Open Access Article
Open Access ArticleReversibly photoswitchable fluorescent proteins: integrating photophysics, photochemistry, bioimaging, and protein engineering
Ryohei Ozaki-Noma
a,
Tetsuichi Wazawa
a,
Kai Lu
ab,
Tomoki Matsuda
b and
Takeharu Nagai
 *acd
*acd
aSANKEN (The Institute of Scientific and Industrial Research), The University of Osaka, Ibaraki, Osaka 567-0047, Japan. E-mail: ng1@sanken.osaka-u.ac.jp
bDepartment of Biosciences, School of Science, Kitasato University, Sagamihara, Kanagawa 252-0373, Japan
cGraduate School of Frontier Biosciences, The University of Osaka, Suita, Osaka 565-0871, Japan
dTransdimensional Life Imaging Division, Institute for Open and Transdisciplinary Research Initiatives, The University of Osaka, Suita, Osaka 565-0871, Japan
First published on 30th December 2025
Abstract
Reversibly photoswitchable fluorescent proteins (rsFPs) represent a unique class of genetically encoded probes that undergo light-driven transitions between non-fluorescent OFF and emissive ON states. Their distinctive switching properties enable repeated, non-destructive control of fluorescence and have become central to advanced bioimaging approaches. In this review, we provide a critical overview of the molecular mechanisms underlying rsFP function, focusing on GFP-like proteins and fluorogen-activating systems that employ external chromophores. We describe switching kinetics, ON/OFF contrast, and fatigue as fundamental performance parameters, and highlight mechanistic insights from spectroscopy, crystallography, and computational studies. The three subclasses of GFP-like rsFPs—negative, positive, and decoupled types—are discussed in detail, alongside external-chromophore systems such as FAST, UnaG, FbFPs, and biliverdin-binding near-infrared proteins. We further survey a wide range of applications, including super-resolution microscopy, functional biosensing, multiplex discrimination, anisotropy-based analyses, diffusion and transport studies, optical data storage, and optogenetic control. Finally, we outline emerging strategies for improving brightness, photostability, spectral diversity, and switching robustness, emphasizing opportunities for rational protein engineering guided by structural and computational approaches. Together, these developments establish rsFPs as versatile, chemically tunable tools that expand the frontiers of fluorescence imaging and quantitative biology.
1. Introduction
Fluorescent proteins have become indispensable tools in biological research. In general, the term “fluorescent proteins” (hereafter “FPs”) refers to proteins that intrinsically emit fluorescence in the visible to near-infrared range. A representative example is the green FP (GFP).1–3 Because the amino acid sequence of a protein is encoded by a gene and expressed through gene expression, the introduction of an exogenous gene encoding an FP into a cell leads to intracellular expression of the corresponding protein.4 This feature has enabled researchers worldwide to design and express a wide variety of FP variants in living systems.FPs are particularly valuable for fluorescence microscopy, as they allow minimally invasive imaging following gene delivery. They are widely used as markers to label specific intracellular compartments or proteins. In addition, FPs serve as building blocks for genetically encoded fluorescent indicators that visualize diverse biological parameters, including ion concentrations, protein interactions, temperature, and macromolecular crowding.5–9 To date, more than 1000 FPs have been identified or engineered (see FPbase: https://www.fpbase.org). While most belong to the GFP-like family, an increasing number of structurally distinct FPs have also been reported.9
Beyond constitutive fluorescence, some FPs exhibit photoinduced changes in absorption, excitation, and emission, either reversible or irreversible. Based on these behaviours, they are classified as photoconvertible, photoactivatable, or reversibly photoswitchable FPs (rsFPs).5 Photoconvertible FPs irreversibly shift their emission to longer wavelengths (typically from green to orange or red) upon irradiation at ∼405 nm. Photoactivatable FPs are initially non-fluorescent under the excitation light used for the activated state and irreversibly become fluorescent upon activation with ∼405 nm light. In contrast, rsFPs reversibly switch between non- or low-fluorescent OFF states and bright ON states under alternating light exposure, enabling repeated imaging cycles without chemical replenishment.
These photoswitching properties have been widely exploited in advanced imaging strategies, particularly in super-resolution (SR) microscopy.10 Beyond SR microscopy, rsFPs have also enabled novel applications such as separation of fluorescence signals from background autofluorescence, discrimination of spectrally overlapping fluorophores, and extension of measurable molecular size ranges in time-resolved fluorescence anisotropy. Collectively, these advances highlight the broad potential of rsFP-based approaches for expanding the limits of optical imaging.
This review summarizes the fundamental mechanisms and photophysical principles underlying rsFP photoswitching, discusses their molecular engineering, and highlights key imaging technologies enabled by these proteins. By providing an integrated perspective, we aim to stimulate the development of next-generation imaging tools and inspire new directions in life science research.
2. Fluorescent proteins
FPs contain chromophores that absorb and emit light in the visible to near-infrared range, in contrast to the ultraviolet fluorescence of aromatic amino acids such as tryptophan, tyrosine, and phenylalanine found in ordinary proteins. These chromophores are generated either autocatalytically from amino acid residues within the FP itself or through the binding of an external biomolecular ligand. GFP-like FPs, for instance, form their chromophores via an intrinsic post-translational cyclisation, dehydration, and oxidation process, enabling fluorescence without the need for additional cofactors.5 In contrast, other FP families require noncovalently bound ligands such as bilirubin, biliverdin, or flavin mononucleotide (FMN) to fluoresce. This section provides an overview of these major classes of FPs. For a comprehensive catalog of reported variants, see FPbase (https://www.fpbase.org/).2.1. GFP-like fluorescent proteins
GFP-like FPs share a characteristic β-barrel structure, often referred to as the “β-can”, which encloses the chromophore (Fig. 1A). The canonical chromophore, p-hydroxybenzylidene-imidazolinone (p-HBI), is autocatalytically formed from a tripeptide motif (typically Ser-Tyr-Gly) through cyclisation, dehydration, and oxidation reactions5 (Fig. 1A(5)). In isolation, such chromophores are virtually non-fluorescent in solution (quantum yield <0.000111). However, when embedded within the rigid β-barrel scaffold, thermal and conformational fluctuations of the chromophore are suppressed, thereby reducing non-radiative decay and resulting in a dramatically enhanced fluorescence quantum yield—for example, 0.93 for mTurquoise2,12 0.60 for EGFP,13 and 0.22 for mCherry.14 | ||
| Fig. 1 Fluorescent protein structures and their chromophores. (A) GFP-like FP structure (pdb ID: 2y0g)15 (left) and its autocatalysed chromophores for GFP-like FP variants (1–9, right). (B) FAST structure derived from Photoactive Yellow Protein (PYP) (pdb ID: 6umz)16 and its external chromophores, HMBR and HBR-3, 5DOM for green and red fluorescence, respectively. (C) UnaG structure (pdb ID: 4i3d)17 and its external chromophore bilirubin. (D) miRFP structure (pdb ID: 5viv)18 and its external chromophore biliverdin. | ||
Although GFP-like proteins have been identified in organisms such as jellyfish, lancelets, and sea anemones, most FPs widely used in research are the products of directed evolution or rational design aimed at improving specific photophysical or biological properties. For bioimaging applications, desirable features include optimized excitation and emission wavelengths, monomeric behaviour to prevent interference with target localisation, high brightness, and resistance to photobleaching. Expanding spectral diversity has been particularly important for multicolour imaging.
The green fluorescence of Aequorea victoria GFP (λ_em = 510 nm; Fig. 1A(5)) can be tuned by modifying the π-electron system of the chromophore. Substitution of Tyr with Trp, His, or Phe results in a shorter π-conjugation length, leading to blue-shifted emission (λ_em = 480, 448, and 424 nm for CFP, BFP,19 and UVFP,20 respectively; Fig. 1A(2–4)). Further reduction of the π-conjugation length, achieved through imidazole ring hydration and protonation of the Tyr residue, produces the deep-ultraviolet FP Sumire21 (λ_em = 414 nm; Fig. 1A(1)). Conversely, in YFP, π–π stacking between the chromophore and Tyr203 extends conjugation, leading to a red-shifted emission22 (λ_em = 525 nm; Fig. 1A(6)).
Most red FPs (RFPs) are derived from Entacmaea quadricolor or Discosoma sp. Despite low sequence homology with GFP, they retain the β-barrel fold (Fig. 1A). Modulation of their π-electron conjugation has yielded numerous variants, such as mOrange (Fig. 1A(7)), mCherry14 (Fig. 1A(8)), and mNeptune23 (Fig. 1A(9)), exhibiting fluorescence from orange to far-red.24
Beyond spectral tuning, engineering efforts have focused on enhancing brightness, folding efficiency, and photostability. Recent developments include highly bright variants such as mGreenLantern25 and mScarlet3.26 Photostable proteins—including StayGold variants,27–30 mScarlet3-H,31 and mGold232—have been successfully applied to SR imaging. For specialized applications such as correlative light and electron microscopy (CLEM), expansion microscopy (ExM), and rapid tissue clearing, chemically robust proteins including HyperfolderYFP,33 mScarlet3-H,31 and mEos4b34 have been developed.
Through extensive molecular engineering, GFP-like FPs now encompass a wide range of spectral and functional variants, greatly expanding the potential of fluorescence-based bioimaging.
2.2. Fluorescent proteins which need external chromophores
Some FPs acquire fluorescence by binding external chromophores that function as cofactors. These proteins are broadly classified as fluorogen-activating systems, in which non-fluorescent fluorogens become emissive only upon binding to a protein scaffold. Representative examples include tetracysteine peptides that bind fluorescein or resorufin derivatives;35 Fluorogen-Activating Proteins (FAPs);36 the Fluorescence-Activating and absorption-Shifting Tag (FAST);37 the bilirubin-binding green FP UnaG;17 flavin-based FPs (FbFPs);38 and biliverdin-binding far-red FPs.39–41Despite their diversity, all share two essential features: (i) a genetically encoded, non-fluorescent apo-protein, and (ii) a fluorogenic cofactor that is non-emissive in solution but becomes fluorescent upon non-covalent binding. While fluorogenic and photoresponsive fluorescent ligands have been reported for covalent labelling systems such as HaloTag,42 SNAP-tag,43 or CLIP-tag,44 these systems fundamentally differ from fluorogen-activating systems in that fluorescence activation relies on covalent labelling rather than reversible fluorogen binding. In fluorogen-activating systems, unbound fluorogens undergo non-radiative decay, whereas binding to the protein restricts intramolecular motions and suppresses these decay pathways, thereby enabling fluorescence emission.
The first demonstration of fluorogen activation employed tetracysteine peptides containing a Cys-Cys-Pro-Gly-Cys-Cys motif that bind fluorescein arsenical hairpin binder (FlAsH–EDT2) and emit green fluorescence.35 Upon binding, the complex formation displaces protective ethanedithiol ligands, generating a rigid arsenic–thiol coordination structure. A red-emitting analogue, ReAsH–EDT2, later extended the colour palette.45 However, nonspecific interactions with endogenous cysteine-rich proteins often caused background fluorescence, limiting the system's applicability in live-cell imaging.
FAPs addressed the limitations of earlier systems by employing a genetically encoded single-chain variable fragment (scFv) that binds synthetic fluorogens with high specificity.36 Reported activatable fluorogens include malachite green derivatives (emission ∼660 nm), thiazole orange derivatives (∼530 nm),46 and dimethylindole red (∼610 nm).47 Because only the bound fluorogen is fluorescent, FAPs provide high signal-to-noise ratios and are well suited for live-cell imaging, pulse-chase experiments, and SR microscopy.
FAST represents a smaller and more versatile alternative to scFv-based FAPs. Engineered from the photoactive yellow protein (PYP) of Halorhodospira halophila, FAST (∼14 kDa) reversibly binds synthetic fluorogens such as 4-hydroxybenzylidene rhodanine (HBR) derivatives with high affinity.37 Variants such as greenFAST and redFAST selectively recognize HMBR and HBR-3,5DOM, respectively.48 The promiscuous pFAST variant further extends the spectral range from blue (473 nm) to red (616 nm).49 FAST fluorescence is oxygen-independent, making it suitable for hypoxic environments, and its reversible, non-covalent ligand binding enables fluorescence recovery and exceptional photostability—features advantageous for SR imaging.49,50
UnaG, a 15.6 kDa FP from the Japanese eel Anguilla japonica, fluoresces upon binding bilirubin, a heme-derived metabolite.17 Binding within the β-barrel stabilizes bilirubin, producing bright green fluorescence (Fig. 1C). Directed evolution has yielded improved variants: eUnaG (Val2Leu), which exhibits approximately twofold greater brightness in bacteria,51 and eUnaG2, carrying nine additional mutations that enhance stability and confer roughly fivefold higher brightness in yeast.52 Beyond use as a fluorescent tag, UnaG-based biosensors have been developed for biomedical applications, as bilirubin serves as a key diagnostic marker, particularly in neonatal jaundice.53
FbFPs originate from Light, Oxygen, Voltage (LOV) domains of phototropins and fluoresce upon binding FMN.38 In their native form, LOV domains undergo a photocycle that involves covalent bond formation between FMN and a conserved cysteine residue. Substitution of this cysteine with alanine abolishes covalent bond formation, suppresses FMN quenching, and halts the photocycle, thereby enhancing fluorescence. FbFPs carrying this substitution have been engineered from multiple species, including Bacillus subtilis, Pseudomonas putida, and Arabidopsis thaliana,54–56 and continued optimisation have further improved their brightness.57,58 Similar to UnaG and FAST, FbFPs are relatively small (12–19 kDa), require no molecular oxygen for chromophore formation, and bind their chromophore noncovalently. These features make FbFPs advantageous as fusion tags compared with GFP-like proteins, allowing imaging under anaerobic conditions.59
Another major class of FAPs utilizes biliverdin IXα as a chromophore (Fig. 1E). These proteins are typically derived from bacterial phytochromes (BphPs), phycobiliproteins, or cyanobacteriochromes (CBCRs), and emit in the far-red to near-infrared range (650–720 nm). The near-infrared FP (iRFP) family—including iRFP670, iRFP682, iRFP703, and iRFP720—was engineered to provide distinct emission spectra suitable for multicolour imaging.60 The subsequent miRFP series (e.g., miRFP670, miRFP703, miRFP720) represents monomeric variants with improved brightness and efficient expression in mammalian cells.39,61 Among the smallest engineered FPs, miRFP670nano (∼17 kDa) and its brighter derivative miRFP670nano3, derived from CBCRs of Nostoc punctiforme,62,63 exhibit superior stability against denaturation and degradation compared to BphP-based iRFPs. These compact monomers can also serve as Förster resonance energy transfer (FRET) donors for miRFP720, enabling red-shifted FRET biosensors that can be used in combination with visible-spectrum sensors and optogenetic tools.
3. Reversibly photoswitchable fluorescent proteins (rsFPs)
rsFPs undergo light-driven transitions between a non-fluorescent OFF state and a bright ON state, followed by thermal relaxation. The transition from ON to OFF is referred to as OFF-switching, whereas the reverse process is called ON-switching. These transitions can be described by pseudo-first-order kinetics,
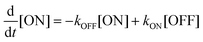 | (1) |
Because [ON] + [OFF] = 1, the general solutions are:
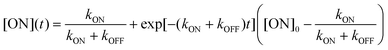 | (2) |
and
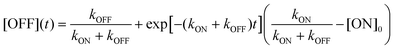 | (3) |
The photoswitching process can be experimentally monitored by recording the time-dependent fluorescence intensity. Provided that the fluorescence intensity is proportional to a[ON] + b[OFF], the photoswitching follows an exponential with time constant 1/(kON + kOFF), where a and b are constants. When illumination strongly favors ON-switching, the photoswitching time approaches 1/kON; conversely, when OFF-switching dominates, the time constant is approximately 1/kOFF.
In many experimental settings, both ON- and OFF-switching may occur simultaneously, for example, when ON- and OFF-switching wavelengths are applied at the same time. In such cases, the same formalism applies, with the understanding that kON and kOFF are determined by the specific wavelengths and light intensities used.
In addition to photoswitching kinetics, two other key properties of rsFPs are ON/OFF contrast and switching fatigue. ON/OFF contrast is defined as the ratio of fluorescence intensity in the ON state to that in the OFF state within an ensemble of rsFPs. During repeated switching cycles, both the fluorescence intensity and the ON/OFF contrast typically decrease. This progressive signal loss, referred to as switching fatigue, is commonly evaluated either by the number of switching cycles or by the time required for the fluorescence intensity to decline to 50% of its initial value.
GFP-like rsFPs are generally classified into three subclasses – negative, positive, and decoupled types – each exhibiting distinct responses to excitation and switching illumination. Their photoswitching behaviour has been investigated in detail through spectroscopic measurements, nuclear magnetic resonance (NMR), crystallographic analyses, and molecular dynamics (MD) simulations, with particular focus on the electronic states of the chromophore, the geometry of its cis/trans isomers, and the associated reaction pathways.64 In contrast, rsFPs that incorporate external chromophores rely primarily on cofactor exchange or reversible covalent bonding to achieve switching. Consequently, their mechanisms differ fundamentally from those of GFP-like rsFPs. The subsequent sections therefore distinguish between the photoswitching modes and mechanisms of GFP-like proteins and those of chromophore-binding variants.
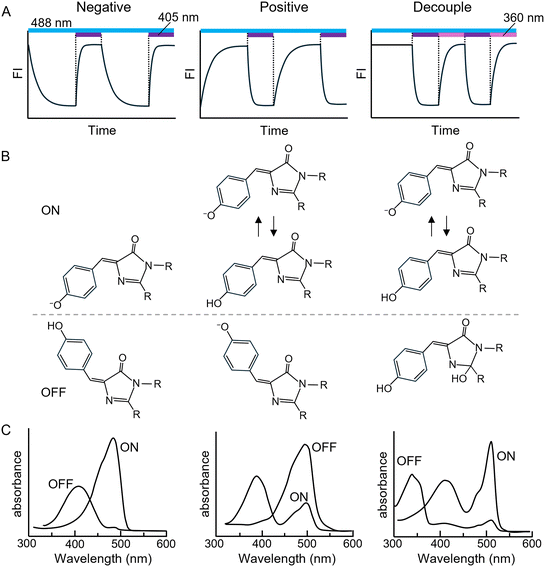
3.1. Photoswitching of GFP-like negative type rsFPs
Negative-type rsFPs (n-rsFPs) exhibit OFF-switching upon irradiation with the excitation light used for fluorescence and ON-switching upon illumination at a different wavelength. Green n-rsFPs such as Dronpa65 and rsEGFP66 emit green fluorescence when excited at 488 nm but simultaneously undergo OFF-switching. Consequently, the fluorescence intensity of an ensemble of n-rsFP molecules decreases over time under continuous 488 nm illumination (Fig. 2A, left). Although this fluorescence decay superficially resembles photobleaching, it can be fully restored by subsequent irradiation at 405 nm (Fig. 2A, left), confirming that the change is reversible photoswitching rather than irreversible photobleaching.The photoswitching mechanism of n-rsFPs is explained by cis–trans isomerisation of the chromophore, accompanied by a change in the protonation state of its phenolic group.67 In typical n-rsFPs such as rsEGFP and Dronpa, the ON state corresponds to the cis-deprotonated form, whereas the OFF state corresponds to the trans-protonated form (Fig. 2B, left). In contrast, in rsGamillus, the ON state is mainly the trans-deprotonated form and the OFF state the cis-protonated form.68
A key feature that enables negative photoswitching behaviour is the spectral separation between the excitation wavelengths of the ON and OFF states (Fig. 2C, left), resulting in distinct wavelength dependencies for ON- and OFF-switching. Cis–trans isomerisation serves as the structural mechanism that differentiates the deprotonated (ON) and protonated (OFF) states by reconfiguring the hydrogen-bond network between the chromophore's phenol/phenolate group and nearby amino acid residues within the β-barrel.69,70
Time-resolved crystallography and absorption spectroscopy, together with computational studies using excited-state quantum mechanics/molecular mechanics (QM/MM) and MD simulations, have revealed multiple intermediate states during photoswitching between the ON and OFF forms.71,72 These studies indicate that isomerisation of the chromophore's phenolic group proceeds via the “hula-twist” and/or “one-bond-flip” mechanisms.73–75
Interestingly, even at cryogenic temperatures, where chromophore mobility is highly constrained, rsEGFP2 still exhibited OFF-switching and adopted a non-fluorescent cis conformational state.76 This observation suggests the possible existence of a photoswitching mechanism independent of cis–trans isomerisation. Further investigation of these mechanisms will likely identify key amino acid residues involved in switching, thereby facilitating the rational design of improved rsFPs (see section 5-1).
3.2. Photoswitching of GFP-like positive type rsFPs
In contrast to n-rsFPs, positive-type rsFPs (p-rsFPs) undergo ON-switching upon irradiation with the excitation light used for fluorescence and OFF-switching upon illumination at another wavelength. For example, when an ensemble of green p-rsFP molecules in the OFF state—such as Padron77 or Kohinoor78—is irradiated with 488 nm excitation light, a time-dependent increase in fluorescence intensity is observed (Fig. 2A, middle). Subsequent illumination at 405 nm, however, induces a decrease in fluorescence intensity (Fig. 2A, middle).The photoswitching mechanism of most p-rsFPs, similar to that of n-rsFPs, involves cis–trans isomerisation of the chromophore accompanied by interconversion between its protonated and deprotonated states (Fig. 2B, middle).79–82 As described above, n-rsFPs achieve negative photoswitching behaviour through stabilisation of either the protonated or deprotonated chromophore in the ON and OFF states (Fig. 2B and C, left). In contrast, p-rsFPs require that fluorescence excitation light both induces ON-switching and directly excites the ON state for emission. This configuration requires two conditions:
(1) the absorption spectrum of the OFF state must overlap the excitation wavelength of the ON state, and
(2) OFF-switching must be driven by light at a wavelength distinct from that used for excitation and ON-switching.
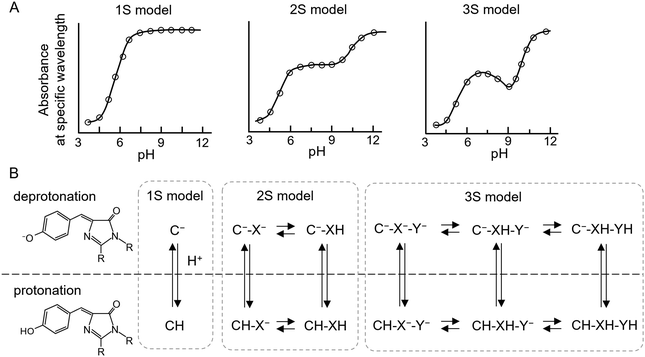
For chromophores that satisfy these criteria, the ON state represents a mixture of protonated and deprotonated species, whereas the OFF state remains fully deprotonated; in this case, the deprotonated form of the ON state is the fluorescent species (Fig. 2B and C, middle). Such a mixed protonation equilibrium can be intrinsically unstable, as small pH fluctuations may significantly alter the balance between the protonated and deprotonated forms according to the single-ionisation-site (1S) model (Fig. 3A and B, left). Nevertheless, several p-rsFPs—such as Kohinoor,78 Padron2,83 and rsZACRO84—exhibit broad absorbance plateaus in the visible range near neutral pH in their titration profiles (Fig. 3A, middle), indicating stable coexistence of protonated and deprotonated species under physiological conditions.
Such behaviour cannot be explained by a single proton-binding equilibrium of the chromophore but instead requires multiple coupled protonation–deprotonation equilibria. For instance, in the ON state of rsZACRO, a red p-rsFP, both the deprotonated and protonated forms of the chromophore coexist in an almost constant ratio (phenolate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phenol = 1
phenol = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.9–2.6) across the pH range 6–9.84 This apparent pH insensitivity of the phenol/phenolate ratio can be described by the association and dissociation of at least two protons—one at the chromophore phenolate and another at a nearby non-chromophoric residue. If the proton-binding sites are located on the chromophore itself (C) and on a neighboring amino acid residue (X), four molecular species—C−–X−, CH–X−, C−–XH, and CH–XH—are presumed to exist in equilibrium near neutral pH (Fig. 3B, middle). Among these, C−–X− and C−–XH species exhibit an absorption peak around 560 nm and are responsible for the red fluorescence, whereas CH–X− and CH–XH species are excited and undergo OFF-switching under blue light irradiation. These CH–X− and CH–XH species are rapidly replenished through dynamic equilibrium with C−–X− and C−–XH, ensuring sustained OFF-switching activity.79
2.9–2.6) across the pH range 6–9.84 This apparent pH insensitivity of the phenol/phenolate ratio can be described by the association and dissociation of at least two protons—one at the chromophore phenolate and another at a nearby non-chromophoric residue. If the proton-binding sites are located on the chromophore itself (C) and on a neighboring amino acid residue (X), four molecular species—C−–X−, CH–X−, C−–XH, and CH–XH—are presumed to exist in equilibrium near neutral pH (Fig. 3B, middle). Among these, C−–X− and C−–XH species exhibit an absorption peak around 560 nm and are responsible for the red fluorescence, whereas CH–X− and CH–XH species are excited and undergo OFF-switching under blue light irradiation. These CH–X− and CH–XH species are rapidly replenished through dynamic equilibrium with C−–X− and C−–XH, ensuring sustained OFF-switching activity.79
Similar behaviour has also been observed not only in p-rsFPs but also in other GFP-like proteins such as avGFP85 and IrisGFP.86 This extended framework is known as the two-ionisation-site (2S) model.87 In both avGFP and IrisGFP, the residue Glu222 has been proposed to participate in the proton association–dissociation equilibrium of the chromophore.
p-rsFPs Kohinoor2.088 also exhibit minimal variation in the relative proportions of protonated and deprotonated chromophore species around neutral pH. However, this behaviour appears to be better described by the association and dissociation of at least three protons, leading to the proposal of the “three-ionisation-site model” (3S model) (Fig. 3A, right). In the 3S model, the proton-binding sites are assumed to reside on the chromophore itself (C) and on two nearby amino acid residues (X and Y), resulting in six molecular species that coexist in equilibrium near neutral pH (Fig. 3B, right). A QM/MM study of Padron0.9,81 which shares a common ancestor with Kohinoor2.0, suggested that a hydrogen-bonding network involving residues Glu144, His193, and Glu211 in the chromophore pocket may contribute to this complex protonation equilibrium.
A p-rsFP following the 1S model may be sufficient to exhibit positive photoswitching under neutral pH conditions.84,89 However, p-rsFPs governed by the 2S or 3S model likely maintain a more stable balance between protonated and deprotonated chromophore species at physiological pH, thereby providing improved photoswitching robustness and substantially higher ON/OFF contrast compared to 1S-type p-rsFPs. Furthermore, these multiple proton networks are dynamic and permit drastic pKa changes upon isomerisation in photoswitching process. Crystallography and ultrafast spectroscopic analyses of Kohinoor have revealed that photoswitching involves multiple excited-state intermediates. These processes are accompanied by changes in intermolecular interactions, including conformational rearrangements and reorganisation of the hydrogen-bonding network in the vicinity of the chromophore and amino acid residue.90
Future identification of key residues and elucidation of the detailed proton-exchange network underlying the 2S or 3S mechanisms will enable the rational design of next-generation high-performance p-rsFPs. Moreover, since several FPs—such as IrisGFP, avGFP, and intriguingly, pHTomato91,92 and cpYGFP93,94—exhibit 2S-type behaviour, engineering studies of these proteins are expected to yield valuable insights for developing novel p-rsFPs. Notably, rsZACRO was developed from the RFP HyperNova,95 which itself displays 2S model behaviour. Such newly engineered p-rsFPs are anticipated to expand the scope of advanced imaging applications, including SR microscopy and multicolour fluorescence techniques (see section 4).
3.3. Photoswitching of GFP-like decouple type rsFPs
Decoupled-type rsFPs utilize distinct irradiation wavelengths for fluorescence excitation, ON-switching, and OFF-switching, whereas p-rsFPs and n-rsFPs employ excitation wavelengths that simultaneously induce fluorescence emission and photoswitching (Fig. 2A). Currently, the only known decoupled-type rsFPs are Dreiklang96 and its variant SPOON,97 which exhibit green fluorescence upon excitation at 500 nm, undergo OFF-switching at 405 nm, and ON-switching at 365 nm (Fig. 2A and C).Unlike the cis–trans isomerisation and protonation–deprotonation mechanisms observed in p- and n-rsFPs, the photoswitching of decoupled-type rsFPs is driven by reversible hydration and dehydration of the imidazolinone ring within the chromophore96 (Fig. 2B). Mutagenesis, absorption spectroscopy, and QM/MM studies of Dreiklang have shown that Glu222 plays a key role in the hydration–dehydration reaction, while Gly65 facilitates the approach of Glu222 toward the chromophore, allowing its participation in this process.96,98,99 Insights from these studies on Dreiklang's chromophore hydration, along with prior mutations introduced into mKalama100 and Sirius20—which eliminate excited-state proton transfer (ESPT) and enhance fluorescence quantum yield—contributed to the semi-rational design of the deep-violet FP Sumire.21 Sumire emits at 414 nm and possesses a chromophore structure analogous to the OFF state of Dreiklang.
In addition to light-induced switching at 405 and 365 nm, both SPOON and Dreiklang exhibit spontaneous ON-switching via thermal relaxation and OFF-switching under 488 nm excitation. Exploiting this property, single-molecule localisation microscopy (SMLM) has been achieved using the spontaneous blinking of SPOON under continuous 488 nm illumination.97
3.4. Switching of negative type rsFPs incorporating external chromophores
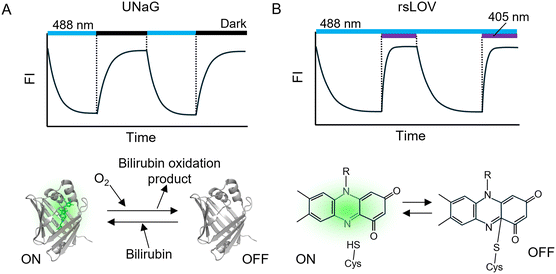
FPs that incorporate external chromophores exhibit switching mechanisms distinct from those of GFP-like rsFPs. In systems such as FAST,37 UnaG,17 and Rep-miRFP,101 switching is primarily driven by cofactor exchange rather than conformational or protonation-state changes of the chromophore. The OFF-switching process is typically initiated by photo-oxidative damage of the chromophore under excitation illumination (Fig. 4A), while the protein scaffold generally remains intact. For ON-switching to occur, the oxidized cofactor must dissociate from the protein. In systems where the chromophore is covalently attached (e.g., smURFP–biliverdin) or forms stable coordination complexes (e.g., FlAsH/ReAsH via arsenic–thiol bonds), this dissociation is largely inhibited.35,102 In contrast, noncovalent systems such as FAST and UnaG permit the damaged chromophore to dissociate more readily.48,103 Genetic engineering has further enabled noncovalent chromophore binding in certain phytochrome-derived systems, exemplified by Rep-miRFP.101ON-switching requires rebinding of a fresh cofactor to regenerate a new fluorescent holo-complex (Fig. 4A). In experimental settings, an excess of free cofactor is typically supplied to promote efficient reconstitution. Proteins such as FAST, UnaG, and Rep-miRFP have demonstrated successful chromophore exchange and fluorescence recovery following photodamage.48,101,103
OFF- and ON-switching in these systems are governed by differences in the binding affinities (Kd) between the protein and its chromophore before and after oxidation. Although direct post-bleaching Kd measurements are not yet available, fluorogen-activating systems such as UnaG–bilirubin complexes exhibit noncovalent interactions (Kd ≈ 98 pM).17 Moreover, photo-oxidized bilirubin derivatives have been detected following blue-light irradiation of holo-UnaG, supporting this exchange-based mechanism.103,104 These processes are distinct from those of DNA-PAINT imaging, where fluorophore exchange is not light-driven but instead relies on transient, reversible interactions between “docking” and “imager” strands (Kn ≈ 1–10 nM).105
YtvA is a FMN-binding FP based on a Light, Oxygen, Voltage (LOV) domain derived from Bacillus subtilis. YtvA106 and its engineered variants rsLOV1 and rsLOV2107 exhibit fluorescence and OFF-switching under 488 nm illumination and ON-switching under 405 nm light, characteristic of negative-type photoswitching (Fig. 4B). The photoswitching mechanism of these proteins differs from that of other rsFPs, involving reversible covalent bond formation between FMN and a cysteine residue within the LOV domain (Fig. 4B), accompanied by changes in chromophore absorption and fluorescence quantum yield. In the ON state, YtvA exhibits an absorption maximum at 450 nm and emits green fluorescence peaking at 500 nm. Upon blue-light irradiation, the LOV domain forms a covalent bond between FMN and the cysteine residue, converting the protein to the OFF state. The ON state is restored either through thermal relaxation or upon UV illumination; however, UV light simultaneously promotes the OFF-switching reaction, leading to a dynamic equilibrium between the two states.106,107
FPs that rely on noncovalent chromophore binding in their ON state—such as FAST, UnaG, miRFP, and YtvA—permit chromophore exchange following photobleaching. This feature confers strong resistance to photobleaching and makes these proteins particularly suitable for SR imaging, which often requires high-intensity excitation.107–109 Emerging protein scaffolds such as phycobiliproteins,40 lipocalins,110 and serpins111 are expected to further expand the toolkit of chromophore-exchanging FPs in the future.
4. Applications of rsFPs
The reversible photoswitching between the ON and OFF states of rsFPs has been applied to a wide range of techniques that advance biological research, most notably in SR imaging.112,113 A fundamental concept in rsFP-based applications is the replacement of conventional fluorophores used in methods that rely on two-state transitions—such as transitions between the ground (S0) and excited (S1) states in fluorescence excitation, or between pre- and post-photobleaching states in bleaching-based techniques (Fig. 5A)—with rsFPs. By utilizing the ON/OFF photoswitching transitions of rsFPs (Fig. 5B) as the equivalent two-state system, rsFPs can be integrated into techniques such as SR imaging, fluorescence lifetime imaging microscopy (FLIM), fluorescence recovery after photobleaching (FRAP), and time-resolved anisotropy measurements.The dwell times of the ON and OFF states of rsFPs are orders of magnitude longer than the excited-state lifetime. Whereas the S1 state typically lasts only a few nanoseconds or less, the ON and OFF states—both corresponding to distinct S0 configurations—persist for seconds or longer (Fig. 5A and B). Consequently, techniques that replace excitation–emission processes with photoswitching can extend the effective measurement time scale by several orders of magnitude. This time-scale extension also enables a substantial reduction in light exposure. For instance, replacing the stimulated emission depletion (STED) process used in SR microscopy with photoswitching allows a dramatic reduction in illumination power density, thereby minimizing phototoxicity to living cells. In addition, incorporating photoswitching kinetics into anisotropy-based measurements, as demonstrated by STARSS, enables rotational correlation analyses of molecular complexes larger than 30 kDa.114 Similarly, substituting photobleaching-based processes with photoswitching can markedly reduce excitation intensity, because photoswitching is a more energy-efficient process than photobleaching. Moreover, unlike photobleaching, rsFP photoswitching is reversible, allowing repeated measurements on the same sample.
The reversible ON–OFF transition of rsFPs has also been exploited for optical data storage applications.66,115 Furthermore, the rsFP Dronpa has been employed as an optogenetic tool for controlling protein–protein interactions and modulating protein activity.116,117 Collectively, rsFPs offer diverse and expanding opportunities across imaging, information storage, and optogenetic applications, as discussed in more detail in the following sections.
4.1. Super-resolution (SR) imaging using rsFPs
SR imaging refers to a class of fluorescence microscopy techniques that achieve spatial resolutions beyond the diffraction limit of light by employing specialized optical configurations and photophysical control of fluorophores. When a point-like fluorescent object is observed through a conventional wide-field fluorescence microscope, the image formed on the detector cannot be smaller than a fundamental lower limit imposed by the wave nature of light. The corresponding size limit on the object plane is known as the Rayleigh criterion, expressed as:
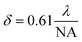 | (4) |
SR fluorescence microscopy methods are broadly classified into stochastic approaches and coordinated illumination/acquisition approaches. Stochastic methods such as STORM, PALM, and SOFI achieve high spatial resolution by exploiting the random blinking, photoactivation, or photobleaching of fluorescent molecules.119,120 Specifically, STORM and PALM rely on the stochastic, temporal isolation of individual fluorophores that switch between fluorescent and non-fluorescent states, enabling high-precision localisation of single emitters within sparsely activated subsets119 (Fig. 6A), whereas SOFI achieves SR imaging by analysing temporal correlations of fluorescence intensity fluctuations without requiring spatial isolation of single emitters.120
In contrast, coordinated illumination/acquisition techniques include STED microscopy,121 saturated structured illumination microscopy (SSIM),122 and super-resolution by polarisation demodulation or excitation polarisation angle narrowing (SPoD-ExPAN)123 (Fig. 6B). These methods achieve SR imaging by spatially engineering excitation and depletion light to confine fluorescence emission below the diffraction limit, by saturating fluorescence to generate high-spatial-frequency emission patterns, or by exploiting polarisation-dependent excitation and emission. Hybrid approaches such as MINFLUX and MINSTED, which combine stochastic fluorophore activation with engineered illumination and precise readout, can achieve localisation precision on the order of a few nanometers.124,125
Both stochastic and engineered illumination/acquisition approaches fundamentally rely on the interconversion between fluorescent and non-fluorescent states of fluorophores. Consequently, rsFPs—by virtue of their reversible ON/OFF photoswitching—enable SR imaging in both modalities while extending the accessible timescale and substantially reducing the illumination intensity required in methods such as STED.
Compared to p- and decoupled-type rsFPs, a larger number of n-rsFPs have been developed, leading to the exploration of diverse SR imaging methods that employ n-rsFPs in both stochastic and coordinated illumination/acquisition approaches. However, in the case of engineered illumination/acquisition techniques, the excitation and OFF-switching wavelengths of n-rsFPs typically exhibit strong spectral overlap. As a result, temporal separation between OFF-switching and fluorescence readout is required, necessitating carefully designed illumination sequences. Moreover, these methods often demand multiple acquisitions or high-intensity excitation to obtain sufficient signal-to-noise ratios, which limits temporal resolution and raises concerns regarding phototoxicity.
In contrast, p- and decoupled-type rsFPs feature distinct spectral bands for OFF-switching and fluorescence excitation. Consequently, SR imaging using these rsFP classes can circumvent the need for temporal separation of illumination or operate at lower excitation intensities while maintaining temporal separation, thereby reducing phototoxicity. For instance, in reversible saturable optical fluorescence transitions (RESOLFT) microscopy employing an n-rsFP such as rsEGFP, a three-step illumination sequence is required for each scanning cycle: (1) ON-switching with a Gaussian beam at 405 nm, (2) OFF-switching with a doughnut-shaped beam at 488 nm, and (3) fluorescence readout with a Gaussian beam at 488 nm (Fig. 6C).
By contrast, when using p-rsFPs such as Padron2, RESOLFT imaging can be performed with a single, combined scan using an overlapped illumination pattern consisting of a doughnut-shaped 405 nm OFF-switching beam and a Gaussian-shaped 488 nm excitation/ON-switching beam84 (Fig. 6D). Furthermore, the p-rsFP Kohinoor has been employed for nonlinear structured illumination microscopy (SIM), achieving fast acquisition speeds.126 Kohinoor and its improved variant Kohinoor2.0 have also been applied to SR imaging at very low illumination intensities using SPoD-OnSPAN, a polarisation-modulated SR imaging method in which cells are simultaneously irradiated with linearly polarized beams for fluorescence excitation/ON-switching and OFF-switching.127,128
4.2. Intracellular functional imaging with rsFPs
Bioimaging enables not only the visualisation of cellular structures, morphology, and biomolecular localisation, but also the measurement of dynamic biological processes such as protein–protein interactions, enzymatic activity, concentrations of functional molecules (e.g., calcium), and physiological parameters (e.g., temperature).5–9 These functional imaging approaches serve as powerful tools for investigating intracellular physiology and signal transduction.Genetically encoded sensors used in functional imaging typically consist of FPs fused to sensing domains that respond to specific biological activities or physicochemical conditions. Upon binding or dissociation of a target molecule, or in response to changes in environmental parameters, the sensing domain undergoes a conformational change. This conformational change is transduced into an alteration of photophysical properties—such as FRET efficiency, fluorescence quantum yield, or molar extinction coefficient—thereby enabling functional imaging through detection of corresponding fluorescence changes.
To visualize cellular physiological parameters such as pH and temperature, some sensors are composed solely of FPs, in which the FP functions simultaneously as both the reporter and the sensing domain. Examples include the pH-sensitive FP pHluorin129 and the genetically encoded temperature indicator B-gTEMP.130
For quantitative bioimaging, it is often preferable to measure parameters that are independent of experimental factors such as intracellular expression levels or optical path variations, rather than relying solely on fluorescence intensity. Fluorescence lifetime represents one such robust quantity, as it is independent of fluorophore concentration. In FRET-based sensors, changes in FRET efficiency directly correspond to variations in the donor's fluorescence lifetime.131 Likewise, single-fluorophore-type sensors that alter quantum yield in response to conformational changes can also be monitored through lifetime variations.132 Consequently, FLIM has emerged as a powerful and quantitative approach for functional imaging.
By analogy with the concept of fluorescence lifetime, the photoswitching kinetics of rsFPs can also be exploited for fluorescence imaging, enabling quantitative measurements of fluorescence signals. Furthermore, FRET can be combined with photoswitching, in which the donor fluorophore is an rsFP—a strategy referred to as photoswitching FRET (psFRET).133 When FRET occurs from the donor rsFP to an acceptor, the photoswitching speed of the donor is reduced due to FRET-mediated quenching of its excited state (Fig. 7A and B). This psFRET approach has been successfully applied to visualize protein–protein interactions.133
In addition, an rsFP can also serve as a FRET acceptor. This approach leverages the distinct absorption spectra of rsFPs in their ON and OFF states, which leads to state-dependent FRET efficiencies. Consequently, the donor fluorescence is modulated according to the photoswitching state of the acceptor rsFP, enabling quantitative fluorescence readouts and the potential implementation of phase-sensitive lock-in detection.134,135
Beyond FRET-based systems, quantitative measurements using photoswitching have also been demonstrated with sensors based on a single rsFP136 (Fig. 7C). For example, the calcium indicator rsGCaMP6s-Q, derived from GCaMP6s, exhibits photoswitching behaviour that is modulated by Ca2+ binding. In the presence of Ca2+, rsGCaMP6s-Q displays negative photoswitching characteristics (Fig. 7D(1)), whereas upon Ca2+ dissociation, the photoswitching kinetics and ON/OFF contrast decrease by severalfold, accompanied by a change from negative to positive photoswitching136 (Fig. 7D(2)).
Unlike switching kinetics, the ON/OFF contrast is independent of both FP concentration and excitation power density, providing a more robust and quantitative fluorescence readout than photoswitching rate alone. This property enables repeated cycling of ON/OFF transitions over multiple illumination sequences, allowing photochromism-enabled absolute quantification (PEAQ) biosensing, which provides an absolute measure of intracellular molecular functions.137
When the photoswitching properties of rsFPs are influenced by a biological parameter, they can serve as quantitative reporters of physiological function. For instance, Dronpa and its variant Dronpa3 exhibit viscosity-dependent changes in OFF-switching kinetics, which have been used to measure intracellular viscosity.138 Photoswitching kinetics also show pH dependence, suggesting the potential for intracellular pH imaging.68,89
Importantly, because photoswitching can be induced using standard wide-field or confocal microscopes, this method offers greater accessibility and experimental simplicity compared to FLIM.133
4.3. Fluorescence signal discrimination with rsFPs
By exploiting the light-dependent fluorescence modulation of rsFPs under designed illumination schemes for photoswitching and excitation, it is possible to distinguish target fluorescence from background noise and achieve high-contrast fluorescence imaging (Fig. 8A). Based on this concept, several lock-in–based imaging modalities have been developed, including optical lock-in detection (OLID),140 synchronously amplified fluorescence image recovery (SAFIRe)141 (Fig. 8B), and out-of-phase imaging after optical modulation (OPIOM).142
In OLID, the rsFP fluorescence signal is selectively extracted from background noise via cross-correlation analysis with a reference signal.140,143 However, the illumination wavelength used for photoswitching is typically shorter than that for fluorescence excitation. For instance, Dronpa emits fluorescence at 517 nm, while OFF- and ON-switching occur under 488 nm and 405 nm light, respectively. Such short-wavelength illumination can also modulate autofluorescence, thereby reducing the signal discrimination capability of OLID.
In contrast, SAFIRe employs intensity-modulated switching light at wavelengths longer than the fluorescence emission wavelength, and extracts fluorescence components in the frequency domain via Fourier transform analysis.141,144 SAFIRe using modBFP—which emits at 455 nm upon 405 nm excitation and can be reversibly activated by 514.5 nm light—demonstrated improved signal-to-noise ratios by effectively suppressing modulation of the autofluorescence background.145
In OPIOM, a single modulated light source is used for both excitation and photoswitching, and rsFP fluorescence is extracted through out-of-phase detection, where the phase-shifted component originates from photoswitching dynamics of the rsFP.142 To maximize the out-of-phase fluorescence signal, the average illumination intensity and modulation frequency must be optimized according to the photophysical properties of the rsFP, including the molecular extinction coefficients for ON/OFF switching and the intrinsic thermal switching kinetics. Using Dronpa3, OPIOM successfully separated rsFP fluorescence signals from autofluorescence in HEK293 cells and zebrafish embryos.142
However, the time resolution of OPIOM in live-cell imaging is limited by the dependence of modulation frequency on the rsFP's thermal switching kinetics. To overcome this limitation, speed-OPIOM was developed, employing dual modulated light sources for excitation and ON/OFF-switching of p- and n-rsFPs (Fig. 8C). These illumination beams are synchronized in antiphase to accelerate photoswitching transitions beyond thermal equilibrium rates, thereby improving temporal resolution.146,147 Speed-OPIOM enables robust extraction of rsFP fluorescence signals even under strong autofluorescence conditions, such as in plant tissues or specimens illuminated by ambient light.148,149
In other implementations, such as multiphoton deactivation and imaging (MADI),151,152 rsFP-specific fluorescence is extracted by subtracting images acquired in the OFF state from those in the ON state. This subtraction effectively isolates the rsFP-derived fluorescence while removing background and non-switchable components.
These optical sectioning concepts based on rsFP photoswitching can be further integrated with SR imaging techniques, including structured illumination microscopy (SIM)154 and RESOLFT microscopy,155,156 thereby extending the achievable spatial resolution beyond the diffraction limit while maintaining optical sectioning capability.
To expand the colour palette, numerous FPs with diverse excitation and emission properties have been developed (Fig. 1). For instance, Sirius,20 bfVFP,157 and Sumire21 have extended the optical window toward shorter wavelengths, whereas FPs such as mNeptune23 and miRFP39 extend the range into the far-red and near-infrared regions. In addition, large Stokes shift FPs (LssFPs) allow signal separation based on differences in excitation rather than emission wavelengths.158–160 Combining these FPs with spectral unmixing techniques further enhances multiplexing capability.161 Nevertheless, the finite optical bandwidth of visible light imposes a physical limit on the number of fluorophores that can be spectrally distinguished, thereby restricting conventional multiplex fluorescence imaging.
To overcome this limitation, fluorescence modulation of rsFPs introduces an orthogonal dimension to excitation and emission wavelengths (Fig. 8A). Strategies such as SAFIRe and Speed-OPIOM, originally designed for suppressing autofluorescence, can also facilitate multiplex signal discrimination and substantially expand multiplexing capacity. For example, SAFIRe successfully distinguished acGFP fluorescence from that of EGFP in NIH 3T3 cells, despite their nearly identical emission spectra.162 Moreover, dual-laser modulated SAFIRe (DM-SAFIRe) modulates rsFP fluorescence via convolution of two modulated light sources—one for ON-switching and the other for OFF-switching and excitation—allowing discrimination of rsFastLime fluorescence from EGFP.163 Interestingly, DM-SAFIRe can also differentiate immobilized from diffusing fluorophores by tuning the modulation frequency relative to molecular diffusion rates, suggesting its applicability to probing protein–protein interactions and oligomerisation states.163
Multiplex imaging can be further extended by discriminating signals from multiple rsFPs themselves. Because each rsFP possesses distinct photoswitching kinetics, their fluorescence signals can be separated according to differential temporal responses under appropriately designed illumination protocols. Speed-OPIOM and its variant HIGHLIGHT (pHase-sensItive imaGing of reversibly pHotoactivatable Labels after Modulation of activatinG ligHT) have enabled independent imaging of four spectrally similar rsFPs by exploiting differences in their resonance frequencies under modulated illumination.164
In addition, several advanced methodologies—such as TMI (temporally multiplexed imaging),165 LIGHTNING (Light-tunable tIme-gated readinG-out of pHotocycles for mulTiplexed fluorescence ImagiNG),166 and NEEMO (intra-exposure excitation modulation)167,168—leverage differences in photoswitching kinetics to achieve rsFP signal separation. In TMI, fluorescence signals are separated based on differences in OFF-switching kinetics of n-rsFPs (Fig. 8D). In LIGHTNING, ON and OFF kinetics are characterized under four illumination conditions (Ilow, IIlow, Ihigh, and IIhigh), and separation is performed using the four corresponding lifetime metrics (τIlow, τIIlow, τIhigh, τIIhigh) (Fig. 8E). In NEEMO, four images (exp1–exp4) are acquired under a programmed excitation/photoswitching sequence, and rsFP signals are separated using the resulting four fluorescence intensities (Fig. 8F).
The distinct photoswitching behaviours of p- and n-rsFPs provide an additional means of achieving multiplex imaging that is orthogonal to conventional spectral separation. Under illumination with 488 nm light, the p-rsFP Padron is converted to its ON state, whereas the n-rsFP rsFastLime is switched to the OFF state, resulting in fluorescence and non-fluorescence, respectively. Conversely, following photoswitching by 405 nm illumination, Padron exhibits little fluorescence, while rsFastLime becomes fluorescent. Consequently, fluorescence signals from these two green rsFPs can be independently imaged despite occupying the same optical emission window—a demonstration successfully achieved in Saccharomyces cerevisiae and Nicotiana benthamiana cells.77,169
Multiplex imaging with rsFPs has also enabled cell-specific visualisation of neurons within complex neural networks in vivo. Neurons possess intricately interwoven axons and dendrites, making it difficult to distinguish individual cells using a single fluorescent colour. Conventional multicolour labelling methods such as Brainbow have addressed this challenge by stochastically expressing different combinations of FPs.170 In contrast, the rsFP-based approach leverages localized photoactivation. In neurons expressing Dronpa, global 488 nm illumination first switches all molecules to the OFF state, after which localized UV or two-photon red-laser excitation selectively switches Dronpa to the ON state within the soma of a single neuron. The activated Dronpa subsequently diffuses throughout the entire cell, including dendrites and axons, enabling selective imaging of the whole neuron. Repeating this procedure for multiple cells and reconstructing the resulting images permits multiplex labelling and identification of individual neurons.171
In summary, multiplex imaging utilizing rsFP photoswitching separates fluorescence signals along an axis orthogonal to excitation and emission wavelengths, effectively expanding the fluorophore identification space into a two-dimensional parameter plane of excitation/emission × photoswitching behaviour. Furthermore, integrating this strategy with lifetime-based172 or other photophysical separation techniques could add additional dimensions to the fluorescence signal space, thereby dramatically enhancing multiplexing capacity for complex biological imaging.
4.4. Anisotropy measurement with rsFPs
Time-resolved fluorescence anisotropy combines fluorescence lifetime measurements with anisotropy analysis to probe molecular dynamics and environmental properties such as viscosity, binding states, and rotational diffusion. However, the size of molecules that can undergo detectable rotational motion within the fluorescence lifetime window—typically in the sub-nanosecond to nanosecond range—is inherently limited (Fig. 9A). Consequently, anisotropy measurements of proteins larger than approximately 30 kDa are often unfeasible.To overcome this limitation, STARSS (Selective Time-Resolved Anisotropy with Reversibly Switchable States) was developed.114 In STARSS, the accessible timescale is extended by exploiting the reversible photoswitching of rsFPs (Fig. 9B). Unlike conventional time-resolved fluorescence anisotropy, STARSS decouples anisotropy detection from fluorescence lifetime by introducing a photocontrollable temporal window defined by the long-lived ON and OFF states of rsFPs (milliseconds to seconds). This extension enables the observation of slow rotational motions and restricted mobility in large macromolecular assemblies such as membrane-bound receptors, protein aggregates, and cytoskeletal scaffolds. As a result, STARSS markedly broadens the applicability of fluorescence anisotropy to biological systems that were previously inaccessible using conventional methods.
Steady-state anisotropy is also frequently employed to detect FRET between identical fluorophores, known as homoFRET, which provides a sensitive means of analysing protein oligomerisation and molecular interactions.173 During homoFRET, differences in dipole orientation between donor and acceptor fluorophores lead to depolarisation of the emitted light, resulting in a measurable decrease in anisotropy. However, accurate quantification requires a FRET-negative reference, which is difficult to obtain when studying proteins that naturally exist in oligomeric states. Traditional approaches have relied on photobleaching to irreversibly eliminate acceptor chromophores, thereby halting FRET and generating a pseudo-control condition.
To overcome the limitations of irreversible photobleaching, photoswitching anisotropy FRET (psAFRET) was developed.174 This method employs rsFPs to reversibly modulate FRET efficiency and measure corresponding changes in anisotropy. By monitoring anisotropy dynamics during the OFF-switching of FPs, psAFRET enables repeated, non-destructive quantification of homoFRET, providing a robust and reversible approach for tracking protein homo-oligomerisation dynamics in living cells.
4.5. Analysis of protein transport with rsFPs
Intracellular protein transport involves a complex interplay of processes such as post-translational modification, protein–protein and protein–organelle interactions, and diffusion. These molecular events often act as triggers for diverse physiological processes, including changes in gene expression through the activation of transcription factors and the reorganisation of intracellular signalling pathways, ultimately leading to alterations in cellular morphology and function.Fluorescence recovery after photobleaching (FRAP) and its related methods—inverse FRAP (iFRAP), fluorescence loss in photobleaching (FLIP), and fluorescence decay after photoactivation (FDAP)—are powerful tools for quantitatively evaluating protein diffusion coefficients in living cells175–177 (Fig. 9C). However, these techniques depend on irreversible fluorescence transitions such as photobleaching or photoconversion, which require high illumination power densities and hinder repeated measurements, as most fluorophores eventually undergo irreversible transitions.
In contrast, the reversible photoswitching of rsFPs enables similar analyses to be conducted at substantially lower illumination intensities than those required for photobleaching or photoconversion. This reversibility facilitates repeated measurements, signal averaging, and time-lapse analyses, although appropriate correction is needed to account for fluorescence modulation introduced by the photoswitching process itself.178 By employing rsFPs in FDAP (Fig. 9C), rapid protein diffusion on the millisecond timescale has been successfully quantified.65,179
These rsFP-based diffusion analyses have been applied to investigate nucleocytoplasmic shuttling and membrane protein transport in mammalian cells,180,181 as well as protein and symplasmic transport within plant tissues.182–184 Collectively, these studies demonstrate that rsFP-based approaches provide a versatile and minimally invasive framework for investigating intracellular transport dynamics across diverse biological systems.
4.6. Optical data storage with rsFPs
Photochromic biomaterials are emerging as promising candidates for high-density, three-dimensional, and potentially biodegradable optical data storage media. Among early examples, bacteriorhodopsin was one of the first biological materials explored as a rewritable storage medium.185–187More recently, rsFPs have attracted attention as alternative photochromic materials due to their bistable optical states and compatibility with optical manipulation and fluorescence readout (Fig. 9D). Notable demonstrations include the encoding of the complete text of 25 Grimm's fairy tales by embedding rsEGFP within a polyacrylamide matrix66 and the recording of a “Pac-Man” video using Dreiklang96—both illustrating the potential of rsFPs for high-capacity, rewritable optical data storage.
Furthermore, crystals of IrisFP, a FP exhibiting both photoswitching and photoconversion properties, have been used to produce three-dimensional optical lettering through two-photon excitation.115 These studies highlight the potential of rsFPs as molecular-scale recording elements for high-density, three-dimensional, and reconfigurable bio-based optical storage systems, offering a unique convergence of biological function, molecular engineering, and photonic control.
4.7. Optical manipulation with rsFP Dronpa
Optical manipulation of protein localisation, oligomeric state, and activity with high spatiotemporal precision provides a powerful means of probing intracellular signalling pathways.188,189 A notable example is Dronpa145N, which exhibits photoswitching-dependent reversible transitions between oligomeric and monomeric states.116 In its ON state, Dronpa145N forms a tetramer, whereas upon cyan-light irradiation it converts to an OFF-state monomer.Building on this property, Dronpa145N was further engineered into pdDronpa, which undergoes light-controlled switching between dimeric and monomeric states.117 This reversible photo-dimerisation enables optical regulation of protein activity by sterically blocking substrate access to active sites through light-induced dimerisation (Fig. 9E). Unlike optogenetic systems based on LOV2 or Cry2, pdDronpa forms its chromophore autocatalytically, eliminating the need for external cofactors and defining a distinct class of self-contained, fluorescent optogenetic tools.
Using pdDronpa, precise optical control has been achieved over a broad range of biological processes, including protein kinase and GTPase activities, gene editing, and transcriptional regulation.190–192 In addition, light-induced oligomerisation of pdDronpa has been harnessed to generate mechanical forces.193 This property has been exploited in photoresponsive hydrogels that exert mechanical stress to regulate cellular migration and differentiation,194,195 as well as in light-controllable drug delivery systems, where the physical properties of hydrogels are modulated by optical stimulation.196
4.8. Novel approaches integrating multiple technologies with rsFPs
Recent integration of diverse application strategies based on rsFPs has given rise to a new wave of innovations that are poised to substantially broaden the scope and capabilities of bioimaging.To monitor protein–protein interactions, bimolecular fluorescence complementation (BiFC) has been adapted for rsFPs.201 In this strategy, an rsFP is split into two fragments that regain both fluorescence and photoswitching capability upon reconstitution.202 This approach enables high-resolution visualisation of interaction sites when combined with SR techniques such as pcSOFI (photochromic stochastic optical fluctuation imaging)203 and RESOLFT.204
Another example, FLINC (Fluorescence Fluctuation Increase by Contact), exploits fluorescence fluctuations of TagRFP that are amplified by its proximity to Dronpa or TagRFP-T. When integrated with pcSOFI, FLINC enabled SR imaging of protein kinase A (PKA) activity.205 This concept was further refined into DrFLINC (Dronpa-chromophore-removed FLINC), which provided a generalizable design for fSR biosensors analogous to FRET-based systems.206
Single-molecule-type fSR probes have also been developed. rsGCaMP, created by introducing photoswitchable mutations into the Ca2+ indicator GCaMP5G, exhibits both calcium sensitivity and reversible photoswitching. Using rsGCaMP in combination with MoNaLISA (a RESOLFT variant), fSR imaging of intracellular Ca2+ dynamics was achieved.207
However, in fSR approaches based on FLINC-type or single-molecule rsFP sensors, only those probes that are functionally activated—through ligand binding or sensing of specific biophysical parameters—exhibit fluorescence fluctuations or signal changes, thereby enabling SR detection. Conversely, in regions or at time points where the probe remains inactive, SR information cannot be obtained. To achieve quantitative and spatially resolved functional measurements, the future development of rsFP-based sensors that couple photoswitching with FRET or fluorescence lifetime mechanisms is expected to further enhance the performance and general applicability of fSR imaging.
To overcome this constraint in SR imaging, multiplexing based on parameters independent of excitation and emission wavelengths has been explored. This strategy enables comprehensive localisation of multiple molecular targets with nanoscale spatial resolution while preserving information about their spatial and functional correlations in living samples. rsFPs offer an effective approach to realize such wavelength-independent multiplexing, as their photoswitching kinetics provide an additional dimension for signal discrimination within the same optical window.
By incorporating rsFP-based multiplexing into SR imaging—or conversely, integrating rsFP-based SR techniques into multiplexed imaging workflows—multiplexed SR imaging can be achieved. For example, STED microscopy employing the p-rsFP Padron together with the n-rsFP rsEGFP enabled two-colour SR imaging, while in vivo STED using rsEGFP, EGFP, and Citrine achieved three-colour SR imaging by exploiting differences in their photoswitching behaviours.209,210 Meanwhile, RESOLFT microscopy combined with fluorescence lifetime–based multiplexing enabled dual-colour SR imaging using two green rsFPs, rsEGFP-N205S and Dronpa-M159T.211
Although these approaches often involve a trade-off in temporal resolution, the integration of rsFP-based multiplexing methods such as TMI, LIGHTNING, NEEMO, and Speed OPIOM with advanced SR imaging modalities is expected to further enhance multiplexing capability, allowing simultaneous, high-resolution visualisation of multiple targets beyond spectral limitations.
In this system, data writing was accomplished by selectively ON-switching rsEGFP within sub-diffraction-limited regions under RESOLFT illumination, followed by photobleaching to fix the written state. Subsequent data readout was performed using RESOLFT microscopy as well, enabling repeated high-resolution visualization of the encoded information.66
In practice, simultaneous detection of two distinct signalling activities—cAMP-dependent protein kinase (PKA) and extracellular signal-regulated kinase (ERK)—was demonstrated using an rsFP-based PKA activity biosensor (rsAKARev)212 in combination with a non-photochromic ERK biosensor (EKARev).213 Both biosensors shared the same optical window for excitation and emission wavelengths, and the incorporation of rsFP-based fluorescence modulation enabled robust signal separation, thereby allowing simultaneous, multiplexed functional imaging within a single sample.212
5. Development of rsFP
A wide variety of rsFPs have been developed to date, enabling a broad range of applications as described above. For all these uses, rsFPs are generally expected to be monomeric—to avoid perturbing the intracellular localisation or function of target proteins—while also exhibiting high brightness and photostability, which are fundamental requirements for any fluorescent probe used in cellular imaging.Beyond these general photophysical properties, the photoswitching characteristics of rsFPs should be carefully optimized according to the intended application. Although high ON/OFF contrast and low photoswitching fatigue are universally desirable, faster ON and OFF switching kinetics are not always advantageous, as the optimal switching speed depends on the specific temporal and experimental requirements of each application.
5.1. Directed evolution of rsFPs
Directed evolution is one of the most powerful methodologies for the development of rsFPs.214 The key steps in directed evolution involve (i) constructing mutant libraries through the introduction of genetic variations, and (ii) screening and isolating variants exhibiting the desired photophysical properties.For mutant library construction, several approaches are available. Random mutagenesis introduces mutations across the entire gene without targeting specific residues.215 In contrast, semi-rational mutagenesis allows mutations to be introduced at defined sites based on structural information. Guided by crystallographic analyses and molecular modelling, substitutions near the phenolic group of the chromophore have proven particularly effective for tuning photoswitching behaviour, as this process is strongly influenced by steric hindrance and hydrophobic/electrostatic interactions surrounding the Tyr residue in the cis and trans chromophore states.74,216
Indeed, many rsFPs—including rsCherry,217 rsFusionRed,218 rsGamillus,68 and rsZACRO84—have been generated through site-directed mutagenesis around the chromophore. To explore all possible amino acid substitutions at a given site, site-saturation mutagenesis can be employed, typically by PCR amplification using primers containing degenerate codons such as NNK (N = A/T/C/G, K = G/T).219 Subsequently, recombination techniques can be used to combine beneficial mutations into a single construct. One useful method is the Staggered Extension Process (StEP), which involves iterative cycles of DNA denaturation, short annealing, and polymerase extension, generating hybrid sequences through random template switching.220
During the screening stage, E. coli is commonly used as a host for expressing mutant libraries. Each bacterial colony on a solid medium corresponds to a unique variant, facilitating the isolation and evaluation of individual clones. Selection of rsFPs requires simultaneous assessment of multiple parameters, including brightness, ON/OFF contrast, and photoswitching kinetics, depending on the intended application. However, the photophysical properties of rsFPs expressed in E. coli colonies do not necessarily reflect their behaviour in other biological contexts such as mammalian cells. Therefore, performing screening directly in the target cell type—for instance, mammalian cells or neurons—is expected to yield rsFPs with characteristics better optimized for specific imaging applications.58
5.2. Rational design
Rational design represents another powerful strategy for engineering rsFPs. In this approach, mutations governing photoswitching behaviour are identified and introduced based on three-dimensional structural information or computational simulations.For example, structural analysis of Dronpa identified the mutation V159G, which likely reduces the energy barrier for photochromic switching and accelerates OFF-switching kinetics.216,221 Similarly, structural characterisation of the excited-state conformation of rsEGFP2 revealed that the V151A substitution improved both photoswitching speed and ON/OFF contrast by alleviating steric hindrance and optimizing the spatial arrangement between the chromophore and surrounding residues.74,222 In IrisFP, a green-to-red photoconvertible and n-rsFP, the mutation M159A, identified through structural studies on photobleaching, prevented irreversible sulfoxidation of Met-159 and thereby improved photoswitching fatigue resistance.223,224
In FPs that utilize external chromophores, photoswitching is governed by chromophore inactivation and subsequent renewal. Thus, introducing mutations at the chromophore-binding site can tune covalent and non-covalent binding modes as well as chromophore affinity, directly influencing photoswitching dynamics. For example, in the near-infrared FP miRFP720, derived from RpBphP2, fluorescence arises from the covalent attachment of biliverdin to Cys-15. Substitution of this residue with serine (C15S) disrupts covalent linkage, enhances chromophore turnover, and facilitates efficient renewal after photooxidation.101
Notably, avGFP-derived FPs can be rationally engineered for photoswitchability by introducing mutations identified during the development of rsEGFP and rsEGFP2.225 For instance, rsFolder was constructed by incorporating mutations T65A, Q69L, and A163S, together with the monomerizing mutation A206K,226 into superfolderGFP,227 yielding a negative-type photoswitchable variant.228 The E222Q mutation in avGFP has also been investigated as a photoswitch-inducing substitution.229
Recently developed avGFP derivatives such as mGold2s32 and HyperfolderYFP,33 which exhibit remarkable resistance to photobleaching and enhanced chemical stability, respectively, provide promising scaffolds for next-generation rsFPs. Combining these robust backbones with photoswitching-enabling mutations is expected to accelerate the rational development of highly photostable and application-tailored rsFPs.
5.3. Fusion with Enhancer
The photophysical properties of rsFPs can be enhanced not only through amino acid substitutions within the β-barrel structure but also by engineering intermolecular interactions with other proteins that act as allosteric modulators. For instance, heterodimerisation of dimerisation-dependent FPs (ddFPs)230,231 or fusion of GFP with nanobodies232 has been shown to alter the spectral characteristics and brightness of the resulting constructs. These binding interactions induce local structural rearrangements around the GFP chromophore, thereby shifting the equilibrium between the protonated and deprotonated states of the chromophore and modifying its spectroscopic properties.This principle has recently been extended to rsFPs, where introducing protein–protein interactions can modulate photoswitching behaviour. In particular, rsGreen variants—rsFPs derived from EGFP—exhibited reduced photoswitching fatigue, faster OFF-switching kinetics, and higher ON/OFF contrast when fused with a GFP nanobody, compared to their unbound counterparts.233 Conversely, fusion of a GFP nanobody to rsEGFP resulted in slower OFF-switching kinetics, which was exploited for temporal multiplex imaging, allowing controlled tuning of switching dynamics to match specific imaging timescales.165
6. Conclusions and perspective
We have reviewed the principal characteristics of GFP-like and fluorogen-activating-based rsFPs, including their photoswitching modes, underlying mechanisms, and key photophysical parameters such as switching kinetics, ON/OFF contrast, and fatigue resistance. These properties have been leveraged across a broad range of measurement modalities, particularly those exploiting two-state fluorophore transitions, such as super-resolution (SR) imaging. In addition, we have summarized the engineering of rsFPs aimed at advancing these applications.In imaging techniques based on transitions between the ground (S0) and excited (S1) states, substituting these processes with the ON/OFF transitions of rsFPs circumvents the intrinsic temporal constraints of conventional approaches. This advantage arises because the lifetimes of the ON and OFF states—both ground states—are several orders of magnitude longer than that of the excited state. Likewise, in modalities that rely on transitions between pre- and post-photobleaching states, replacing these irreversible processes with reversible photoswitching enables repeated measurements. Notable examples include: (i) the reduction of illumination power in RESOLFT imaging compared to STED microscopy by substituting the depletion beam with photoswitching light; (ii) overcoming molecular-size limitations in time-resolved fluorescence anisotropy via STARSS; and (iii) repeated quantification of molecular mobility using FDAP. Collectively, these examples demonstrate how rsFP-based state transitions can significantly extend both the temporal and functional reach of fluorescence imaging.
The integration of rsFPs into diverse modalities has also led to the emergence of hybrid imaging techniques, such as functional super-resolution (fSR) microscopy, which visualizes physiological activities at nanoscale resolution. Continued integration of rsFPs with complementary imaging approaches is expected to drive further innovation. For instance, the modulation of rsFP signals can be combined to enhance image contrast in photoacoustic imaging.234 Additionally, combining polarisation imaging with rsFP-based SR techniques enables narrowing of the excitation polarisation angle, thereby significantly enhancing polarisation contrast and signal-to-noise ratio.127,235 Furthermore, exploiting fluorescence signal separation of rsFPs offers promising strategies for expanding the diversity of fluorescent barcodes, while novel biosensing methods such as bioluminescence-assisted switching and fluorescence imaging (BASFI) provide new opportunities to probe protein–protein interactions.236
The performance of rsFPs—in terms of switching speed, ON/OFF contrast, brightness, and fatigue resistance—has been greatly advanced through directed evolution and structure-guided rational design. Nevertheless, the current palette of rsFPs still lacks diversity in both spectral coverage and photoswitching kinetics. While numerous green n-rsFPs have been developed, other spectral classes and switching types remain underrepresented, limiting applications that demand multiplexing or specific switching dynamics. Although near-infrared fluorescent negative rsFPs are being developed and applied to SR imaging,101,237 blue rsFPs with photoswitching performance sufficient for practical applications have yet to be achieved. For emerging imaging modalities, it is increasingly necessary to establish comprehensive rsFP libraries spanning a wide range of parameters, including excitation/emission spectra, switching wavelengths, and kinetics.
In addition to diversification of emission wavelengths and photoswitching characteristics, improvements in brightness and resistance to photoswitching fatigue remain strongly demanded for applications. To date, directed evolution approaches, including the screening of random mutagenesis libraries, have made substantial contributions to the development of rsFPs. However, given the vast protein sequence space, such approaches are inherently labor-intensive and time-consuming. Consequently, rational design driven by three-dimensional structural analyses, spectroscopic analyses, and computational simulations is an attractive strategy. Moreover, understanding photoswitching mechanisms through these analyses not only facilitates rsFP engineering but also inspires new applications and alternative strategies for improving rsFP performance beyond protein engineering. For example, ultrafast spectroscopic studies have suggested that the photoswitching process of rsFPs is not a simple two-state transition between ON and OFF states, but instead involves multiple intermediates in the excited states.64 In addition, cryo-crystallographic analyses have indicated the existence of photoswitching mechanisms that do not rely on cis–trans isomerisation.76 These insights may enable novel photoswitching schemes in rsFPs that require more than two photons, for example, sequential transitions such as OFF1 → OFF2 → ON state. Such multi-step switching processes in rsFPs could contribute to further improvements in spatial resolution in SR imaging.238 Furthermore, applications employing light irradiation for triplet states in the excited state have been reported to suppress photoswitching fatigue.239 These clearly highlight how deeper mechanistic understanding of photophysical behaviour is driving the development of novel applications and approaches for controlling rsFP.
In addition to the rational design, artificial intelligence (AI)-driven protein design have opened powerful new directions for rsFP development.240 Notably, large-scale protein language models trained on evolutionary sequence data (e.g., ESM-3) have successfully generated artificial FPs.241 In parallel, de novo, physics- and structure-based design approaches have achieved the rational creation of FAPs with β-barrel scaffolds capable of binding small-molecule fluorogens such as DFHBI.242 These methods are expected to complement conventional directed evolution and rational design by:
(i) rapidly generating functional candidates from vast sequence spaces;
(ii) integrating structural and physicochemical constraints to propose sequences optimized for specific photophysical properties (e.g., colour, brightness, kinetics, and stability); and
(iii) accelerating the experimental–computational design cycle.
By bridging AI-driven sequence generation with experimental screening, these emerging approaches are poised to close the existing gaps in rsFP diversity—particularly in excitation, emission, and photoswitching spectra—thereby accelerating the discovery of high-performance, next-generation rsFP probes for advanced bioimaging.
Author contributions
Ryohei Ozaki-Noma: conceptualisation (equal), visualisation (lead), writing – original draft (lead). Tetsuichi Wazawa: conceptualisation (supporting), writing – original draft (supporting). Kai Lu: writing – original draft (supporting). Tomoki Matsuda: conceptualisation (supporting), writing – original draft (supporting). Takeharu Nagai: conceptualisation (equal), writing – review and editing (lead).Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Acknowledgements
This work was in part supported by grants from Japan Science and Technology Agency (JPMJCR15N3 and JPMJTR254C to T. N.; JPMJCR20E4 to T. M.), Japan Society for the Promotion of Science (23115003, 18H05410, 21K19225, 23H05421, and 25H02457 to T. N.; 19K05226 and 22K04891 to T. W.; 23KJ1492 to R. O. N.), The Naito Foundation (to T. N.), The Sumitomo Foundation (to T. N.), and the Research Program of “Dynamic Alliance for Open Innovation Bridging Human, Environment (to T. N.).References
- O. Shimomura, F. H. Johnson and Y. Saiga, J. Cell. Comp. Physiol., 1962, 59, 223–239 CrossRef CAS.
- F. H. Johnson, O. Shimomura, Y. Saiga, L. C. Gershman, G. T. Reynolds and J. R. Waters, J. Cell. Comp. Physiol., 1962, 60, 85–103 CrossRef CAS.
- D. C. Prasher, V. K. Eckenrode, W. W. Ward, F. G. Prendergast and M. J. Cormier, Gene, 1992, 111, 229–233 CrossRef CAS PubMed.
- M. Chalfie, Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, Science, 1994, 263, 802–805 CrossRef CAS PubMed.
- K. Nienhaus and G. U. Nienhaus, J. Phys.: Condens. Matter, 2016, 28, 443001 CrossRef.
- M. Wang, Y. Da and Y. Tian, Chem. Soc. Rev., 2023, 52, 1189 RSC.
- E. C. Greenwald, S. Mehta and J. Zhang, Chem. Rev., 2018, 118, 11707–11794 CrossRef CAS.
- T. Wazawa, R. Ozaki-Noma, K. Lu, S. Fukushima, T. Matsuda and T. Nagai, Biophys. Physicobiol., 2025, 22, e220008 CAS.
- E. A. Rodriguez, R. E. Campbell, J. Y. Lin, M. Z. Lin, A. Miyawaki, A. E. Palmer, X. Shu, J. Zhang and R. Y. Tsien, Trends Biochem. Sci., 2017, 42, 111–129 CrossRef CAS PubMed.
- S. W. Hell, Science, 2007, 316, 1153–1158 CrossRef CAS PubMed.
- H. Niwa, S. Inouye, T. Hirano, T. Matsuno, S. Kojima, M. Kubota, M. Ohashi and F. I. TsujiNiwa, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13617–13622 CrossRef CAS.
- J. Goedhart, D. von Stetten, M. Noirclerc-Savoye, M. Lelimousin, L. Joosen, M. A. Hink, L. van Weeren, T. W. J. Gadella Jr and A. Royant, Nat. Commun., 2012, 3, 751 CrossRef.
- B. P. Cormack, R. H. Valdivia and S. Falkow, Gene, 1996, 173, 33–38 CrossRef CAS.
- N. C. Shaner, R. E. Campbell, P. A. Steinbach, B. N. G. Giepmans, A. E. Palmer and R. Y. Tsien, Nat. Biotechnol., 2004, 22, 1567–1572 CrossRef CAS.
- A. Royant and M. Noirclerc-Savoye, J. Struct. Biol., 2011, 174, 385–390 CrossRef CAS PubMed.
- M. G. Romei, C.-Y. Lin and S. G. Boxer, J. Photochem. Photobiol., A, 2020, 401, 112738 CrossRef CAS PubMed.
- A. Kumagai, R. Ando, H. Miyatake, P. Greimel, T. Kobayashi, Y. Hirabayashi, T. Shimogori and A. Miyawaki, Cell, 2013, 153, 1602–1611 CrossRef CAS PubMed.
- M. Baloban, D. M. Shcherbakova, S. Pletnev, V. Z. Pletnev, J. C. Lagarias and V. V. Verkhusha, Chem. Sci., 2017, 8, 4546–4557 RSC.
- R. Heim, D. C. Prasher and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 12501–12504 CrossRef CAS PubMed.
- W. Tomosugi, T. Matsuda, T. Tani, T. Nemoto, I. Kotera, K. Saito, K. Horikawa and T. Nagai, Nat. Methods, 2009, 6, 351–353 CrossRef PubMed.
- K. Sugiura and T. Nagai, Commun. Biol., 2022, 5, 1172 CrossRef PubMed.
- M. Ormö, A. B. Cubitt, K. Kallio, L. A. Gross, R. Y. Tsien and S. J. Remington, Science, 1996, 273, 1392–1395 CrossRef PubMed.
- M. Z. Lin, M. R. McKeown, H.-L. Ng, T. A. Aguilera, N. C. Shaner, R. E. Campbell, S. R. Adams, L. A. Gross, W. Ma, T. Alber and R. Y. Tsien, Chem. Biol., 2009, 16, 1169–1179 CrossRef.
- A. Miyawaki, D. M. Shcherbakova and V. V. Verkhusha, Curr. Opin. Struct. Biol., 2012, 22, 679–688 CrossRef PubMed.
- B. C. Campbell, E. M. Nabel, M. H. Murdock, C. Lao-Peregrin, P. Tsoulfas, M. G. Blackmore, F. S. Lee, C. Liston, H. Morishita and G. A. Petsko, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 30710–30721 CrossRef.
- T. W. J. Gadella Jr, L. van Weeren, J. Stouthamer, M. A. Hink, A. H. G. Wolters, B. N. G. Giepmans, S. Aumonier, J. Dupuy and A. Royant, Nat. Methods, 2023, 20, 541–545 CrossRef PubMed.
- M. Hirano, R. Ando, S. Shimozono, M. Sugiyama, N. Takeda, H. Kurokawa, R. Deguchi, K. Endo, K. Haga, R. Takai-Todaka, S. Inaura, Y. Matsumura, H. Hama, Y. Okada, T. Fujiwara, T. Morimoto, K. Katayama and A. Miyawaki, Nat. Biotechnol., 2022, 40, 1132–1142 CrossRef PubMed.
- H. Zhang, G. D. Lesnov, O. M. Subach, W. Zhang, T. P. Kuzmicheva, A. V. Vlaskina, V. R. Samygina, L. Chen, X. Ye, A. Y. Nikolaeva, A. Gabdulkhakov, S. Papadaki, W. Qin, V. Borshchevskiy, M. M. Perfilov, A. S. Gavrikov, M. Drobizhev, A. S. Mishin, K. D. Piatkevich and F. V. Subach, Nat. Methods, 2024, 21, 657–665 CrossRef PubMed.
- E. Ivorra-Molla, D. Akhuli, M. B. L. McAndrew, W. Scott, L. Kumar, S. Palani, M. Mishima, A. Crow and M. K. Balasubramanian, Nat. Biotechnol., 2024, 42, 1368–1371 CrossRef.
- R. Ando, S. Shimozono, H. Ago, M. Takagi, M. Sugiyama, H. Kurokawa, M. Hirano, Y. Niino, G. Ueno, F. Ishidate, T. Fujiwara, Y. Okada, M. Yamamoto and A. Miyawaki, Nat. Methods, 2024, 21, 648–656 CrossRef PubMed.
- H. Xiong, Q. Chang, J. Ding, S. Wang, W. Zhang, Y. Li, Y. Wu, P. Lin, C. Yang, M. Liu, G. Fang, Y. Yang, J. Xie, D. Qi, T. Jiang, W. Fu, F. Hu, Y. Chen, R. Yue, Y. Li, Y. Cui, M. Li, S. Fan, Y. Yang, Y. Xu, D. Li, F. Zhang, H. Zhao, C. Wu, Q. Zheng, K. D. Piatkevich and Z. Fu, Nat. Methods, 2025, 22, 1288–1298 CrossRef.
- J. Lee, S. Lai, S. Yang, S. Zhao, F. A. Blanco, A. C. Lyons, R. Merino-Urteaga, J. F. Ahrens, N. A. Nguyen, H. Liu, Z. Liu, G. G. Lambert, N. C. Shaner, L. Chen, K. F. Tolias, J. Zhang, T. Ha and F. St-Pierre, Nat. Commun., 2025, 16, 3241 CrossRef PubMed.
- B. C. Campbell, M. G. Paez-Segala, L. L. Looger, G. A. Petsko and C. F. Liu, Nat. Methods, 2022, 19, 1612–1621 CrossRef PubMed.
- M. G. Paez-Segala, M. G. Sun, G. Shtengel, S. Viswanathan, M. A. Baird, J. J. Macklin, R. Patel, J. R. Allen, E. S. Howe, G. Piszczek, H. F. Hess, M. W. Davidson, Y. Wang and L. L. Looger, Nat. Methods, 2015, 12, 215–218 CrossRef PubMed.
- B. A. Griffin, S. R. Adams and R. Y. Tsien, Science, 1998, 281, 269–272 CrossRef PubMed.
- C. Szent-Gyorgyi, B. F. Schmidt, Y. Creeger, G. W. Fisher, K. L. Zakel, S. Adler, J. A. J. Fitzpatrick, C. A. Woolford, Q. Yan, K. V. Vasilev, P. B. Berget, M. P. Bruchez, J. W. Jarvik and A. Waggoner, Nat. Biotechnol., 2008, 26, 235–240 CrossRef PubMed.
- M.-A. Plamont, E. Billon-Denis, S. Maurin, C. Gauron, F. M. Pimenta, C. G. Specht, J. Shi, J. Quérard, B. Pan, J. Rossignol, K. Moncoq, N. Morellet, M. Volovitch, E. Lescop, Y. Chen, A. Triller, S. Vriz, T. Le Saux, L. Jullien and A. Gautier, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 497–502 CrossRef PubMed.
- A. M. Buckley, J. Petersen, A. J. Roe, G. R. Douce and J. M. Christie, Curr. Opin. Chem. Biol., 2015, 27, 39–45 CrossRef PubMed.
- D. M. Shcherbakova, M. Baloban, A. V. Emelyanov, M. Brenowitz, P. Guo and V. V. Verkhusha, Nat. Commun., 2016, 7, 12405 CrossRef PubMed.
- E. A. Rodriguez, G. N. Tran, L. A. Gross, J. L. Crisp, X. Shu, J. Y. Lin and R. Y. Tsien, Nat. Methods, 2016, 13, 763–769 CrossRef PubMed.
- O. S. Oliinyk, A. A. Shemetov, S. Pletnev, D. M. Shcherbakova and V. V. Verkhusha, Nat. Commun., 2019, 10, 279 CrossRef PubMed.
- G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina, C. Zimprich, M. G. Wood, R. Learish, R. F. Ohana, M. Urh, D. Simpson, J. Mendez, K. Zimmerman, P. Otto, G. Vidugiris, J. Zhu, A. Darzins, D. H. Klaubert, R. F. Bulleit and K. V. Wood, ACS Chem. Biol., 2008, 3, 373–382 CrossRef PubMed.
- A. Keppler, S. Gendreizig, T. Gronemeyer, H. Pick, H. Vogel and K. Johnsson, Nat. Biotechnol., 2003, 21, 86–89 CrossRef.
- A. Gautier, A. Juillerat, C. Heinis, I. R. Corrêa Jr, M. Kindermann, F. Beaufils and K. Johnsson, Chem. Biol., 2008, 15, 128–136 CrossRef.
- S. R. Adams, R. E. Campbell, L. A. Gross, B. R. Martin, G. K. Walkup, Y. Yao, J. Llopis and R. Y. Tsien, J. Am. Chem. Soc., 2002, 124, 6063–6076 CrossRef.
- E. E. Rastede, M. Tanha, D. Yaron, S. C. Watkins, A. S. Waggoner and B. A. Armitage, Photochem. Photobiol. Sci., 2015, 14, 1703–1712 CrossRef PubMed.
- N. I. Shank, K. J. Zanotti, F. Lanni, P. B. Berget and B. A. Armitage, J. Am. Chem. Soc., 2009, 131, 12960–12969 CrossRef.
- A. G. Tebo, B. Moeyaert, M. Thauvin, I. Carlon-Andres, D. Böken, M. Volovitch, S. Padilla-Parra, P. Dedecker, S. Vriz and A. Gautier, Nat. Chem. Biol., 2021, 17, 30–38 CrossRef PubMed.
- H. Benaissa, K. Ounoughi, I. Aujard, E. Fischer, R. Goïame, J. Nguyen, A. G. Tebo, C. Li, T. Le Saux, G. Bertolin, M. Tramier, L. Danglot, N. Pietrancosta, X. Morin, L. Jullien and A. Gautier, Nat. Commun., 2021, 12, 6989 CrossRef PubMed.
- M. Venkatachalapathy, V. Belapurkar, M. Jose, A. Gautier and D. Nair, Nanoscale, 2019, 11, 3626–3632 RSC.
- S. Ko, J. Kwon and S.-H. Shim, Front. Mol. Biosci., 2021, 8, 647590 CrossRef PubMed.
- J. Zahradník, D. Dey, S. Marciano, L. Kolářová, C. I. Charendoff, A. Subtil and G. Schreiber, ACS Synth. Biol., 2021, 10, 3445–3460 CrossRef.
- Y. Itoh, M. Hattori, T. Wazawa, Y. Arai and T. Nagai, ACS Sens., 2021, 6, 889–895 CrossRef PubMed.
- A. Losi, E. Polverini, B. Quest and W. Gärtner, Biophys. J., 2002, 82, 2627–2634 CrossRef.
- U. Krauss, A. Losi, W. Gärtner, K.-E. Jaeger and T. Eggert, Phys. Chem. Chem. Phys., 2005, 7, 2804–2811 RSC.
- S. Chapman, C. Faulkner, E. Kaiserli, C. Garcia-Mata, E. I. Savenkov, A. G. Roberts, K. J. Oparka and J. M. Christie, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 20038–20043 CrossRef CAS.
- J. M. Christie, K. Hitomi, A. S. Arvai, K. A. Hartfield, M. Mettlen, A. J. Pratt, J. A. Tainer and E. D. Getzoff, J. Biol. Chem., 2012, 287, 22295–22304 CrossRef CAS.
- S. Babakhanova, E. E. Jung, K. Namikawa, H. Zhang, Y. Wang, O. M. Subach, D. A. Korzhenevskiy, T. V. Rakitina, X. Xiao, W. Wang, J. Shi, M. Drobizhev, D. Park, L. Eisenhard, H. Tang, R. W. Köster, F. V. Subach, E. S. Boyden and K. D. Piatkevich, Protein Sci., 2022, 31, 728–751 CrossRef CAS.
- T. Drepper, T. Eggert, F. Circolone, A. Heck, U. Krauss, J.-K. Guterl, M. Wendorff, A. Losi, W. Gärtner and K.-E. Jaeger, Nat. Biotechnol., 2007, 25, 443–445 CrossRef CAS PubMed.
- D. M. Shcherbakova and V. V. Verkhusha, Nat. Methods, 2013, 10, 751–754 CrossRef CAS.
- D. M. Shcherbakova, N. C. Cammer, T. M. Huisman, V. V. Verkhusha and L. Hodgson, Nat. Chem. Biol., 2018, 14, 591–600 CrossRef CAS.
- O. S. Oliinyk, A. A. Shemetov, S. Pletnev, D. M. Shcherbakova and V. V. Verkhusha, Nat. Commun., 2019, 10, 279 CrossRef PubMed.
- O. S. Oliinyk, M. Baloban, C. L. Clark, E. Carey, S. Pletnev, A. Nimmerjahn and V. V. Verkhusha, Nat. Methods, 2022, 19, 740–750 Search PubMed.
- L. Tang and C. Fang, Int. J. Mol. Sci., 2022, 23, 6459 CrossRef CAS PubMed.
- R. Ando, H. Mizuno and A. Miyawaki, Science, 2004, 306, 1370–1373 Search PubMed.
- T. Grotjohann, I. Testa, M. Leutenegger, H. Bock, N. T. Urban, F. Lavoie-Cardinal, K. I. Willig, C. Eggeling, S. Jakobs and S. W. Hell, Nature, 2011, 478, 204–208 CrossRef CAS.
- M. Andresen, A. C. Stiel, S. Trowitzsch, G. Weber, C. Eggeling, M. C. Wahl, S. W. Hell and S. Jakobs, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 13005–13009 CrossRef CAS.
- H. Shinoda, K. Lu, R. Nakashima, T. Wazawa, K. Noguchi, T. Matsuda and T. Nagai, Cell Chem. Biol., 2019, 26, 1469–1479 CrossRef CAS PubMed.
- H. Mizuno, T. K. Mal, M. Wälchli, A. Kikuchi, T. Fukano, R. Ando, J. Jeyakanthan, J. Taka, Y. Shiro, M. Ikura and A. Miyawaki, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 9227–9232 CrossRef CAS.
- N.-E. Christou, I. Ayala, K. Giandoreggio-Barranco, M. Byrdin, V. Adam, D. Bourgeois and B. Brutscher, Biophys. J., 2019, 117, 2087–2100 CrossRef CAS PubMed.
- J. Woodhouse, G. N. Kovacs, N. Coquelle, L. M. Uriarte, V. Adam, T. R. M. Barends, M. Byrdin, E. de la Mora, R. B. Doak, M. Feliks, M. Field, F. Fieschi, V. Guillon, S. Jakobs, Y. Joti, P. Macheboeuf, K. Motomura, K. Nass, S. Owada, C. M. Roome, C. Ruckebusch, G. Schirò, R. L. Shoeman, M. Thepaut, T. Togashi, K. Tono, M. Yabashi, M. Cammarata, L. Foucar, D. Bourgeois, M. Sliwa, J.-P. Colletier, I. Schlichting and M. Weik, Nat. Commun., 2020, 11, 741 Search PubMed.
- Y.-H. Zhu, X.-X. Liu, Q. Fang, X.-Y. Liu, W.-H. Fang and G. Cui, J. Phys. Chem. Lett., 2023, 14, 2588–2598 CrossRef CAS PubMed.
- A. Fadini, C. D. M. Hutchison, D. Morozov, J. Chang, K. Maghlaoui, S. Perrett, F. Luo, J. C. X. Kho, M. G. Romei, R. M. L. Morgan, C. M. Orr, V. Cordon-Preciado, T. Fujiwara, N. Nuemket, T. Tosha, R. Tanaka, S. Owada, K. Tono, S. Iwata, S. G. Boxer, G. Groenhof, E. Nango and J. J. van Thor, J. Am. Chem. Soc., 2023, 145, 15796–15808 CrossRef CAS.
- N. Coquelle, M. Sliwa, J. Woodhouse, G. Schirò, V. Adam, A. Aquila, T. R. M. Barends, S. Boutet, M. Byrdin, S. Carbajo, E. De la Mora, R. B. Doak, M. Feliks, F. Fieschi, L. Foucar, V. Guillon, M. Hilpert, M. S. Hunter, S. Jakobs, J. E. Koglin, G. Kovacsova, T. J. Lane, B. Lévy, M. Liang, K. Nass, J. Ridard, J. S. Robinson, C. M. Roome, C. Ruckebusch, M. Seaberg, M. Thepaut, M. Cammarata, I. Demachy, M. Field, R. L. Shoeman, D. Bourgeois, J.-P. Colletier, I. Schlichting and M. Weik, Nat. Chem., 2018, 10, 31–37 CrossRef CAS PubMed.
- J. Chang, M. G. Romei and S. G. Boxer, J. Am. Chem. Soc., 2019, 141, 15504–15508 Search PubMed.
- A. M. R. Mantovanelli, O. Glushonkov, V. Adam, J. Wulffelé, D. Thédié, M. Byrdin, I. Gregor, O. Nevskyi, J. Enderlein and D. Bourgeois, J. Am. Chem. Soc., 2023, 145, 14636–14646 Search PubMed.
- M. Andresen, A. C. Stiel, J. Fölling, D. Wenzel, A. Schönle, A. Egner, C. Eggeling, S. W. Hell and S. Jakobs, et al., Nat. Biotechnol., 2008, 26, 1035–1040 CrossRef CAS PubMed.
- D. K. Tiwari, Y. Arai, M. Yamanaka, T. Matsuda, M. Agetsuma, M. Nakano, K. Fujita and T. Nagai, Nat. Methods, 2015, 12, 515–518 CrossRef CAS PubMed.
- A. Walter, M. Andresen, S. Jakobs, J. Schroeder and D. Schwarzer, Phys. Chem. B, 2015, 119, 5136–5144 CrossRef CAS PubMed.
- E. Fron, M. Van der Auweraer, J. Hofkens and P. Dedecker, J. Phys. Chem. B, 2013, 117, 16422–16427 CrossRef CAS.
- T. Brakemann, G. Weber, M. Andresen, G. Groenhof, A. C. Stiel, S. Trowitzsch, C. Eggeling, H. Grubmüller, S. W. Hell, M. C. Wahl and S. Jakobs, J. Biol. Chem., 2010, 285, 14603–14609 CrossRef CAS.
- A. R. Faro, P. Carpentier, G. Jonasson, G. Pompidor, D. Arcizet, I. Demachy and D. Bourgeois, J. Am. Chem. Soc., 2011, 133, 16362–16365 CrossRef PubMed.
- T. Konen, D. Stumpf, T. Grotjohann, I. Jansen, M. Bossi, M. Weber, N. Jensen, S. W. Hell and S. Jakobs, ACS Nano, 2021, 15, 9509–9521 CrossRef CAS.
- R. Ozaki-Noma, T. Wazawa, T. Kakizuka, H. Shidara, K. Takemoto and T. Nagai, ACS Nano, 2025, 19, 7188–7201 Search PubMed.
- G. J. Palm and A. Wlodawer, Methods Enzymol., 1999, 302, 378–394 CAS.
- S. Gayda, K. Nienhaus and G. U. Nienhaus, Biophys. J., 2012, 103, 2521–2531 Search PubMed.
- R. Bizzarri, R. Nifosì, S. Abbruzzetti, W. Rocchia, S. Guidi, D. Arosio, G. Garau, B. Campanini, E. Grandi, F. Ricci, C. Viappiani and F. Beltram, Biochemistry, 2007, 46, 5494–5504 CrossRef CAS PubMed.
- T. Wazawa, R. Noma, S. Uto, K. Sugiura, T. Washio and T. Nagai, Microscopy, 2021, 70, 340–352 CrossRef CAS.
- A. C. Bourges, B. Moeyaert, T. Y. H. Bui, F. Bierbuesse, W. Vandenberg and P. Dedecker, Chem. Commun., 2023, 59, 8810–8813 RSC.
- A. Fatima, Y. He, J. N. Iuliano, G. M. Greetham, P. Malakar, C. Hall, H. A. Woroniecka, B. C. Richardson, J. B. French, A. Lukacs, P. J. Tonge and S. R. Meech, Chem. Sci., 2025, 16, 16955–16969 Search PubMed.
- Y. Li and R. W. Tsien, Nat. Neurosci., 2012, 15, 1047–1053 CrossRef PubMed.
- Y. Shen, M. Rosendale, R. E. Campbell and D. Perrais, J. Cell Biol., 2014, 207, 419–432 CrossRef PubMed.
- H. Masuda, Y. Takenaka, A. Yamaguchi, S. Nishikawa and H. Mizuno, Gene, 2006, 372, 18–25 CrossRef PubMed.
- H. Shinoda, Y. Ma, R. Nakashima, K. Sakurai, T. Matsuda and T. Nagai, Cell Chem. Biol., 2018, 25, 330–338 CrossRef PubMed.
- H. Shidara, T. Shirai, R. Ozaki-Noma, S. Jitsuki, T. Nagai and K. Takemoto, Commun. Biol., 2024, 7, 945 CrossRef PubMed.
- T. Brakemann, A. C. Stiel, G. Weber, M. Andresen, I. Testa, T. Grotjohann, M. Leutenegger, U. Plessmann, H. Urlaub, C. Eggeling, M. C. Wahl, S. W. Hell and S. Jakobs, Nat. Biotechnol., 2011, 29, 942–947 CrossRef PubMed.
- Y. Arai, H. Takauchi, Y. Ogami, S. Fujiwara, M. Nakano, T. Matsuda and T. Nagai, ACS Chem. Biol., 2018, 13, 1938–1943 CrossRef PubMed.
- F. Lacombat, P. Plaza, M.-A. Plamont and A. Espagne, J. Phys. Chem. Lett., 2017, 8, 1489–1495 CrossRef PubMed.
- B. L. Grigorenko, I. V. Polyakov, A. I. Krylov and A. V. Nemukhin, J. Phys. Chem. B, 2019, 123, 8901–8909 CrossRef PubMed.
- H.-w. Ai, N. C. Shaner, Z. Cheng, R. Y. Tsien and R. E. Campbell, Biochemistry, 2007, 46, 5904–5910 CrossRef PubMed.
- K. Lu, T. Wazawa, T. Matsuda, D. M. Shcherbakova, V. V. Verkhusha and T. Nagai, Commun. Biol., 2024, 7, 473 CrossRef PubMed.
- J.-H. Machado, R. Ting, J. Y. Lin and E. A. Rodriguez, RSC Chem. Biol., 2021, 2, 1221–1226 RSC.
- J. Kwon, J.-S. Park, M. Kang, S. Choi, J. Park, G. T. Kim, C. Lee, S. Cha, H.-W. Rhee and S.-H. Shim, Nat. Commun., 2020, 11, 273 CrossRef PubMed.
- E. Lee, S.-H. Shim and M. Cho, Chem. Sci., 2018, 9, 8325–8336 Search PubMed.
- J. Schnitzbauer, M. T. Strauss, T. Schlichthaerle, F. Schueder and R. Jungmann, Nat. Protoc., 2017, 12, 1198–1228 CrossRef PubMed.
- A. Losi, W. Gärtner, S. Raffelberg, F. C. Zanacchi, P. Bianchini, A. Diaspro, C. Mandalari, S. Abbruzzettia and C. Viappiani, Photochem. Photobiol. Sci., 2013, 12, 231–235 CrossRef PubMed.
- C. Gregor, S. C. Sidenstein, M. Andresen, S. J. Sahl, J. G. Danzl and S. W. Hell, Sci. Rep., 2018, 8, 2724 Search PubMed.
- F. M. Pimenta, G. Chiappetta, T. Le Saux, J. Vinh, L. Jullien and A. Gautier, Sci. Rep., 2017, 7, 12316 CrossRef PubMed.
- J. Kwon, M. S. Elgawish and S.-H. Shim, Adv. Sci., 2022, 9, e2101817 CrossRef.
- S. Ghosh, C.-L. Yu, D. J. Ferraro, S. Sudha, S. K. Pal, W. F. Schaefer, D. T. Gibson and S. Ramaswamy, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 11513–11518 CrossRef PubMed.
- K. Y. Manoilov, A. Ghosh, S. C. Almo and V. V. Verkhusha, J. Mol. Biol., 2022, 434, 167359 CrossRef PubMed.
- R. Nifosì, B. Storti and R. Bizzarri, Riv. Nuovo Cimento, 2024, 47, 91–178 CrossRef.
- N. A. Jensen, I. Jansen, M. Kamper and S. Jakobs, Nanoscale Photonic Imaging. Topics in Applied Physics, 2020, vol. 134, pp. 241–261 Search PubMed.
- A. Volpato, D. Ollech, J. Alvelid, M. Damenti, B. Müller, A. G. York, M. Ingaramo and I. Testa, Nat. Biotechnol., 2023, 41, 552–559 Search PubMed.
- V. Adam, H. Mizuno, A. Grichine, J. Hotta, Y. Yamagata, B. Moeyaert, G. U. Nienhaus, A. Miyawaki, D. Bourgeois and J. Hofkens, J. Biotechnol., 2010, 149, 289–298 Search PubMed.
- X. X. Zhou, H. K. Chung, A. J. Lam and M. Z. Lin, Science, 2012, 338, 8140–8814 Search PubMed.
- X. X. Zhou, L. Z. Fan, P. Li, K. Shen and M. Z. Lin, Science, 2017, 355, 836–842 CrossRef.
- S. J. Sahl, S. W. Hell and S. Jakobs, Nat. Rev. Mol. Cell Biol., 2017, 18, 685–701 CrossRef.
- I. M. Khater, I. R. Nabi and G. Hamarneh, Patterns, 2020, 1, 100038 CrossRef.
- T. Dertinger, R. Colyer, G. Iyer, S. Weiss and J. Enderlein, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 22287–22292 Search PubMed.
- S. W. Hell and J. Wichmann, Opt. Lett., 1994, 19, 780–782 Search PubMed.
- M. G. L. Gustafsson, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 13081–13086 Search PubMed.
- N. Hafi, M. Grunwald, L. S. van den Heuvel, T. Aspelmeier, J.-H. Chen, M. Zagrebelsky, O. M. Schütte, C. Steinem, M. Korte, A. Munk and P. J. Walla, Nat. Methods, 2014, 11, 579–584 CrossRef.
- F. Balzarotti, Y. Eilers, K. C. Gwosch, A. H. Gynnå, V. Westphal, F. D. Stefani, J. Elf and S. W. Hell, Science, 2017, 355, 606–612 Search PubMed.
- M. Weber, M. Leutenegger, S. Stoldt, S. Jakobs, T. S. Mihaila, A. N. Butkevich and S. W. Hell, Nat. Photonics, 2021, 15, 361–366 CrossRef.
- H.-W. Lu-Walther, W. Hou, M. Kielhorn, Y. Arai, T. Nagai, M. M. Kessels, B. Qualmann and R. Heintzmann, PLoS One, 2016, 11, e0165148 CrossRef PubMed.
- T. Wazawa, Y. Arai, Y. Kawahara, H. Takauchi, T. Washio and T. Nagai, Microscopy, 2018, 67, 89–98 CrossRef.
- T. Wazawa, T. Washio and T. Nakai, Neuromethods, 2020, 154, 229–244 CrossRef.
- G. Miesenböck, D. A. De Angelis and J. E. Rothman, Nature, 1998, 394, 192–195 CrossRef.
- K. Lu, T. Wazawa, J. Sakamoto, C. Q. Vu, M. Nakano, Y. Kamei and T. Nagai, Nano Lett., 2022, 22, 5698–5707 CrossRef.
- H. Wallrabe and A. Periasamy, Curr. Opin. Biotechnol., 2005, 16, 19–27 CrossRef PubMed.
- C. Zhong, S. Arai and Y. Okada, Cells Rep. Methods, 2024, 4, 100902 CrossRef.
- K. H. Rainey and G. H. Patterson, Proc. Natl. Acad. Sci. U. S. A., 2018, 116, 864–873 CrossRef.
- F. V. Subach, L. Zhang, T. W. J. Gadella, N. G. Gurskaya, K. A. Lukyanov and V. V. Verkhusha, Chem. Biol., 2010, 17, 745–755 CrossRef PubMed.
- S. Mao, R. K. P. Benninger, Y. Yan, C. Petchprayoon, D. Jackson, C. J. Easley, D. W. Piston and G. Marriott, Biophys. J., 2008, 94, 4515–4524 CrossRef.
- B. Adelizzi, V. Gielen, T. Le Saux, P. Dedecker and L. Jullien, ACS Sens., 2021, 6, 1157–1165 CrossRef PubMed.
- F. Bierbuesse, A. C. Bourges, V. Gielen, V. Mönkemöller, W. Vandenberg, Y. Shen, J. Hofkens, P. V. Berghe, R. E. Campbell, B. Moeyaert and P. Dedecker, Nat. Commun., 2022, 13, 1850 CrossRef.
- Y.-T. Kao, X. Zhu and W. Min, Proc. Natl. Acad. Sci. U. S. A., 2011, 109, 3220–3225 CrossRef.
- J. E. Aubin, J. Histochem. Cytochem., 1979, 27, 36–43 Search PubMed.
- G. Marriott, S. Mao, T. Sakata, J. Ran, D. K. Jackson, C. Petchprayoon, T. J. Gomez, E. Warp, O. Tulyathan, H. L. Aaron, E. Y. Isacoff and Y. Yan, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17789–17794 Search PubMed.
- C. I. Richards, J.-C. Hsiang and R. M. Dickson, J. Phys. Chem. B, 2010, 114, 660–665 CrossRef PubMed.
- J. Querard, T.-Z. Markus, M.-A. Plamont, C. Gauron, P. Wang, A. Espagne, M. Volovitch, S. Vriz, V. Croquette, A. Gautier, T. Le Saux and L. Jullien, Angew. Chem., Int. Ed., 2015, 54, 2633–2637 CrossRef.
- G. Abbandonato, B. Storti, G. Signore, F. Beltram and R. Bizzarri, Microsc. Res. Tech., 2016, 79, 929–937 CrossRef CAS PubMed.
- J.-C. Hsiang, A. E. Jablonski and R. M. Dickson, Acc. Chem. Res., 2014, 47, 1545–1554 CrossRef CAS PubMed.
- A. E. Jablonski, R. B. Vegh, J.-C. Hsiang, B. Bommarius, Y.-C. Chen, K. M. Solntsev, A. S. Bommarius, L. M. Tolbert and R. M. Dickson, J. Am. Chem. Soc., 2013, 135, 16410–16417 Search PubMed.
- J. Quérard, R. Zhang, Z. Kelemen, M.-A. Plamont, X. Xie, R. Chouket, I. Roemgens, Y. Korepina, S. Albright, E. Ipendey, M. Volovitch, H. L. Sladitschek, P. Neveu, L. Gissot, A. Gautier, J.-D. Faure, V. Croquette, T. Le Saux and L. Jullien, Nat. Commun., 2017, 8, 969 Search PubMed.
- R. Chouket, A. Pellissier-Tanon, A. Lemarchand, A. Espagne, T. Le Saux and L. Jullien, Chem. Sci., 2020, 11, 2882–2887 Search PubMed.
- R. Zhang, R. Chouket, M.-A. Plamont, Z. Kelemen, A. Espagne, A. G. Tebo, A. Gautier, L. Gissot, J.-D. Faure, L. Jullien, V. Croquette and T. Le Saux, Light: Sci. Appl., 2018, 7, 97 CrossRef.
- Z. Kelemen, R. Zhang, L. Gissot, R. Chouket, Y. Bellec, V. Croquette, L. Jullien, J.-D. Faure and T. Le Saux, ACS Omega, 2020, 5, 15105–15114 Search PubMed.
- M. Ingaramo, A. G. York, E. J. Andrade, K. Rainey and G. H. Patterson, Nat. Commun., 2015, 6, 8184 CrossRef.
- X. Zhu, Y.-T. Kao and W. Min, J. Phys. Chem. Lett., 2012, 3, 2082–2086 CrossRef CAS.
- Y.-T. Kao, X. Zhu, F. Xu and W. Min, Biomed. Opt. Express, 2012, 3, 1955–1963 Search PubMed.
- T. Kubo, K. Temma, K. Sugiura, H. Shinoda, K. Lu, N. I. Smith, T. Matsuda, T. Nagai and K. Fujita, ACS Photonics, 2021, 8, 2666–2673 Search PubMed.
- K. Temma, R. Oketani, T. Kubo, K. Bando, S. Maeda, K. Sugiura, T. Matsuda, R. Heintzmann, T. Kaminishi, K. Fukuda, M. Hamasaki, T. Nagai and K. Fujita, Nat. Methods, 2024, 21, 889–896 CrossRef CAS PubMed.
- P. Hoyer, G. de Medeiros, B. Balázs, N. Norlin, C. Besir, J. Hanne, H.-G. Kräusslich, J. Engelhardt, S. J. Sahl, S. W. Hell and L. Hufnagel, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 3442–3446 CrossRef CAS.
- A. Bodén, D. Ollech, A. G. York, A. Millett-Sikking and I. Testa, Nat. Methods, 2024, 21, 882–888 CrossRef.
- A. Roldán-Salgado, L. Muslinkina, S. Pletnev, N. Pletneva, V. Pletnev and P. Gaytán, Protein Sci., 2022, 31, 688–700 Search PubMed.
- T. Kogure, S. Karasawa, T. Araki, K. Saito, M. Kinjo and A. Miyawaki, Nat. Biotechnol., 2006, 24, 577–581 Search PubMed.
- H.-w. Ai, K. L. Hazelwood, M. W. Davidson and R. E. Campbell, Nat. Methods, 2008, 5, 401–403 CrossRef CAS PubMed.
- K. D. Piatkevich, J. Hulit, O. M. Subach, B. Wu, A. Abdulla, J. E. Segall and V. V. Verkhusha, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5369–5374 CrossRef CAS.
- J. Seo, Y. Sim, J. Kim, H. Kim, I. Cho, H. Nam, Y.-G. Yoon and J.-B. Chang, Nat. Commun., 2022, 13, 2475 CrossRef CAS PubMed.
- A. E. Jablonski, J.-C. Hsiang, P. Bagchi, N. Hull, C. I. Richards, C. J. Fahrni and R. M. Dickson, J. Phys. Chem. Lett., 2012, 3, 3585–3591 CrossRef CAS PubMed.
- Y.-C. Chen, A. E. Jablonski, I. Issaeva, D. Bourassa, J.-C. Hsiang, C. J. Fahrni and R. M. Dickson, J. Am. Chem. Soc., 2015, 137, 12764–12767 CrossRef CAS PubMed.
- A. Pellissier-Tanon, R. Chouket, R. Zhang, A. Lahlou, A. Espagne, A. Lemarchand, V. Croquette, L. Jullien and T. Le Saux, ChemPhysChem, 2022, 23, e202200295 Search PubMed.
- Y. Qian, O. T. Celiker, Z. Wang, B. Guner-Ataman and E. S. Boyden, Cell, 2023, 186, 5656–5672 CrossRef CAS.
- R. Chouket, A. Pellissier-Tanon, A. Lahlou, R. Zhang, D. Kim, M.-A. Plamont, M. Zhang, X. Zhang, P. Xu, N. Desprat, D. Bourgeois, A. Espagne, A. Lemarchand, T. Le Saux and L. Jullien, Nat. Commun., 2022, 13, 1482 CrossRef CAS.
- H. Valenta, S. Hugelier, S. Duwé, G. L. Gerfo, M. Müller, P. Dedecker and W. Vandenberg, Biophys. J., 2021, 1, 100026 CAS.
- H. Valenta, F. Bierbuesse, R. Vitale, C. Ruckebusch, W. Vandenberg and P. Dedecker, Talanta, 2024, 269, 125397 CrossRef CAS PubMed.
- M. Lummer, F. Humpert, M. Wiedenlübbert, M. Sauer, M. Schüttpelz and D. Staiger, Mol. Plant, 2013, 6, 1518–1530 CrossRef CAS.
- J. Livet, T. A. Weissman, H. Kang, R. W. Draft, J. Lu, R. A. Bennis, J. R. Sanes and J. W. Lichtman, Nature, 2007, 450, 56–62 CrossRef CAS PubMed.
- S. Aramaki and K. Hatta, et al., Dev. Dyn., 2006, 235, 2192–2199 Search PubMed.
- Z. Tan, C.-H. Hsiung, J. Feng, Y. Zhang, Y. Wan, J. Chen, K. Sun, P. Lu, J. Zang, W. Yang, Y. Gao, J. Yin, T. Zhu, Y. Lu, Z. Pan, Y. Zou, C. Liao, X. Li, Y. Ye, Y. Liu and X. Zhang, Cell, 2025, 188, 6987–7005 CrossRef CAS.
- F. T. S. Chan, C. F. Kaminski and G. S. K. Schierle, ChemPhysChem, 2011, 12, 500–509 CrossRef CAS PubMed.
- N. Ojha, K. H. Rainey and G. H. Patterson, Nat. Commun., 2020, 11, 21 Search PubMed.
- T. Matsuda and T. Nagai, Microscopy, 2014, 63, 403–408 CrossRef CAS PubMed.
- I. Nemet, P. Ropelewski and Y. Imanishi, Photochem. Photobiol. Sci., 2015, 14, 1787–1806 CrossRef CAS.
- T. Matsuda, A. Miyawaki and T. Nagai, Nat. Methods, 2008, 5, 339–345 CrossRef CAS.
- F. Mueller, T. Morisaki, D. Mazza and J. G. McNally, Biophys. J., 2012, 102, 1656–1665 CrossRef CAS.
- D. M. Chudakov, T. V. Chepurnykh, V. V. Belousov, S. Lukyanov and K. A. Lukyanov, Traffic, 2006, 7, 1304–1310 CrossRef CAS PubMed.
- O. Y. Kwon, I. C. Kwon, H. K. Song and H. Jeon, Biochim. Biophys. Acta, 2008, 1780, 1403–1407 CrossRef CAS PubMed.
- M. Chisari, D. K. Saini, V. Kalyanaraman and N. Gautam, J. Biol. Chem., 2007, 282, 24092–24098 CrossRef CAS PubMed.
- M. Lummer, F. Humpert, C. Steuwe, K. Caesar, M. Schüttpelz, M. Sauer and D. Staiger, Traffic, 2011, 12, 693–702 CrossRef CAS.
- M. Lummer, F. Humpert, M. Wiedenlübbert, M. Sauer, M. Schüttpelz and D. Staiger, Mol. Plant, 2013, 6, 1518–1530 CrossRef CAS.
- N. Gerlitz, R. Gerum, N. Sauer and R. Stadler, Plant J., 2018, 94, 751–766 CrossRef CAS PubMed.
- J. Tallent, Q. W. Song, Z. Li, J. Stuart and R. R. Birge, Opt. Lett., 1996, 21, 1339–1341 CrossRef CAS.
- Y. S. Bae, J. Yang, S. Jin, S. Y. Lee and C. H. Park, Biotechnol. Prog., 1999, 15, 971–973 CrossRef CAS.
- T. Fischer, M. Neebe, T. Juchem and N. A. Hampp, IEEE Trans. Nanobiosci., 2003, 2, 1–5 CrossRef PubMed.
- D. Tischer and O. D. Weiner, Nat. Rev. Mol. Cell Biol., 2014, 15, 551–558 CrossRef CAS.
- K. Zhang and B. Cui, Trends Biotechnol., 2015, 33, 92–100 CrossRef CAS PubMed.
- X. X. Zhou, X. Zou, H. K. Chung, Y. Gao, Y. Liu, L. S. Qi and M. Z. Lin, ACS Chem. Biol., 2018, 13, 443–448 CrossRef CAS PubMed.
- A. L. Patel, E. Yeung, S. E. McGuire, A. Y. Wu, J. E. Toettcher, R. D. Burdine and S. Y. Shvartsman, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 25756–25763 CrossRef CAS.
- J. Ju, H. N. Lee, L. Ning, H. Ryu, X. X. Zhou, H. Chun, Y. W. Lee, A. I. Lee-Richerson, C. Jeong, M. Z. Lin and J. Seong, Cell Rep., 2022, 40, 111080 Search PubMed.
- R. Jöhr, M. S. Bauer, L. C. Schendel, C. Kluger and H. E. Gaub, Nano Lett., 2019, 19, 3176–3181 CrossRef PubMed.
- X. Wu, W. Huang, W.-H. Wu, B. Xue, D. Xiang, Y. Li, M. Qin, F. Sun, W. Wang, W.-B. Zhang and Y. Cao, Nano Res., 2018, 11, 5556–5565 CrossRef CAS.
- J. Zhang, S. H. D. Wong, X. Wu, H. Lei, M. Qin, P. Shi, W. Wang, L. Bian and Y. Cao, Adv. Mater., 2021, 33, e2105765 CrossRef PubMed.
- Y. T. Nguyen, H. J. Lee and N. Kim, Bioconjugate Chem., 2025, 36, 1247–1256 Search PubMed.
- A. Nickerson, T. Huang, L.-J. Lin and X. Nan, PLoS One, 2014, 9, e100589 CrossRef.
- Z. Liu, D. Xing, Q. P. Su, Y. Zhu, J. Zhang, X. Kong, B. Xue, S. Wang, H. Sun, Y. Tao and Y. Sun, Nat. Commun., 2014, 5, 4443 CrossRef CAS.
- R. Yan, B. Wang and K. Xu, Curr. Opin. Chem. Biol., 2019, 51, 92–97 Search PubMed.
- K. Lu, C. Q. Vu, T. Matsuda and T. Nagai, Int. J. Mol. Sci., 2019, 20, 5784 CrossRef CAS.
- H. Ren, Q. Ou, Q. Pu, Y. Lou, X. Yang, Y. Han and S. Liu, Biomolecules, 2024, 14, 859 CrossRef CAS PubMed.
- Y. R. Lee, J.-H. Park, S.-H. Hahm, L.-W. Kang, J. H. Chung, K.-H. Nam, K. Y. Hwang, I. C. Kwon and Y. S. Han, Mol. Imaging Biol., 2010, 12, 468–478 Search PubMed.
- F. Hertel, G. C. H. Mo, S. Duwé, P. Dedecker and J. Zhang, Cell Rep., 2016, 14, 390–400 CrossRef CAS.
- S. Wang, M. Ding, X. Chen, L. Chang and Y. Sun, Biomed. Opt. Express, 2017, 8, 3119–3131 CrossRef CAS PubMed.
- G. C. H. Mo, B. Ross, F. Hertel, P. Manna, X. Yang, E. Greenwald, C. Booth, A. M. Plummer, B. Tenner, Z. Chen, Y. Wang, E. J. Kennedy, P. A. Cole, K. G. Fleming, A. Palmer, R. Jimenez, J. Xiao, P. Dedecker and J. Zhang, Nat. Methods, 2017, 14, 427–434 CrossRef CAS PubMed.
- W. Lin, G. C. H. Mo, S. Mehta and J. Zhang, J. Am. Chem. Soc., 2021, 143, 14951–14955 CrossRef CAS PubMed.
- K. Mishra, J. P. Fuenzalida-Werner, F. Pennacchietti, R. Janowski, A. Chmyrov, Y. Huang, C. Zakian, U. Klemm, I. Testa, D. Niessing, V. Ntziachristos and A. C. Stiel, Nat. Biotechnol., 2021, 40, 598–605 CrossRef PubMed.
- F. Lavoie-Cardinal, N. A. Jensen, V. Westphal, A. C. Stiel, A. Chmyrov, J. Bierwagen, I. Testa, S. Jakobs and S. W. Hell, ChemPhysChem, 2014, 15, 655–663 CrossRef CAS PubMed.
- K. I. Willig, A. C. Stiel, T. Brakemann, S. Jakobs and S. W. Hell, Nano Lett., 2011, 11, 3970–3973 CrossRef CAS.
- K. I. Willig, W. Wegner, A. Müller, V. Clavet-Fournier and H. Steffens, Cell Rep., 2021, 35, 109192 CrossRef CAS.
- I. Testa, E. D'Este, N. T. Urban, F. Balzarotti and S. W. Hell, Nano Lett., 2015, 15, 103–106 CrossRef CAS PubMed.
- T. Roebroek, W. Vandenberg, F. Sipieter, S. Hugelier, C. Stove, J. Zhang and P. Dedecker, Nat. Commun., 2021, 12, 2005 Search PubMed.
- N. Komatsu, K. Aoki, M. Yamada, H. Yukinaga, Y. Fujita, Y. Kamioka and M. Matsuda, Mol. Biol. Cell, 2011, 22, 4647–4656 CrossRef CAS PubMed.
- L. S. Vidal, M. Isalan, J. T. Heap and R. Ledesma-Amaro, RSC Chem. Biol., 2023, 4, 271–291 RSC.
- E. O. McCullum, B. A. R. Williams, J. Zhang and J. C. Chaput, Methods Mol. Biol., 2010, 634, 103–109 Search PubMed.
- S. L. C. Moors, S. Michielssens, C. Flors, P. Dedecker, J. Hofkens and A. Ceulemans, J. Chem. Theory Comput., 2008, 4, 1012–1020 Search PubMed.
- A. C. Stiel, M. Andresen, H. Bock, M. Hilbert, J. Schilde, A. Schönle, C. Eggeling, A. Egner, S. W. Hell and S. Jakobs, Biophys. J., 2008, 95, 2989–2997 CrossRef CAS PubMed.
- F. Pennacchietti, E. O. Serebrovskaya, A. R. Faro, I. I. Shemyakina, N. G. Bozhanova, A. A. Kotlobay, N. G. Gurskaya, A. Bodén, J. Dreier, D. M. Chudakov, K. A. Lukyanov, V. V. Verkhusha, A. S. Mishin and I. Testa, Nat. Methods, 2018, 15, 601–604 CrossRef CAS PubMed.
- E. M. Williams, J. N. Copp and D. F. Ackerley, Methods Mol. Biol., 2014, 1179, 83–101 CrossRef.
- A. M. Aguinaldo and F. H. Arnold, Methods Mol. Biol., 2003, 231, 105–110 CAS.
- A. C. Stiel, S. Trowitzsch, G. Weber, M. Andresen, C. Eggeling, S. W. Hell, S. Jakobs and M. C. Wahl, Biochem. J., 2007, 402, 35–42 CrossRef CAS.
- V. Adam, K. Hadjidemetriou, N. Jensen, R. L. Shoeman, J. Woodhouse, A. Aquila, A.-S. Banneville, T. R. M. Barends, V. Bezchastnov, S. Boutet, M. Byrdin, M. Cammarata, S. Carbajo, N. E. Christou, N. Coquelle, E. De la Mora, M. El Khatib, T. M. Chicano, R. B. Doak, F. Fieschi, L. Foucar, O. Glushonkov, A. Gorel, M. L. Grünbein, M. Hilpert, M. Hunter, M. Kloos, J. E. Koglin, T. J. Lane, M. Liang, A. Mantovanelli, K. Nass, G. N. Kovacs, S. Owada, C. M. Roome, G. Schirò, M. Seaberg, M. Stricker, M. Thépaut, K. Tono, K. Ueda, L. M. Uriarte, D. You, N. Zala, T. Domratcheva, S. Jakobs, M. Sliwa, I. Schlichting, J.-P. Colletier, D. Bourgeois and M. Weik, ChemPhysChem, 2022, 23, e202200192 CrossRef CAS.
- C. Duan, V. Adam, M. Byrdin, J. Ridard, S. Kieffer-Jaquinod, C. Morlot, D. Arcizet, I. Demachy and D. Bourgeois, J. Am. Chem. Soc., 2013, 135, 15841–15850 CrossRef CAS.
- C. Duan, M. Byrdin, M. El Khatib, X. Henry, V. Adam and D. Bourgeois, Methods Appl. Fluoresc., 2015, 3, 014004 CrossRef.
- T. Grotjohann, I. Testa, M. Reuss, T. Brakemann, C. Eggeling, S. W. Hell and S. Jakobs, eLife, 2012, 1, e00248 CrossRef PubMed.
- D. A. Zacharias, J. D. Violin, A. C. Newton and R. Y. Tsien, Science, 2002, 296, 913–916 CrossRef CAS.
- J.-D. Pédelacq, S. Cabantous, T. Tran, T. C. Terwilliger and G. S. Waldo, Nat. Biotechnol., 2005, 24, 79–88 CrossRef.
- M. El Khatib, A. Martins, D. Bourgeois, J.-P. Colletier and V. Adam, Sci. Rep., 2016, 6, 18459 CrossRef CAS.
- R. Bizzarri, M. Serresi, F. Cardarelli, S. Abbruzzetti, B. Campanini, C. Viappiani and F. Beltram, J. Am. Chem. Soc., 2010, 132, 85–95 CrossRef CAS.
- S. C. Alford, A. S. Abdelfattah, Y. Ding and R. E. Campbell, Chem. Biol., 2012, 19, 353–360 Search PubMed.
- S. C. Alford, Y. Ding, T. Simmen and R. E. Campbell, ACS Synth. Biol., 2012, 1, 569–575 CrossRef CAS PubMed.
- A. Kirchhofer, J. Helma, K. Schmidthals, C. Frauer, S. Cui, A. Karcher, M. Pellis, S. Muyldermans, C. S. Casas-Delucchi, M. C. Cardoso, H. Leonhardt, K.-P. Hopfner and U. Rothbauer, Nat. Struct. Mol. Biol., 2010, 17, 133–138 CrossRef CAS.
- T. Roebroek, S. Duwé, W. Vandenberg and P. Dedecker, Int. J. Mol. Sci., 2017, 18, 2015 CrossRef PubMed.
- A. C. Stiel and V. Ntziachristos, Nat. Methods, 2024, 21, 1996–2007 CrossRef CAS PubMed.
- L. J. Münker, M. Hohgardt, A. Albrecht, D. Pfennig, J. S. Tegtmeier, A. Holz, M. Zagrebelsky, M. Korte and P. J. Walla, npj Imaging, 2025, 3, 31 CrossRef.
- L. Zhang, F. Xu, Z. Chen, X. Zhu and W. Min, J. Phys. Chem. Lett., 2013, 4, 3897–3902 CrossRef CAS.
- D. Stumpf, N. Jensen, C. Mittelheisser, J. Keller-Findeisen, A. I. Chizhik, M. Kamper, T. Diekmann, F. Habenstein, I. Jansen, J. Enderlein, M. Sliwa, K. Inamdar, S. W. Hell and S. Jakobs, Proc. Natl. Acad. Sci. U. S. A., 2025, 122, e2504748122 CrossRef CAS.
- J. G. Danzl, S. C. Sidenstein, C. Gregor, N. T. Urban, P. Ilgen, S. Jakobs and S. W. Hell, Nat. Photonics, 2016, 10, 122–128 CrossRef CAS.
- G. Marín-Aguilera, F. Pennacchietti, A. Volpato, A. Papalini, A. Kulkarni, N. Bagheri, G. Minet, J. Widengren and I. Testa, Nat. Commun., 2025, 16, 10843 CrossRef.
- J. Yao and X. Wang, Med. Novel Technol. Devices, 2025, 26, 100366 CrossRef.
- T. Hayes, R. Rao, H. Akin, N. J. Sofroniew, D. Oktay, Z. Lin, R. Verkuil, V. Q. Tran, J. Deaton, M. Wiggert, R. Badkundri, I. Shafkat, J. Gong, A. Derry, R. S. Molina, N. Thomas, Y. A. Khan, C. Mishra, C. Kim, L. J. Bartie, M. Nemeth, P. D. Hsu, T. Sercu, S. Candido and A. Rives, Science, 2025, 387, 850–858 CrossRef CAS PubMed.
- J. Dou, A. A. Vorobieva, W. Sheffler, L. A. Doyle, H. Park, M. J. Bick, B. Mao, G. W. Foight, M. Y. Lee, L. A. Gagnon, L. Carter, B. Sankaran, S. Ovchinnikov, E. Marcos, P.-S. Huang, J. C. Vaughan, B. L. Stoddard and D. Baker, Nature, 2018, 561, 485–491 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |