replaced or cleaned chemically when the TMP reached up to 45 kPa to prevent the membrane fouling.
1.20. In order to maintain anaerobic conditions in each reactor, the synthetic wastewater was fed to the reactor after deoxygenation by flushing with argon gas. The pH of the influent varied in the range of 7.5–8.0 without intended control. The injection rate was adjusted to maintain the initial HRT at 48 h (the first 10 days was set to 96 h for anammox bacteria in seed sludge to adapt to the new environment), and then the HRT was shortened by increasing the injection rate when the removal of ammonium and nitrite was effective (>90%) and stable. The membrane module was replaced or cleaned chemically when the TMP reached up to 45 kPa to prevent the membrane fouling.
| Substance | Concentration | Unit |
|---|---|---|
| (NH4)2SO4 | 594 | mg L−1 |
| NaNO2 | 746 | mg L−1 |
| KHCO3 | 500 | mg L−1 |
| CaCl2·2H2O | 180 | mg L−1 |
| MgSO4·7H2O | 120 | mg L−1 |
| KH2PO4 | 27 | mg L−1 |
| Trace elements | 1 | mL L−1 |
2.3. Chemical analyses
The concentrations of ammonium, nitrite and nitrate were measured according to standard APHA method after filtering samples through a 0.45 μm syringe filter. The MLSS of sludge and suspended solids (SS) of effluent were also determined according to standard methods.23 A digital portable DO meter (HQ40d, Hach, America) and a microelectrode (5307, Unisense, Denmark) were employed to detect DO level and N2O emission in liquid, respectively.2.4. DNA extraction and Illumina MiSeq sequencing
A 1 mL biomass sample from each reactor was collected and harvested by centrifugation for DNA extraction on days 60 and 120 using a PowerSoil™ DNA Isolation Kit (MO BIO Laboratories, USA) according to the manufacturer’s protocol. Based on the main existing form of sludge, the biomass sample in cMBR was collected from suspended sludge while cake layer biofilm was employed for nMBR.Agarose gel electrophoresis (1%), staining with ethidium bromide solution, was adopted to assess extracted DNA quality. The following PCR reactions were processed on an ABI GeneAmp® 9700 (Applied Biosystems, USA) and the PCR product was gel purified. Afterwards, Illumina MiSeq sequencing was carried out by Shanghai Majorbio Bio-pharm Technology Co., Ltd (Shanghai, China) using DNA samples on day 120.
2.5. Fluorescence in situ hybridization (FISH)
FISH analysis was applied to verify the existence and distribution of anammox bacteria in mature sludge. The sludge samples were collected from both reactors on day 120. The EUB338Mix (EUB338, EUB338-II and EUB338-III) probe (FITC-labeled) was used to monitor almost all bacteria (green) while anammox bacteria (red) were targeted using an AMX820 probe (Cy3-labeled). An Olympus BX53 confocal scanning laser microscope (Olympus, Japan) was employed for image acquisitions. Fluorescence images obtained were also processed in a semi-quantitative analysis using the software Image-Pro Plus 6.0. The concrete steps of FISH were conducted as described elsewhere.242.6. Real-time quantitative PCR (qPCR)
Amx809F and Amx1066R (Table 2) were used as the primer set to target the 16S rRNA gene for quantifying anammox bacteria.25 The concrete steps of qPCR were carried out according to the previous study22 and the relevant curves were added as ESI (Fig. S1 and S2†).| Primer name | Target | Sequence (5′–3′) | Target site | Annealing temperature (°C) |
|---|---|---|---|---|
| Amx809F | Anammox | GCCGTAAACGATGGGCACT | 809–826 | 60 |
| Amx1066R | Anammox | AACGTCTCACGACACGAGCTG | 1047–1066 | 60 |
3. Results and discussion
3.1. Enrichment and reactor performance
