Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploring cucurbit[6]uril–peptide interactions in the solid state: crystal structure of cucurbit[6]uril complexes with glycyl-containing dipeptides†
Oksana
Danylyuk

Institute of Physical Chemistry, Polish Academy of Sciences, Kasprzaka 44/52, 01-224 Warsaw, Poland. E-mail: odanylyuk@ichf.edu.pl
First published on 15th June 2017
Abstract
Macrocyclic host cucurbit[6]uril forms supramolecular complexes with dipeptides sequenced as Gly-X (X is either an aromatic amino acid residue Phe, Tyr, and Trp or Gly) in the solid state. Despite exclusion complexation, the interaction between guest dipeptide and host cucurbit[6]uril is multipoint. The ammonium and amide nitrogen atoms of dipeptide participate in hydrogen bonding with a host carbonyl rim and water molecules or chloride anions trapped inside the macrocyclic cavity. The structural study reveals the stabilizing role of the aromatic residues in the supramolecular assembly due to their complementarity with the outer surface of cucurbit[6]uril. In the absence of an aromatic side chain, the calcium ions can stabilize and guide the supramolecular assembly between the macrocycle and Gly-Gly dipeptide.
Cucurbit[n]urils are supramolecular macrocyclic hosts with hydrophobic cavities and two identical carbonyl rimmed portals. Interest in cucurbit[n]urils as containers and supramolecular receptors is increasingly growing due to their excellent recognition and complexation properties in aqueous media.1 One of the most intriguing aspects of their host–guest chemistry is the recognition of biologically important molecules, such as amino acids, small peptides and even proteins.2 Supramolecular approaches that involve cucurbit[n]uril macrocyclic hosts to modify and control protein assembly are particularly attractive and may have important implications in chemical biology.3 Investigation of supramolecular interactions of cucurbit[n]urils with biomolecules is necessary for providing mechanistic insight into their molecular recognition and self-assembly properties and designing new protocols to control and regulate protein activity.4 On the other hand, the interaction of these macrocyclic hosts with amino acids and peptides is important in the context of cucurbituril-based drug delivery, as it might affect the stability of cucurbituril–drug complexes, for example accelerating drug release from the host cavity by competitive binding. Moreover, the knowledge about cucurbit[n]uril binding preference towards specific biomolecules (or residues) could be beneficial for drug targeting. Despite remarkable progress in the study of cucurbit[n]uril interactions with amino acids, oligopeptides and even proteins, the X-ray structural investigations of these assemblies are rather scarce,5 which may be explained by difficulties in obtaining suitable crystals for X-ray single crystal diffraction, as well as particularly challenging and time-consuming supramolecular crystallography arising from small crystals, poor diffraction, flexibility of the framework manifested in high degrees of structural disorder and often air sensitivity. Despite these difficulties, the structural study of supramolecular systems is worth the effort as the crystal structure provides unequivocal proof for the formation of supramolecular assembly and direct insight into molecular recognition, elucidating interaction mode (geometry, stoichiometry, and non-covalent interactions) between hosts and guests.
In this work, we would like to present our results on the structural study of supramolecular complexes between the macrocyclic host cucurbit[6]uril (CB6) and a series of dipeptides. For our study, we selected glycine and aromatic amino acid containing dipeptides sequenced as Gly-X, where X is either an aromatic amino acid residue (Phe, Tyr, Trp) or Gly. Glycine is the simplest of all twenty amino acids with only hydrogen on its side chain. The lack of a complex side chain allows high flexibility in the proteins and that is why glycine is often found in the loop regions, where the polypeptide chain makes a sharp turn.6 Despite its simplicity, glycine is a major building component of many structural proteins, such as collagen,7 some spider silks,8 silkworm silk9 and marine mussel bioadhesives.10 Aromatic amino acid residues are commonly found at the important binding sites of natural complexes of peptides and proteins.11 The aromatic groups provide a large binding surface/interface and enable several types of intermolecular interactions, such as π-stacking, C–H⋯π interactions, and hydrophobic binding.
