Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Developing an in vitro model of haematoma for study of intracerebral haemorrhage†
Siobhan
Crilly
 *ab,
Victor Sebastian
Tapia
ab,
Carlo
Bawn
*ab,
Victor Sebastian
Tapia
ab,
Carlo
Bawn
 c and
Annalisa
Tirella
de
c and
Annalisa
Tirella
de
aDivision of Neuroscience, School of Biological Sciences, Faculty of Biology, Medicine and Health, Manchester Academic Health Science Centre, The University of Manchester, Oxford Road, Manchester, M13 9PT, UK. E-mail: Siobhan.crilly@manchester.ac.uk
bGeoffrey Jefferson Brain Research Centre, The Manchester Academic Health Science Centre, Northern Care Alliance & University of Manchester, UK
cDepartment of Chemistry, School of Natural Sciences, Chemistry Building, University of Manchester, Oxford Road, M13 9PL, UK
dDivision of Pharmacy and Optometry, School of Biological Sciences, Faculty of Biology, Medicine and Health, Manchester Academic Health Science Centre, The University of Manchester; Oxford Road, Manchester, M13 9PT, UK
eBIOtech centre, Department of Industrial Engineering, University of Trento, Via Sommarive 9, 38122 Trento, Italy
First published on 22nd April 2024
Abstract
Intracerebral haemorrhage (ICH) is a devastating neurovascular attack with limited treatment options. Alternative, pre-clinical modelling approaches are required to identify and trial therapeutic drug compounds. In this study we have used alginate hydrogels to model blood insult in vitro. Human whole blood was mixed with alginate and encapsulated into hydrogel beads. Beads were then incorporated in a second layer of alginate containing hyaluronic acid/chitosan nanoparticles to mimic the mechanical properties of brain tissue and create a model haematoma. Beads and model haematomas were characterised to profile size, volume, mechanical properties, release capacity and storage stability over time. Beads and model haematomas stimulate a pro-inflammatory phenotype in human monocytic and macrophage-like cells, however have no pathogenic effect on brain endothelial and neuronal cell survival or function. In conclusion, we have developed an effective strategy to model ICH in vitro, to investigate the human immune response to blood insult.
Introduction
Intracerebral haemorrhage (ICH) occurs when blood vessels in the brain spontaneously rupture and release an insult of blood into the brain parenchyma.1 This devastating neurological damage causes widespread cell death and inflammation resulting in disability and mortality, and there are no specific therapies for patients. Primary injury associated with the rupture of blood into the brain causes swelling, increased intracranial pressure, oedema and compression of surrounding brain structures. After the bleeding stops the haematoma condenses, red blood cells lyse, and there is a release of toxic blood compounds and breakdown products into the brain tissue.2 Best clinical practice includes surgically removing the haematoma to reduce the mass effect for better clinical outcomes3 but this is only possible in ∼10% of patients.4 There has been little investigation into how the extravasated blood damages cells when left to degrade and reabsorb.Commonly, modelling intracerebral haemorrhage pre-clinically relies on the use of animals to recreate the complex, multi-system disease and investigate behavioural outcomes.5 Despite improving our understanding of disease mechanisms, pre-clinical animal studies have been limited in translation of outcomes to the clinic.6 As such, there is an urgent need for novel, complementary models7 to address specific questions about blood toxicity and the localised human immune response.
Human-relevant data has historically come from clinical observations, patient peripheral blood samples, or analysis of post-mortem tissue. Such approaches limit the information available to specific time frames and holds little relevance for understanding the immediate pathology caused by extravasated blood. Traditionally, in vitro models of ICH recreate injury by adding haemoglobin,8 hemin,8,9 glutamate10 or hydrogen peroxide to damage cells and the advantages and limitations have been reviewed elsewhere.11 This approach does not recreate the full complement of factors in the blood that contribute to primary cell damage and stimulate secondary inflammatory injury.12 Neurotoxic factors from blood include thrombin, haemoglobin, free iron, fibrinogen, complement, leukocytes, platelets, oxyhaemoglobin, methemoglobin and hemin.13 Adding whole blood to cell culture introduces practical problems due to opacity and reduction of available nutrients and oxygen in the media with increasing blood doses. To overcome these problems, in this study we have used alginate-based hydrogels to encapsulate human blood to control injury in a cell culture system.
Alginate is a polysaccharide derived from brown algae that has been widely used in biomedical applications and tissue engineering due to structural similarities with extracellular matrices (ECM).14 Alginate is composed of different sequences of β-D-mannuronic acid (M) and α-L-guluronic acid (G) monomers determined by the species of source algae used. Alginate hydrogels are also used as drug delivery systems, enabling the release of both small chemicals and biomolecules in a controlled manner dependant on crosslinking methods.15 Alginate hydrogels form easily with the addition of divalent cations, such as Ca2+, upon which G monomers are bound into ‘egg box’ structure.16 The G/M ratio is often used to determine gelation, crosslinking density and the resulting mechanical properties of the formed hydrogels. In this study, we investigated 4 types of alginate and different concentrations of CaCl2 for crosslinking, to encapsulate whole blood and characterise breakdown product release for ICH modelling in vitro. This is the first study, to our knowledge, that investigates whether alginate-based hydrogels can provide controllable release of blood breakdown compounds in vitro that mimic the cellular toxicity following an ICH in a culture system.
To further model characteristics of the brain tissue, alginate hydrogels containing hyaluronic acid (HA) were characterised to identify mechanical properties that mimic the cerebral ECM. HA was selected as a crucial component of the brain ECM, providing a backbone structure for HA-binding lecticans.17 In addition to providing biomechanical integrity, HA alters tissue hydration and plays a key role in synaptic plasticity,18 stroke recovery19 and inflammation.20 We incorporated HA into the model haematoma to promote hydration of the gel, and to provide structure that would mimic the soft, damaged, oedemic brain tissue following a haemorrhage that surrounds a haematoma.21 In polymeric form, HA in hydrogel absorbs water and weakens the network, making the hydrogel unstable within 24–48 hours in cell culture.22 In this study we have fabricated nanoparticles from HA and chitosan (CS), to stabilise the hydrogel whilst maintaining HA in the network as an essential component of the ECM. HA forms a corona around the nanoparticle23,24 and ensures hydrogel stability by reducing hydration.
This methodology is highly adaptable and offers an entirely human, in vitro platform for interrogating the mechanistic pathology of ICH, and screening neuroprotective compounds that may have translational benefit for the clinic.