The initial nitrogen loading rate (NLR) which was applied in the cMBR was 67.5 mg N per L per d and the influent ammonium and nitrite concentrations were maintained around 123.3 ± 4.0 and 152.4 ± 3.7 mg L−1 respectively as shown in Fig. 2. During days 0 to 12, the input ammonium was barely consumed while much more nitrite was consumed, indicating no significant anammox phenomenon was detected. The effluent nitrate concentration ranged from 0 to 5 mg L−1, which might be attributed to endogenous denitrification since no organics were fed as in other relevant research.26,27 After 30 days, the removal efficiencies of ammonium and nitrite were about 44.8 ± 0.7% and 52.8 ± 1.0%, leading to a NRR of about 63 mg N per L per d. The occurrence of the anammox process could be confirmed by ammonium (removal efficiency varied between 44.8 and 87.6%) and nitrite (removal efficiency varied between 52.7 and 91.0%) being consumed synchronously in conjunction with nitrate production in the range of 9.0–35.8 mg N per L. The increasing ammonium and nitrite removal efficiencies reflected a tendency toward gradual increase in anammox activity.During days 56 to 60, both ammonium and nitrite removal efficiencies were above 90%, suggesting a successful anammox start-up. However when the NLR was further increased to 203.2 mg N per L per d from days 64 to 72, the nitrogen removal performance suddenly decreased to 66.0 ± 4.8% and 60.4 ± 9.1% for ammonium and nitrite respectively. Literature shows that a residual nitrite concentration of 50 mg L−1 could inhibit anammox bacteria partially while a 100 mg L−1 concentration could inactivate anammox bacteria completely.28 As the cMBR did not present the capacity for accommodating higher nitrogen concentrations of nMBR, the NLR was adjusted back to 137.2 mg N per L per d again through regulating the HRT to the initial 48 h to avoid damage by nitrite. After that, the performance of cMBR improved gradually again. On day 108, the ammonium and nitrite removal efficiencies were steadily maintained at 94.0 ± 1.5% indicating the success of anammox activity recovery.
The initial nitrogen compound concentrations together with the NLR of nMBR were set to the same values as those for cMBR while endogenous denitrification was also found during days 0 to 12 which led to low ammonium and high nitrite removal efficiencies. From days 16 to 28, the removal of ammonium and nitrite improved rapidly to 64.8 ± 20.2% and 79.6 ± 12.0% respectively, revealing significant evidence of anammox activity, and then nMBR achieved stable ammonium and nitrite nitrogen removal above 90% on day 44. Similarly, in order to supply efficient nutrients and increase anammox growth capacity, the NLR was also increased to 203.2 mg N per L per d on day 64. On the following day, the nitrite concentration of the effluent increased to 38.4 mg N per L and the total nitrogen removal efficiency dropped to 67.7%, which was consistent with the phenomenon observed by Isaka et al.29 However, unlike the inferior performance of cMBR, after accommodation for a few days, anammox removal in nMBR rose significantly again. During days 84 to 100, the nitrite removal efficiency remained close to 100%, while ammonium removal efficiency varied from 88% to 95%.
No inhibition of nitrogen removal was observed in nMBR until the NLR was increased by 2 times (from around 141.7 to the maximum 283.3 mg N per L per d) on day 104, when the ammonium and nitrite removal efficiencies speedily declined to 29.9 ± 1.2% and 39.7 ± 0.8% respectively. Afterwards, the anammox activity in the system was recovered within 4 days. On day 128, the nMBR exhibited 2-times the NRR (nMBR: 245.4 mg N per L per d, cMBR: 122.6 mg N per L per d), showing the enormous advantage of adopting a non-woven fabric for improving the NRR. Compared with other relevant research demonstrated in Table 3 (start up from activated or anoxic sludge), the nMBR could accomplish the start-up of the anammox process successfully in a shorter time with a higher NRR and nitrogen removal efficiency. In addition, inoculation with mixed sludge rather than the total anammox sludge used in many other studies would decrease the financial pressure during start-up and operation due to the scarcity and rarity of anammox sludge.
