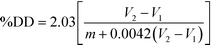Anti-viral RNAi nanoparticles protect shrimp against white spot disease
Shai
Ufaz
a,
Adi
Balter
a,
Chen
Tzror
a,
Shai
Einbender
a,
Ori
Koshet
a,
Janna
Shainsky-Roitman
a,
Zvi
Yaari
b and
Avi
Schroeder
 *b
*b
aViAqua Therapeutics, Technion – Israel Institute of Technology, Haifa 3200003, Israel
bDepartment of Chemical Engineering, Technion – Israel Institute of Technology, Haifa 32000, Israel. E-mail: avids@technion.ac.il; Tel: +972 778871953
First published on 8th November 2017
Abstract
Nearly 20% of cultured shrimp die every year due to viral diseases. In this study, we evaluated the capacity of nanoparticulate RNA interference (RNAi) to down-regulate genes in Penaeus vannamei shrimp and protect shrimp against white spot syndrome virus (WSSV, i.e. white spot disease). Using a reporter target gene, Rab7, we show that the length of the administered dsRNA correlates with gene knockdown. We found that 250 bp-long dsRNA strands knocked down gene expression most effectively, followed by 125 and 70 bp-long strands. The 21 bp long strands did not downregulate the gene. We also show gene downregulation to be concentration dependent. Even at low RNA concentrations of 0.01 μg per gram-body-weight gene knockdown exceeded 80%. Knockdown levels were similar in multiple organs, including the gills, gut, hepato-pancreas, pleopods and muscle. Gene knockdown lasted for one month, after which gene expression recuperated. To protect the RNA molecules from enzymatic degradation, we complexed the RNA with the cationic polysaccharide chitosan, forming 90–200 nm particles that facilitated efficient RNAi in vivo. In a double-blinded viral challenge test, RNAi targeting viral-protein 28 (VP28) protected the shrimp against WSSV infection. Survival of animals treated with RNAi nanoparticles exceeded 95% compared to no survival in the untreated controls. Nanoparticulate RNAi is an effective modality for protecting against viral diseases.
Design, System, ApplicationWe describe the molecular design of a RNA-chitosan nanoparticle for treating viral diseases in aquaculture. Specifically, we show that RNA-interference downregulates viral gene expression in shrimp for nearly one month after administering a single dose of RNA-nanoparticles. This targeted treatment protected shrimp against a fatal disease – white spot syndrome virus, which impacts the global food chain and food safety. We demonstrate that designing the molecular building blocks plays an important role in the structure and function of the self-assembled supra-molecular nanoparticle. For example, chemically-modified chitosan polymers complexed with double-stranded RNA formed 150–200 nm nanoparticles, while the same polymer complexed with long single-strand RNA formed particles of half the size. This study highlights the promise of nanotechnologies for treating pandemics. |
1. Introduction
Shrimp is a primary dietary source in both developing and developed countries.1–4 To meet this large demand, shrimp farming has intensified over the past decades. At the same time, the impact of viral diseases has surged, destroying nearly 20% of the global produce annually.2White spot syndrome virus (WSSV, the virus that induces white spot disease – WSD), is one of the most lethal infections in shrimp and has no existing treatment. Countries effected by WSSV include the Philippines, China, Indonesia, Iran, Brazil, Australia, India and many others.5
WSSV is a 300 kb dsDNA virus that is approximately 350 nm long and 130 nm in diameter.5 The surface of the viral capsid is decorated with proteins that facilitate its invasion through the shrimp's stomach, gills, cuticular epidermis and hepatopancreas.6–8 The virus utilizes clathrin mediated endocytosis to penetrate cells, eventually escaping from early endosomes via Rab7 (endogenous to the shrimp) and viral protein-28 (VP28) pathways.8,9 Infected shrimp present white spots on their body, and usually die three to seven days post infection.8
RNA interference (RNAi) is an effective gene downregulation mechanism induced by RNA.10 In this process, mRNA is degraded through a RNA-induced silencing complex (RISC) by introducing fragments of complementary RNA to the treated cell.11,12 Recent Phase III clinical trials with RNAi have proven effective in treating disease.13,14 We sought to use RNAi to downregulate genes that facilitate the viral progression within the shrimp's body. Specifically, we targeted Rab7 and VP28.
RNA is susceptible to enzymatic degradation in the water or before entering cells inside the shrimp's body.15 Nanoparticulate RNA formulations are playing a major role in facilitating RNAi in vivo.16,17 For a nanoparticle formulation to be effective it must protect the RNA payload from enzymatic degradation and release the RNA at a therapeutic dose intracellularly.12,18–23 RNA nanoparticles usually contain a cationic component, such as polymers or lipids, that complex electrostatically with the negatively-charged RNA payload to form nanoparticles.15
We used chitosan, a polysaccharide that is sourced from the shrimp's exoskeleton, as our cationic molecular template. To adjust the charge density on the chitosan we modified the number of primary amine groups on the molecule, resulting in a cationic polymer capable of complexing with RNA into nanoparticles.
Chitosan is a biodegradable and biocompatible co-polymer of glucosamine and N-acetyl-D-glucosamine derived from chitin. Specifically, chitosan is a deacetylated chitin polymer, which, in contrast to chitin, has free amine groups.24,25 At acidic pH, below its pKa (∼6.5), the primary amines in chitosan become positively charged. These protonated amines enable the chitosan to bind to the negatively-charged dsRNA and condense into particles.26 The degree of deacetylation on the polymer backbone plays an important role in controlling its affinity to the nucleic acid.26–28
In this study we used chemically-modified chitosan-based nanoparticles to carry anti-Rab7 and anti-VP28 RNA and evaluated their ability to downregulate gene expression in vivo.