The first study on the interaction of CB6 with small peptides in aqueous solution was performed by Buschmann and co-workers in 2005.12 The dipeptides were found to form exclusion complexes with the N-terminal ammonium groups binding to the CB6 portal. Early work of Kim and Inoue showed the complexation of dipeptides containing Phe by CB7.13 Later, it was found that CB7 effectively recognizes peptide sequences Phe-Gly over Gly-Phe and Tyr-Gly over Gly-Tyr as well as Trp-Gly over Gly-Trp.14CB8 has been shown to selectively recognize and bind aromatic tripeptides Trp-Gly-Gly and Phe-Gly-Gly in aqueous solution.15 The authors also reported very interesting crystal structures of CB8 complexes with these tripeptides elucidating the structural basis for selective recognition and unprecedented peptide sequence selectivity. Namely, the crystal structure of a CB8 complex with Trp-Gly-Gly revealed the inclusion of an indole ring into the host macrocyclic cavity and the ion–dipole interactions between the N-terminal nitrogen and carbonyl rim of CB8. The crystal structure of a CB8 complex with Phe-Gly-Gly has shown the dimerization of the tripeptide through inclusion of two Phe residues interacting via π-stacking inside the host cavity. It should be mentioned that these two crystal structures are the first and only structural study on cucurbituril interactions with small peptides. Interestingly, Urbach and co-workers also succeeded in the crystallization of a human insulin complex with CB7.16 The beautiful crystal structure showed that binding occurs at the N-terminal Phe residue and that the N-terminus unfolds to enable binding. Kim and co-workers showed that CB7 prevents the fibrillation of insulin and β-amyloid by capturing Phe residues, which are crucial to the hydrophobic interactions formed during amyloid fibrillation.17
In order to better understand the versatility and capability of cucurbit[n]urils as host molecules towards bioactive guests, particularly amino acids and small peptides, we continue to explore their host–guest supramolecular chemistry in the solid state. Herein, four supramolecular complexes of CB6 with glycyl-containing dipeptides have been crystallized and their crystal structures have been established via single crystal X-ray diffraction. The structural aspects of the host–guest complexation, main non-covalent interactions that guide the assembly, the role of the aromatic residues and the metal ion in the self-assembly and resulting supramolecular architecture in the solid state have been elucidated.
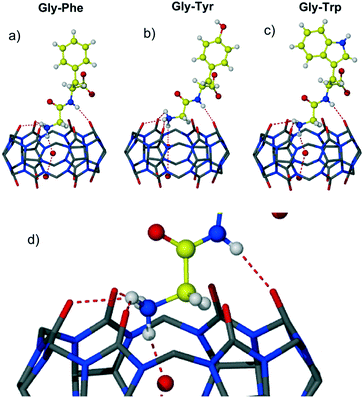
Three complexes 1–3 of CB6 with aromatic dipeptides Gly-Phe, Gly-Tyr and Gly-Trp crystallized from aqueous solution of CB6, the corresponding dipeptide and magnesium chloride used to improve solubility of the macrocycle. Magnesium cations have not been found in the final crystalline assemblies. An attempt to introduce calcium ions into the CB6-aromatic dipeptide systems as solubilizing and/or structure-directing agents resulted in the crystallization of the known CB6 coordination polymer with calcium ions.18 Crystal structure analysis revealed that all three complexes crystallize in the chiral monoclinic space group P21 and are isostructural. The dipeptides are in a zwitterionic form and are complexed exo with respect to the host cavity. An interesting trend found in these three complexes is the slight decrease in the unit cell volume upon enlarging the aromatic residue Phe → Tyr → Trp (3180, 3159 and 3097 Å3). This trend might suggest more effective packing of CB6 complexes with larger aromatic systems, thus leading to more efficient interaction between the macrocycle and aromatic Gly-X dipeptide in the solid state. The careful crystal structure analysis revealed that the main host–guest binding motif in all three complexes involves a highly polar CB6 rim and two nitrogen atoms of dipeptide – terminal ammonium and amido nitrogen of the peptide bond, Fig. 1. The main structural features, non-covalent interactions and supramolecular organization in the resulting assemblies will be discussed on the example of CB6 complex 1 with Gly-Phe.
The asymmetric unit of complex 1 comprises one CB6 molecule, one dipeptide Gly-Phe as a zwitterion and fourteen water molecules. The dipeptide is complexed at one of the CB6 rims leaving the second identical rim not involved in the host–guest interaction. The CB6 macrocyclic cavity is occupied with two water molecules. The dipeptide adopts conformation suitable for the interaction with the CB6 carbonyl rim via both ammonium and amido nitrogen atoms. The terminal ammonium of Gly-Phe lies in the plane of six carbonyl oxygen atoms of the host and donates hydrogen bonds to three oxygen atoms (one of the H-bonds is bifurcated, N–HO distances are 2.76, 2.88 and 3.03 Å). The fourth hydrogen bond is realized towards water molecules included deeply in the macrocyclic cavity (2.79 Å). The amido nitrogen of the dipeptide also forms hydrogen bond with carbonyl oxygen of the host (N–H⋯O distance 2.88 Å). The aromatic group of the Phe residue is directed away from the host portal and is positioned almost perpendicular to the CB6 rim; the dihedral angle between the guest phenyl group and plane defined by six carbonyl oxygen atoms of CB6 is 86°. The carboxylate and carbonyl groups of Gly-Phe are involved in rich hydrogen bonding with water molecules.
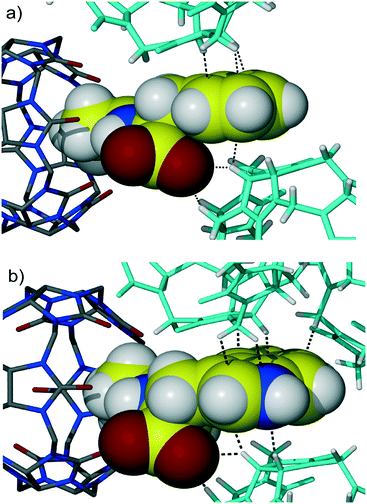
Further symmetry expansion shows that the aromatic group of the Phe residue is in close contact with the outer surface of two CB6 macrocycles in the crystal lattice. The aromatic moiety of the dipeptide contributes to the overall assembly through C–H⋯π interactions between backbones of adjacent CB6 and the aromatic system of Gly-Phe, Fig. 2a. There are one methine and one methylene carbon atoms from one CB6 and one methine carbon atom from another CB6 in close contact with the Phe aromatic residue enabling C–H⋯π interactions. Additionally, there are two C–H⋯O weak hydrogen bonds between methine carbon atoms of CB6 and a carboxyl oxygen atom of the dipeptide (C–H⋯O distance is 3.22 and 3.23 Å). When comparing similar host–guest organization in CB6 complex 3 with Gly-Trp, it is evident that there are more contact points between the CB6 outer surfaces and large indole system of Trp, Fig. 2b. Moreover, the third CB6 takes part in the interaction with an enlarged aromatic moiety, in this way, making overall supramolecular packing more efficient.
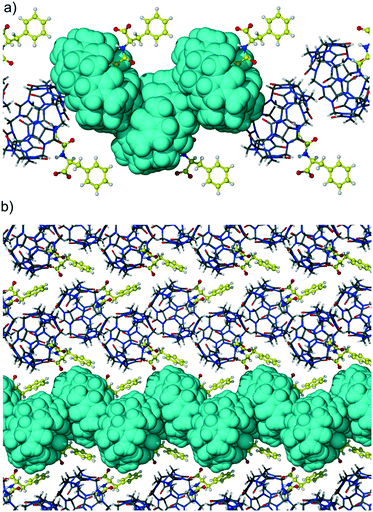
The second portal of CB6 is not engaged in the interaction with dipeptide and is closed with the backbone of the adjacent macrocycle in the T-shaped geometry. Such a ‘side to portal’ arrangement of cucurbiturils is quite favorable because of the multiple CH2 (methylene) and CH (methine) weak hydrogen bonds with carbonyl oxygen atoms of the adjacent macrocycle. The T-shaped arrangement is realized in many cucurbituril crystalline hydrates as well as in some of their host–guest and even coordination complexes.19 These multipoint interactions are believed to be responsible for the solid-state assembly of CBs, their excellent thermal and chemical stability, and low water solubility. The supramolecular organization of adjacent CB6 molecules in complex 1 is depicted in Fig. 3a which represents zigzag strands of CB6-dipeptide units running along the b direction. The host molecules are arranged in a ‘side to portal’ manner and guest dipeptides protrude from other portals of CB6s. The adjacent zigzag strands are arranged in a parallel way and stitched together by C–H⋯π interactions between CB6 and aromatic groups of dipeptide, Fig. 3b.
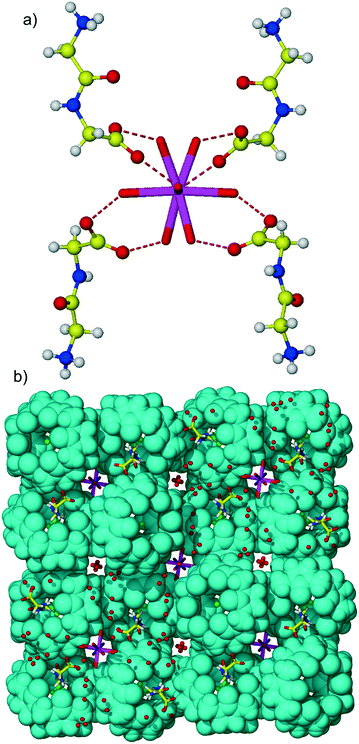
The crystallization protocol used for obtaining CB6 complexes 1–3 with aromatic dipeptides was not successful in the case of the Gly-Gly guest. It is not surprising as this simple dipeptide differs significantly from the previously employed Gly-X aromatic dipeptides – it is achiral and lacks an aromatic side chain that obviously contributes significantly to the stabilization of the CB6-dipeptide assembly. The change of the salt used for enhancing CB6 aqueous solubility from magnesium chloride to calcium chloride resulted in the crystallization of the desired compound 4. The CB6 complex with Gly-Gly crystallized in the tetragonal space group I41/a. X-ray analysis revealed that calcium ions are embedded in the crystal lattice of the complex and are active participants in the overall supramolecular assembly. It should be mentioned that metal ions are frequently found in the protein structures and are often necessary for protein stability, structure and function.20 Moreover, the presence of metal ions is often necessary for protein crystallization.
The asymmetric unit of multi-component complex 4 comprises half of the CB6 molecule lying over an inversion centre, one Gly-Gly dipeptide in the zwitterionic form, one quarter of a calcium cation positioned on the 4-fold rotoinversion axis, half of a chloride anion and water molecules. The crystal structure is characterized by substantial disorder of several components: the Gly-Gly dipeptide was modeled as disordered over two positions, water molecules constituting the coordination sphere of the calcium ion are disordered because of their position close to the rotoinversion axis, chloride anions occupying the macrocyclic cavity are also disordered, and additionally, disordered lattice water molecules are present. The calcium cation is coordinated by disordered aqua ligands, the coordination number of the metal ion is six, and the coordination polyhedron of calcium is the distorted tetragonal bipyramid. The metal–oxygen distances are in the range of 2.26–2.49 Å and typical to those found in the literature.21
Different from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CB6 complexes with aromatic Gly-X dipeptides, here, Gly-Gly is complexed at each portal of the macrocycle giving 1
1 CB6 complexes with aromatic Gly-X dipeptides, here, Gly-Gly is complexed at each portal of the macrocycle giving 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
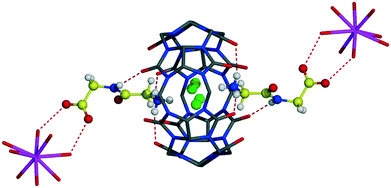
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 host–guest assembly, Fig. 4. The host–guest main binding motif is again the interaction of terminal ammonium and amide nitrogen atoms of dipeptide with carbonyl oxygen atoms of CB6via ion–dipole and hydrogen bonding. Surprisingly, the CB6 cavity is taken by chloride anions. We have already observed similar trapping of chloride anions by a CB6 cavity in a solid-state complex with tryptophan.5c The inclusion of chloride anions is stabilized by charge-assisted N–H⋯Cl hydrogen bonding with ammonium functions of two Gly-Gly dipeptides closing each portal of CB6 (N–H⋯Cl distances are in the range of 2.94–3.12 Å). The carboxylate group of dipeptide is involved in hydrogen bonding with aqua ligands of a calcium cation thus providing stabilization to the overall supramolecular assembly, Fig. 5a. Each aqua-coordinated calcium interacts with four Gly-Gly dipeptides, donating two hydrogen bonds to each carboxylate group. Such interaction mode generates tubular organization of the multi-component assembly in the solid state, Fig. 5b. The tube formed of CB6 and Gly-Gly molecules assembles around aqua-coordinated calcium cations.
2 host–guest assembly, Fig. 4. The host–guest main binding motif is again the interaction of terminal ammonium and amide nitrogen atoms of dipeptide with carbonyl oxygen atoms of CB6via ion–dipole and hydrogen bonding. Surprisingly, the CB6 cavity is taken by chloride anions. We have already observed similar trapping of chloride anions by a CB6 cavity in a solid-state complex with tryptophan.5c The inclusion of chloride anions is stabilized by charge-assisted N–H⋯Cl hydrogen bonding with ammonium functions of two Gly-Gly dipeptides closing each portal of CB6 (N–H⋯Cl distances are in the range of 2.94–3.12 Å). The carboxylate group of dipeptide is involved in hydrogen bonding with aqua ligands of a calcium cation thus providing stabilization to the overall supramolecular assembly, Fig. 5a. Each aqua-coordinated calcium interacts with four Gly-Gly dipeptides, donating two hydrogen bonds to each carboxylate group. Such interaction mode generates tubular organization of the multi-component assembly in the solid state, Fig. 5b. The tube formed of CB6 and Gly-Gly molecules assembles around aqua-coordinated calcium cations.
To conclude, the solid-state complexes of CB6 with dipeptides sequenced as Gly-X (X is Phe, Tyr, Trp or Gly) show a number of interesting structural features. Despite exclusion complexation, the interaction of the CB6 macrocycle with dipeptides is multipoint. It involves ion–dipole interaction, hydrogen bonding between ammonium and amido nitrogen atoms of the dipeptide with the polar carbonyl rim of CB6, hydrogen bonding of the ammonium nitrogen of the dipeptide with water molecules or chloride anions trapped inside the macrocyclic cavity. The crystal structure analysis revealed the stabilizing role of aromatic residues in the supramolecular assembly due to their complementarity with the outer surface of the CB6 skeleton. In the absence of an aromatic side chain, the metal ion can stabilize and guide the supramolecular assembly between CB6 and Gly-Gly dipeptide. The role of calcium ions is mainly in engaging the carboxylate group of dipeptide via hydrogen bonding with aqua ligands. Although much work remains to be done to explore the structural supramolecular chemistry of cucurbituril macrocycles with different peptides, the present work suggests potential design strategies for CB6 assemblies with peptide chains – the preference of aromatic residues to be surrounded with outer skeletons of CB6 when the size of the macrocycle does not enable the formation of the inclusion complex and the use of the metal ion as a structure-directing agent in the absence of other structure stabilizing elements.
Experimental
Crystallization
The dipeptides Gly-Gly, Gly-Phe, Gly-Tyr and Gly-Trp were purchased from Sigma Aldrich. CB6 was synthesized according to a literature procedure [A. Day et al., J. Org. Chem., 2001, 66, 8094].Complexes 1–3
CB6·10H2O (10 mg, 8.5 μmol) and MgCl2·6H2O (173 mg, 0.85 mmol) were dissolved in distilled water (1 ml) upon gentle heating. The solution of dipeptide Gly-Phe, Gly-Tyr or Gly-Trp (2 equivalents) in 1 ml of water was added to the solution of the macrocycle. The diffraction quality crystals were formed after several days.Complex 4
CB6·10H2O (10 mg, 8.5 μmol) and CaCl2·2H2O (12.5 mg, 0.085 mmol) were dissolved in distilled water (1 ml) upon gentle heating. The solution of dipeptide Gly-Gly (2 equivalents) in 1 ml of water was added to the solution of the macrocycle. The diffraction quality crystals were formed after several days.Single crystal X-ray diffraction
The crystals were selected under Paratone-N oil, mounted on the nylon loops and positioned in the cold stream on the diffractometer. Uniformity of the samples was checked by unit cell determination of several crystals for each sample. The X-ray data for complexes 1, 2 and 4 were collected on a Nonius KappaCCD diffractometer using MoKα radiation (λ = 0.71073 Å). The data were processed with HKL2000.22 The X-ray data for complex 3 were collected on a SuperNova Agilent diffractometer using CuKα radiation (λ = 1.54184 Å). The data were processed with CrysAlisPro.23 Structures were solved by direct methods and refined using SHELXL.24 The figures were prepared using X-Seed25/POV-Ray.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 337 reflections measured, 15
337 reflections measured, 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 276 unique, 929 parameters, R = 0.083, wR = 0.205 (R = 0.094, wR = 0.213 for all data), GooF = 1.09. CCDC 1548093.
276 unique, 929 parameters, R = 0.083, wR = 0.205 (R = 0.094, wR = 0.213 for all data), GooF = 1.09. CCDC 1548093.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 148 reflections measured, 16
148 reflections measured, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 170 unique, 1151 parameters, R = 0.081, wR = 0.181 (R = 0.122, wR = 0.210 for all data), GooF = 1.11. CCDC 1548094.
170 unique, 1151 parameters, R = 0.081, wR = 0.181 (R = 0.122, wR = 0.210 for all data), GooF = 1.11. CCDC 1548094.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 272 reflections measured, 11
272 reflections measured, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 unique, 1182 parameters, R = 0.065, wR = 0.179 (R = 0.067, wR = 0.182 for all data), GooF = 1.03. CCDC 1548096.
900 unique, 1182 parameters, R = 0.065, wR = 0.179 (R = 0.067, wR = 0.182 for all data), GooF = 1.03. CCDC 1548096.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 959 reflections measured, 7167 unique, 673 parameters, R = 0.077, wR = 0.208 (R = 0.091, wR = 0.223 for all data), GooF = 1.03. CCDC 1548095.
959 reflections measured, 7167 unique, 673 parameters, R = 0.077, wR = 0.208 (R = 0.091, wR = 0.223 for all data), GooF = 1.03. CCDC 1548095.
References
- J. W. Lee, S. Samal, N. Selvapalam, H.-J. Kim and K. Kim, Acc. Chem. Res., 2003, 36, 621 CrossRef CAS PubMed; E. Masson, X. Ling, R. Joseph, L. Kyeremeh-Mensah and X. Lu, RSC Adv., 2012, 2, 1213 RSC; X.-L. Ni, X. Xiao, H. Cong, L.-L. Liang, K. Cheng, X.-J. Cheng, N.-N. Ji, Q.-J. Zhu, S.-F. Xue and Z. Tao, Chem. Soc. Rev., 2013, 42, 9480 RSC; X.-L. Ni, X. Xiao, H. Cong, Q.-J. Zhu, S.-F. Xueand and Z. Tao, Acc. Chem. Res., 2014, 47, 1386 CrossRef PubMed; K. I. Assaf and W. M. Nau, Chem. Soc. Rev., 2015, 44, 394 RSC.
- S. J. Barrow, S. Kasera, M. J. Rowland, J. del Barrio and O. A. Scherman, Chem. Rev., 2015, 115, 12320 CrossRef CAS PubMed.
- F. de Greef, M. Merkx and L. Brunsveld, Angew. Chem., Int. Ed., 2016, 55, 8899 CrossRef CAS PubMed; S. Sankaran, M. C. Kiren and P. Jonkheijm, ACS Nano, 2015, 9(4), 3579 CrossRef PubMed.
- C. Hou, Z. Huang, Y. Fang and J. Liu, Org. Biomol. Chem., 2017, 15, 4272 CAS.
- J.-M. Yi, Y.-Q. Zhang, H. Cong, S.-F. Xue and Z. Tao, J. Mol. Struct., 2009, 933, 112 CrossRef CAS; P. Thuery, Inorg. Chem., 2011, 50(21), 10558 CrossRef PubMed; O. Danylyuk and V. P. Fedin, Cryst. Growth Des., 2012, 12(2), 550 Search PubMed; Z.-Z. Gao, R.-L. Lin, D. Bai, Z. Tao, J.-X. Liu and X. Xiao, Sci. Rep., 2017, 7, 44717 CrossRef PubMed; P.-H. Shan, S.-C. Tu, R.-L. Lin, Z. Tao, J.-X. Liu and X. Xiao, CrystEngComm, 2017, 19, 2168 RSC.
- F. Krieger, A. Möglich and T. Kiefhaber, J. Am. Chem. Soc., 2005, 127(10), 3347 CrossRef PubMed.
- M. D. Shoulders and R. T. Raines, Annu. Rev. Biochem., 2009, 78, 929 CrossRef CAS PubMed.
- J. D. van Beek, S. Hess, F. Vollrath and B. H. Meier, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(16), 10266 CrossRef CAS PubMed.
- T. Asakura, T. Ito, M. Okudaira and T. Kameda, Macromolecules, 1999, 32(15), 4940 CrossRef CAS.
- K. J. Coyne, X.-X. Qin and J. H. Waite, Science, 1997, 277(5333), 1830 CrossRef CAS PubMed.
- B. Ma, T. Elkayam, H. Wolfson and R. Nussinov, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(10), 5772 CrossRef CAS PubMed.
- H.-J. Buschmann, L. Mutihac, R.-C. Mutihac and E. Schollmeyer, Thermochim. Acta, 2005, 430(1–2), 79 CrossRef CAS.
- M. V. Rekharsky, H. Yamamura, C. Inoue, M. Kawai, I. Osaka, R. Arakawa, K. Shiba, A. Sato, Y. Ho Ko, N. Selvapalam, K. Kim and Y. Inoue, J. Am. Chem. Soc., 2006, 128, 14871 CrossRef CAS PubMed.
- M. V. Rekharsky, H. Yamamura, Y. H. Ko, N. Selvapalam, K. Kim and Y. Inoue, Chem. Commun., 2008, 2236 RSC.
- L. M. Heitmann, A. B. Taylor, P. J. Hart and A. R. Urbach, J. Am. Chem. Soc., 2006, 128(38), 12574 CrossRef CAS PubMed.
- J. M. Chinai, A. B. Taylor, L. M. Ryno, N. D. Hargreaves, C. A. Morris, P. J. Hart and A. R. Urbach, J. Am. Chem. Soc., 2011, 133(23), 8810 CrossRef CAS PubMed.
- H. H. Lee, T. S. Choi, S. J. C. Lee, J. W. Lee, J. Park, Y. H. Ko, W. J. Kim, K. Kim and H. I. Kim, Angew. Chem., Int. Ed., 2014, 53, 7461 CrossRef CAS PubMed.
- W. A. Freeman, W. L. Mock and N.-Y. Shih, J. Am. Chem. Soc., 1981, 103, 7367 CrossRef CAS.
- D. Bardelang, K. A. Udachin, D. M. Leek, J. C. Margeson, G. Chan, C. I. Ratcliffe and J. A. Ripmeester, Cryst. Growth Des., 2011, 11, 5598 Search PubMed; O. Danylyuk, V. P. Fedin and V. Sashuk, CrystEngComm, 2013, 15, 7414 RSC; O. Danylyuk, V. P. Fedin and V. Sashuk, Chem. Commun., 2013, 49, 1859 RSC.
- T. Dudev and C. Lim, Chem. Rev., 2014, 114, 538 CrossRef CAS PubMed.
- H. Zheng, M. Chruszcz, P. Lasota, L. Lebioda and W. Minor, J. Inorg. Biochem., 2008, 102, 1765 CrossRef CAS PubMed.
- Z. Otwinowski and W. Minor, Methods Enzymol., 1997, 276, 307 CAS.
- Agilent Technologies, CrysAlisPro, Version 1.171.36.32 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112 CrossRef CAS PubMed.
- L. J. Barbour, J. Supramol. Chem., 2001, 1, 189–191 CrossRef CAS.
Footnote |
| † CCDC 1548093–1548096. For crystallographic data in CIF or other electronic format see DOI: 10.1039/c7ce00881c |
| This journal is © The Royal Society of Chemistry 2017 |