| Reactor type | Source sludge | Membrane module | Start-up conditions | Maximal NRR (mg N per L per d) | Ammonium removal efficiency (%) | Nitrite removal efficiency (%) | Start-up time (day) | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| Ammonium (mg L−1) | Nitrite (mg L−1) | ||||||||
| a A non-woven fabric membrane was used as the external membrane module in this study.b A hollow fiber membrane module was employed as the curtain shape in this study.c NRR was considered as the removal of ammonium and nitrite in this study.d Start-up date was defined as the end of the unstable phase in this study.e The average total nitrogen removal efficiency in the stable stage was 73.6% in this study and the MSBR was a membrane sequencing batch reactor with a submerged hollow fiber membrane module for biomass retention. | |||||||||
| nMBR | Activated sludge | Non-woven fabrica | 40 | 53 | 1047.5 | 90.9 | 95.0 | 64 | 17 |
| nMBR | Anammox sludge | Non-woven fabric | 25 | 25 | 1600 | >80 | >90 | — | 13 |
| MBR | Activated sludge | Hollow fiber membraneb | 50 | 50 | 80 | >90 | >90 | 50 | 10 |
| MBR | Activated sludge | Hollow fiber membrane | 50 | 50 | 345.2c | ∼100 | ∼100 | 59 | 10 |
| MBR | Anoxic sludge | Hollow fiber sheet | 25 | 25 | 218.5 | >95 | >95 | 25d | 21 |
| MBR | Anammox sludge | Hollow fiber membrane | 1680 | 1680 | 1600 | — | ≥99 | — | 11 |
| MSBR | Anammox sludge | Hollow fiber membrane | 75.3 | 83.7 | 710 | >80 | >90e | — | 14 |
| cMBR | Mixed sludge | Hollow fiber membrane | 126 | 151.3 | 124.2 | >95 | >95 | 56 | This study |
| nMBR | Mixed sludge | Non-woven fabric | 126 | 151.3 | 245.4 | >95 | >95 | 44 | This study |
Fig. 3 illustrates the values of consumed nitrite/ammonium ratio and produced nitrate/consumed ammonium ratio. The values of consumed nitrite/ammonium ratio in the two reactors during the anammox stage varied from 1.20 to 1.30, which was a little lower than the theoretical value of 1.32 as described in eqn (1). This could partly be attributed to the influent nitrite/ammonium ratio of 1.20 aimed at avoiding the damage of high residual nitrite on anammox bacteria. The existence of nitrification and denitrification also contributed to this phenomenon. The values of the produced nitrate/consumed ammonium ratio in the two reactors during the anammox stage also varied in a normal range of 0.24 to 0.29, approaching the theoretical value of 0.26. Corresponding with the lower nitrogen removal efficiency in cMBR from day 64, the values of nitrite removal/ammonium removal in cMBR also were lower than those in nMBR, particularly during days 64 to 92. Such long-term different nitrogen loading might further lead to an effect on biomass or microorganism growth.
 | ||
| Fig. 3 The values of nitrite removal/ammonium removal in cMBR (■) and nMBR (●) and the values of nitrate production/ammonium removal in cMBR (□) and nMBR (○) from days 32 to 130. | ||
3.2. Biomass growth and retention
The main purpose for adopting a MBR configuration was to minimize the loss of biomass through effluent flow. During the whole experiment, the effluent SS from the cMBR was less than 15 mg L−1, whereas nMBR showed a higher biomass loss (25–35 mg L−1). On day 60, the effluent SS in nMBR dropped below 19 mg L−1 because the suspended sludge turned into attached biofilm moderately, preserving more precious anammox bacteria, and was maintained at less than 13 mg L−1 from day 90 (Fig. S3†).Apart from the function of membrane-like separation, the non-woven fabric modules also served as biofilm carriers. Thus, anammox bacteria in the suspended sludge were attached to the non-woven fabric due to the significant attachment propensity of anammox bacteria.13 The shear stress, which generated from water flow crossing the flat non-woven membrane, also directed biomass to approach the fabric and finally attach on the membrane loosely.30 With the extension of this process, the loose biofilm structure became denser. The suction from the effluent pump might also facilitate this phenomenon.
After 130 days of operation, nearly all biomass in cMBR existed in the form of suspended sludge with a little sludge adhering on the membrane surface, and the MLSS in cMBR gradually climbed to over 6650 ± 50 mg L−1. On the other hand, the biomass in nMBR mainly presented as attached biofilm on the non-woven fabric while a small number of residual free living bacteria were observed, resulting in a difficulty in MLSS evaluation. At the end of the operating stage, a dry weight of 46.48 g of dominant attached growth biomass and 254 ± 20 mg L−1 of MLSS were detected, representing approximately 8000 ± 20 mg L−1 of total MLSS in nMBR. In addition, the attached biomass on the non-woven membrane could act as a permeable reactive barrier for nitrogen removal and ammonium-rich wastewater would be disposed of again by the microorganisms in the cake layer before being discharged. Analysis of the nitrogen concentration showed that around half of the ammonium (ca. 49%) and nitrite (ca. 53%) in the sludge mixture of nMBR were removed during their transport to the effluent through the biofilm.