2. Materials and methods
dsRNA production
Rab7, a primary gene that is involved in shrimp viral infection cascades, was chosen as the target gene for optimizing the silencing effect. Inhibition of Rab7 protein leads to increased immunity against the WSSV virus. dsRNA was produced in-house using in vivo bacterial expression system. Rab7 gene was cloned into a E. coli expression vector in forward and reverse orientation downstream to the T7 promoter. The two fragments were separated by a 100 bp non coding loop region. For cloning a recombinant plasmid containing an inverted repeat of stem and loop a synthetic fragment containing Litopenaeus vannamei Ras-related protein (Rab7, accession # JQ581679) partial sequence and non-coding addition for the loop was synthesized (Loop, sequence below). This fragment was used as template for amplification of sense 250 bp Rab7 sequence + loop region using the primers: XbaI Rab7 250 Forward GAAACTCTAGATGGGTAACAAGATTGATCTGGAG and BamHI Rab7 250 Reverse AGCCGGATCCtagcttacga. The PCR product was cloned into pET9a (Novagen) using XbaI + BamHI restriction sites. The antisense Rab7 was PCR amplified using the same template with the following primers: BamHI 250 forward GCATAGGATCCTGGGTAACAAGATTGATCTGGAG and PstI 250 reverse GAGTACTGCAGCATCCTGTTTAGCCTTGTTGTCA and was cloned using BamHI, PstI to generate the final plasmid – pET9a-Rab7 250 RNAi. The recombinant plasmid (pET9a-Rab7 250) containing an inverted repeat of stem loop Rab7 was transformed by heat shock method into the ribonuclease III (Rnase III) mutant E. coli strain HT115. Expression of stem loop Rab7 was induced by adding 0.4 mM isopropyl-β-D thiogalactoside (IPTG) into the bacterial culture. The culture was harvested 4 hours after IPTG induction. Bacterial single-stranded RNA (ssRNA) and loop region of stem loop Rab7 were digested with ribonuclease A (Rnase A). Then, the dsRNA was extracted by TRIzol RNA Isolation Reagents (ThermoFisher Scientific) according to the manufacturer's instructions. Concentration of dsRNA-Rab7 were determined by UV-spectrophotometry at wavelength 260 nm and agarose gel electrophoresis. The same procedure using different primers (below) was used for production of 125 and 70 bp dsRNA fragments.Rab7 silencing levels in shrimp, using either in vitro or bacteria production of dsRNA were similar.
Sample collection, RNA extraction and cDNA synthesis
Gill or other organs samples from each shrimp were collected and placed immediately in liquid nitrogen until use. The samples were homogenized using glass beads in 1 ml TRIzol reagent using an Argos pestle motor mixer and total RNA was extracted by TRIzol RNA Isolation Reagents (ThermoFisher Scientific) according to the manufacturer's instructions, followed by Dnase treatment (TURBO DNA-free, Ambion). First strand cDNA was synthesized using RevertAid First Strand cDNA Synthesis according to the manufacturer's instructions.RNA quantification by quantitative PCR
Quantification of the Rab7 gene (Accession # FJ811529) by quantitative PCR was performed as described by (Vatanavicharn et al., 2014). The primers sequence for qRT analysis was selected from Rab7 fragment that is not part of the fragment chosen for the dsRNA production to avoid errors caused by the dsRNA delivery. 2 μl of the cDNA was used in a 10 μl reaction prepared using qPCRBIO Probe Mix Lo-ROX (PCR Biosystems) and PrimeTime qPCR Assays (IDT) containing the following primers and probe: Rab7 Forward GGGATACAGCTGGTCAAGAAA; Rab7 Reverse CGAGAGACTTGAAGGTATTGGG and FAM labeled probe – CGAGGAGCTGATTGTTGTGTTCTCGT (500 nM primers and 250 nM probe).PCR was performed in CFX96 Touch Real-Time PCR Detection System using the default thermal cycling conditions. Real-time RT-PCR Ct values obtained for Rab7 mRNA were normalized against Ct values obtained for EF1α mRNA (Accession # GU136229) using the following primers and probe: EF1a Forward GTGGAGACCTTCCAACAGTATG, EF1a Reverse CCTTCTTGTTGACCTCCTTGAT and FAM labeled probe TGCGTGACATGAAGCAGACGG. A mean delta Ct value ± SD was determined for each treatment (minimum of 5 shrimp each) and the quantification was relative to a non-treated control that was set to 1.
Tail muscle injection of dsRNA
To deliver dsRNA by injection, 10 μL saline solution containing each dsRNA formulation in the tested concentration was injected into the tail muscle of the 3rd abdominal segment of each shrimp using a 50 μl Hamilton glass syringe equipped with a 26-guage needle.N-Acetylation of chitosan
1.5 g chitosan (HMC+ Chitoscience Chitosan 80/20, Mw 40–150 kDa) were dissolved in 4% (v/v) acetic acid solution (50 ml), and acetic anhydride in 50 ml absolute ethanol was added. As the amount of acetic anhydride determines the extent of the N-acetylation reaction and therefore the deacetylation degree (DD) of the final product, different amounts of acetic anhydride were used. After stirring at 40 °C for 12 h, the reaction was stopped by pH neutralization with NaOH (5 M) to pH 9. The resulting precipitate was centrifuged (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 10 min at 4 °C) and washed thoroughly with deionized water at pH 9. This procedure was repeated 3 times. Then, the precipitate was washed from solvents and impurities by dialysis (SpectrumLabs, 12–14 kDa pore size) in 5 L of deionized water for 72 h. The water was replaced 8 times during this period. The resulting N-acetylated chitosan was recovered from the dialysis bag using centrifugation (15
000 g for 10 min at 4 °C) and washed thoroughly with deionized water at pH 9. This procedure was repeated 3 times. Then, the precipitate was washed from solvents and impurities by dialysis (SpectrumLabs, 12–14 kDa pore size) in 5 L of deionized water for 72 h. The water was replaced 8 times during this period. The resulting N-acetylated chitosan was recovered from the dialysis bag using centrifugation (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 15 min at 4 °C) and freeze-drying for 48 h using lyophilizer (Labconco) (Scheme 1).
000 g for 15 min at 4 °C) and freeze-drying for 48 h using lyophilizer (Labconco) (Scheme 1).
Potentiometric determination of deacetylation degree
Potentiometric titration was used to determine the deacetylation degree (DD) of the N-acetylated chitosan. Briefly, 0.2 g of the synthesized chitosan was dissolved in 30 ml of 0.1 M hydrochloric acid. After 2 hours of continuous stirring, 25 ml of deionized water was added and the stirring continued for additional 1 hour. When the chitosan was completely dissolved, the solution was titrated with a 0.1 M sodium hydroxide solution. From the titration of this solution, a curve with two inflexion points was obtained. The amount of the acid consumed between these two points was considered to correspond to the amount of the free amino groups in the solution (Tolaimate, 2000). The titration was performed with a pH meter (EUTECH instruments).Degree of deacetylation (DD) of chitosan was calculated using the following formula:
Preparation of chitosan-dsRNA nanoparticles
Formulations with different polymer/dsRNA mass ratios were prepared by a self-assembly method while keeping the amount of dsRNA constant (20 μg mL−1). Chitosan or its N-acetylated derivative was dissolved to a concentration of 4% (w/v) in 0.2 M sodium acetate buffer (pH 4.6). Working solutions with concentrations of 0.01–0.1% were made from the stock by dilution with the same buffer. Equal volumes of Rab7 dsRNA solution (100 μg ml−1 in 0.2 M sodium acetate buffer) and chitosan solutions were quickly mixed under high vortexing for 30 s and incubated for 1 hour at RT. The assembled nanoparticles were used without further purification. Since the dsRNA concentration remains constant for all nanoparticle solutions, manipulating the chitosan concentration changes the polymer/dsRNA mass ratio.Characterization of nanoparticles
Particle size and surface charge (zeta potential) of the chitosan/dsRNA nanoparticles were determined using a Zetasizer Nano ZSP (Malvern Instruments, UK). Particle size measurements were performed at a 173° angle and a temperature of 25 °C. The size is expressed as the z-average hydrodynamic diameter obtained by a cumulative analysis of the correlation function using the viscosity and refractive index of water in the calculations. The surface charge of the nanoparticles distributed in sodium acetate buffer (pH 4.6 unless otherwise is stated) was measured using a disposable folded capillary cell (Malvern Instruments, UK).Production of ssRNA
The Rab7-ssRNA was synthesized in vitro. For generating the DNA template for the ssRNA synthesis, a synthetic fragment containing Litopenaeus vannamei Ras-related protein (Rab7, accession # JQ581679) partial sequence was synthesized. This fragment was used as a template for amplification of sense and antisense 250 b Rab7 sequence using the following primers:Rab7 for TGGGTAACAAGATTGATCTGGAG; T7 rab7 Rev GGATCCTAATACGACTCACTATAGGCATCCTGTTTAGCCTTGTTGTC.