Results
Differences in alginate composition affect inflammation in culture
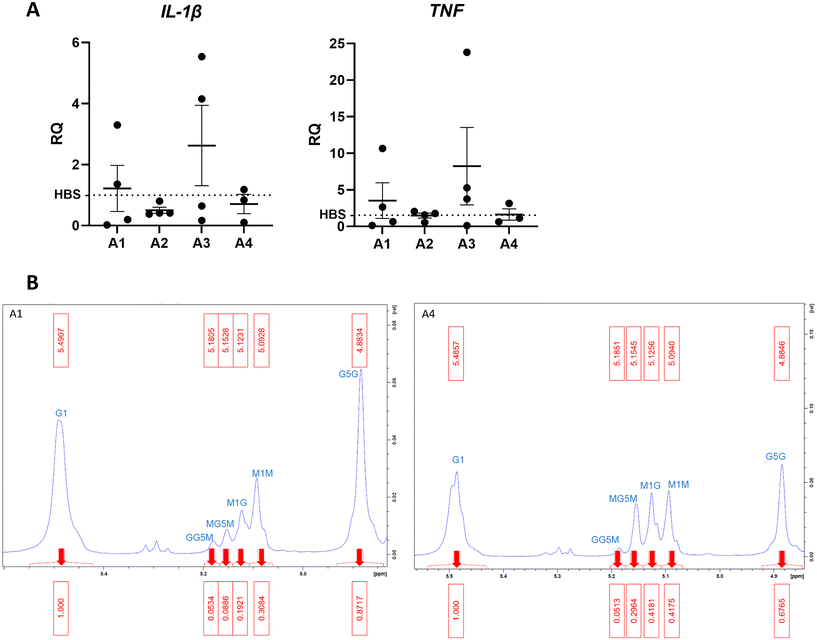
To investigate the effect of different alginate compositions on inflammation, beads generated from 10 mg mL−1 alginate hydrogel (AB) were firstly formed using 4 different alginate types varying in G/M ratio and molecular weight (MW). Different alginate-only beads were cultured for 6 hours with THP-1 cells to determine inflammatory stimulation (Fig. 1A). A panel of primers were used to detect differential gene expression from the HEPES buffered saline (HBS) control indicative of proinflammatory phenotypes. Phosphate buffered solutions cannot be used as phosphate anions degrade alginate gels by sequestering Ca2+ ions from the matrix. Reverse transcriptase-qPCR analysis of gene transcription was performed to determine whether alginates stimulated a pro-inflammatory response in THP-1 cells. Tumour necrosis factor (TNF) is a pro-inflammatory cytokine involved in regulating macrophage function and cell survival. Interleukin-1β (IL-1β) is a potent pro-inflammatory cytokine that is stimulated in response to injury and drives downstream transcription of other pro-inflammatory molecules. Data showed that A3 was proinflammatory and A1 produced a variable 2-fold increase of TNF transcription however this was not significant from the HBS control. A2 did not produce an inflammatory response however, was excluded from further experiments because of practical difficulty with filter sterilisation due to high viscosity. A4 consistently showed the least inflammatory activation (Fig. 1A) Interleukin-6, arginase 1, interleukin-10, nitric oxide synthase-2 and CC motif chemokine ligand 18 were all undetected confirming the absence of an anti-inflammatory phenotype.To confirm whether the G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M ratio is as different as reported in A1 and A4, proton nuclear magnetic resonance (H1-NMR) analysis was performed. The ratio is known to influence physiochemical and mechanical properties of alginate hydrogels.25 H1-NMR shows ratio G
M ratio is as different as reported in A1 and A4, proton nuclear magnetic resonance (H1-NMR) analysis was performed. The ratio is known to influence physiochemical and mechanical properties of alginate hydrogels.25 H1-NMR shows ratio G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M is 2.1 in A1 and 1.3 in A4 (Fig. 1B). The high M content of A4 is desirable to make soft hydrogels with properties that mimic brain tissue (∼1 kPa).26
M is 2.1 in A1 and 1.3 in A4 (Fig. 1B). The high M content of A4 is desirable to make soft hydrogels with properties that mimic brain tissue (∼1 kPa).26
Synthesising a hyaluronic acid-alginate layer
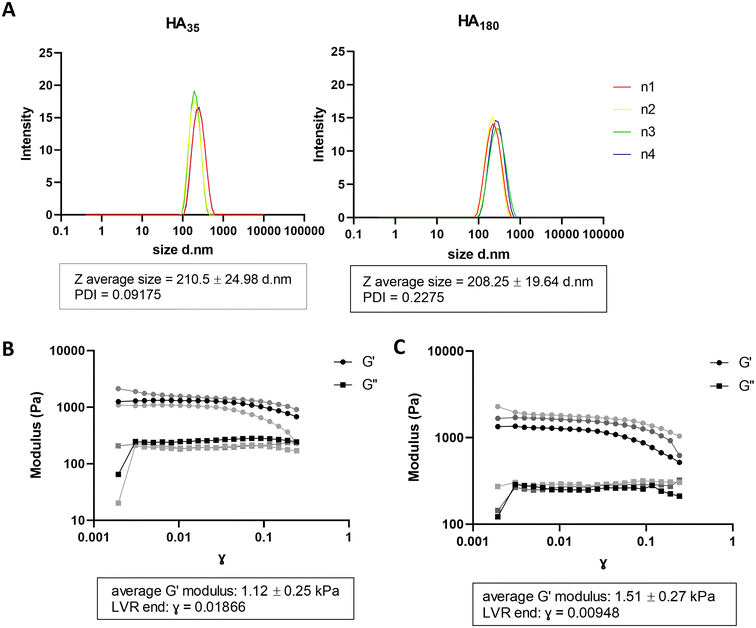
In physiological conditions, the haematoma is surrounded by damaged tissue and oedema is observed by CT scan,27 likely to contain disrupted ECM and cell matter. This provides a physical layer between the healthy tissue and contact with the blood in the haematoma. In order to generate a model of haematoma that mimics human disease physiology, we sought to introduce hyaluronic acid (HA) to alginate hydrogels to recreate the layer of water-dense damaged brain ECM between the haematoma and cells in culture. Hyaluronic acid has been previously reported to be unstable in water after 24–48 hours due to water absorbance.22 To mitigate instability in water, hyaluronic acid/chitosan (HA/CS) nanoparticles were prepared and mixed with A4 for hydrogel formulation.24 HA/CS nanoparticles were made using two MW of hyaluronic acid: 35 kDa (HA35) and 180 kDa (HA180). HA35/CS and H180/CS nanoparticles were consistent in Z-average size (∼210 nm) and polydispersity index (PDI < 0.25) (Fig. 2A). Of note, HA35/CS nanoparticles showed more uniformity (PDI = 0.10) than HA180/CS (PDI = 0.23). Alginate HA/CS gels were prepared by mixing 10 mg mL−1 A4 solution with 1 mg mL−1 HA/CS nanoparticles solution at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio, and hydrogels obtained after incubation with 75 mM CaCl2 (aq.) for 10 min at RT. CaCl2 concentration was optimised to generate alginate hydrogel containing HA/CS nanoparticles that had a stiffness close to 1.1 kPa, the reported stiffness of grey matter.26 Rheological analysis of the elastic modulus (G′) and viscous modulus (G′′) was used to determine material stiffness. Data revealed that A4-HA35/CS hydrogel has an average elastic modulus of 1.12 kPa ± 0.25 s.d (Fig. 2B) compared to A4-HA180/CS hydrogel which is 1.51 kPa ± 0.27 s.d (Fig. 2C). Therefore, A4-HA35/CS hydrogels were selected for the stiffness most closely fit with grey matter and used for the rest of the experiments. Data from further experiments with A4-HA180/CS hydrogels is available in ESI Fig. 1.†
1 volume ratio, and hydrogels obtained after incubation with 75 mM CaCl2 (aq.) for 10 min at RT. CaCl2 concentration was optimised to generate alginate hydrogel containing HA/CS nanoparticles that had a stiffness close to 1.1 kPa, the reported stiffness of grey matter.26 Rheological analysis of the elastic modulus (G′) and viscous modulus (G′′) was used to determine material stiffness. Data revealed that A4-HA35/CS hydrogel has an average elastic modulus of 1.12 kPa ± 0.25 s.d (Fig. 2B) compared to A4-HA180/CS hydrogel which is 1.51 kPa ± 0.27 s.d (Fig. 2C). Therefore, A4-HA35/CS hydrogels were selected for the stiffness most closely fit with grey matter and used for the rest of the experiments. Data from further experiments with A4-HA180/CS hydrogels is available in ESI Fig. 1.†