3.3. Illumina MiSeq sequencing analysis
The pyrosequencing-based analysis of 16S rRNA genes was used to assess the compositions of the bacterial communities. Good’s coverage values were 91.34% and 91.60% for cMBR and nMBR, respectively, which indicated that the number of sequences was sufficient to characterize the microbial communities. The composition of bacterial communities at the phylum level in both reactors is shown in Fig. 4(a). The top 9 phyla observed in cMBR and nMBR were Actinobacteria, Armatimonadetes, Bacteroidetes, Chlorobi, Chloroflexi, GOUTA4, Nitrospirae, Planctomycetes and Proteobacteria.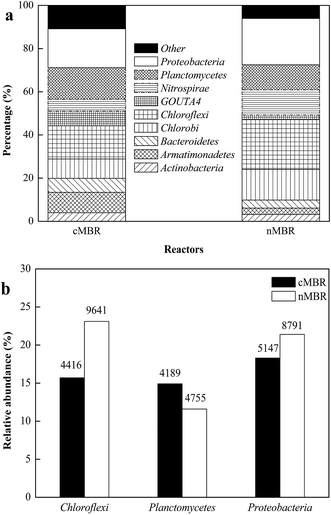 | ||
| Fig. 4 (a) Compositions of different communities at the phylum level in cMBR and nMBR; (b) relative abundance of different communities at the phylum level in cMBR and nMBR. | ||
Planctomycetes was the most important division (Fig. 4(b)), comprising approximately 14.5% (4189 reads) in cMBR and 11.8% (4755 reads) in nMBR. Anammox bacteria are mainly identified as Planctomycetes.31–33 However, with regard to species, Illumina MiSeq sequencing analysis cannot indicate specifically what these microorganisms are, as most of them are unclassified Planctomycetes. Only two of the 102 detected Planctomycetes operational taxonomic units (OTUs) were identified to be the recognized anammox bacteria species (Candidatus Kuenenia and Candidatus Jettenia). In addition, not all the detected Planctomycetes could be defined as anammox bacteria, and thus anammox bacteria as Planctomycetes had a quite lower abundance than expected.
Similar to previous findings,32,34 there were some other phyla in the reactors coexisting with anammox bacteria (i.e. Planctomycetes). The most abundant phylum was Proteobacteria whose relative abundances were 18.0% (5147 reads) in cMBR and 21.5% (8971 reads) in nMBR. Previous studies indicated that several species of β-Proteobacteria (38 OTUs, 3233 reads in cMBR; 54 OTUs, 5530 reads in nMBR) could embody low anammox activity to convert ammonium and nitrite to N2 using nitrogen dioxide as an electron acceptor, for instance autotrophic aerobic ammonium-oxidizing bacteria (AAOB), especially Nitrosomonas eutropha and N. europaea.32,34 Another abundant phylum was Chloroflexi bacteria (4416, 9641 reads in cMBR, nMBR respectively), which has been found frequently in anammox reactors, playing an important role in sludge granulation and biofilm formation.13,34,35 The numbers of Chloroflexi bacteria coincided well with experimental results of significant biofilm formation in nMBR while such a phenomenon in cMBR was not clear.
Fig. 5 depicts the phylogenetic tree based on 16S rRNA gene fragments of almost all bacteria in cMBR and nMBR. As part of the Illumina MiSeq sequencing analysis, the phylogenetic tree reflects the main bacteria communities and their relatives in public databases. Bootstrap values (>50%) are indicated at branch points. The number 0.01 is the scale bar, which represents 0.01 nucleotide substitutions per nucleotide position. Candidatus Kuenenia (7, 2 reads in cMBR, nMBR respectively) and Candidatus Jettenia (9, 86 reads in cMBR, nMBR respectively) were the only definitely detected anammox bacteria genera, which showed a relatively high genetic similarity with the Blastocatella genus, which is affiliated to Actinobacteria as one of the dominant bacterial communities in soil. Overall, these results suggest that Illumina MiSeq sequencing, used with the aim of detecting the categories and quantities of universal bacteria based on 16S rRNA, was not effective or sensitive enough for this anammox culture.32,36 This might be attributed to sensitive anammox genes being mainly 250 bp whilst Illumina MiSeq sequencing mainly focuses on genes of about 300–400 bp. Therefore, FISH and qPCR analysis were adopted for further detection of the proportion and quantity of anammox bacteria to improve the sensitivity and specificity.
 | ||
| Fig. 5 Phylogenetic tree of clones obtained from DNA samples of cMBR and nMBR used in Illumina MiSeq sequencing analysis. | ||
3.4. FISH analysis
Sludge samples were collected from cMBR (suspended growth sludge) and nMBR (attached-growth biofilm) on day 120 for FISH analysis to investigate the spatial distribution and proportion of anammox bacteria. As shown in Fig. S4,† the red color, which represents anammox bacteria, was observed in both sludge samples. Through relative abundance analysis using Image-Pro Plus 6.0, the results also revealed that anammox bacteria accounted for about 58% of the total bacteria in nMBR which was significantly higher than those (51%) in cMBR. Although the results are estimated values with certain limitations, they showed that anammox bacteria had been the dominant community in the reactors and that non-woven fabric is more adaptable for the proliferation of anammox bacteria. The observations, together with the findings of qPCR analysis, corresponded well to the performance of nitrogen removal in reactors: the more anammox bacteria, the higher the nitrogen removal efficiency.3.5. qPCR analysis
To further investigate the benefit of non-woven fabric in terms of proliferating anammox bacteria, qPCR analysis was performed to enumerate anammox bacteria precisely. The qPCR enumeration results on day 60 demonstrated improved proliferation for nMBR with more preferable anammox cells enumeration of 1.24 × 107 copies per mL compared to cMBR (6.79 × 106 copies per mL). In the case of assuming all nitrogen was removed by anammox bacteria, the estimated per cell nitrogen removal rate in cMBR was also much higher than that in nMBR due to the orders of magnitude difference in cell enumeration. In other words, the low per cell nitrogen removal rate in nMBR indicated that nMBR had higher potential for improving nitrogen removal by anammox bacteria, which could be attributed to the use of the non-woven fabric membrane.Afterwards, the numbers of anammox bacteria continued to ascend speedily and reached 5.32 × 108 copies per mL in cMBR and 1.07 × 109 copies per mL in nMBR on day 120. Nevertheless, the estimated per cell nitrogen removal rate declined again considering the change in the orders of magnitude. Variation in the activity of anammox bacteria is a common phenomenon in the stationary phase of bacteria community growth,37 meaning that insufficient substrate supply could become the main restriction factor. Once higher nitrogen concentration was employed to satisfy the need of anammox bacteria, a higher NRR and more anammox bacteria could be obtained.
3.6. N2O emission
Considering the existence of nitrifying bacteria and denitrifying bacteria in the seed sludge, N2O, which is a typical by-product of nitrification and denitrification, could still be produced in the reactors during the cultivation of anammox bacteria. The initial liquid N2O emissions from cMBR and nMBR were measured as about 56.8 and 58.0 μmol L−1 (Fig. 6(a)), respectively, which could be attributed to endogenous denitrification consuming the bacteria themselves as a carbon source as well as using the nitrite/nitrate existing in the influent or transformation by the anammox process.26,27 Many studies also found that anammox bacteria (affiliated to Planctomycetes, 4189 and 4755 reads in cMBR and nMBR respectively), nitrifying and denitrifying bacteria (affiliated to Proteobacteria, 5147 and 8971 reads in cMBR and nMBR respectively) coexist in anammox reactors.32,34 Hence, the occurrence of denitrification was entirely possible. | ||
| Fig. 6 (a) The total liquid N2O emission in cMBR and nMBR; (b) the liquid N2O emission per consumed NRR (1 mg N per L per d) in cMBR and nMBR. | ||
During the 60 day period the emission of liquid N2O in nMBR fell from 58.0 to 41.6 μmol L−1 gradually. Meanwhile, the emission in cMBR also declined, but significantly less (46.1 μmol L−1), suggesting that anammox bacteria replaced the nitrifying and denitrifying bacteria to become the dominant bacteria in the two MBRs with more significant tendency in nMBR. From day 70, the HRT of nMBR moderately was shortened to 12 h, resulting in the NLR doubling to 283.3 mg N per L per d approximately. As a result, liquid N2O emission in nMBR increased dramatically to 60.5 μmol L−1 on day 120 associated with a transient decrease of anammox activity to accommodate the new conditions, as excess nitrogen was provided to residual nitrifying bacteria and denitrifying bacteria. The results could also be proved by the presence of Proteobacteria, 18.0% of relative abundance (5147 reads) in cMBR and 21.5% (8971 reads) in nMBR, such as Nitrosococcus oceani and Nitrosococcus halophilus (γ-Proteobacteria), Nitrosomonas and Nitrosospira (β-Proteobacteria).38
The arithmetical unit was changed to μmol d per g N (N2O emission/NRR) (Fig. 6(b)) to compare the N2O emission in cMBR and nMBR for treating commensurate nitrogen. The downward trend in both two reactors during the whole cultivation demonstrated the gradual purified anammox process and strengthened anammox activity. Being consistent with the performance of nitrogen removal, the N2O emission indexes of nMBR were lower than those of cMBR, reflecting the diminishment of N2O emission being superior to that in cMBR.