In vitro transcription was performed using T7 RiboMAX Express RNAi System (Promega) according to the manufacturer protocol. The reaction was incubated for 1 hour at 37 °C, followed by DNase treatment and ethanol precipitation. The RNA was resuspended in nuclease free water and its concentration was determined by UV-spectrophotometry at wavelength 260 nm and agarose gel electrophoresis.
Antisense fragment:
CATCCTGTTTAGCCTTGTTGTCATTGGTCAATTTGATCTGGTCTGGAAACTCATTGTACAGCTCCACCTCTGACTCCTGAGCAAGAGCATTGCGAGCAATGGTCTGGAAAGCTAGCTCCACATTAATAGCTTCCTTTGCACTAGTTTCAAAGTAGGGAACTTCATTTTTACTATGACACCATTGTTGTGCTCGCTTCGTCGATACCGCCCTATTCTCCAGATCAATCTTGTTACCCA.
RNAse assay
Complex stability and ability to protect the dsRNA from degradation was examined using the RNAse A enzymatic degradation assay. Briefly, free dsRNA and H55/dsRNA and H42/dsRNA complexes were first exposed to RNAse A (Promega, Cat# A7970, Madison, WI) for an hour at 37 °C (20 ng RNAse A per 1 μg complexed RNA). Then, the enzyme was inactivated by RNAse inhibitor (80 units per 1 μg of complexed RNA, Ribolock RNAse inhibitor, 20 U μl−1, Thermo Fisher Scientific, Waltham, MA) and the dsRNA was released from the nanoparticles by incubation of 1 hour at 37 °C with Chitosanase (Sigma). The free dsRNA was precipitated with cold isopropanol. The pellets were then re-dissolved in 30 μl of DEPC-water and applied to a 1.5% agarose gel electrophoresis for 30 min at 100 V.WSSV challenge test
CCATGGGCAGCAGCCATCATCATCATCATCACGGATCCGGCAGGTATCACAACACTGTGACCAAGACCATCGAAACCCACACAGACAATATCGAGACAAACATGGATGAAAACCTCCGCATTCCTGTGACTGCTGAGGTTGGATCAGGCTACTTCAAGATGACTGATGTGTCCTTTGACAGCGACACCTTGGGCAAAATCAAGATCCGCAATGGAAAGTCTGATGCACAGATGAAGGAAGAAGATGCGGATCTTGTCATCACTCCCGTGGAGGGCCGAGCACTCGAAGTGACTGTGGGGCAGAATCTCACCTTTGAGGGAACATTCAAGGTGTGGAACAACACATCAAGAAAGATCAACATCACTGGTATGCAGATGGTGCCAAAGATTAACCCATCAAAGGCCTTTGTCGGTAGCTCCAACACCTCCTCCTTCACCCCCGTCTCTATTGATGAGGATGAAGTTGGCACCTTTGTGTGTGGTACCACCTTTGGCGCACCAATTGCAGCTACCGCCGGTGGAAATCTTTTCGACATGTACGTGCACGTCACCTACTCTGGCACTGAGACCGAGTAAGCGGCCGCAACTCGAGAACG.
This fragment was used as template for amplification of sense 250 bp VP28 sequence using the primers: XbaI VP28-3 for GACTATCTAGACATTCAAGGTGTGGAACAACAC and NcoI VP28-3 Rev ACGTACCATGGCTCGGTCTCAGTGCCAGAGT. The PCR product was cloned into pET9a based plasmid (containing a loop fragment between its PstI and NcoI sites) using XbaI + NcoI restriction sites. The antisense VP28 was PCR amplified using the same template with the following primers: BamHI VP28-3 For ACATAGGATCCCATTCAAGGTGTGGAACAACAC and PstI VP28-3 Rev ACAGACTGCAGCTCGGTCTCAGTGCCAGAGT and was cloned using BamHI, PstI to generate the final plasmid – pET9a-VP28 250 RNAi. The recombinant plasmid (pET9a-VP28 250 RNAi) containing an inverted repeat of stem loop VP28 was transformed by heat shock method into the ribonuclease III (RNase III) mutant E. coli strain HT115. Expression of stem loop VP28 was induced by adding 0.4 mM isopropyl-β-D thiogalactoside (IPTG) into the bacterial culture.29 The culture was harvested 4 hours after IPTG induction. Bacterial single-stranded RNA (ssRNA) and loop region of stem loop VP28 were digested with ribonuclease A (RNase A). Then, the dsRNA-VP28 was extracted by TRIzol RNA Isolation Reagents (ThermoFisher Scientific) according to the manufacturer's instructions. Concentration of dsRNA-VP28 was determined by UV-spectrophotometry at 260 nm and agarose gel electrophoresis.
Production of VP28 125 bp fragment was performed in vitro. For production of the VP28 125 bp fragment, the synthetic VP28 DNA fragment was used as a template for amplification antisense 125 bp VP28 sequence using the following primers:
T7 VP28 125 Forward:
GGATCCTAATACGACTCACTATAGGGGCAGAATCTCACCTTTGAGG.
VP28 Rev: GTTGGAGCTACCGACAAAGG
In vitro transcription was performed using T7 RiboMAX Express RNAi System (Promega) according to the manufacturer's protocol. The reaction was incubated for 1 h at 37 °C, followed by DNase treatment and ethanol precipitation. The RNA was resuspended in nuclease-free water and its concentration was determined by UV-spectrophotometry at 260 nm and agarose gel electrophoresis.
Treatment 1: injection of free (uncomplexed) VP28 250 bp fragment.