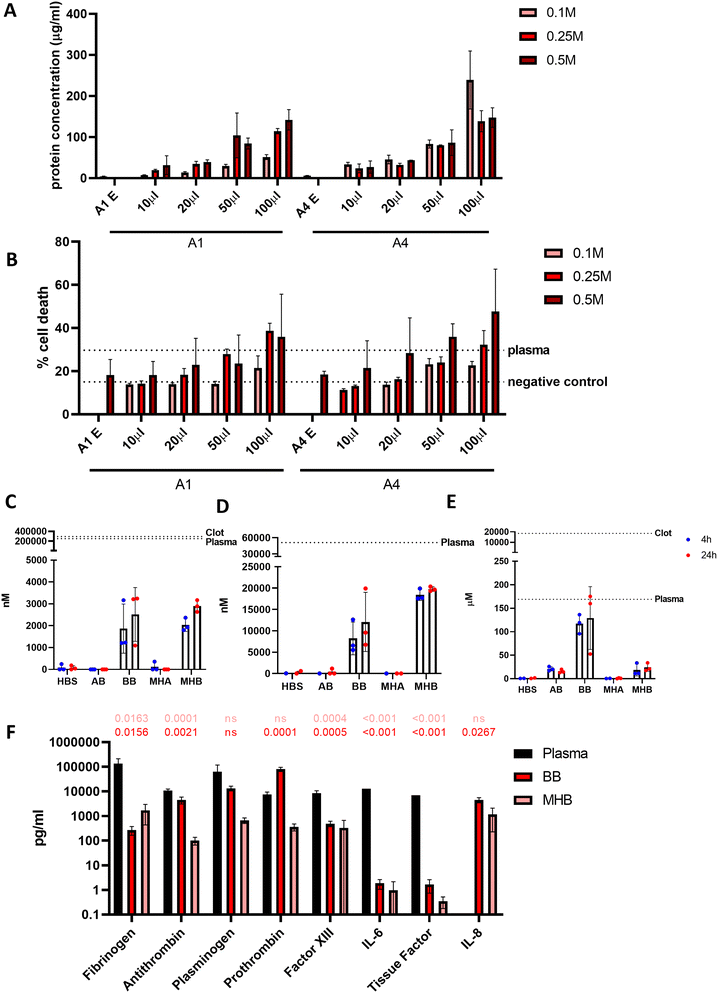
Alginate blood beads are consistent in fabrication, but release blood in a short time period
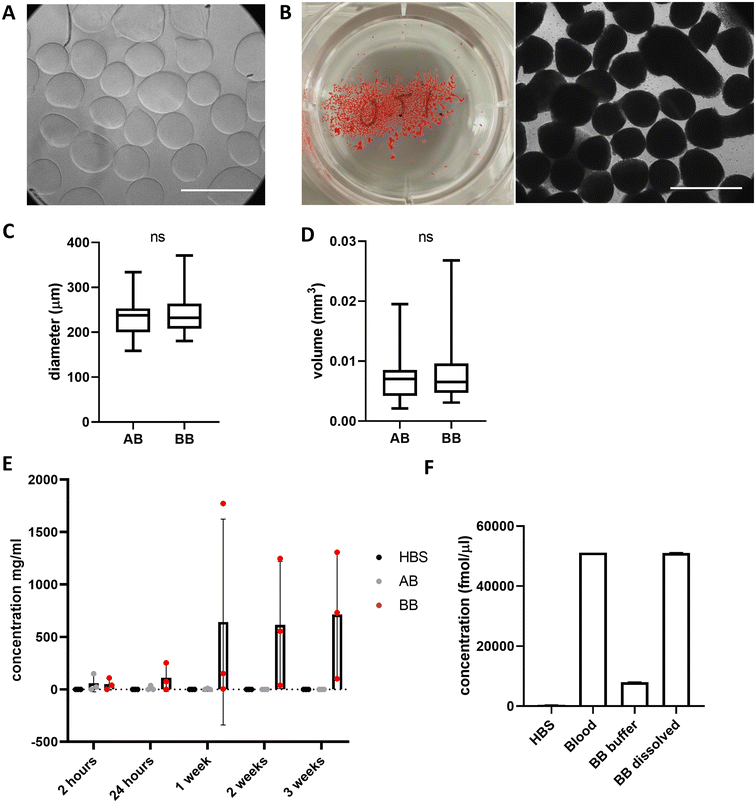
Beads generated from 10 mg mL−1 alginate hydrogel mixed with whole blood to form blood beads (BB) showed no difference in size or volume from alginate-only beads (AB) (Fig. 3A–D). On average, beads are 250 μm in diameter and have a volume of 0.007 mm3. Constant values were observed with different ratios of blood![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) alginate for bead preparation however protein release was accelerated at higher ratios. Blood beads were made 50
alginate for bead preparation however protein release was accelerated at higher ratios. Blood beads were made 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 v/v blood and 10 mg mL−1 alginate for all further experiments as beads had good stability and blood content. Blood beads showed evidence of degradation and protein release into the buffer after 24 hours at 4 °C (Fig. 3E). This varied between preparation repeats however did not reach levels of significance. After 3 weeks stored at 4 °C, blood beads had released ∼15% (Fig. 3F) of the hemin levels that were detected in the dissolved blood beads, implying that although blood beads are initially leaking protein from breakdown products, hemin can still release after 3 weeks to initiate a pathogenic response.
50 v/v blood and 10 mg mL−1 alginate for all further experiments as beads had good stability and blood content. Blood beads showed evidence of degradation and protein release into the buffer after 24 hours at 4 °C (Fig. 3E). This varied between preparation repeats however did not reach levels of significance. After 3 weeks stored at 4 °C, blood beads had released ∼15% (Fig. 3F) of the hemin levels that were detected in the dissolved blood beads, implying that although blood beads are initially leaking protein from breakdown products, hemin can still release after 3 weeks to initiate a pathogenic response.
Generating a model haematoma adds stability to beads
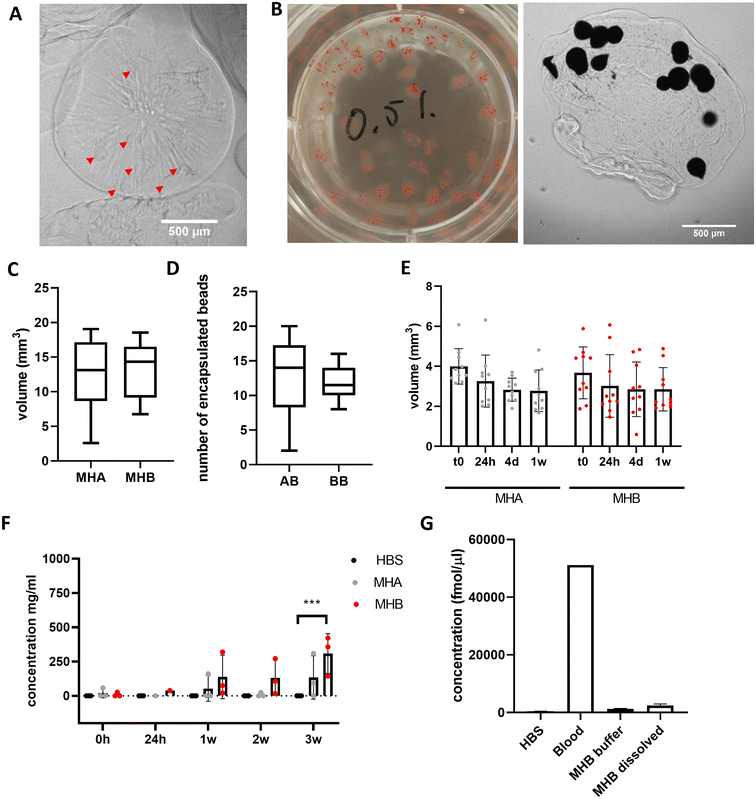
Alginate-only beads and blood beads were incorporated into A4 HA35/CS hydrogel, final concentration of 5 mg mL−1 ALG4 and 0.5 mg mL−1 HA35/CS nanoparticles, prior to extrusion and gelation in a 75 mM CaCl2 solution (aq.) at RT to form model haematomas (controls with alginate-only beads = MHA, and with blood beads = MHB) (Fig. 4A and B). Model haematomas are on average 2 mm in diameter with a volume of 15 mm3 (Fig. 4C) and on average contain the same number of beads regardless of blood, however there is a much larger range of number of alginate-only beads encapsulated within the hyaluronic acid layer (Fig. 4D). This may be due to difficulty identifying alginate-only beads from contrast images. To confirm limited swelling of the hyaluronic acid -containing hydrogel we determined that there is no significant change in volume over 1 week in storage at 4 °C (Fig. 4E). By measuring the protein content of the storage buffer as an indication of blood breakdown release, data revealed that MHB can be stored for ∼3 weeks before significant protein detection (Fig. 4F). Mean protein concentrations at 3 weeks were half of detected levels for blood beads without the A4 HA35/CS hydrogel layer (308 mg mL−1 compared to 712 mg mL−1, Fig. 1E). Detection of signal in control model haematoma samples at 3 weeks perhaps indicates a breakdown in the polysaccharide in the HA35/CS hydrogel and interference in the BCA detection assay. To ensure model consistency, model haematomas were formed within 24 hours of blood donation and bead formation and were used for further investigation and cell culture within 2 weeks of fabrication. Analysis of hemin concentration in the buffer and dissolved MHB after 3 weeks reveals that MHB only had about 2% of the total hemin detected in whole blood samples (Fig. 4G) whereas the blood beads had equal amounts of hemin as whole blood (Fig. 1F). This implies a faster breakdown of the haemoglobin and loss of hemin than in blood beads.Beads release toxic compounds in a dose dependent manner
Blood beads were added in increasing volumes to wells for a 24 h incubation at 37 °C to show a dose-dependent effect on protein release (Fig. 5A). This resulted in dose-dependent cell death in macrophage-like THP-1 cells when cultured for 24 hours (Fig. 5B). The largest influence on cell death was caused by the number of blood beads, and to a lesser extent, the concentration of CaCl2 used to crosslink the alginate regardless of alginate type (A1 and A4). Blood beads made with A4 and 0.25 M CaCl2 were selected for further experiments and dosed at 50 μL into 200 μL wells.In order to determine what factors released from the beads in culture are causing toxicity, further analysis was performed to analyse the contents of the media that may be damaging the cells. Reactive oxygen species (ROS) are released from the haematoma and from surrounding damaged cells. After 24 hours at 37 °C detectable ROS levels from blood beads and model haematomas with blood beads were 100× less than undiluted plasma at 2.52 μM and 2.89 μM respectively (Fig. 5C). Hemin is commonly used to damage cells in culture and previous studies in the literature have shown 100 μM of hemin results in 50% neuronal cell death.8Fig. 5D shows detection was greater from the model haematomas with blood beads than blood beads, and there was less of a difference dependent on time of incubation. Hemin released from the model haematomas with blood beads was ∼20 μM, almost half of the levels detected in plasma. Blood beads released ∼10 μM. Haemoglobin, another toxic compound released from the haematoma when red blood cells lyse, was detectable at the same levels as plasma after 24 hours (Fig. 5E). Our study shows that beads release 130 μM haemoglobin after 24 hours in culture, and plasma contains as much as 170 μM. Data suggests that there is a faster degradation of haemoglobin in model haematomas with blood beads than in blood beads, and this results in the quick release of hemin which breaks down and is undetectable after storage for 3 weeks.
Human fibrinolysis LEGENDplex assay analysis revealed that when incubated at 37 °C for 6 hours, blood beads and model haematomas with blood beads released significantly less fibrinogen, anti-thrombin, factor XIII, IL-6 and tissue factor than found in donor plasma (Fig. 5F). Levels of plasminogen were not significantly different from plasma, and prothrombin released from blood beads was significantly higher than plasma controls. IL-8 (CXCL8) was not detected in plasma however, was detected to be released from the models. IL-8 is a chemokine that promotes angiogenesis and proinflammatory roles that damage the endothelial cells, implying perhaps donor leukocytes survive and are active within the hydrogel. These data imply that blood beads exhibit fibrinolytic and thrombotic factors which may contribute to the toxic effects observed in culture however this is less than detected physiologically.
Characterising the cellular response to toxic factors
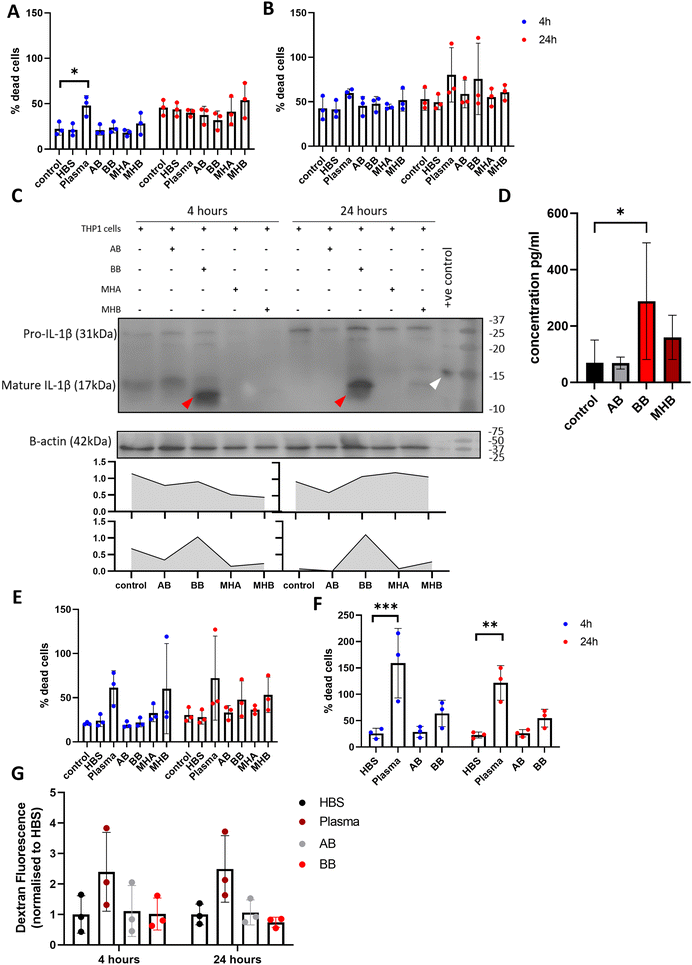
To investigate the effect of beads in culture with different cell types, 50 μL blood beads and 3× model haematomas with blood beads were added to monocytic THP-1 cells in suspension, stimulated macrophage-like THP-1 cells, SH-SY5Y neuronal cells and brain endothelial hCMEC/D3 cells. Blood beads and model haematomas with blood beads had no effect on cell death of monocytic THP-1 cells or macrophage-like THP-1 cells at either time point (Fig. 6A and B). IL-1β is activated and released in a two-step process and is a potent down-stream pro-inflammatory cytokine. Western blot analysis showed that monocytic THP-1 cells produced pro- and mature IL-1β in response to blood beads (Fig. 6C), however this was undetected by ELISA and therefore unreleased from cells. Macrophage-like THP-1 cells release IL-1β detected by ELISA when treated with blood beads, and a lesser extent with model haematomas with blood beads (Fig. 6D). In literature, it has been observed before that heme,28 red blood cells and haemoglobin29 all induce IL-1β release from phorbal-12-myristate-13-acetate (PMA) – stimulated THP-1 cells. This implies that perhaps the blood insult is the first priming step for IL-1β maturation, however a second activation stimulus is required for release, such as the PMA stimulation of cells.30 These results support the conclusion that blood beads and model haematomas with blood beads is a good model to study the immune response to haemorrhage. Blood beads and model haematomas with blood beads had no effect on cell death of SH-SY5Y neuronal cells or hCMEC/D3 cells at either time point (Fig. 6E and F). SH-SY5Y cells did not produce or release any IL-1β nor was IL-6 and TNFα detected in any of the cell types. To determine whether blood beads impacted brain vascular cell function, we conducted a barrier permeability assay on hCMEC/D3 cultures (Fig. 6G). Incubation with blood beads had no effect on the permeability of the cells showing that the barrier function of these cells is not impaired.Discussion
In this study we have shown that blood can be encapsulated into alginate beads and stored for up to 24 hours. When introduced to cell culture, blood beads were proven to mimic the toxicity of blood on cells in a dose-dependent manner. This model presents a novel method of investigating pathophysiology of blood extravasation and the associated immune response that can be applied with medical relevance for pre-clinical modelling of ICH. Application of such a model would allow for investigation of pathology in a human system, to screen compounds for anti-inflammatory or cyto-protective effects.When considering the use of this model in the pre-clinical ICH pipeline, it is important to compare with the current in vitro models commonly used. Hemin and haemoglobin, as breakdown products of RBCs within the haematoma, have been previously used to model ICH in vitro. There is limited understanding of pathology when trying to recreate ICH in vitro as dosing cell culture with individual factors does not include the full neurotoxicity of whole blood. In this study we have shown that after 4 hours of incubation at 37 °C, blood beads released 8 μM hemin and 117 μM of haemoglobin. Model haematomas with blood beads released 20 μM of hemin and 11 μM of haemoglobin. Previously, researchers have shown that similar doses of hemin have had considerable deleterious effect on cell survival and are capable of stimulating ROS release from cells (Table 1). In our study we showed that blood beads stimulated vastly more ROS release than in previous studies,31 released 10× the concentration of haemoglobin used in literature8 and was capable of initiating IL-1β release without a liposaccharide (LPS) priming step, which highlights the discrepancies between using single neurotoxic factors and whole blood at initiating a translational injury response that recreates human disease.
| Neurotoxic factor | Dose | Cell type | Outcome | Ref. |
|---|---|---|---|---|
| Hemin | 20 μM for 2 hours | THP-1 | 40% reduction in cell viability | Imoto et al. 201831 |
| 8× increase in ROS production | ||||
| Hemin | 100 μM for 14–18 hours | Primary mouse neurons | 50% reduction in cell viability | Zille et al. 20178 |
| Haemoglobin | 1.5 μM for 24–28 hours | Primary mouse neurons | 25% reduction in cell viability | Zille et al. 20178 |
| Hemin | 5 pg mL−1 LPS for 24 hours | Macrophage-like THP-1 | No cytotoxic effect | Carrasco-Pozo et al. 202032 |
| 10 μM hemin | Reduced glycolysis, HIF-1α, cell metabolism and phagocytosis | |||
| Hemin | 0.01–1 μg mL−1 LPS for 3 hours | Human primary macrophages | 1000 pg mL−1 IL-1β release | Nurmi et al. 201733 |
| 30 μM hemin | Macrophage-like THP-1 | NLRP3 inflammasome activation | ||
| 4 μM ATP for 1 hour | Mouse bone marrow-derived macrophages | No change in ROS | ||
| Increased autophagy |
Formulation of the beads in this study was simplified for selection however, there are many parameters that can be further altered to investigate the influence on size, volume, permeability, and stability of the beads and subsequent outcomes in culture. In principle, high G/M ratios and high crosslinking cation concentration lead to a high crosslinking density and a stiff hydrogel network which slows the diffusion of loaded molecules. Design criteria in this study focused on the use of non-animal derived polymers, generating mechanical properties to match brain tissue, and encapsulating and releasing human blood. Within whole blood, the pathogenic molecules are a range of sizes and alginate hydrogels typically have a pore size of ∼5 nm.34 We selected Ca2+ as crosslinking cations based on previous knowledge in modelling mechanics of soft tissues in vitro.35,36 The size of fabricated blood beads can be increased by using a larger extrusion nozzle, decreasing the vibration frequency and increasing the flow rate during encapsulation.37 Alginate type and concentration influences the diffusion of large molecules through the hydrogel matrix.25 We used alginates with different G/M ratios to best match the stiffness of soft tissues.38 With this strategy we demonstrated that high M alginates (A4) form softer hydrogels with properties similar to brain tissue stiffness and that are also suitable for diffusion of blood components.
We investigated the use of hyaluronic acid-coated chitosan nanoparticles for the inclusion of hyaluronic acid in alginate hydrogels as a component of the ECM, to overcome well known issues of network instability with polymeric hyaluronic acid. We used two different molecular weights of hyaluronic acid and found a slight increase in hydrogel stiffness with nanoparticles made with high MW hyaluronic acid (∼1.5 kPa A4-HA180/CS vs. ∼1.1 kPa A4-HA35/CS). Of note, while using HA35/CS alginate-based hydrogels, volumes remained consistent for the duration of the experiment. As expected, adding a higher volume of blood beads to cell culture resulted in a higher amount of blood breakdown products available for diffusion.
Further use of this model with different cell types, in different culture settings, or with 3D cultures may reveal new insights into the pathology of blood extravasation in the brain and help reveal different proinflammatory and cell damage/death pathways that are activated following ICH. We can use this model to investigate therapies that target specific cell types in a human system, prior to further pre-clinical testing. Investigation into patient-specific responses or using genetic analysis of primary patient-derived cell cultures may identify specific products of injury that may be used as disease biomarkers or for targetable therapies. Further characterisation of this model can reveal insights into the mechanisms behind haematoma breakdown and resorption, by investigating culture with microglia and neuronal stem cells. Broadly, the encapsulation of blood into alginate can be employed in other culture systems to investigate the cellular response to haematoma, whilst providing a reversible and controllable blood insult.
By adding models to the pre-clinical toolkit for studying ICH, we hope to close the gap between animal models and clinical trial to improve translational outcomes and benefit patients.
Materials and methods
Reagents and materials
| Reagents | Recipe |
| HEPES buffered saline (HBS) | 150 mM NaCl |
| 20 mM HEPES | |
| pH 7.4 | |
| Materials | Supplier |
| Sodium alginate #71238 | Sigma |
| Sodium alginate-LN | PROALG |
| Sodium alginate-LT | PROALG |
| Sodium alginate-MC | PROALG |
| Hyaluronic acid MW 35 kDa (HA35) | Medipol SA |
| Hyaluronic acid MW 180 kDa (HA180), | Medipol SA |
| Chitosan MW 36 kDa #9012-76-4 | Sigma |
Preparation of alginate
HBS was made fresh and sterilised. A 10 mg mL−1 alginate solution was prepared using the following alginate types:| Notation | Species | Commercial source | Country of origin | Reported G/M ratio |
| A1 | Laminaria hyperborea | Sigma 71238 Lot# BCCD8789 | Norway | 2.3 |
| A2 | Lessonia nigrescens (bertorona) | LN – PROALG | Chile | =1 |
| A3 | Lessonia trabeculata | LT – PROALG | Chile | >1 |
| A4 | Macrocystis pyrifera | MC – PROALG | Chile | <1 |
After complete hydration, each solution was warmed to 37 °C and filter sterilised using 0.22 μm polyethersulfone filter (SLGP033RS, Millipore).
Encapsulation of blood in alginate beads
All experiments involving human samples were conducted following the guidelines of ethical involvement of human participants published by the University of Manchester (UK) and with ethical approval from the University of Manchester Research Ethics Committee (#16181–28064). Peripheral blood samples were freely repeatedly donated by 5 healthy volunteers who gave informed consent, in tubes containing 10% anticoagulant citrate dextrose (ACD) (Sigma C3821) and 0.1% PPACK Dihydrochloride (Millipore 520222) and immediately processed for encapsulation. Equal volumes of 10 mg mL−1 alginate solution was mixed with blood or HBS for controls, and loaded into a 10 mL syringe and encapsulated into beads as previously described.37 In brief, blood/alginate suspension was extruded at 8.9 mL min−1 through a 150 μm nozzle. Droplets were generated using a vibration frequency of 5500 Hz and the stream was disrupted by electrostatic potential of 1.0 kV. Droplets were collected into a rotating, sterile CaCl2 bath (speed 100 rpm) and incubated for 10 minutes to allow for ionic crosslinking. Alginate hydrogel beads were then poured through a 50 μm cell strainer and washed twice with HBS. Beads were stored at 10% volume in HBS at 4 °C (2.5 mL of beads and 25 mL of HBS). All blood beads (BB) in experiments were compared to alginate-only beads (AB) formulated at the same time.Making hyaluronic acid/chitosan nanoparticles
Hyaluronic acid (HA) MW 35 kDa (HA35) and 180 kDa (HA180), (Medipol SA) was dissolved in dH2O to 1.5 mg mL−1 and stored at 4 °C. Chitosan (CS) MW 36 kDa (Sigma 9012-76-4) was made to 0.69 mg mL−1 in 4.6 mM HCl (aq.) and adjusted to pH 5.0. Hyaluronic acid coated chitosan nanoparticles (HAMW/CS) were made as previously described.23 In brief, 250 μL of CS was added to 250 μL of dH2O in a round bottomed tube and stirred at 1000 rpm for 10 min under sterile conditions. Following this, 500 μL was then added to 500 μL of HA solution and stirred at 1000 rpm for 30 min at RT. Dynamic light scattering (DLS) using a Malvern Zetasizer Nano ZS and software (Malvern Instruments Ltd, Malvern, UK) was used to determine hydrodynamic diameter and polydispersity index (PDI) at 298 ± 0.1 K, recorded at a backscattering angle of 173 °C.Protein release assay
Samples for assay were collected from either the storage buffer (HBS) or dissolved alginate samples at the appropriate time point. AB and BB were dissolved using 100 mM HEPES and 500 mM trisodium citrate dehydrate adjusted to pH 7.3. Protein concentration was determined using the Pierce BCA protein assay kit (Thermo Scientific 23225) according to kit protocol. Colourmetric detection of absorbance at 562 nm was performed using the Biotek plate reader and Gen5 software. Experimental replicates (n) were defined as beads formed from different blood donors.Blood breakdown release assays
BB and MHB were incubated in HBS at 37 °C for 4 and 24 hours. Buffer samples were collected and assayed for ROS release (Abcam ab238535), haemoglobin release (Sigma MAK115), hemin release (Abcam ab65332), fibrinolytic and thrombotic factor release using the LEGENDplex panels (BioLegend 740760 and 740908 respectively), all according to kit instructions. Control whole blood/plasma samples were used from the same donation. Detection of absorbance/fluorescence was performed using the Biotek plate reader and Gen5 software. LEGENDplex samples were analysed using the BD FACSVerse flow cytometer and data analysed using the open source LEGENDplex software (https://www.biolegend.com/en-us/legendplex?tab=software). Experimental replicates (n) were defined as beads formed from different blood donors.Proton nuclear magnetic resonance (H1-NMR) analysis of alginates
Prior to 1H-NMR analysis, sodium alginate (A1 and A4) was partially degraded by acid hydrolysis as described in standard test method ASTM F2259-10.39 In brief, 0.1% w/v alginate solution was prepared under sterile conditions and adjusted to pH 5.6, then pH 3.8 each time heated to 100 °C for 1 h. Final pH was adjusted to 7.4 and samples were freeze-dried at −80 °C, 0.01 mbar (Scanvac Coolsafe Pro55 with Vacuubrand Vacuum pump). Samples (10 mg) were fully hydrated at RT in pure deuterium oxide (D2O, 7789-20-0, Acros Organics) and freeze-dry repeated. A 10 mg mL−1 alginate in D2O solution was prepared and 700 μL was tested. 1H NMR scans were performed using the Bruker Avance III 400 MHz spectrometer (Bruker) at 78 °C with 30° pulse proton angle, relaxation delay 1 s and acquisition time 3.984 s using a 5 mm prodigy cryo-probe. The total number of scans was 16. Obtained spectra were processed using Bruker Topspin (version 3.6.3). H1-NMR traces were annotated as previously described40 and the area under the curve was used to calculate the G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M ratio using eqn (1).39
M ratio using eqn (1).39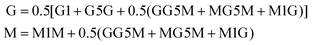 | (1) |
Rheology
Rheological tests were performed to measure the elastic (G′) and viscous (G′′) moduli of alginate-based hydrogels. Oscillatory rheology tests were performed using a Discovery HR-2 rheometer (TA Instruments, UK) equipped with a 20 mm parallel plate geometry. Frequency sweeps were performed at 0.5% strain, 1–10 rad s−1 frequency range at 37 °C with the gap (plate-to-plate) adjusted to the height of each sample (5 mm). Retrieved data were analysed using DIN 51810-2 recommended settings in RheoWin Data Manager (version 4.87.0002) to determine linear viscoelastic range (LVR).Model haematomas
In sterile conditions, AB or BB were well mixed and 2 mL of each transferred to a tube. Beads were left to settle and HBS removed. 1 mL of 10 mg mL−1 A4 and 1 mL HA/CS nanoparticles were added and gently mixed until a homogenous suspension was obtained. The solution was loaded into a 2 mL syringe and extruded through a 30G gauge needle into a rotating 75 mM CaCl2 solution bath, allowing crosslinking at RT. After 10 minutes, MH were poured through a 50 μm cell strainer to remove the CaCl2 and washed twice with HBS. Model haematomas were stored in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 volume of HBS at 4 °C.
10 volume of HBS at 4 °C.
Imaging
Images were acquired using a fluorescent inverted microscope (Leica DMI6000, Leica Microsystems, UK) coupled with a 5.5 Neo sCMOS camera (Andor, UK), and equipped with 2.5× (PLAN 2.5×/0.07, Leica) and 10× objectives (PL 10×/0.3 PH1, Leica). Images were acquired and analysed using μManager software (v.1.46, Vale Lab, UCSF, USA) and ImageJ.Cell culture
The human leukaemia monocytic cell line, THP-1, was cultured in RPMI-1640 medium supplemented with 2 mM L-glutamine, 100 μg mL−1 penicillin–streptomycin (Pen-Strep, Sigma), and 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Gibco). Cells were grown in T75 flasks and incubated in a humidified 5% CO2 (v/v) atmosphere at 37 °C and routinely split for optimal culture numbers. Cells between passages 11 and 23 were plated 1 million cells per mL to a 96 well plate the day of loading haemorrhages. THP-1 cells between passages 11 and 19 were plated 1 mil mL−1 to a 96 well plate and differentiated using 250 nM of phorbal-12-myristate-13-acetate (PMA) overnight. Media was changed in the morning when plating the models to PMA-free supplemented RPMI.Immortalised human neuroblastoma cell line SH-SY5Y, were cultured between passages 13 and 20 were raised in DMEM supplemented with 100 μg mL−1 penicillin–streptomycin (Pen-Strep, Sigma) and 10% (v/v) FBS. Cells were trypsinized with a solution of 0.5% (w/v) trypsin and 0.2% (w/v) EDTA (Sigma) to split cultures. Cells were plated 0.0375 million cells per mL to a 96 well plate and allowed to adhere overnight. Media was changed the following morning prior to adding the models.
Immortalised human cerebral microvascular endothelial cell line, hCMEC/D3, was purchased from Merck (UK). Cells were cultured between passages 8 and 10, and plated on pre-coated surfaces with rat tail collagen type I (Merck), (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in PBS) at 37 °C for 1 h. Cultures were maintained in complete Endothelial Cell Growth Medium MV (PromoCell, C-22020) supplemented with 1 ng mL−1 recombinant human basic fibroblast growth factor (Merck), 100 units per ml penicillin, and 100 μg mL−1 penicillin–streptomycin (Pen-Strep, Sigma). Cells were seeded in the upper compartment of collagen-coated inserts (24-well, 0.4 μm pores, PET translucent, CellQART) at a density of 0.5 million cells per ml in 100 μL 24 h prior to experiments.
100 in PBS) at 37 °C for 1 h. Cultures were maintained in complete Endothelial Cell Growth Medium MV (PromoCell, C-22020) supplemented with 1 ng mL−1 recombinant human basic fibroblast growth factor (Merck), 100 units per ml penicillin, and 100 μg mL−1 penicillin–streptomycin (Pen-Strep, Sigma). Cells were seeded in the upper compartment of collagen-coated inserts (24-well, 0.4 μm pores, PET translucent, CellQART) at a density of 0.5 million cells per ml in 100 μL 24 h prior to experiments.
Estimation of blood content
For AB/BB treatment, 50 μL media was removed from 200 μL in 96 well plates and equal volume of homogenous bead solution added. At 10% v/v density this was estimated to contain ∼5 μL of blood held within the beads. For MHA/MHB treatment, 3× MH were isolated using a micro spatula spoon and added to the wells. MHB were made with 2 mL BB at 10% v/v equalling 200 μL blood per batch. 2 mL A4-HA35/CS solution resulted in ∼100 MH assuming equal distribution of blood beads equals 2 μL of blood held in each MHB.Cell death, ELISA and microscopy
At 4 and 24 hours following incubation, 100 μL of media was removed and analysed for hIL-1B using ELISA kit and instructions (Biotechne DY201). Cell death was analysed using a lactate dehydrogenase (LDH) release assay (CytoTox 96 Promega G1780) on supernatant samples following bead incubation as per the kit instructions. Colourmetric detection of absorbance was performed using the Biotek plate reader and Gen5 software. Experimental replicates (n) were defined as experiments performed on different passages of cells.Dextran permeability of hCMEC/D3 monolayers
Plasma, AB and BB were added to the lower compartment of the well for 24 h. Following treatments, media was changed to phenol-red free Opti-MEM medium (Gibco) and fluorescein isothiocyanate–dextran 70 kDa (Sigma) was added to the upper compartment at 1 mg mL−1 in a total volume of 100 μL. 30 min later media was collected from the lower compartment. Fluorescence of the media samples was measured using a Fluorstar reader with excitation at 492 nm and emission at 520 nm (BMG Labtech). Fluorescence was normalised to the buffer control group. Experimental replicates (n) were defined as experiments performed on different passages of hCMEC/D3 cells.Quantitative PCR
Cells were pelleted and RNA extracted using Trizol and chloroform/isopropanol DNA precipitation. Complementary DNA sequences were generated from 800 ng of RNA and reverse transcriptase from the High-Capacity RNA-to-cDNA kit (Applied Biosystems). Gene expression analysis was performed using RT-qPCR with sample cDNA and SYBRGreen qPCR Master mix (Thermo Fisher Scientific). Primers used for IL-1β and TNF, are listed below and compared to relative quantities of house keeper genes HPRT1 and GAPDH. Data was acquired using the Applied Biosystems StepOne software and quantified using 2−ΔΔCT.41 Experimental replicates (n) were defined from different cell passages.| Primers | Sequence |
| Fw_hs-HPRT1 | CAGGCGAACCTCTCGGCTTT |
| Rv_hs-HPRT1 | GGGTCGCCATAACGGAGCC |
| Fw_hs-GAPDH | TCGGAGTCAACGGATTTGGTC |
| Rv_hs-GAPDH | AGTTGAGGTCAATGAAGGGT |
| Fw_hs-IL1B | ACGATGCACCTGTACGATCACT |
| Rv_hs-IL1B | CACCAAGCTTTTTTGCTGTGAGT |
| Fw_hs-TNF | TGCACTTTGGAGTGATCGGC |
| Rv_hs-TNF | AGCTTGAGGGTTTGCTACAAC |
Western blot
Cells were incubated with beads and model haematomas for 4 and 24 hours before being pelleted, supernatant removed and dissociated using a 1% Triton X lysis buffer and protease inhibitor. Protein was loaded into 12% resolving gel and separated by SDS-PAGE before blotting using a semi-dry transfer system (BIO-Rad). Primary antibody (goat anti-IL-1β, RnD Systems) was added 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 overnight at 4 °C and a secondary HRP antibody (anti-goat, Agilent Technologies) (1
1000 overnight at 4 °C and a secondary HRP antibody (anti-goat, Agilent Technologies) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) used to visualise bands with ECL detecting reagent (Amersham) in the G
5000) used to visualise bands with ECL detecting reagent (Amersham) in the G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Box with GeneSys software (Syngene). Membranes were washed and incubated with anti-β actin (Sigma A3854) 1
Box with GeneSys software (Syngene). Membranes were washed and incubated with anti-β actin (Sigma A3854) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 as a loading control and imaged again. Gel bands were quantified using ImageJ gel analysis tool and area under the curve.
000 as a loading control and imaged again. Gel bands were quantified using ImageJ gel analysis tool and area under the curve.
Statistical analysis
All data was analysed using GraphPad Prism. Box and whisker plots show the median value and extend to 25th and 75th percentile, whiskers show the minimum and maximum values. Analysis of multiple groups was performed using a one-way or two-way ANOVA with appropriate multiple comparisons. Error bars show s.d. Analysis of fibrinolytic factor release was performed using unpaired Student's T-tests.Abbreviations
| A1–4 | Alginate samples 1–4 |
| AB | Alginate-only beads |
| BB | Blood beads |
| MHA | Model haematomas made with alginate-only beads and A4 HA35/CS gels |
| MHB | Model haematomas made with blood beads and A4 HA35/CS gels |
| HA35 | 35 kDa Hyaluronic acid |
| HA180 | 180 kDa Hyaluronic acid |
| CS | Chitosan |
| HBS | HEPES buffered solution |
Author contributions
SC is responsible for the project conceptualisation, methodology, investigation, data acquisition, analysis, validation and visualisation and the writing of the original manuscript. VST is responsible for the data presented in Fig. 1A and 6G. CB is responsible for the NMR data acquisition and analysis in Fig. 1B. AT is responsible for the conceptualisation of the project, provision of resources and supervision for SC. All authors were involved in the reviewing and editing of the final manuscript.Data availability
All raw data pertaining to this study is available without restriction from the authors. Data acquired using models fabricated with A4-HA180/CS hydrogels is available as ESI Fig. 1.†Conflicts of interest
The authors declare no competing interests.Acknowledgements
This study was funded by the NC3Rs (NC/V002082/1) to SC. The authors would like to thank the University of Manchester NMR core facility for use of the Bruker BioSpin spectrometer, Dr Katharina Edkins for use of the Discovery HR-2 rheometer and Dr Gareth Howell and the University of Manchester Genomic Technologies Core Facility for cell sorting for LEGENDplex assays.References
- M. Zille, T. D. Farr, R. F. Keep, C. Römer, G. Xi and J. Boltze, Novel targets, treatments, and advanced models for intracerebral haemorrhage, EBioMedicine, 2022, 76, 103880 CrossRef CAS PubMed.
- R. F. Keep, Y. Hua and G. Xi, Intracerebral haemorrhage: mechanisms of injury and therapeutic targets, Lancet Neurol., 2012, 11(8), 720–731 CrossRef CAS PubMed.
- A. L. de Oliveira Manoel, Surgery for spontaneous intracerebral hemorrhage, Crit. Care, 2020, 24(1), 45 CrossRef PubMed.
- A. R. Parry-Jones, C. Sammut-Powell, K. Paroutoglou, E. Birleson, J. Rowland and S. Lee, et al. An Intracerebral Hemorrhage Care Bundle Is Associated with Lower Case Fatality, Ann. Neurol., 2019, 86(4), 495–503 CrossRef PubMed.
- C. L. MacLellan, G. Silasi, A. M. Auriat and F. Colbourne, Rodent models of intracerebral hemorrhage, Stroke, 2010, 41(10 suppl 1), S95–SS8 Search PubMed.
- S. E. Withers, A. R. Parry-Jones, S. M. Allan and P. R. Kasher, A Multi-Model Pipeline for Translational Intracerebral Haemorrhage Research, Transl. Stroke Res., 2020, 1–14 Search PubMed.
- M. Selim, D. Hanley, J. Broderick, J. N. Goldstein, B. A. Gregson and G. Falcione, et al. Basic and Translational Research in Intracerebral Hemorrhage: Limitations, Priorities, and Recommendations, Stroke, 2018, 49(5), 1308–1314 CrossRef PubMed.
- M. Zille, S. S. Karuppagounder, Y. Chen, P. J. Gough, J. Bertin and J. Finger, et al. Neuronal death after hemorrhagic stroke in vitro and in vivo shares features of ferroptosis and necroptosis, Stroke, 2017, 48(4), 1033–1043 CrossRef PubMed.
- L. Goldstein, Z. P. Teng, E. Zeserson, M. Patel and R.F, Regan, Hemin induces an iron–dependent, oxidative injury to human neuron–like cells, J. Neurosci. Res., 2003, 73(1), 113–121 CrossRef CAS PubMed.
- H. Parfenova, S. Basuroy, S. Bhattacharya, D. Tcheranova, Y. Qu and R. F. Regan, et al. Glutamate induces oxidative stress and apoptosis in cerebral vascular endothelial cells: contributions of HO-1 and HO-2 to cytoprotection, Am. J. Physiol., 2006, 290(5), C1399–CC410 CrossRef CAS PubMed.
- B. Syed, A. Nirwane and Y. Yao, In vitro models of intracerebral hemorrhage, Brain Hemorrh., 2022, 3(3), 105–107 CrossRef PubMed.
- J. Lok, W. Leung, S. Murphy, W. Butler, N. Noviski and E. H. Lo, Intracranial hemorrhage: mechanisms of secondary brain injury, in Intracerebral Hemorrhage Research, Springer, 2011, pp. 63–69 Search PubMed.
- J. A. Stokum, G. J. Cannarsa, A. P. Wessell, P. Shea, N. Wenger and J. M. Simard, When the blood hits your brain: the neurotoxicity of extravasated blood, Int. J. Mol. Sci., 2021, 22(10), 5132 CrossRef CAS PubMed.
- K. Y. Lee and D. J. Mooney, Alginate: properties and biomedical applications, Prog. Polym. Sci., 2012, 37(1), 106–126 CrossRef CAS PubMed.
- D. M. Hariyadi and N. Islam, Current status of alginate in drug delivery, Adv. Pharmacol. Pharm. Sci., 2020, 2020, 8886095 CAS.
- G. T. Grant, E. R. Morris, D. A. Rees, P. J. Smith and D. Thom, Biological interactions between polysaccharides and divalent cations: the egg–box model, FEBS Lett., 1973, 32(1), 195–198 CrossRef CAS.
- G. Jensen, J. L. Holloway and S. E. Stabenfeldt, Hyaluronic acid biomaterials for central nervous system regenerative medicine, Cells, 2020, 9(9), 2113 CrossRef CAS PubMed.
- A. Dityatev, A. Irintchev and F. Morellini, Extracellular matrix molecules: synaptic plasticity and learning, Elsevier Ltd, Encyclopedia of Neuroscience, 2009, pp. 149–156 Search PubMed.
- L. S. Sherman, S. Matsumoto, W. Su, T. Srivastava and S. A. Back, Hyaluronan synthesis, catabolism, and signaling in neurodegenerative diseases, Int. J. Cell Biol., 2015, 2015, 368584 Search PubMed.
- J. E. Rayahin, J. S. Buhrman, Y. Zhang, T. J. Koh and R. A. Gemeinhart, High and low molecular weight hyaluronic acid differentially influence macrophage activation, ACS Biomater. Sci. Eng., 2015, 1(7), 481–493 CrossRef CAS PubMed.
- Y. Chen, S. Chen, J. Chang, J. Wei, M. Feng and R. Wang, Perihematomal edema after intracerebral hemorrhage: an update on pathogenesis, risk factors, and therapeutic advances, Front. Immunol., 2021, 12, 740632 CrossRef CAS PubMed.
- M. Rubert, M. Alonso-Sande, M. Monjo and J. Ramis, Evaluation of alginate and hyaluronic acid for their use in bone tissue engineering, Biointerphases, 2012, 7(1), 44 CrossRef CAS.
- A. Almalik, H. Benabdelkamel, A. Masood, I. O. Alanazi, I. Alradwan and M. A. Majrashi, et al. Hyaluronic acid coated chitosan nanoparticles reduced the immunogenicity of the formed protein corona, Sci. Rep., 2017, 7(1), 1–9 CrossRef CAS PubMed.
- A. Almalik, R. Donno, C. J. Cadman, F. Cellesi, P. J. Day and N. Tirelli, Hyaluronic acid-coated chitosan nanoparticles: Molecular weight-dependent effects on morphology and hyaluronic acid presentation, J. Controlled Release, 2013, 172(3), 1142–1150 CrossRef CAS PubMed.
- B. Enobakhare, D. Bader and D. Lee, Concentration and M/G ratio influence the physiochemical and mechanical properties of alginate constructs for tissue engineering, J. Appl. Biomater. Biomech., 2006, 4(2), 87–96 CAS.
- T. Kaster, I. Sack and A. Samani, Measurement of the hyperelastic properties of ex vivo brain tissue slices, J. Biomech., 2011, 44(6), 1158–1163 CrossRef CAS PubMed.
- S. Urday, L. A. Beslow, D. W. Goldstein, A. Vashkevich, A. M. Ayres and T. W. Battey, et al. Measurement of perihematomal edema in intracerebral hemorrhage, Stroke, 2015, 46(4), 1116–1119 CrossRef PubMed.
- D. Kour, M. Ali, P. Khajuria, K. Sharma, P. Ghosh and S. Kaur, et al. Flurbiprofen inhibits heme induced NLRP3 inflammasome in Berkeley sickle cell disease mice, Front. Pharmacol., 2023, 14, 1123734 CrossRef CAS PubMed.
- L. Qin, Z. Fengyong, Z. Jiamin, Y. Qixiu, L. Geming and X. Rongwei, et al. NLRP3 Inflammasome activation regulates aged RBC clearance, Inflammation, 2018, 41, 1361–1371 CrossRef CAS PubMed.
- S. P. Cullen, C. J. Kearney, D. M. Clancy and S. J. Martin, Diverse activators of the NLRP3 inflammasome promote IL-1β secretion by triggering necrosis, Cell Rep., 2015, 11(10), 1535–1548 CrossRef CAS PubMed.
- S. Imoto, M. Kono, T. Suzuki, Y. Shibuya, T. Sawamura and Y. Mizokoshi, et al. Haemin-induced cell death in human monocytic cells is consistent with ferroptosis, Transfus. Apher. Sci., 2018, 57(4), 524–531 CrossRef PubMed.
- C. Carrasco-Pozo, K. N. Tan and V. M. Avery, Hemin prevents increased glycolysis in macrophages upon activation: protection by microbiota-derived metabolites of polyphenols, Antioxidants, 2020, 9(11), 1109 CrossRef CAS PubMed.
- K. Nurmi, I. Kareinen, J. Virkanen, K. Rajamäki, V.-P. Kouri and K. Vaali, et al. Hemin and cobalt protoporphyrin inhibit NLRP3 inflammasome activation by enhancing autophagy: a novel mechanism of inflammasome regulation, J. Innate Immun., 2017, 9(1), 65–82 CrossRef CAS PubMed.
- T. Boontheekul, H.-J. Kong and D. J. Mooney, Controlling alginate gel degradation utilizing partial oxidation and bimodal molecular weight distribution, Biomaterials, 2005, 26(15), 2455–2465 CrossRef CAS PubMed.
- L. Shah, A. Latif, K. J. Williams and A. Tirella, Role of stiffness and physico-chemical properties of tumour microenvironment on breast cancer cell stemness, Acta Biomater., 2022, 152, 273–289 CrossRef CAS PubMed.
- R De La Rosa J, J Wubetu, N Tirelli and A. Tirella, Colorectal tumor 3D in vitro models: Advantages of biofabrication for the recapitulation of early stages of tumour development, Biomed. Phys. Eng. Expr., 2018, 4(4), 045010 CrossRef PubMed.
- B. Almari, D. Brough, M. Harte and A. Tirella, Fabrication of Amyloid-β-Secreting Alginate Microbeads for Use in Modelling Alzheimer's Disease, J. Visualized Exp., 2019,(149), e59597 CAS.
- X. Yang, H. Sui, H. Liang, J. Li and B. Li, Effects of M/G Ratios of Sodium Alginate on Physicochemical Stability and Calcium Release Behavior of Pickering Emulsion Stabilized by Calcium Carbonate, Front. Nutr., 2022, 8, 818290 CrossRef PubMed.
- ASTM, International. Standard test method for determining the chemical composition and sequence in alginate by proton nuclear magnetic resonance (1H NMR) spectroscopy, West Conshohocken, PA, 2012 Search PubMed.
- C. Zhao, A. Latif, K. J. Williams and A. Tirella, The characterization of molecular weight distribution and aggregation by asymmetrical flow field-flow fractionation of unmodified and oxidized alginate, React. Funct. Polym., 2022, 175, 105292 CrossRef CAS.
- K. J. Livak and T. Schmittgen, Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method, Methods, 2001, 25(4), 402–408 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm00039k |
| This journal is © The Royal Society of Chemistry 2024 |