3.7. Membrane fouling
During the entire operation period, the hollow fiber module in cMBR was replaced once on day 80 when the TMP reached 45 kPa (Fig. 7), whereas the TMP development of the non-woven module in nMBR was only 25 kPa and the module was not replaced or cleaned during the 128 days of operation, indicating less serious membrane fouling. Some researchers have reported that a bigger pore size filter could cause more membrane fouling issues, particularly pore blocking in micro-filtration in terms of the same biomass concentration in MBRs.19,39,40 Nevertheless, the different separation modes of hollow fiber membrane (micro-filtration) and non-woven fabric (macro-filtration) meant the discipline was not appropriate in this situation. Such a distinction was also the main reason for the large TMP gap. As the practical pore sizes of the hollow fiber membrane and non-woven fabric decreased gradually with the formation of a loose cake layer, especially in nMBR, the increasing trend of membrane fouling corresponding with TMP became lower and lower (Fig. 7). With a lower MLSS of suspended sludge (e.g. cMBR: 6650 ± 50 mg L−1, nMBR: 254 ± 20 mg L−1 on day 130), the TMP in nMBR was lower than that in cMBR during the whole cultivation. The TMP growth rate of nMBR also slowed down gradually while the decrease rate of suspended sludge slowed down. Thus, the use of a non-woven module could mitigate membrane fouling, indicating notable lower operation cost and competitive advantage compared to polymer membranes.4. Conclusions
This study compared the performance of two MBRs during anammox bacteria cultivation. Compared to cMBR, nMBR exhibited the advantage of accelerating the start-up of the anammox process corresponding to a higher NRR of approximately 245.3 mg N per L per d. The non-woven membrane was beneficial to the formation of biofilm, which contributed to anammox bacteria proliferation, leading to a qPCR enumeration result of 1.07 × 109 copies per mL. The reduction of N2O emission was also observed during the period of operation. Moreover, nMBR could lead to less serious membrane fouling due to less TMP development. Overall, nMBR could be a promising, labor-saving and money-saving technology for anammox process start-up. In view of the start-up time and engineering cost, the method of this paper might be a more suitable choice for further municipal and industrial applications of anammox technology.Acknowledgements
The authors gratefully acknowledge the support from the National Natural Science Foundation of China (No. 21477063, 51108251 and 21177075), Taishan Scholar Foundation of Shandong Province, Research Award Fund for Outstanding Middle-aged and Young Scientist of Shandong Province (No. BS2012HZ007), Natural Science Foundation for Distinguished Young Scholar of Shandong Provincial (No. JQ201216) and Jinan Science and Technology Project (201401364).References
- M. S. Jetten, I. Cirpus, B. Kartal, L. van Niftrik, K. T. van de Pas-Schoonen, O. Sliekers, S. Haaijer, W. van der Star, M. Schmid, J. van de Vossenberg, I. Schmidt, H. Harhangi, M. van Loosdrecht, J. Gijs Kuenen, H. Op den Camp and M. Strous, Biochem. Soc. Trans., 2005, 33, 119–123 CrossRef CAS PubMed.
- A. Joss, D. Salzgeber, J. Eugster, R. Konig, K. Rottermann, S. Burger, P. Fabijan, S. Leumann, J. Mohn and H. Siegrist, Environ. Sci. Technol., 2009, 43, 5301–5306 CrossRef CAS.
- W. R. van der Star, W. R. Abma, D. Blommers, J. W. Mulder, T. Tokutomi, M. Strous, C. Picioreanu and M. C. van Loosdrecht, Water Res., 2007, 41, 4149–4163 CrossRef CAS PubMed.
- W. R. Abma, C. E. Schultz, J. W. Mulder, W. R. van der Star, M. Strous, T. Tokutomi and M. C. van Loosdrecht, Water Sci. Technol., 2007, 55, 27–33 CrossRef CAS.
- N. Chamchoi, S. Nitisoravut and J. E. Schmidt, Bioresour. Technol., 2008, 99, 3331–3336 CrossRef CAS PubMed.
- U. Imajo, T. Tokutomi and K. Furukawa, Water Sci. Technol., 2004, 49, 155–163 CAS.
- M. S. M. Jetten, M. Strous, K. T. van de Pas-Schoonen, J. Schalk, U. G. J. M. van Dongen, A. A. van de Graaf, S. Logemann, G. Muyzer, M. C. M. van Loosdrecht and J. G. Kuenen, FEMS Microbiol. Rev., 1998, 22, 421–437 CrossRef CAS PubMed.
- M. Strous, J. J. Heijnen, J. G. Kuenen and M. S. M. Jetten, Appl. Microbiol. Biotechnol., 1998, 50, 589–596 CrossRef CAS.
- Z. Li, X. Xu, B. Shao, S. Zhang and F. Yang, Chem. Eng. J., 2014, 254, 9–16 CrossRef CAS PubMed.
- T. Wang, H. Zhang, F. Yang, S. Liu, Z. Fu and H. Chen, Bioresour. Technol., 2009, 100, 2501–2506 CrossRef CAS PubMed.
- W. R. L. van der Star, A. I. Miclea, U. G. J. M. van Dongen, G. Muyzer, C. Picioreanu and M. C. M. van Loosdrecht, Biotechnol. Bioeng., 2008, 101, 286–294 CrossRef CAS PubMed.
- R. J. W. Meulepas, C. G. Jagersma, J. Gieteling, C. J. N. Buisman, A. J. M. Stams and P. N. L. Lens, Biotechnol. Bioeng., 2009, 104, 458–470 CrossRef CAS PubMed.
- F. Meng, G. Su, Y. Hu, H. Lu, L.-N. Huang and G.-H. Chen, Water Res., 2014, 58, 82–91 CrossRef CAS PubMed.
- C. Trigo, J. L. Campos, J. M. Garrido and R. Méndez, J. Biotechnol., 2006, 126, 475–487 CrossRef CAS PubMed.
- G. Gonzalez-Gil, L. Seghezzo, G. Lettinga and R. Kleerebezem, Biotechnol. Bioeng., 2001, 73, 125–134 CrossRef CAS PubMed.
- T. Fujii, H. Sugino, J. D. Rouse and K. Furukawa, J. Biosci. Bioeng., 2002, 94, 412–418 CrossRef CAS.
- S.-Q. Ni, P.-H. Lee, A. Fessehaie, B.-Y. Gao and S. Sung, Bioresour. Technol., 2010, 101, 1792–1799 CrossRef CAS PubMed.
- N. Lee, G. Amy, J.-P. Croue and H. Buisson, Water Res., 2004, 38, 4511–4523 CrossRef CAS PubMed.
- W. Guo, H.-H. Ngo and J. Li, Bioresour. Technol., 2012, 122, 27–34 CrossRef CAS PubMed.
- P. Bai, X. Chen, H. Yin, J. Wang and G.-H. Chen, Desalin. Water Treat., 2010, 18, 297–301 CrossRef CAS.
- Y. Tao, D.-W. Gao, Y. Fu, W.-M. Wu and N.-Q. Ren, Bioresour. Technol., 2012, 104, 73–80 CrossRef CAS PubMed.
- L.-F. Ren, S.-Q. Ni, C. Liu, S. Liang, B. Zhang, Q. Kong and N. Guo, Environ. Sci. Pollut. Res., 2015, 22, 2925–2934 CrossRef CAS PubMed.
- Water Environment Federation and American Public Health Association (APHA), Washington, DC, USA, 2005.
- M. Schmid, U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. Jetten, J. W. Metzger, K.-H. Schleifer and M. Wagner, Syst. Appl. Microbiol., 2000, 23, 93–106 CrossRef CAS.
- I. Tsushima, T. Kindaichi and S. Okabe, Water Res., 2007, 41, 785–794 CrossRef CAS PubMed.
- N. Chamchoi and S. Nitisoravut, Chemosphere, 2007, 66, 2225–2232 CrossRef CAS PubMed.
- A. Dapena-Mora, S. W. H. van Hulle, J. Luis Campos, R. Méndez, P. A. Vanrolleghem and M. Jetten, J. Chem. Technol. Biotechnol., 2004, 79, 1421–1428 CrossRef CAS PubMed.
- M. S. M. Jetten, M. Wagner, J. Fuerst, M. van Loosdrecht, G. Kuenen and M. Strous, Curr. Opin. Biotechnol., 2001, 12, 283–288 CrossRef CAS.
- K. Isaka, T. Sumino and S. Tsuneda, J. Biosci. Bioeng., 2007, 103, 486–490 CrossRef CAS PubMed.
- Y. Liu and J.-H. Tay, Water Res., 2002, 36, 1653–1665 CrossRef CAS.
- B.-l. Hu, P. Zheng, C.-j. Tang, J.-w. Chen, E. van der Biezen, L. Zhang, B.-j. Ni, M. S. Jetten, J. Yan and H.-Q. Yu, Water Res., 2010, 44, 5014–5020 CrossRef CAS PubMed.
- X.-R. Li, B. Du, H.-X. Fu, R.-F. Wang, J.-H. Shi, Y. Wang, M. S. Jetten and Z.-X. Quan, Syst. Appl. Microbiol., 2009, 32, 278–289 CrossRef CAS PubMed.
- M. Strous, E. Pelletier, S. Mangenot, T. Rattei, A. Lehner, M. W. Taylor, M. Horn, H. Daims, D. Bartol-Mavel, P. Wincker, V. Barbe, N. Fonknechten, D. Vallenet, B. Segurens, C. Schenowitz-Truong, C. Médigue, A. Collingro, B. Snel, B. E. Dutilh, H. J. M. Op den Camp, C. van der Drift, I. Cirpus, K. T. van de Pas-Schoonen, H. R. Harhangi, L. van Niftrik, M. Schmid, J. Keltjens, J. van de Vossenberg, B. Kartal, H. Meier, D. Frishman, M. A. Huynen, H.-W. Mewes, J. Weissenbach, M. S. M. Jetten, M. Wagner and D. le Paslier, Nature, 2006, 440, 790–794 CrossRef PubMed.
- S. Qiao, Y. Kawakubo, Y. Cheng, T. Nishiyama, T. Fujii and K. Furukawa, Biodegradation, 2009, 20, 117–124 CrossRef CAS PubMed.
- S. Cho, Y. Takahashi, N. Fujii, Y. Yamada, H. Satoh and S. Okabe, Chemosphere, 2010, 78, 1129–1135 CrossRef CAS PubMed.
- M. C. Schmid, A. B. Hooper, M. G. Klotz, D. Woebken, P. Lam, M. M. Kuypers, A. Pommerening-Roeser, H. J. Op Den Camp and M. S. Jetten, Environ. Microbiol., 2008, 10, 3140–3149 CrossRef CAS PubMed.
- T.-F. C. Mah and G. A. O’Toole, Trends Microbiol., 2001, 9, 34–39 CrossRef CAS.
- S. Avrahami, R. Conrad and G. Braker, Appl. Environ. Microbiol., 2002, 68, 5685–5692 CrossRef CAS.
- A. Lim and R. Bai, J. Membr. Sci., 2003, 216, 279–290 CrossRef CAS.
- S. Hong, T. Bae, T. Tak, S. Hong and A. Randall, Desalination, 2002, 143, 219–228 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra16802c |
| This journal is © The Royal Society of Chemistry 2015 |