Treatment 2: injection of free (uncomplexed) VP28 125 bp fragment.
Treatment 3: injection of H55/VP28 250 bp fragment.
Survival analysis determined the time to death, over a significant period of time, between 3 different treatments. Means of survival from the different groups are compared by ANOVA. Both statistical methods are processed using R Program.
3. Results and discussion
RNAi is an effective gene regulation modality that is facilitated by dsRNA having an anti-sense strand that is complementary to a target mRNA.10 The specificity of RNAi towards a target gene grants it major advantages over other therapeutic modalities. In this study, we evaluated the capacity of nanoparticulate RNAi to downregulate gene expression in shrimp against WSSV.To test the effect of RNA dose on gene expression in shrimp we chose to target the Rab7 gene, which is associated with endosomal escape. Specifically, Rab7 binds the WSSV capsid and facilitates its release from endosomes.30,31 To be safe, one must verify that the chosen RNA sequence has no known coding homology in nature, specifically not in shrimp or human (Table 1).
| Primer name | Sequence |
|---|---|
| XbaI Rab7 250 forward | GAAACTCTAGATGGGTAACAAGATTGATCTGGAG |
| BamHI Rab7 250 reverse | AGCCGGATCCTAGCTTACGA |
| BamHI 250 forward | GCATAGGATCCTGGGTAACAAGATTGATCTGGAG |
| PstI 250 reverse | GAGTACTGCAGCATCCTGTTTAGCCTTGTTGTCA |
| XbaI for Rab7 70 | GATCTCTAGAGAGGTGGAGCTGTACAATGAG |
| PstI for loop | CGTCTGCAGCGCATCTTGC |
| BamHI for Rab7 70 | GATCGGATCCGAGGTGGAGCTGTACAATGAG |
| PstI Rab7 rev | GACTACTGCAGCATCCTGTTTAGCCTTGTTGTC |
| BamHI Rab7 125 | GAATCGGATCCTAATGTGGAGCTAGCTTTCCAG |
| PstI rab7s rev | GAGTACTGCAGCATCCTGTTTAGCCTTGTTGTCA |
| PstI loop rev | tacgtCTGCAGCGCATCTTG |
| XbaI rab7 125 for | GAATCTCTAGATAATGTGGAGCTAGCTTTCCAG |
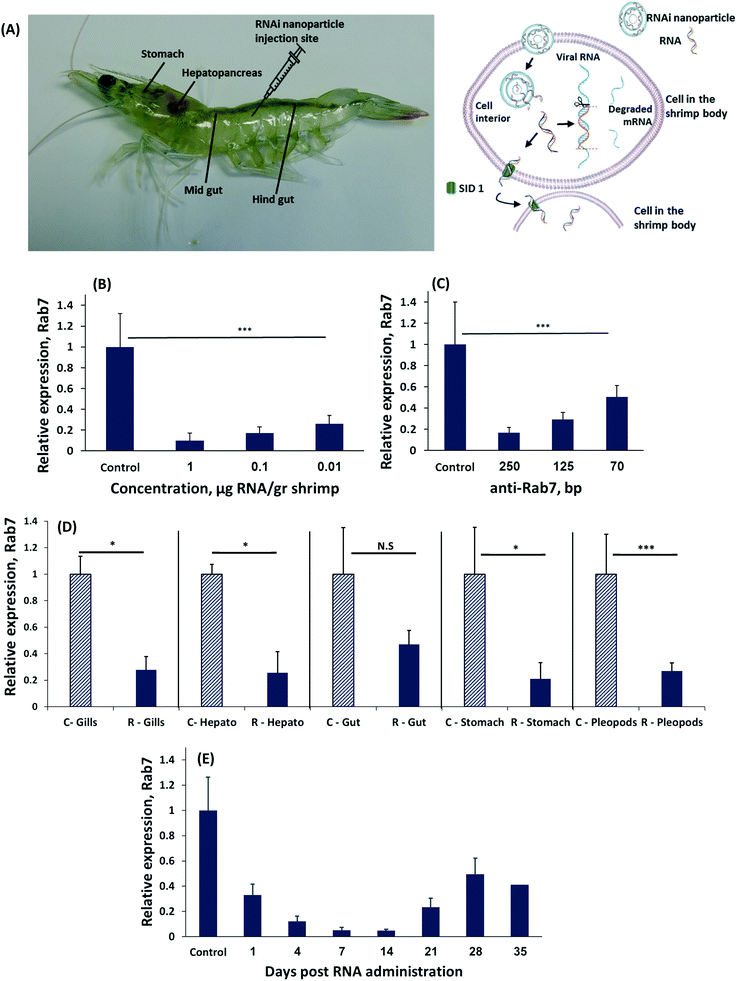
We induced RNAi by administering different concentrations of dsRNA to Penaeus vannamei shrimp by an intramuscular injection (Fig. 1A).32 Forty-eight hours later the shrimp were sacrificed and the level of Rab7 expression in the shrimp's tissues was quantified using real-time (rt)PCR. A dose-dependent effect of the dsRNA on Rab7 expression was recorded (Fig. 1B). Animals administered 0.01 μg-dsRNA per g-shrimp had a 75 ± 8% Rab7 downregulation compared to 90 ± 7% at 1 μg-dsRNA per g-shrimp. These knockdown results are an improvement over the state-of-the-art (∼2 μg-RNA per g-shrimp33) and are owed to the design of the dsRNA strand that targets the 3′ region of the Rab7 gene.34,35
Interestingly, the length of the RNA strand had a significant effect on gene knockdown. At a concentration of 0.1 μg-dsRNA per g-shrimp the 70 bp-long dsRNA strands downregulated Rab7 by 50 ± 11%, while the same dose of 250 bp strands knocked-down Rab7 by 90 ± 5% (Fig. 1C). In mammals, delivering 20 to 25 bp-long dsRNA is considered optimal for inducing RNAi.34 However, in plants and crustaceans there is evidence that longer RNA strands are more effective.22,33,36–39 While the explanation for this phenomenon remains a matter of scientific investigation, it is known these long RNA strands are diced into 21 bp-long RNA fragments intracellularly in order to then enter the RISC complex.40 In the cytoplasm the various diced RNA fragments can target a greater portion of the Rab7 gene, thereby improving specificity to the target gene.41
To be effective against a viral infection, gene knockdown must persist throughout the body. We measured the level of Rab7 expression in different organs after a local administration of dsRNA to the shrimp's tail muscle. All organs had significant Rab7 knockdown (Fig. 1D) exceeding 70% in the stomach, pleopods, hepatopancreas and gills. A slightly lower level of knockdown was recorded in the gut, due to the rapid turnover of the tissues in the gastrointestinal tract.42 These data indicate that RNAi occurs in the entire body, even after a local administration of dsRNA. The mechanism by which the dsRNA is trafficked throughout the shrimp's body differs from RNAi in mammals. In mammals, dsRNA must be administered into each targeted cell individually.43 In shrimp, however, a transcellular trafficking mechanism exists in a manner that one cell introduces the siRNA to the neighboring cells. Therefore, a local administration of the RNA treatment is sufficient to treat the entire body.19
The duration of gene knockdown is important for determining the period the shrimp will be protected from a viral infection. To test how long the target gene was downregulated we injected shrimp with 0.1 μg-anti-Rab7-dsRNA per g-shrimp and measured expression over time (Fig. 1E). Gene knockdown lasted for 35 days, after which Rab7 expression resumed.
RNA is susceptible to biodegradation in the aqueous environments shrimps are farmed in. To stabilize the therapeutic RNA payload we formulated it into chitosan-based nanoparticles. Different chitosan molecules, including chitosan and partially N-acetylated chitosan derivatives (having less primary amine groups on the polymer backbone), were synthesized and compared for their ability to deliver dsRNA and subsequently mediate gene silencing.
The physical properties of a nanoparticle influence its biodistribution and cellular fate.44 Specifically, the particle size, surface charge and affinity to the RNA payload greatly influence its activity in vivo.45
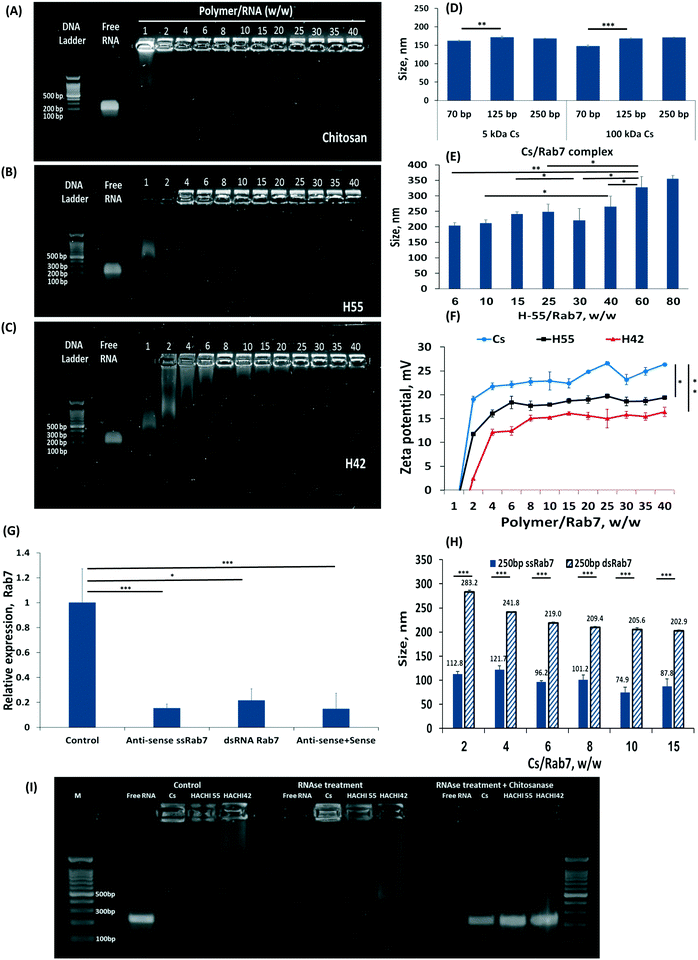
Increasing the percent of amine groups on the chitosan backbone increases the cationic charge along the molecule, thereby strengthening the affinity to the complexed RNA payload.27 Nanoparticles were prepared by self-assembly of chitosan-based polymers and anti-Rab7 dsRNA. A gel electrophoresis migration assay was used to distinguish between the complexed and free RNA. Free RNA migrated along the gel when charge is applied while the stable RNA-chitosan complexes retard its electrophoretic mobility and remained in the loading well (a bright spot can be noticed in the upper part of the gel, Fig. 2A–C). As the degree of acetylation increased, the dsRNA binding affinity decreased and a greater ratio of polymer/RNA was needed to stabilize the RNA complex (Fig. 2A–C). Specifically, chitosan (naturally having approx. 82.4% amine groups) binds RNA at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 wt ratio (Fig. 2A). Acetylated chitosan, having less amine groups, either H55 (with 55% of the polymer aminated) or H42, required increasing the polymer/RNA ratio to complex the RNA (Fig. 2B and C). Fig. 2A–C presents the effect of the deacetylation degree on the dsRNA binding efficacy for chitosan and H42 and H55 derivatives, respectively. The polymer/anti-Rab7 dsRNA mass ratio for complete dsRNA binding increased from 2 to 25 as the degree of amination decreased from 82.4% (chitosan, Fig. 2A) to 42% (H42, Fig. 2C).
1 wt ratio (Fig. 2A). Acetylated chitosan, having less amine groups, either H55 (with 55% of the polymer aminated) or H42, required increasing the polymer/RNA ratio to complex the RNA (Fig. 2B and C). Fig. 2A–C presents the effect of the deacetylation degree on the dsRNA binding efficacy for chitosan and H42 and H55 derivatives, respectively. The polymer/anti-Rab7 dsRNA mass ratio for complete dsRNA binding increased from 2 to 25 as the degree of amination decreased from 82.4% (chitosan, Fig. 2A) to 42% (H42, Fig. 2C).
The size of the RNA-nanoparticle affects its biodistribution and cellular fate. We measured the effect the lengths of the dsRNA construct, and the chitosan polymer, have on particle size. We found that at similar polymer/RNA weight ratios, varying the length of the RNA strand, from 70 to 250 bp, or the length of the chitosan polymer, either 5 or 150 kDa (Fig. 2D). The explanation for this phenomenon is that there are two main parameters that affect particle size: the charge density along the cationic polymer and the polymer/RNA mass ratio. Varying the length of the dsRNA strand (while maintaining its total mass in the particle), or the length of the complexing polymer did not affect particle size. Thereby, as long as the mass ratio between the dsRNA and the complexing polymer are held constant, the particle size will not be affected. This was confirmed by increasing the mass ratio of the H55/RNA particles (Fig. 2E). To form stable RNA-H55 particles, a greater mass ratio was needed (2B). Increasing the polymer/RNA mass ratio increased particle size (Fig. 2E).
The surface charge of the particles can mediate cellular uptake by adhering to negatively-charged groups in the tissue or on the cell surface.46 As expected, increasing the polymer/RNA mass ratio not only increases particle size but also increases the zeta potential measured on the particle surface (Fig. 2F). Partially N-acetylated chitosan derivatives (H55 and H42) had lower zeta-potential values compared to chitosan for all mass ratios tested. For example, the zeta-potential of H42- and H55-based nanoparticles was approximately 15 and 17 mV at polymer/Rab7 mass ratio of 40, while chitosan-based nanoparticles measured a zeta-potential value of 26 mV.
For many therapeutic application a particle size of 100 nm has been shown to grant an advantage in penetrating tissues and cells.47 The particle size we achieved using dsRNA/polymer complexes were approx. 160 nm for chitosan and 200 to 260 nm for H55. We sought to further reduce these particle sizes. We hypothesized the rigidity of the molecular building blocks within the particle may prevent more efficient folding.48,49 Specifically, dsRNA is an extremely rigid molecule compared to single-strand (ss)RNA.50,51 To test this hypothesis, we complexed anti-Rab7 ssRNA at similar mass ratios and found that these particles are nearly half the size of the dsRNA complexes, i.e. <100 nm (Fig. 2H). Furthermore, the long, 250 bp, ssRNA induced efficient RNAi owed to complementary RNA duplexes, and stem-and-loop structures, formed in the long strand that allow its introduction into the RNAi cascade, Fig. 2G.10
The ability of a carrier to protect its payload from nuclease degradation is an important property for efficient gene delivery. To assess the capacity of the particles to protect RNA from biodegradation, we exposed free RNA as well as RNA/H55 and RNA/H42 complexes to RNase A (Fig. 2I). Free RNA was fully digested within 30 minutes while the particulate RNA remained protected from the RNase until released from the formulation. The stability of the H55 particles was greatest, rendering to release the RNA from the formulation by enzymatically degrading the chitosan backbone with chitosanase, after which the RNA was susceptible to RNase in the solution (Fig. 2I).
Taken together, these results demonstrate that increasing the charge density along the complexing polymer enables formulating stable particles at low polymer/RNA ratios. Self-assembly of dsRNA and chitosan is dependent on the degree of deacetylation of the chitosan derivative. As the percent of cationic amine groups on the polymer increased, the affinity to RNA increases. At the same time, pure chitosan has been shown to adhere too strongly to DNA/RNA payloads, preventing their therapeutic effective release at the target site.26,52 Therefore, we chose to continue our viral-challenge studies with H55 rather than cationic chitosan.
We tested the capacity of RNA-nanoparticles to protect 2 g Penaeus vannamei juvenile shrimp from a lethal WSSV infection. Prior to the experiment, 1000 shrimp were acclimatized in a 3-ton tank at 30 ppt salinity, 4 ppm dissolved oxygen, 28 °C, for 5 days, while being fed a normal diet of 5% body weight. Three treatment groups of 80 shrimp each included animals administered 125, 250 bp anti-Rab7 RNA, or H55/250 bp nanoparticulate-RNA. In addition, two control groups were included: one infected by the virus without treatment (positive control) and one not infected (negative control). Thus, the juvenile shrimp were divided into 5 groups of 4 replicates of 20 animals each. The shrimp groups were held in 60 L tanks (see Materials and methods).
The shrimp were injected either 125, 250 bp anti-Rab7 RNA, H55/250 bp nanoparticulate-RNA, or with 2% NaCl (control). Sixty-five hours post treatment the shrimp were challenged with WSSV. For this, the shrimp feed was supplemented with WSSV-infected muscle having approx. 107 WSSV copies per gr. Shrimp survival was recorded twice a day, and tissue was obtained for qPCR analysis from the surviving and dead shrimp.
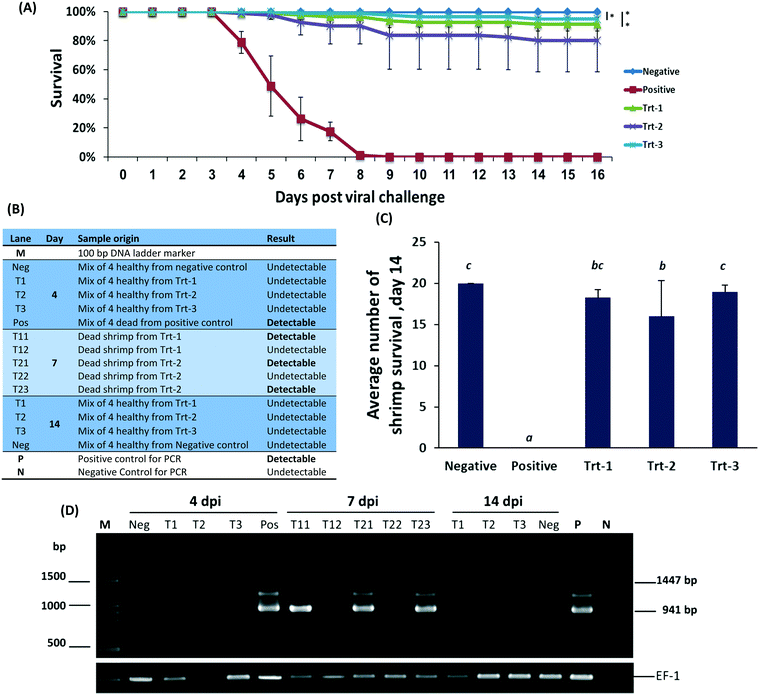
On day 2 post infection shrimp began dying in the WSSV positive control group and continued dying at an average rate of four to six animals per day. The mortality pattern was very similar in all replicates in this treatment. One hundred percent mortality was observed on day 7 post infection. No mortality was observed in the negative control group (Fig. 3B). All RNA treatments showed immunization, reaching 99% survival in some of the groups (Fig. 3A) without body-weight loss or signs of distress. All treatment groups were statistically different from the positive control (p < 0.05). The nanoparticulate RNA group also did not have a significant difference from the uninfected negative control (P < 0.05). These data indicate that nanoparticulate dsRNA is effective in protecting shrimp from WSSV infection.9,53–55
To assess the level of WSSV infection, pleopods and gills were evaluated for WSSV infection by PCR, Fig. 3C. On day 4, WSSV was undetectable in the RNA treatments and negative control but was detected in the positive control. Samples taken on day 7 had a positive indication of the virus in animals treated by free RNA but not in animals treated with the nanoparticulate RNA. On day 14, all the remaining animals were undetectable for WSSV. These results, confirmed by quantitative PCR, demonstrate that the protective effect of the treatment remains active at least two weeks post the viral challenge. Furthermore, the animals treated with the nanoparticulate treatment exceeded all other groups.
In conclusion, we report here that our findings about antiviral activity of nanoparticulate RNAi for targeting WSSV can be applied to make global improvements in food production processes and food safety. Gene knockdown was demonstrated in healthy and disease models, and achieved significant protection against a viral challenge. RNAi is an extremely promising modality for treating diseases. In the case of WSSV, no other therapeutic agent has been found to be capable of treating this lethal infection. RNAi is biologically targeted and therefore has a known and safe activity profile.56 While the data presented herein are promising regarding the ability to treat disease, implementing RNAi in aquaculture will require overcoming several challenges: a) administration – this paper describes an anti-viral RNAi therapeutic proof-of-concept by injecting the nanoparticles to the shrimp one-by-one, large scale systems should aim for an oral delivery of the RNA nanoparticles to the entire pond simultaneously; b) cost effective – while prices of RNA are dropping significantly, the cost of manufacturing and quality assurance of the particles (including RNA and complexing materials) should be less than 10% of the cost of final product; c) environmental and food safety – RNA strands must be optimized not to affect other organisms and the particles should be composed of safe and biodegradable and biocompatible materials.
The future of disease management depends greatly on novel RNAi-based therapeutics. RNAi triggers a natural, non-GMO, non-antibiotic gene-knockdown mechanism. This goal is being facilitated be the development of new and promising nanomaterials that deliver the RNA intracellularly.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors wish to thank Mrs Rebekah Roiter for editing this manuscript.References
- M. Lafourcade, T. Larrieu, S. Mato, A. Duffaud, M. Sepers, I. Matias, V. De Smedt-Peyrusse, V. F. Labrousse, L. Bretillon, C. Matute, R. Rodriguez-Puertas, S. Laye and O. J. Manzoni, Nat. Neurosci., 2011, 14, 345–350 CrossRef CAS PubMed.
- F.-F. A. A. O. O. T. U. Nations, GLOBEFISH – Update on world seafood markets, Rome, Italy, 2016 Search PubMed.
- A. G. J. Tacon, Thematic Review of Feeds and Feed Management Practices in Shrimp Aquaculture, Aquatic Farms Ltd. for the World Bank, NACA, WWF and FAO Consortium Program on Shrimp Farming and the Environment, Kaneohe, HI, USA, 2002 Search PubMed.
- Overview of the U.S. Seafood Supply Universities of Oregon State, Cornell, Delaware, Rhode Island, Florida, and California, and the Community Seafood Initiative, 2015, http://www.seafoodhealthfacts.org/seafood-choices/overview-us-seafood-supply Search PubMed.
- A. Sanchez-Paz, Vet. Res., 2010, 41, 43 CrossRef PubMed.
- P. S. Chang, C. F. Lo, Y. C. Wang and G. H. Kou, Dis. Aquat. Org., 1996, 27, 131–139 CrossRef.
- W. Wu, L. Wang and X. Zhang, Virology, 2005, 332, 578–583 CrossRef CAS PubMed.
- B. Verbruggen, L. K. Bickley, R. van Aerle, K. S. Bateman, G. D. Stentiford, E. M. Santos and C. R. Tyler, Viruses, 2016, 8, E23 CrossRef PubMed.
- J. Xu, F. Han and X. Zhang, Antiviral Res., 2007, 73, 126–131 CrossRef CAS PubMed.
- A. Fire, S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Nature, 1998, 391, 806–811 CrossRef CAS PubMed.
- S. M. Hammond, E. Bernstein, D. Beach and G. J. Hannon, Nature, 2000, 404, 293–296 CrossRef CAS PubMed.
- A. Sagi, R. Manor and T. Ventura, Genes, 2013, 4, 620–645 CrossRef CAS PubMed.
- D. Adams, A. Gonzalez-Duarte, W. O'Riordan, C. Yang, T. Yamashita, A. Kristen, I. Tournev, H. Schmidt, T. Coelho, J. Berk, K. Lin, P. Dyck, P. Gandhi, M. Sweetser, J. Chen, J. Gollob and O. Suhr, Patisiran, an investigational RNAi therapeutic for patients with hereditary transthyretin-mediated (hATTR) amyloidosis with polyneuropathy: Results from the Phase 3 APOLLO study, Paris, 2017 Search PubMed.
- P. R. Cullis and M. J. Hope, Mol. Ther., 2017, 25, 1467–1475 CrossRef CAS PubMed.
- A. Schroeder, C. G. Levins, C. Cortez, R. Langer and D. G. Anderson, J. Intern. Med, 2010, 267, 9–21 Search PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS PubMed.
- R. Kedmi and D. Peer, Nanomedicine, 2009, 4, 853–855 CrossRef CAS PubMed.
- A. Schroeder, J. E. Dahlman, G. Sahay, K. T. Love, Y. Wang and D. G. Anderson, J. Controlled Release, 2012, 160, 172–176 CrossRef CAS PubMed.
- A. M. Jose, Genesis, 2015, 53, 395–416 CrossRef CAS PubMed.
- B. C. Yoo, F. Kragler, E. Varkonyi-Gasic, V. Haywood, S. Archer-Evans, Y. M. Lee, T. J. Lough and W. J. Lucas, Plant Cell, 2004, 16, 1979–2000 CrossRef CAS PubMed.
- A. M. Jose and C. P. Hunter, Annu. Rev. Genet., 2007, 41, 305–330 CrossRef CAS PubMed.
- J. Robalino, T. Bartlett, E. Shepard, S. Prior, G. Jaramillo, E. Scura, R. W. Chapman, P. S. Gross, C. L. Browdy and G. W. Warr, J. Virol., 2005, 79, 13561–13571 CrossRef CAS PubMed.
- Z. Yaari, D. da Silva, A. Zinger, E. Goldman, A. Kajal, R. Tshuva, E. Barak, N. Dahan, D. Hershkovitz, M. Goldfeder, J. S. Roitman and A. Schroeder, Nat. Commun., 2016, 7, 13325 CrossRef CAS PubMed.
- M. Rinaudo, Prog. Polym. Sci., 2006, 31, 603–632 CrossRef CAS.
- S. Aiba, Int. J. Biol. Macromol., 1989, 11, 249–252 CrossRef CAS PubMed.
- A. Schroeder, C. G. Levins, C. Cortez, R. Langer and D. G. Anderson, J. Intern. Med., 2010, 267, 9–21 CrossRef CAS PubMed.
- J. E. Dahlman, C. Barnes, O. Khan, A. Thiriot, S. Jhunjunwala, T. E. Shaw, Y. Xing, H. B. Sager, G. Sahay, L. Speciner, A. Bader, R. L. Bogorad, H. Yin, T. Racie, Y. Dong, S. Jiang, D. Seedorf, A. Dave, K. S. Sandu, M. J. Webber, T. Novobrantseva, V. M. Ruda, A. K. Lytton-Jean, C. G. Levins, B. Kalish, D. K. Mudge, M. Perez, L. Abezgauz, P. Dutta, L. Smith, K. Charisse, M. W. Kieran, K. Fitzgerald, M. Nahrendorf, D. Danino, R. M. Tuder, U. H. von Andrian, A. Akinc, D. Panigrahy, A. Schroeder, V. Kotelianski, R. Langer and D. G. Anderson, Nat. Nanotechnol., 2014, 9(8), 648–655 CrossRef CAS PubMed.
- A. Schroeder, J. E. Dahlman, G. Sahay, K. T. Love, S. Jiang, A. A. Eltoukhy, C. G. Levins, Y. Wang and D. G. Anderson, J. Controlled Release, 2012, 160, 172–176 CrossRef CAS PubMed.
- N. Krinsky, M. Kaduri, J. Shainsky-Roitman, M. Goldfeder, E. Ivanir, I. Benhar, Y. Shoham and A. Schroeder, PLoS One, 2016, 11, e0165137 Search PubMed.
- K. Sritunyalucksana, W. Wannapapho, C. F. Lo and T. W. Flegel, J. Virol., 2006, 80, 10734–10742 CrossRef CAS PubMed.
- C. Ongvarrasopone, M. Chanasakulniyom, K. Sritunyalucksana and S. Panyim, Mar. Biotechnol., 2008, 10, 374–381 CrossRef CAS PubMed.
- C. S. Kim, Z. Kosuke, Y. K. Nam, S. K. Kim and K. H. Kim, Fish Shellfish Immunol., 2007, 23, 242–246 CrossRef CAS PubMed.
- D. V. Nguyen, O. Christiaens, P. Bossier and G. Smagghe, Rev. Fish. Sci., 2016, 0, 1–12 Search PubMed.
- A. Birmingham, E. Anderson, K. Sullivan, A. Reynolds, Q. Boese, D. Leake, J. Karpilow and A. Khvorova, Nat. Protoc., 2007, 2, 2068–2078 CrossRef CAS PubMed.
- J. K. Lam, M. Y. Chow, Y. Zhang and S. W. Leung, Mol. Ther.--Nucleic Acids, 2015, 4, e252 CrossRef CAS PubMed.
- C. A. Schumpert, J. L. Dudycha and R. C. Patel, BMC Biotechnol., 2015, 15, 91 CrossRef PubMed.
- J. Toxopeus, A. H. Warner and T. H. MacRae, Cell Stress Chaperones, 2014, 19, 939–948 CrossRef CAS PubMed.
- O. Sharabi, R. Manor, S. Weil, E. D. Aflalo, Y. Lezer, T. Levy, J. Aizen, T. Ventura, P. B. Mather, I. Khalaila and A. Sagi, Endocrinology, 2016, 157, 928–941 CrossRef CAS PubMed.
- R. G. Feijo, R. Maggioni, P. C. Cunha Martins, K. L. de Abreu, J. M. Oliveira-Neto, C. Guertler, E. B. Justino, L. M. Perazzolo and L. F. Marins, Dis. Aquat. Org., 2015, 114, 89–98 CrossRef CAS PubMed.
- M. Karlikow, B. Goic and M. C. Saleh, Dev. Comp. Immunol., 2014, 42, 85–92 CrossRef CAS PubMed.
- A. L. Jackson, J. Burchard, J. Schelter, B. N. Chau, M. Cleary, L. Lim and P. S. Linsley, RNA, 2006, 12, 1179–1187 CrossRef CAS PubMed.
- F. Ren, Q. Shi, Y. Chen, A. Jiang, Y. T. Ip, H. Jiang and J. Jiang, Cell. Res., 2013, 23, 1133–1146 CrossRef CAS PubMed.
- J. Soutschek, A. Akinc, B. Bramlage, K. Charisse, R. Constien, M. Donoghue, S. Elbashir, A. Geick, P. Hadwiger, J. Harborth, M. John, V. Kesavan, G. Lavine, R. K. Pandey, T. Racie, K. G. Rajeev, I. Rohl, I. Toudjarska, G. Wang, S. Wuschko, D. Bumcrot, V. Koteliansky, S. Limmer, M. Manoharan and H. P. Vornlocher, Nature, 2004, 432, 173–178 CrossRef CAS PubMed.
- J. A. Champion, Y. K. Katare and S. Mitragotri, J. Controlled Release, 2007, 121, 3–9 CrossRef CAS PubMed.
- G. Sahay, W. Querbes, C. Alabi, A. Eltoukhy, S. Sarkar, C. Zurenko, E. Karagiannis, K. Love, D. Chen, R. Zoncu, Y. Buganim, A. Schroeder, R. Langer and D. G. Anderson, Nat. Biotechnol., 2013, 31, 653–658 CrossRef CAS PubMed.
- R. Kedmi, N. Ben-Arie and D. Peer, Biomaterials, 2010, 26, 6867–6875 CrossRef PubMed.
- A. Schroeder, D. A. Heller, M. W. Winslow, J. E. Dahlman, G. W. Pratt, R. Langer, T. Jacks and D. G. Anderson, Nat. Rev. Cancer, 2011, 12, 39–50 CrossRef PubMed.
- H. Dietz, S. M. Douglas and W. M. Shih, Science, 2009, 325, 725–730 CrossRef CAS PubMed.
- G. I. Livshits, J. Ghabboun, N. Borovok, A. B. Kotlyar and D. Porath, Adv. Mater., 2014, 26, 4981–4985 CrossRef CAS PubMed.
- K. Hayashi, H. Chaya, S. Fukushima, S. Watanabe, H. Takemoto, K. Osada, N. Nishiyama, K. Miyata and K. Kataoka, Macromol. Rapid Commun., 2016, 37, 486–493 CrossRef CAS PubMed.
- P. Kebbekus, D. E. Draper and P. Hagerman, Biochemistry, 1995, 34, 4354–4357 CrossRef CAS PubMed.
- P. L. Ma, M. Lavertu, F. M. Winnik and M. D. Buschmann, Biomacromolecules, 2009, 10, 1490–1499 CrossRef CAS PubMed.
- T. Rattanarojpong, S. Khankaew, P. Khunrae, R. Vanichviriyakit and K. Poomputsa, J. Biotechnol., 2016, 229, 44–52 CrossRef CAS PubMed.
- M. Sarathi, M. C. Simon, C. Venkatesan, J. Thomas, M. Ravi, N. Madan, S. Thiyagarajan and A. S. Sahul Hameed, J. Fish Dis., 2010, 33, 603–607 CrossRef CAS PubMed.
- F. Zhu and X. Zhang, Mar. Biotechnol., 2012, 14, 63–68 CrossRef CAS PubMed.
- M. E. Davis, J. E. Zuckerman, C. H. Choi, D. Seligson, A. Tolcher, C. A. Alabi, Y. Yen, J. D. Heidel and A. Ribas, Nature, 2010, 464, 1067–1070 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |