High-speed cell partitioning through reactive machine learning-guided inkjet printing
Eric
Cheng
 a,
Glenn
Chang
a,
Glenn
Chang
 b,
Haley
MacDonald
b,
Haley
MacDonald
 b,
Miguel
Ramirez
b,
Miguel
Ramirez
 b,
Pamela A.
Hoodless
b,
Pamela A.
Hoodless
 bc,
Robin
Coope
bc,
Robin
Coope
 d,
Adi
Steif
d,
Adi
Steif
 *bcd and
Karen C.
Cheung
*bcd and
Karen C.
Cheung
 *aef
*aef
aDepartment of Electrical and Computer Engineering, University of British Columbia, Vancouver, V6T 1Z4, Canada. E-mail: kcheung@ece.ubc.ca
bDepartment of Basic and Translational Research, BC Cancer Research Institute, Vancouver, V5Z 1L3, Canada
cDepartment of Medical Genetics, University of British Columbia, Vancouver, V6T 1Z3, Canada. E-mail: adi.steif@ubc.ca
dCanada's Michael Smith Genome Sciences Centre, Vancouver, V5Z 4S6, Canada
eSchool of Biomedical Engineering, University of British Columbia, Vancouver, V6T 2B9, Canada
fCentre for Blood Research, University of British Columbia, Vancouver, V6T 1Z3, Canada
First published on 8th August 2025
Abstract
Partitioning cells in open nanowells permits high confidence in single cell occupancy and enables flexibility in the development of different molecular assays. A challenge for this approach however is to print cells sufficiently quickly to enable experiments of adequate statistical power in a reasonable time. To address this, we developed a single cell dispensing instrument leveraging inkjet technology with continuous real-time optical feedback and machine learning algorithms for high-throughput single cell isolation. The Isolatrix enables rapid partitioning of cells into open substrates such as nanowell arrays, permitting high-throughput application of custom genomic assays such as direct-transposition single cell whole genome sequencing (scWGS). We trained the classifier on manually labelled data with a range of cell sizes and applied the instrument to generate scWGS profiles from cell lines and primary mouse tissue. Comparison to existing predictive workflows demonstrated that this reactive approach, featuring machine learning classification of events post-dispensing, gives up to a 9.69 times increase in isolation speed. Validation via fluorescent imaging of cell lines confirmed a classification accuracy of 98.7%, at a rate of 0.52 seconds per single cell, under tuned spotting parameters. Genomic analysis showed low background contamination and high coverage uniformity across the genome, enabling detection of chromosomal copy number alterations. With data tracing capabilities and a convenient user interface, we expect the Isolatrix to enable large-scale profiling of a range of genomic data modalities.
Introduction
Single cell genomics has emerged as a transformative tool for understanding cellular heterogeneity in health and disease.1–3 By enabling segregation and analysis of individual cells, this approach uncovers insights into genetic and phenotypic diversity,4 rare mutations,5 and identification of distinct cellular subpopulations that are obscured through bulk sequencing.6,7 Profiling cell-to-cell heterogeneity is particularly important in cancer studies,8,9 where somatic mutation and epigenetic rewiring may lead to competing tumour sub-clones that can contribute to treatment resistance and metastatic progression.10–12Commercial instruments for emulsion-based microfluidic single cell isolation, whereby a cell suspension is randomly partitioned in aqueous droplets within an oil emulsion, include the widely-adopted 10× Genomics Chromium platform. Emulsion-based instruments have popularized profiling of transcript expression, chromatin accessibility, and targeted protein abundance. While these microfluidic systems are widely used due to their ability to rapidly isolate and encapsulate thousands of cells,13,14 the approach has limitations.
First, random encapsulation combined with an inability to verify cell count within individual droplets results in the inclusion of doublets or multiplets, which can confound downstream analyses.15 Although bioinformatic tools have been developed to address these artifacts, they are often inconsistent and struggle with identifying homotypic doublets.
The closed architecture of these emulsion-based instruments limits flexibility in adopting or modifying protocols. Examples of such emerging single cell library preparation protocols include direct-tagmentation whole genome sequencing,16,17 proteomics,18–20 metabolomics,21,22 and various multiomics protocols.23
Cells that are fragile or tightly linked can be difficult to dissociate and therefore tend to be missing or underrepresented in emulsion-based libraries (e.g. adipocytes, hepatocytes).24–26 Large cells or debris from primary tissue can clog microfluidic channels, leading to failed runs.27,28 This has presented, for example, challenges in the study of various liver diseases including hepatocellular carcinoma (HCC), the most common form of liver cancer.29 Large-scale single cell atlas studies on the liver have used different approaches to overcome the challenge of profiling hepatocytes. Some have ignored this majority cell type altogether,30 while others acknowledge they are substantially underrepresented31 or show representation primarily from a single donor liver.32 Other studies overcome the cell dissociation challenge by profiling nuclei instead of cells,33,34 but this can reduce sequencing yield, limit the profiling of some biomolecules and organelles, and preclude interrogation of cell size and morphology.
Piezoelectric inkjet dispensing is an established technology for spotting droplets, tunable in the range of 150–400 picolitres.35,36 These can be delivered at variable frequencies up to thousands of droplets per second37 or the inkjet can be operated in drop-on-demand mode.38 The flexibility of the technique provides utility in a range of applications from everyday printing to manufacturing multiplexed biomolecular assays.39–41 Additionally, emerging clinical applications of bioprinting in fields such as tissue regeneration or wound healing underscore the potential of inkjet printing for delivery of bioinks.42,43 Recent integration of machine learning models into bioprinters has demonstrated optimization of printing parameters, enhancing control and accuracy.44,45 Inkjet printheads have also demonstrated gentle handling of a number of cell types due to the low shear stress imparted on the particles during dispensing, achieving high dispensed cell viability.46,47
Inkjet-based spotting, being a low-volume dispensing modality, can permit reactions in open wells with nanolitre volumes, where both cells and reagents can be sequentially added. It should be noted that miniaturization preserves the robustness of reactions, maintaining the relative concentration of reagents and input deoxyribonucleic acid (DNA) from conventional bulk reactions. A single cell with 6 pg of DNA in a ∼100 nl well is proportional to a polymerase chain reaction (PCR) based bulk library construction with 6 ng of DNA in a ∼100 μl reaction.16 Spotting into open nanowells enables sequential reagent addition, permitting implementation of novel protocols, so emerging methods like single cell whole genome sequencing,16 proteomics,48 or whole transcript ribonucleic acid (RNA) sequencing49,50 may be deployed at scale. Another feature of open wells is the ability to selectively build libraries only on wells of interest,16 as well as selectively recover cells, either individually or in defined segments of a well plate or nanowell chip.51
Extant commercial inkjet-based single cell dispensing systems, such as the CellenONE (Cellenion) and F.Sight (Cytena) use deterministic image-based algorithms to assess cell size, shape, and position within the nozzle, aiming to predict single cell capture. They assess a defined region within the nozzle immediately before each dispensing event to determine if a single cell should be deposited on the target substrate or into a waste receptacle. While this approach enables high single cell occupancy on the substrate, it has limitations. These include the setting of strict gating criteria which can lead to the exclusion of genuine single cells that fall outside predefined parameters, in addition to potentially discarding viable single cells due to conditions aimed at minimizing the risk of doublet capture. Importantly, while these instruments can dispense into standard well-plates within reasonable time frames, their cell isolation speeds present challenges for high-throughput dispensing into nanowell arrays, as hardware latency associated with travelling back and forth between the waste receptacle and target wells compounds across large numbers of cells, significantly impacting overall throughput at scale. We hypothesized that improved optics and image analysis combined with a reactive dispensing protocol to capture all dispensing events could result in faster cell partitioning, albeit with the trade-off of variable dispense volumes per well.
To this end, we developed the Isolatrix, a new inkjet-based cell dispensing instrument which features a novel printing algorithm and machine learning classification of events post-dispensing for rapid and accurate single cell isolation, by depositing single cells into nanowells, onto glass slides, or into other discrete compartments. By dispensing empty droplets onto the same target location until an event is identified, the Isolatrix printing algorithm reduces cell isolation time. This is a reactive approach to enumerate spotting events in contrast to the predictive approach used by competing instruments, and enables higher cell portioning rates while preserving the advantages of direct visual confirmation of partitioning events.
Single cell isolation instrument
Design criteria
The design criteria for this instrument were driven primarily by improving the throughput of single cell whole genome sequencing using the direct library preparation (DLP+) protocol.16 The DLP+ workflow involves cell lysis, heat-inactivated protease treatment, and direct DNA transposition within high-density nanowell arrays. A single DLP+ sequencing experiment typically involves isolation of 1000–3000 cells within 72 × 72 Takara SmartChip™ nanowell arrays.52 The latter is a 41 mm square aluminium plate with 460 μm diameter wells drilled ∼600 μm deep at a pitch of 542 μm, giving a volume of 100 nl per well. DLP+ in SmartChips has proven to be robust and scalable,53 but the substantive operator-time needed to spot cells has remained a significant challenge.The goals of the Isolatrix instrument design were as follows:
1. Enable single cell throughput of <1 second per cell
2. Provide high confidence in single cell dispense calling
3. Ensure compatibility with common open well plates, slides, and nanowells
The first design goal, motivated by operator experience, was to be able to spot ∼1000 cells in the order of minutes, i.e. one second per cell or less. This was selected to address the need to ease scheduling, minimize cell exposure to ambient conditions resulting in a change in cell state or cell death,54,55 and minimize the risk of cell aggregation from sedimentation.56–58 The second design goal emphasizes the importance of verifying single cell spotting events with high confidence, so as to minimize unidentified cell doublets and multiplets in constructed libraries. The third design goal was to enable a diverse range of workflows by ensuring instrument compatibility with different target substrates, including the Takara SmartChip™ nanowell array, microscope slides, and standard microplates.
Single cell instrument hardware
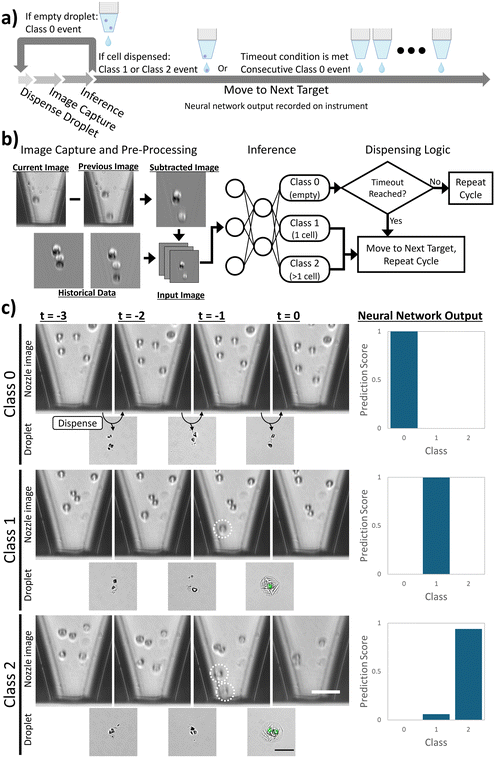
The Isolatrix is designed around the MicroFab MJ-AL-02-080 piezoelectrically actuated inkjet printhead. This printhead is constructed from a tapered glass capillary with an 80 μm orifice and can operate in drop-on-demand mode enabling a machine vision system to guide cell dispensing. The glass nozzle allows imaging of the inner channel of the printhead to monitor and control cell dispensing events. The imaging system, as shown in Fig. 1a, is fixed in line with the inkjet printhead and continuously monitors cell motions in response to droplet ejection. The optics consist of a ThorLabs MVL12X12Z 12× zoom lens configuration and a FLIR Blackfly S BFS-U3-32S4M greyscale scientific camera with a resolution of 0.26 pixels per μm and a field of view of 538 × 200 μm into the nozzle's channel adjacent to the orifice. A light emitting diode (LED) light source is positioned 100 mm behind the printhead and is synchronized with droplet ejection and image capture. A Zaber LSQ Series XY gantry system with a microstep size of 0.496 μm enables positioning of the substrate beneath the inkjet nozzle (Fig. 1b).Dispensing algorithm
The instrument employs a novel cell printing strategy (Fig. 2a) leveraging machine vision-based feedback with a trained neural network to achieve single-cell isolation rates beyond random capture.59 During printing, the printhead is positioned over a target and droplets are dispensed under observation by the machine learning model. Cell encapsulation in a suspension is governed by the Poisson distribution, so the majority of the droplets will be empty. If a cell dispensing event is detected, the system records the event and moves to the next well, otherwise, it repeats the ejection and analysis cycle as highlighted in Fig. 2b. To facilitate inference of the captured cell motions within the nozzle images, the network multiplexes historical data by combining images from prior droplets into a single input incorporating cell motion data to identify cell encapsulation events.60 Each image is subtracted from the preceding one to remove static features, such as debris on the outer capillary surface. The resulting subtracted images are concatenated into a single composite image, which is then used as input to the neural network.A timeout condition prevents overfilling or reagent dilution by skipping wells after a user-defined number of consecutive empty droplet events.
As analysis is performed on images captured after the droplet has been ejected, the Isolatrix approach aims to identify post-dispense events, with an example of the three possible classes given in Fig. 2c. The droplet ejection cycle was measured to be 25 ms which includes the droplet actuation, image capture, neural network inference and user interface (UI) processing. Conversely, the time required for the gantry to travel 542 μm from well to well was measured to be 140 ms on average. Therefore, to enable high-throughput cell dispensing, the algorithm minimizes machine movement actions by continuously dispensing into the current target and traversing the grid in a raster pattern.
Cell dispensing event classifier
A neural network was trained to classify droplets into three classes: class 0 are empty droplets, class 1 are droplets containing a single cell, and class 2 are droplets containing multiple cells. A dataset of dispensing event images was captured and manually labeled, using a variety of cell lines including 184-hTERT, A549, MCF-7, OCI-AML3 and Huh-7. The image set consists of 6197 class 0 images, 5317 class 1 images and 1559 class 2 images with an 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 split for training and validation. All models were trained with the same learning rate (0.0001), batch size (64) and dropout rate (0.5) while varying only architectural hyperparameters such as the number of layers, filters and nodes.
15 split for training and validation. All models were trained with the same learning rate (0.0001), batch size (64) and dropout rate (0.5) while varying only architectural hyperparameters such as the number of layers, filters and nodes.
Two neural networks were deployed on the instrument equipped with an AMD Ryzen 9 3950X CPU and a NVIDIA GeForce RTX 3080 GPU with frozen weights. A convolutional neural network (CNN)61–63 was trained, comprising four 2D convolutional layers with progressively increasing filter counts (16, 32, 64, 64), followed by a 64-unit dense layer and a softmax output layer for three-class classification (Fig. 3a). With 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 895
895![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 395 trainable parameters, the model achieved a weighted F1 score of 0.84 on the validation dataset and an inference time of 4.6 ms per event.
395 trainable parameters, the model achieved a weighted F1 score of 0.84 on the validation dataset and an inference time of 4.6 ms per event.
A ResNet model64,65 was trained on an expanded dataset (class 0: 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 033, class 1: 7257, class 2: 2501) and deployed on the instrument. This model consisted of a series of eight blocks of convolutional, pooling and feedforward connections with increasing channel depth from 64 to 512, before flattening and mapping to a fully connected layer with 64 units outputting to the three classes (Fig. 3b). In total, there were 7
033, class 1: 7257, class 2: 2501) and deployed on the instrument. This model consisted of a series of eight blocks of convolutional, pooling and feedforward connections with increasing channel depth from 64 to 512, before flattening and mapping to a fully connected layer with 64 units outputting to the three classes (Fig. 3b). In total, there were 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 689
689![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 992 trainable parameters in the ResNet model resulting in an inference time of 9.72 ms per image. The validation dataset had a weighted F1 score of 0.94.
992 trainable parameters in the ResNet model resulting in an inference time of 9.72 ms per image. The validation dataset had a weighted F1 score of 0.94.
Dispensed single cell calling
To facilitate manual inspection for cell calling (e.g. for network training) or to review the algorithm's predictions, a UI (Fig. 4) aggregates and displays neural network class probabilities, corresponding nozzle images, and, if available, fluorescent microscopy images of the well contents. It displays the neural network's output in real time using color-coded markers to represent cell count predictions for each well, simplifying the selection of target wells for downstream workflows (Fig. 4a). In general, only wells containing single cells are selected to undergo library construction, along with positive and negative controls. Wells with only class 0 events (empty droplets) can be selected as no cell controls (NCCs) to allow estimates of the level of background contamination in the cell suspension fluid, while no template control (NTC) wells contain neither cells nor cell suspension fluid and can be used to assess contamination in the library reagents. Positive cell controls (PCCs) from a well-characterized cell line can be included alongside genomic DNA (gDNA) wells to identify issues related to input sample quality. The neural network's output and corresponding nozzle images are displayed with UI buttons to label the selected well (Fig. 4b).Results
Workflow to verify dispensed single cell purity
An experiment was conducted to verify droplet classification with orthogonal imaging. While dispensing into the nanowell chip allows for the capture of single cells to be used in one-pot chemistry workflows such as the DLP+ protocol, the high aspect ratio of the aluminum wells presents challenging imaging conditions when differentiating between single or multiple cells. This makes verification of the single cell purity of the dried-down contents within the well via microscopy alone insufficient, necessitating inspection of additional data such as the captured nozzle images. Dispensing droplets onto a glass slide enables fluorescent imaging and facilitates review to differentiate droplets containing single or multiple cells. Each dispensed position can be associated with a readout from the instrument. The slide-imaged droplets (Fig. 5) showed excellent agreement with nozzle images; 100% of 100 manually inspected droplets had the correct cell count as seen in the corresponding nozzle images. Manually verified dispensing events were used for network training.Validation of timeout conditions and comparison to existing instrumentation using a karyotypically normal cell line
The Isolatrix was evaluated in a whole genome single cell sequencing experiment using the DLP+ method and the 184-hTERT normal immortalized breast epithelial cell line, which is a female cell line with a generally normal karyotype.16 The goals of this experiment were to enable evaluation of sequencing quality metrics between 5-droplet and 10-droplet timeout conditions, as well as a direct comparison with the current instrument used for DLP+ implementation, the CellenONE. In comparing the 5-droplet and 10-droplet timeout conditions, we sought to determine whether the increased volume of cell suspension fluid (phosphate buffered saline, PBS) and ambient DNA per well would have an adverse impact on sequencing data quality and contaminating reads. The DLP+ workflow utilizes nanowell chips pre-spotted with primers containing unique row and column barcodes which allows for DNA fragments to be associated with their respective cell of origin after sequencing.For this experiment, the Isolatrix dispensed 664 single cells in 14.7 minutes or, on average, 1.46 seconds per cell. The library-constructed single cells were verified via fluorescence microscopy of the chip and the captured nozzle images. Within this, 313 cells were dispensed under the 5-droplet timeout condition, while 291 cells were dispensed under the 10-droplet timeout condition (Fig. 6a), requiring print times of 535 s and 347 s respectively. This corresponds to a throughput of 1.71 seconds per single cell for the 5-droplet timeout condition and 1.19 seconds per single cell for the 10-droplet timeout condition. In comparison, the CellenONE instrument deposited 458 single cells in 38.5 minutes, with a throughput of 5.04 seconds per single cell (Fig. 6a).
This library was sequenced on an Illumina NovaSeq 6000 and aligned to the hg38 reference genome using Minimap2.66 Sequencing quality metrics, copy number profiles, and copy number quality scores were derived as previously described.16 Cells dispensed by the Isolatrix demonstrated comparable sequencing yield to those dispensed by the CellenONE, with no significant difference in total mapped reads observed under both the 5-droplet timeout condition (mean = 604![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 630; Wilcoxon test, p-value = 0.18) and the 10-droplet timeout condition (mean = 556
630; Wilcoxon test, p-value = 0.18) and the 10-droplet timeout condition (mean = 556![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 175; Wilcoxon test, p-value = 0.26), compared to the CellenONE (mean = 562
175; Wilcoxon test, p-value = 0.26), compared to the CellenONE (mean = 562![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 426) (Fig. 6b and d). Additional quality control metrics including coverage depth (CellenONE: 0.016; 5-droplet: 0.017; 10-droplet: 0.016) and total reads (CellenONE: 643
426) (Fig. 6b and d). Additional quality control metrics including coverage depth (CellenONE: 0.016; 5-droplet: 0.017; 10-droplet: 0.016) and total reads (CellenONE: 643![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 791; 5-droplet: 680
791; 5-droplet: 680![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 828.5; 10-droplet: 633
828.5; 10-droplet: 633![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 643.2), were also comparable across all conditions. While a significant difference was observed in total mapped reads between NCC wells with increasing numbers of droplets, particularly under the 10-droplet condition (mean: 2089.5) compared to the 5-droplet condition (mean: 1126.2), this background signal remains negligible relative to wells containing cells (mean: 573
643.2), were also comparable across all conditions. While a significant difference was observed in total mapped reads between NCC wells with increasing numbers of droplets, particularly under the 10-droplet condition (mean: 2089.5) compared to the 5-droplet condition (mean: 1126.2), this background signal remains negligible relative to wells containing cells (mean: 573![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151.7) (Fig. 6d). The increase in background reads is consistent with the higher average number of droplets dispensed per well under the 10-droplet timeout condition (mean: 5.10) versus the 5-droplet condition (mean: 2.73), increasing the chance of low-level contamination. Nevertheless, the background signal remains orders of magnitude lower than signal from wells with cells and is not expected to impact downstream copy number analysis or overall data interpretation.
151.7) (Fig. 6d). The increase in background reads is consistent with the higher average number of droplets dispensed per well under the 10-droplet timeout condition (mean: 5.10) versus the 5-droplet condition (mean: 2.73), increasing the chance of low-level contamination. Nevertheless, the background signal remains orders of magnitude lower than signal from wells with cells and is not expected to impact downstream copy number analysis or overall data interpretation.
Chromosomal copy number was inferred using HMMcopy.67 Copy number quality from cells dispensed by the Isolatrix was significantly higher than that of the CellenONE (mean = 0.63) under both the 5-droplet timeout condition (mean = 0.77; Wilcoxon test, p-value <0.001) and the 10-droplet timeout condition (mean = 0.70; Wilcoxon test, p-value = 0.03) (Fig. 6c and e).
To illustrate how variations in copy number quality scores affect confidence in chromosomal copy number calls, cells R06-C53 and R21-C29, both dispensed by the Isolatrix, exhibit high-quality copy number profiles (Fig. 6f). As 184-hTERT is a normal diploid cell line, a consistent copy number of two is observed across most chromosomes in these cells. Notably, R21-C29 displays an unexpected copy number gain on chromosome 8, despite maintaining an otherwise diploid profile, demonstrating the Isolatrix's ability to dispense high-quality cells suitable for identifying rare genomic variations at the chromosomal level. In contrast, the copy number profile of cell R35-C03, also dispensed by the Isolatrix, is characterized by increased noise and high variability, representing a low-quality copy number profile that fails to accurately recapitulate the expected diploid copy number of two.
A potential spatial bias in copy number quality was observed, where the bottom-right region of the chip dispensed by the CellenONE exhibited lower copy number quality. However, total mapped reads were not affected in this region, suggesting that the reduction in copy number quality may reflect a true biological or technical variance (e.g. due to longer dispensing time) rather than issues with sequencing or library construction.
Isolation of primary dissociated mouse liver tissue
To assess the performance of the Isolatrix on primary tissue samples, we dispensed cells from a primary mouse liver sample for scWGS using the DLP+ protocol. Hepatocytes are typically large and fragile cells, exhibiting a propensity to burst under shear stress,68 presenting challenges for consistent single cell handling. Unlike cell lines, dissociation of primary tissue can include substantial cellular debris, resulting in elevated background contamination. Given low levels of background contamination in the 184-hTERT experiment, an input cell concentration of 600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL and a timeout condition of 20 droplets was used for this dispensing run. Staining the sample enabled differentiation between fluorescent live cells and non-fluorescent extracellular debris via microscopy imaging of the nanowell chip post-dispensing.
000 cells per mL and a timeout condition of 20 droplets was used for this dispensing run. Staining the sample enabled differentiation between fluorescent live cells and non-fluorescent extracellular debris via microscopy imaging of the nanowell chip post-dispensing.
A grid of 30 × 59 wells was selected for dispensing, yielding 381 live single cells, as verified by nozzle images and fluorescent signal within the wells. The total print time was 554 seconds. Lower than expected single cell yield was observed due to the presence of debris within the primary dissociated sample being misclassified as single cells. Sequencing was performed on an Illumina NextSeq 2000 instrument and aligned to the mm10 reference genome using Minimap2.66 On average, each single cell yielded 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 553
553![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 884 total reads (SD: 1
884 total reads (SD: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 191
191![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 486) (Fig. 7a). As a positive control, human genomic DNA (gDNA) was also dispensed and mapped to the hg38 reference genome. The majority of single live cell reads (mean = 76.6%, SD = 27.4%) and gDNA reads (mean = 93.7%, SD = 1.35%) successfully mapped to their respective references (Fig. 7b).
486) (Fig. 7a). As a positive control, human genomic DNA (gDNA) was also dispensed and mapped to the hg38 reference genome. The majority of single live cell reads (mean = 76.6%, SD = 27.4%) and gDNA reads (mean = 93.7%, SD = 1.35%) successfully mapped to their respective references (Fig. 7b).
Copy number profiles were inferred using HMMcopy.67 As expected, due to the challenging nature of dispensing primary liver tissue, only 80 out of the 381 dispensed cells produced high-quality copy number profiles (mean copy number quality = 0.45) (Fig. 7c and d). In contrast, all gDNA control wells achieved high copy number quality (mean = 1.00), indicating that the limitations were specific to the liver sample rather than the dispensing platform or library preparation (Fig. 7C). Despite the limited number of high-quality cells, the liver cells displayed diploid copy number profiles with one X and one Y chromosome, consistent with a healthy male mouse (Fig. 7e). Notably, a rare focal copy number gain was detected at the start of chromosome 14 in 22 of these high-quality live cells (Fig. 7f).
Neural network performance optimization
The DLP+ experiment validated the instrument's variable volume print strategy which allows optimization to improve the achievable single-cell throughput and yield. Through empirical testing with the hTERT cell line, a working input cell concentration was determined to be in the range of 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml. This corresponds to an expected value of 0.112 cells per droplet. Further increasing the input cell concentration would theoretically improve single cell yield, but practical limits, such as the increased risk of clogging, militates against higher concentrations. We note that appropriate choice of this parameter is sample and cell type dependent. Samples with smaller cell size may permit higher input concentrations without compromising droplet formation or inducing nozzle clogging, while samples with extracellular debris or large cell size could benefit from lower cell concentrations. Bioinformatics analysis of the 184-hTERT experiment also suggests that the timeout condition can be increased with minimal negative impact on ambient contamination and chemistry compatibility. As such, for the mouse liver cell experiment the timeout condition was increased to 20 droplets. Increasing the timeout condition will lead to improvements in single cell yield by reducing the number of empty wells. Using these conditions, the neural networks were deployed on the instrument and their performance was benchmarked with the resulting confusion matrix shown in Fig. 8a.
000 cells per ml. This corresponds to an expected value of 0.112 cells per droplet. Further increasing the input cell concentration would theoretically improve single cell yield, but practical limits, such as the increased risk of clogging, militates against higher concentrations. We note that appropriate choice of this parameter is sample and cell type dependent. Samples with smaller cell size may permit higher input concentrations without compromising droplet formation or inducing nozzle clogging, while samples with extracellular debris or large cell size could benefit from lower cell concentrations. Bioinformatics analysis of the 184-hTERT experiment also suggests that the timeout condition can be increased with minimal negative impact on ambient contamination and chemistry compatibility. As such, for the mouse liver cell experiment the timeout condition was increased to 20 droplets. Increasing the timeout condition will lead to improvements in single cell yield by reducing the number of empty wells. Using these conditions, the neural networks were deployed on the instrument and their performance was benchmarked with the resulting confusion matrix shown in Fig. 8a.
A test print was conducted with the CNN, dispensing HEK cells into a 64 × 12 grid within the nanowell chip. An input cell concentration of 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL and a timeout of 20 droplets was used. Manual inspection of captured nozzle images identified 408 wells containing isolated single cells, corresponding to 53.1% single cell occupancy. The weighted F1 score of the test print was 0.899. The total grid print time was recorded on the instrument to take 188.75 seconds, achieving a single cell throughput of 0.46 seconds per cell.
000 cells per mL and a timeout of 20 droplets was used. Manual inspection of captured nozzle images identified 408 wells containing isolated single cells, corresponding to 53.1% single cell occupancy. The weighted F1 score of the test print was 0.899. The total grid print time was recorded on the instrument to take 188.75 seconds, achieving a single cell throughput of 0.46 seconds per cell.
The experiment was repeated using the trained ResNet dispensing SiHa cells in an expanded grid of 31 × 61 wells (1924 wells total) simulating a full-scale DLP+ print run using the same concentration and timeout conditions. Manual inspection confirmed 1370 wells with isolated single cells, corresponding to a 71.2% single-cell occupancy. From this test print, the weighted F1 score was 0.986, and the grid was printed in 11 minutes and 43.65 seconds, resulting in a benchmarked throughput of 0.52 seconds per single cell, although general performance may vary depending on experimental conditions.
The CNN demonstrated a slightly higher single cell throughput compared to the ResNet due to the faster inference time per image associated with smaller model size (comparison of model metrics provided in Table S1). However, its lower accuracy limited the achievable single cell yield per chip. A limitation of the trained CNN was its tendency to misclassify class 0 empty droplets as class 1 or class 2 events containing cells. This increased the number of erroneously empty wells within the printed area and reduced overall single cell per well yield. The ResNet demonstrated high accuracy in distinguishing between empty droplets and cell dispensing events, leading to fewer empty wells due to misclassifications which resulted in higher overall single cell occupancy. This improvement is critical for enhancing the efficiency of single cell isolation in large-scale experiments.
While the confusion matrices in Fig. 8 capture the accuracy of individual droplet dispensing events, we next evaluated the performance for well occupancy. For example, an individual well with a correct class prediction for the final droplet (e.g. class 1) could still contain more than one cell if a previous droplet deposited in the same well was a class 1 event misclassified as class 0. Of all the wells which the neural network labeled as belonging to class 1, 1345 out of 1538 wells (87.4%) contain a genuine single cell, as verified via nozzle and fluorescence microscopy images with an overall accuracy of 98.7% across all classified events. Of the 1538 wells, 142 wells were empty but were erroneously classified as class 1 (9.3%). Finally, 51 wells contained multiple cells (3.33%). For single cell sequencing applications, empty wells do not significantly negatively impact downstream results, as they do not produce a signal. The contamination of multiple cells makes up only 3.3% of the total identified single-cell containing wells. This may obviate the need for manual inspection and verification of the single cell purity of the wells selected for library construction, a time-consuming step in the current DLP+ workflow. We note a major advantage of inkjet dispensing systems is the ability to log comprehensive metadata, allowing for retrospective review to verify the single cell purity of the sample, should, for example, an unusual or unexpected cell population be identified in downstream informatic analysis. The number of droplets dispensed into each single cell containing well from the ResNet test print was counted and plotted in the histogram in Fig. 8b. With a timeout condition of 20 droplets, the distribution has a weighted mean of 6.29 droplets before a single cell was dispensed.
Methods
DLP+ single cell whole genome sequencing experiments
The cell suspension was stained with carboxyfluorescein succinimidyl ester (CSFE) fluorescent dye, as previously described,16 and dispensed into individual wells of a SmartChip™ nanowell array (Takara). The cell suspension fluid was allowed to evaporate and the chip imaged to validate the content of each well. Imaging was performed on a Nikon Eclipse Ti inverted microscope with the chip flipped over on the stage. A 10× extra-long working objective lens was focused on the bottom of the nanowell and each well was imaged under epifluorescence. Selected target wells are processed for downstream sequencing library construction.A head-to-head experiment compared cell dispensing using the new Isolatrix instrument with the current implementation of the DLP+ workflow, which uses the CellenONE platform, to assess compatibility and evaluate the impacts of the variable volume dispensing algorithm. The experiment was conducted on a single nanowell chip segmented into three regions. A region was designated for gDNA positive controls, a region for conducting current DLP+ workflow with CellenONE dispensing, and a region dispensed into using the Isolatrix. This region was further subdivided into two regions testing two different timeout conditions of a maximum of 5 and 10 droplets, to evaluate the trade-offs between single cell yield and background contamination. The number of wells for each cell dispensing region were selected with the aim of capturing an approximately equal number of single cells per condition. The experiment used a cell suspension from the 184-hTERT normal immortalized human breast epithelial cell line, with a measured average cell diameter of 16.4 μm and an input cell concentration of 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL. Within the chip's cell dispensing area, periodic wells were skipped (pitch spacing = 8 wells) to allow for no template control (NTC) wells to assess contamination within the reagents. A subset of non-cell wells which reached the timeout conditions were randomly selected and libraries constructed for no cell control (NCC) wells to assess the background DNA contamination level. A gDNA solution was dispensed using the instrument in a checkerboard pattern at the bottom segment of the chip in order to assess any cross-contamination of the dispensed droplets between wells.
000 cells per mL. Within the chip's cell dispensing area, periodic wells were skipped (pitch spacing = 8 wells) to allow for no template control (NTC) wells to assess contamination within the reagents. A subset of non-cell wells which reached the timeout conditions were randomly selected and libraries constructed for no cell control (NCC) wells to assess the background DNA contamination level. A gDNA solution was dispensed using the instrument in a checkerboard pattern at the bottom segment of the chip in order to assess any cross-contamination of the dispensed droplets between wells.
Reagent dispensing for the DLP+ protocol was performed at Canada's Michael Smith Genome Sciences Centre (GSC) using a sciFLEXARRAYER S3 instrument, as previously described.16 Libraries were sequenced on an Illumina NovaSeq 6000 with paired-end 150 bp reads.
Bioinformatic analysis
Demultiplexed paired FASTQ files were obtained for each well from the sequencing core at Canada's Michael Smith Genome Sciences Centre. For each FASTQ pair, DNA reads were processed using FASTP (v0.23.4)69 to trim adapters and poly-G artefacts. The resulting trimmed reads were aligned to human reference genome (hg38) using Minimap2 (v2.26)66 with the short-read preset parameter (−x sr). The total number of mapped reads per well was quantified using the Samtools (v1.19)70 flagstat command, and coverage depth was measured with Picard CollectWgsMetrics (v2.27.5).71 Copy number profiles and quality metrics for each cell were generated using HMMcopy (v0.0.80)67 as previously described.16 Cells were classified as high quality if the copy number quality score was greater than 0.75 and the total number of mapped reads exceeded 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Figures were generated using tidyverse (v2.0.0)72 and ggplot2 (v3.4.4), and statistical significance was calculated using ggpubr (v0.6.0).73 A link to the full analysis pipeline can be found in the Code availability section.
000. Figures were generated using tidyverse (v2.0.0)72 and ggplot2 (v3.4.4), and statistical significance was calculated using ggpubr (v0.6.0).73 A link to the full analysis pipeline can be found in the Code availability section.
Mouse husbandry
All mice used in this study were maintained in accordance with the University of British Columbia's Animal Care Committee's standards under specific pathogen-free conditions. Up to four mice were housed per cage and were maintained on a regular chow diet and water ad libitum on a 12 hour light–dark cycle. Adult mice used for dissociation experiments were B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (Strain #: 00799, The Jackson Laboratory). All mice were viable and fertile with normal phenotype. All animal procedures were performed in accordance with and approved by the University of British Columbia's Animal Care Committee's standards (Research Ethics Board Certificate numbers: A24-0215, A24-0204, A20-021, A20-022).Primary mouse liver sample preparation
An adult mouse liver was dissected, washed in cold PBS and divided into pieces (UBC Research Ethics Board Certificate: A24-0215, A24-0204, A20-021, A20-022). Tissue was enzymatically digested with Liberase (Millipore-Sigma) at 37 °C shaking 2000 rpm for 30 minutes, followed by dissociation using the gentleMACS Dissociator (Miltenyi Biotec). The disaggregated sample was passed through a 70 μm strainer and enriched for hepatocytes by slow centrifugation at 50 g for 4 min at 4 °C followed by a density gradient using a 25% Percoll (Sigma-Aldrich) solution and centrifugation at 1250g for 20 min at 4 °C. Cells were resuspended and red blood cell lysis performed using ammonium chloride (Stemcell). Cells were washed three times and then stained with CFSE (ThermoFisher) before dispensing.Single cell dispensing validation experiment using glass slides
The flexure spring on the substrate holder was adapted to hold a standard 3′′ × 1′′ × 1 mm microscope slide. A grid of 20 × 72 targets was dispensed at the same pitch spacing of the nanowell chip. A CSFE stained HEK cell line sample was prepared and dispensed under normal conditions. The PBS suspension medium left salt outlines of the dispensed droplets, enabling clear localization of each droplet under brightfield microscopy (Nikon Eclipse Ti-E). At a timeout of a maximum of 20 consecutive droplets, the outline of each dispensed position did not overlap, maintaining the distinct feature of each printed target on the glass slide. The slide was imaged under fluorescence imaging (488 nm excitation, 530 nm emission) to identify any cells within the droplet and brightfield imaging to resolve the outline of each dispensed target. The images were composited together and compared with the predictions of the instrument.Discussion
The Isolatrix is a new single cell inkjet dispensing instrument that enables rapid, high-throughput cell isolation under real-time monitoring. Compared to existing instruments, the dispensing algorithm results in 9.69× faster isolation times with consistent dispensing at less than one second per cell. The trained neural network minimizes labelling errors such as doublets and ensures high confidence in isolated sample purity. As such, the reactive printing algorithm enhances throughput and scalability by improving capture efficiency. Throughput can be further improved upon by replacing the current motorized stage with a faster alternative, such as with an air-bearing system. Notably, parameters such as droplet frequency and image analysis times are a function of the instrument's computational performance and can also benefit from hardware improvements. Direct visual confirmation via captured nozzle images enhances precision over emulsion-based microfluidic systems while maintaining compatibility with a broad range of cell types and sample plates. The incorporation of information from the captured nozzle and microscopy images of each cell, post-isolation, offers robust validation that spotted cells are indeed singleton. This combination enables the analysis of thousands of cells per experiment, providing a scalable and information-driven approach to single cell workflows.Compared to current emulsion-based platforms where a lack of capacity for sequential reagent addition limits available protocols, Isolatrix dispensing into open nanowells is compatible with a variety of single cell omics protocols. The dispensing algorithm, speed, and compatibility with sequencing library chemistry was first validated with a single cell whole genome sequencing experiment performed in nanowell chips. Further testing using a dissociated primary mouse liver sample demonstrated the compatibility of the low shear stress environment within the inkjet printhead in maintaining the integrity of the fragile sample. Future work can focus on optimizing the tissue dissociation to reduce debris or expansion of the neural network's detection capabilities to differentiate debris from cells. This experiment also validated the use of a higher droplet timeout condition to improve single cell capture efficiency. We note that given the stability of DNA, the DLP+ protocol permits complete evaporation of the deposited cell suspension fluid prior to sequencing library preparation. The instrument is compatible with humidity control to prevent evaporation and future implementation of protocols that target less stable biomolecules (e.g. RNA) can take advantage this. The system's performance was tuned, identifying parameters such as the input cell concentration and timeout conditions which can improve throughput and single cell occupancy. The neural networks demonstrated robust generalization across different cell types. The neural network's output correlated well with the dispensed outcome as validated by fluorescent imaging, and the integrated IUI and dispensing summary facilitates data traceability and provides high confidence in the single cell purity of the dispensed samples.
Conclusions
The Isolatrix is an inkjet-based single cell dispensing instrument that uses machine vision feedback and neural network classification for rapid and accurate cell isolation. This allows for high-throughput experiments with high single-cell confidence as validated using the DLP+ whole genome sequencing workflow. The open-well format opens the door for future support for scaled implementation of a range of single cell transcriptomics, epigenomics, and proteomics workflows, where the precise isolation and rapid processing of cells are critical for capturing transient signals. The instrument's flexibility can further enable ease of development for future multi-omics protocols, filling critical knowledge gaps and driving new discoveries in the transformative field of single cell biology.Author contributions
EC designed and built the instrument and carried out the cell dispensing experiments. GC and HM contributed to bioinformatic workflow development and genomic data analysis. MR and PAH contributed to dissociation protocol development and sample preparation for the mouse liver experiments. EC, GC, RC, AS, and KCC wrote the manuscript. RC contributed to instrument development and supervision. AS and KCC acquired funding for this project and oversaw and supervised the study.Conflicts of interest
KCC and EC are inventors on a patent application covering elements of the dispensing technology (CA3149667/US20220155331A1/EP4010681A4/CN114341617A/WO2021022374A1).Data availability
Supplementary information is available. See DOI: https://doi.org/10.1039/D5LC00514K.Genomic data generated in this study is available through the Sequence Read Archive under accession number PRJNA1299548. Data related to the Isolatrix instrument is available upon request.
Code availability
The genomic data analysis pipeline is available at: https://svn.bcgsc.ca/bitbucket/projects/SL/repos/scwgs_pipeline/browse.Acknowledgements
The authors gratefully acknowledge funding support from Genome British Columbia (PIF0001), the Natural Sciences and Engineering Research Council of Canada (I2IPJ 544495-19, RGPIN-2020-04798), and the Canada Foundation for Innovation (40044). AS is a Michael Smith Foundation for Health Research Scholar (SCH-2022-2625) and acknowledges additional funding support from the BC Cancer Foundation (1NSRG052). The authors are grateful to work and live on land that is the traditional, ancestral, and unceded territory of the Coast Salish Peoples, including the territories of the Musqueam, Squamish, and Tsleil-Waututh First Nations.References
- G. D. Evrony, A. G. Hinch and C. Luo, Applications of Single-Cell DNA Sequencing, Annu. Rev. Genomics Hum. Genet., 2021, 22(1), 171–197 CrossRef PubMed.
- H. O. Lee and W. Y. Park, Single Cell Genomics for Tumor Heterogeneity, Adv. Exp. Med. Biol., 2021, 1187, 205–214 CrossRef CAS PubMed.
- D. Wang and S. Bodovitz, Single cell analysis: the new frontier in 'omics', Trends Biotechnol., 2010, 28(6), 281–290 CrossRef CAS PubMed.
- K. Watanabe, et al., Genetic mapping of cell type specificity for complex traits, Nat. Commun., 2019, 10(1), 3222 CrossRef CAS PubMed.
- L. A. Miles, et al., Single-cell mutation analysis of clonal evolution in myeloid malignancies, Nature, 2020, 587(7834), 477–482 CrossRef CAS PubMed.
- L. G. Martelotto, et al., Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples, Nat. Med., 2017, 23(3), 376–385 CrossRef CAS PubMed.
- H. Wu, et al., Evolution and heterogeneity of non-hereditary colorectal cancer revealed by single-cell exome sequencing, Oncogene, 2017, 36(20), 2857–2867 CrossRef CAS PubMed.
- A. P. Patel, et al., Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma, Science, 2014, 344(6190), 1396–1401 CrossRef CAS PubMed.
- I. Tirosh, et al., Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq, Science, 2016, 352(6282), 189–196 CrossRef CAS PubMed.
- J. L. Schillo, et al., Single-cell genomics analysis reveals complex genetic interactions in an in vivo model of acquired BRAF inhibitor resistance, NAR Cancer, 2024, 6(1), zcad061 CrossRef PubMed.
- D. Ramazzotti, et al., Longitudinal cancer evolution from single cells, bioRxiv, 2020, preprint, DOI:10.1016/j.jocs.2021.101523.
- L. M. LaFave, R. E. Savage and J. D. Buenrostro, Single-Cell Epigenomics Reveals Mechanisms of Cancer Progression, Annu. Rev. Cancer Biol., 2022, 6(1), 167–185 CrossRef.
- N. Shembekar, et al., Droplet-based microfluidics in drug discovery, transcriptomics and high-throughput molecular genetics, Lab Chip, 2016, 16(8), 1314–1331 RSC.
- R. Zilionis, et al., Single-cell barcoding and sequencing using droplet microfluidics, Nat. Protoc., 2017, 12(1), 44–73 CrossRef CAS PubMed.
- T. P. Lagus and J. F. Edd, High-throughput co-encapsulation of self-ordered cell trains: cell pair interactions in microdroplets, RSC Adv., 2013, 3(43), 20512–20522 RSC.
- E. Laks, et al., Clonal Decomposition and DNA Replication States Defined by Scaled Single-Cell Genome Sequencing, Cell, 2019, 179(5), 1207–1221.e22 CrossRef CAS PubMed.
- H. Zahn, et al., Scalable whole-genome single-cell library preparation without preamplification, Nat. Methods, 2017, 14(2), 167–173 CrossRef CAS PubMed.
- S. Stanisheuski, et al., Thermal inkjet makes label-free single-cell proteomics accessible and easy, Front. Chem., 2024, 12, 1428547 CrossRef CAS PubMed.
- Z. Yang, et al., AM-DMF-SCP: Integrated Single-Cell Proteomics Analysis on an Active Matrix Digital Microfluidic Chip, JACS Au, 2024, 4(5), 1811–1823 CrossRef CAS PubMed.
- J. Woo, et al., High-throughput and high-efficiency sample preparation for single-cell proteomics using a nested nanowell chip, Nat. Commun., 2021, 12(1), 6246 CrossRef CAS PubMed.
- J. Wu, et al., One-Shot Single-Cell Proteome and Metabolome Analysis Strategy for the Same Single Cell, Anal. Chem., 2024, 96(14), 5499–5508 CrossRef CAS PubMed.
- T. Hu, et al., Single-cell spatial metabolomics with cell-type specific protein profiling for tissue systems biology, Nat. Commun., 2023, 14(1), 8260 CrossRef CAS PubMed.
- J. M. Fulcher, et al., Parallel measurement of transcriptomes and proteomes from same single cells using nanodroplet splitting, Nat. Commun., 2024, 15(1), 10614 CrossRef CAS PubMed.
- K. Yamashita, et al., An improved technique for dissociating hepatocytes from fixed mouse liver for cytochemistry, Stain Technol., 1985, 60(1), 29–37 CrossRef CAS PubMed.
- E. C. Welch, H. Yu and A. Tripathi, Optimization of a Clinically Relevant Chemical-Mechanical Tissue Dissociation Workflow for Single-Cell Analysis, Cell. Mol. Bioeng., 2021, 14(3), 241–258 CrossRef CAS PubMed.
- R. Sekiguchi and B. Hauser, Preparation of Cells from Embryonic Organs for Single-Cell RNA Sequencing, Curr. Protoc. Cell Biol., 2019, 83(1), e86 CrossRef PubMed.
- G. P. Dunn, et al., Emerging insights into the molecular and cellular basis of glioblastoma, Genes Dev., 2012, 26(8), 756–784 CrossRef CAS PubMed.
- N. L. Calistri, et al., Microfluidic active loading of single cells enables analysis of complex clinical specimens, Nat. Commun., 2018, 9(1), 4784 CrossRef PubMed.
- F. Bray, et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, Ca-Cancer J. Clin., 2018, 68(6), 394–424 CrossRef PubMed.
- P. Ramachandran, et al., Resolving the fibrotic niche of human liver cirrhosis at single-cell level, Nature, 2019, 575(7783), 512–518 CrossRef CAS PubMed.
- R. Carlessi, et al., Single-nucleus RNA sequencing of pre-malignant liver reveals disease-associated hepatocyte state with HCC prognostic potential, Cell Genomics, 2023, 3(5), 100301 CrossRef CAS PubMed.
- S. A. MacParland, et al., Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations, Nat. Commun., 2018, 9(1), 4383 CrossRef PubMed.
- R. Carlessi, et al., Single-nucleus RNA sequencing of pre-malignant liver reveals disease-associated hepatocyte state with HCC prognostic potential, Cell Genomics, 2023, 3(5), 100301 CrossRef CAS PubMed.
- M. L. Richter, et al., Single-nucleus RNA-seq2 reveals functional crosstalk between liver zonation and ploidy, Nat. Commun., 2021, 12(1), 4264 CrossRef CAS PubMed.
- T.-M. Liou, C.-Y. Chan and K.-C. Shih, Effects of actuating waveform, ink property, and nozzle size on piezoelectrically driven inkjet droplets, Microfluid. Nanofluid., 2009, 8(5), 575–586 CrossRef.
- H.-C. Wu, et al., Study of Micro-Droplet Behavior for a Piezoelectric Inkjet Printing Device Using a Single Pulse Voltage Pattern, Mater. Trans., 2004, 45(5), 1794–1801 CrossRef CAS.
- J. Dou, et al., Study on the dynamic characteristics of piezoelectric inkjet print head with basic excitation methods, Microsyst. Technol., 2019, 25(8), 2985–2992 CrossRef.
- R. Bernasconi, et al., Piezoelectric Drop-On-Demand Inkjet Printing of High-Viscosity Inks, Adv. Eng. Mater., 2021, 24(1), 2100733 CrossRef.
- L. Li, et al., All Inkjet-Printed Amperometric Multiplexed Biosensors Based on Nanostructured Conductive Hydrogel Electrodes, Nano Lett., 2018, 18(6), 3322–3327 CrossRef CAS PubMed.
- J. T. Kirk, et al., Multiplexed inkjet functionalization of silicon photonic biosensors, Lab Chip, 2011, 11(7), 1372–1377 RSC.
- A. Memic, et al., Bioprinting technologies for disease modeling, Biotechnol. Lett., 2017, 39(9), 1279–1290 CrossRef CAS PubMed.
- E. Pagan, et al., A handheld bioprinter for multi-material printing of complex constructs, Biofabrication, 2023, 15(3), 035012 CrossRef CAS PubMed.
- N. Hakimi, et al., Handheld skin printer: in situ formation of planar biomaterials and tissues, Lab Chip, 2018, 18(10), 1440–1451 RSC.
- J. Shin, et al., Optimized 3D Bioprinting Technology Based on Machine Learning: A Review of Recent Trends and Advances, Micromachines, 2022, 13(3), 363 CrossRef PubMed.
- J. Shin, et al., Machine Learning-Enhanced Optimization for High-Throughput Precision in Cellular Droplet Bioprinting, Adv. Sci., 2025, 2412831 CrossRef PubMed.
- M. Yumoto, et al., Evaluation of the effects of cell-dispensing using an inkjet-based bioprinter on cell integrity by RNA-seq analysis, Sci. Rep., 2020, 10(1), 7158 CrossRef CAS PubMed.
- W. L. Ng and V. Shkolnikov, Optimizing cell deposition for inkjet-based bioprinting, Int. J. Bioprint., 2024, 2135 CrossRef CAS.
- A. Leduc, et al., Massively parallel sample preparation for multiplexed single-cell proteomics using nPOP, Nat. Protoc., 2024, 19(12), 3750–3776 CrossRef CAS PubMed.
- F. Salmen, et al., High-throughput total RNA sequencing in single cells using VASA-seq, Nat. Biotechnol., 2022, 40(12), 1780–1793 CrossRef CAS PubMed.
- S. Haile, et al., Frontiers | A Scalable Strand-Specific Protocol Enabling Full-Length Total RNA Sequencing From Single Cells, Front. Genet., 2021, 12 DOI:10.3389/fgene.2021.665888.
- V. Akbari, et al., Parent-of-origin detection and chromosome-scale haplotyping using long-read DNA methylation sequencing and Strand-seq, Cell Genomics, 2023, 3(1), 100233 CrossRef CAS PubMed.
- O. Lucas, et al., Characterizing the evolutionary dynamics of cancer proliferation in single-cell clones with SPRINTER, Nat. Genet., 2025, 57(1), 103–114 CrossRef CAS PubMed.
- M. J. Williams, et al., Luminal breast epithelial cells of BRCA1 or BRCA2 mutation carriers and noncarriers harbor common breast cancer copy number alterations, Nat. Genet., 2024, 56(12), 2753–2762 CrossRef CAS PubMed.
- F. Guerrera, et al., The Influence of Tissue Ischemia Time on RNA Integrity and Patient-Derived Xenografts (PDX) Engraftment Rate in a Non-Small Cell Lung Cancer (NSCLC) Biobank, PLoS One, 2016, 11(1), e0145100 CrossRef PubMed.
- L. Opitz, et al., Impact of RNA degradation on gene expression profiling, BMC Med. Genomics, 2010, 3(1), 36 CrossRef PubMed.
- E. Cheng, et al., Investigation of the hydrodynamic response of cells in drop on demand piezoelectric inkjet nozzles, Biofabrication, 2016, 8(1), 015008 CrossRef PubMed.
- S. Parsa, et al., Effects of surfactant and gentle agitation on inkjet dispensing of living cells, Biofabrication, 2010, 2(2), 025003 CrossRef PubMed.
- D. Chahal, A. Ahmadi and K. C. Cheung, Improving piezoelectric cell printing accuracy and reliability through neutral buoyancy of suspensions, Biotechnol. Bioeng., 2012, 109(11), 2932–2940 CrossRef CAS PubMed.
- K. C. Cheung and E. Cheng, Facilitating controlled particle deposition from a droplet dispenser, WO2021022374, The University of British Columbia, CA, 2021.
- H. Ismail Fawaz, et al., Deep learning for time series classification: a review, Data Min. Knowl. Discov., 2019, 33(4), 917–963 CrossRef.
- K. Piaskowski and D. Belter, Fast Object Detector Based on Convolutional Neural Networks, Computational Modeling of Objects Presented in Images. Fundamentals, Methods, and Applications, 2019 Search PubMed.
- S. Nishimoto, et al., Predicting the future direction of cell movement with convolutional neural networks, PLoS One, 2019, 14(9), e0221245 CrossRef CAS PubMed.
- M. Motamedi, et al., Scalable CNN Synthesis for Resource-Constrained Embedded Platforms, IEEE Internet Things J., 2022, 9(3), 2267–2276 Search PubMed.
- J. Xu, et al., RegNet: Self-Regulated Network for Image Classification, IEEE Trans. Neural Netw. Learn. Syst., 2023, 34(11), 9562–9567 Search PubMed.
- M. Wenninger, et al., Timage - A Robust Time Series Classification Pipeline, International Conference on Artificial Neural Networks, 2019 Search PubMed.
- H. Li, New strategies to improve minimap2 alignment accuracy, Bioinformatics, 2021, 37(23), 4572–4574 CrossRef CAS PubMed.
- G. Ha, et al., Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer, Genome Res., 2012, 22(10), 1995–2007 CrossRef CAS PubMed.
- Y. Tanaka, et al., Evaluation of effects of shear stress on hepatocytes by a microchip-based system, Meas. Sci. Technol., 2006, 17(12), 3167–3170 CrossRef CAS.
- S. Chen, Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp, iMeta, 2023, 2(2), e107 CrossRef CAS PubMed.
- P. Danecek, et al., Twelve years of SAMtools and BCFtools, GigaScience, 2021, 10(2), giab008 CrossRef PubMed.
- Picard toolkit, Broad Institute, 2019 Search PubMed.
- H. Wickham, et al., Welcome to the Tidyverse, J. Open Source Softw., 2019, 4(43), 1686 CrossRef.
- A. Kassambara, ggpubr: 'ggplot2' Based Publication Ready Plots, 2023 Search PubMed.
| This journal is © The Royal Society of Chemistry 2025 |