 Open Access Article
Open Access ArticleThe outputs of molecular sensors detectable by human senses
Maria S.
Rubel
 ab,
Tatiana
Zemerova
ab,
Tatiana
Zemerova
 a and
Dmitry M.
Kolpashchikov
a and
Dmitry M.
Kolpashchikov
 *cd
*cd
aLaboratory of DNA-nanosensoric Diagnostic, ChemBio Cluster, ITMO University, Saint Petersburg 191002, Russia
bAmyloid Biology Laboratory, St. Petersburg State University, St. Petersburg, 199034, Russia
cChemistry Department, University of Central Florida, Orlando, FL 32816-2366, USA. E-mail: Dmitry.Kolpashchikov@ucf.edu
dBurnett School of Biomedical Sciences, University of Central Florida, Orlando, FL 32816, USA
First published on 23rd January 2025
Abstract
Molecular sensors respond to the presence of biological analytes by producing signals that are either directly perceivable by human sensory systems or converted into electric signals, which require electronic devices for communicating the signals to humans. Here, we review the outputs of molecular sensors detectable directly by human senses. According to the literature, sensors with visual outputs dominate. Undeservedly unnoticed, sensors that release gases might be particularly useful since the gas output can be detected with the several human senses in a quantifiable format. Relatively new sensors with tactile outputs can be accessed by visually impaired people. Molecular sensors communicating their outputs directly to human senses bypassing electronic devices may contribute to the development of point-of-care testing technologies, as well as providing the direct communication of molecular nanorobots with humans.
1. Introduction
Sensing biological molecules is important in medical diagnostics, environmental monitoring, and forensic applications, as well as in fundamental research.1 A common trend in developing tests is making them compatible with point-of-care or home testing, having the qualities manifested by the ASSURED criteria (affordable, sensitive, specific, user-friendly, rapid, robust, equipment-free, and deliverable to end-users) announced by the World Health Organization (WHO).2 The COVID-19 pandemic emphasized the importance of fast and affordable testing. Additionally, a multiplexing capability that enables cost- and time-efficient utilization of available resources is valuable for laboratory-based diagnostics.3The success of affordable and easy to use home tests can be demonstrated by the glucometer, an essential device used to test blood glucose levels of diabetic or hypoglycemia patients.4 In a glucometer, the enzyme glucose oxidase acts as a glucose-specific molecular sensor that produces electrons as an output signal. This technology has empowered patients to monitor their blood sugar level conveniently and effectively.
According to the International Union of Pure and Applied Chemistry (IUPAC), a chemical sensor is a device that converts chemical information, ranging from the concentration of a specific sample component to total composition analysis, into an analytically useful signal.5 The most common signals include light absorbance (colorimetric) or emission (e.g., fluorescence and chemiluminescence), and electric (current). The nature of signal outputs may affect the compliance to the ASSURED criteria.2 For example, tests employing color changes as outputs could be more affordable than those producing fluorescence signals since they require neither light-emitting nor light-sensing devices.
We are interested in outputs of molecular sensors in the context of molecular robots,6,7 nanodevices equipped with sensing, computing, and actuating functions. Molecular robots, if created, should be able to sense complex sets of biological markers, analyse them using sophisticated algorithms, and autonomously initiate environmental changes, such as therapeutic actions.8 Molecular robots could also communicate directly with humans through one of the five senses: taste, smell, vision, hearing, or touch, without the need for an electronic (computer) mediator. This feature article focuses on outputs of molecular sensors (Scheme 1), where sensing, signal transduction, and signaling components consist of a discrete number of molecules. These sensors can serve as building blocks for molecular robots. However, sensors that produce electric and magnetic outputs and thus require specialized equipment are outside of the scope of this minireview. This work deliberately excluded machine learning and artificial intelligence, because they rely on computational support, and have been covered in a series of recent reviews on this subject.9–11 Such support can indeed increase the sensitivity but more data and systematic assessments are still needed to accurately characterize the size of the effect.
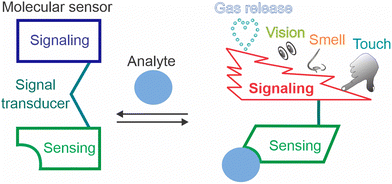 | ||
| Scheme 1 The components of molecular sensors. The signal output can be sensed by human senses directly without instruments. | ||
This article starts with the description of sensors with the most common visually detectable outputs. We then discuss outputs that can be sensed by touch, smell, gas release, taste, and hearing. The discussion also highlights the potential and prospects of these biosensors in diagnostics.
2. Visual outputs
Optical molecular sensors are extensively used both for equipment-free qualitative/semi-quantitative analysis and equipment-based quantitative analysis. This section covers selected and most recent examples of molecular sensors that change color, sample transparency, or light emission intensity (fluorescence or chemiluminescence) and, therefore, produce an output that can be detected visually.2.1. Organic dyes as molecule sensors
The ability of organic dyes to change color in response to a pH change has been used in education,12 research,13 and industry.14 A well-known example is phenolphthalein (PP in Fig. 1(A)). The triarylmethane lactone structure of PP loses two protons at high pH to form a conjugated quinone-like structure of PP2− responsible for the pink color.15 Takezawa et al. demonstrated the use of nanostructure-encapsulated PP for the detection of adamantane derivatives (Fig. 1(B)).16 In this strategy, the nanostructure-bound PP− is colorless even at pH of 10.0 as the nanostructure stabilizes its colorless lactone PP− form. Binding at the adamantane derivatives displaces PP− into the solution thus converting it into pink PP2−. Recently, Zhang et al. proposed a similar strategy for reporting metal ions (e.g., Fe3+) using the Rhodamine B dye encapsulated in a porous coordination cage.17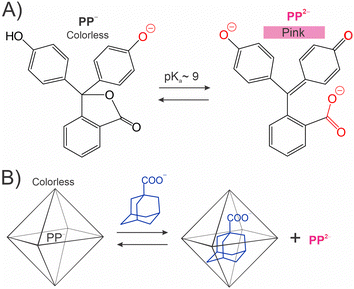 | ||
| Fig. 1 pH indicators as molecule sensors. (A) Phenolphthalein (PP).15 (B) Detection of adamantane derivatives using a caged PP.16 | ||
pH indicators can detect the progress of nucleic acid amplification reactions. For example, Tanner et al. sensed the decrease in pH from 8 to 6.5 during loop mediated amplification (LAMP) using several dyes (e.g., phenol red and cresol red).18 It was found that the amount of DNA input, amplicon length, sample preparation, and even atmospheric conditions impact the color change, thus compromising the robustness of the method.19
The color of organic dyes can be changed in response to binding ions such as K+, Na+, Ca2+ or as a result of RedOx reactions.20,21 For example, Lian et al. synthesized oligo-arene compounds that display yellow or pink hues upon binding with Hg2+ or Ag+ ions, achieving a practically significant limit of detection (LOD) of 28 nM for Hg2+.22
Noteworthily, the concentrations of dyes used for visual detection are typically measured in the micromolar to submicromolar range (0.25–10 μM). Correspondingly, the LOD below the submicromolar range cannot be reached without adding a signal amplification step, which aligns with the sensing capabilities of both a human eye and a standard spectrophotometer. This LOD level enables detection of practically significant concentrations of small molecules and ions but falls short to detect proteins or nucleic acids in biological samples.
2.2. Aggregation of nanoparticles
The color change caused by aggregation of gold nanoparticles (GNPs) is one of the most common strategies for biosensing with visual output. GNPs have been used in enzyme-linked immunosorbent assay (ELISA), lateral flow assays, polymerase chain reaction (PCR), immuno-PCR,23 among other formats.24 The most commercially successful applications use GNPs as tags for antibodies, oligonucleotides or glycans.25 This format typically achieves a moderate LOD of 5 nM or higher.26 For example, the lateral flow urine pregnancy test detects human chorionic gonadotropin at concentrations of ∼25 IU L−1, which corresponds to approximately 75 nM.27To achieve multicolor/multiplex detection, gold nanorods can be used. The plasmonic properties of gold nanorods can be fine-tuned by varying the aspect ratio, allowing light absorption at different wavelengths.28 Other nanoparticles can be used for signal visualization including silver, carbon, and selenium. Ozefe et al. reported a smartphone-assisted Hepatitis C virus detection using a magnetic levitation assay that separates virus-free magnetic particles from those bound to viral particles.29 The LOD was about 10 fold lower than that of an ELISA assay.29
Most approaches based on analyte-specific GNP aggregation produce aggregates of a poorly controlled structure and sedimentation rate. In contrast, Guo et al. developed a GNP-base molecular sensor that takes advantage of a controlled association of only two GNPs attached to two DNA sensing strands. A nucleic acid analyte brings the particles in proximity due to the formation of a DNA three-way junction complex (Fig. 2).26 They used asymmetrically modified GNPs with the DNA probe attached only at a restricted area of each of the two particles. This led to a controlled positioning of the particles after the complex formation. Such a design improved color stability, reduced the LOD to ∼1.0 pM and increased the dynamic range.26 This approach is yet to be applied for other types of nanoparticles.
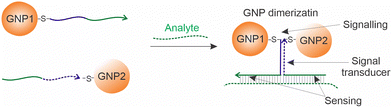 | ||
| Fig. 2 Analyte-induced GNP dimerization as reported by Guo et al.26 A nucleic acid analyte brings two nanoparticles in proximity, which causes an absorption spectral shift. | ||
Common limitations of the GNP aggregation-based techniques include sedimentation of the GNP aggregates, accompanied by the color loss, which leads to false negative signals. The analyte-independent aggregation can lead to false-positive signals, relatively low sensitivity, and a narrow dynamic range.
2.3. Enzyme-free chemical reactions
Avoiding protein enzymes in test systems can be advantageous, since enzymes are unstable and/or require specific storage conditions. Enzyme-free chemical reactions resulting in visually detectable products can be exemplified by the Tollens’ reagent for detection of aldehydes and reducing sugars (Fig. 3(A)). Recently, Xiang et al. demonstrated that GNPs facilitate this reaction under alkaline conditions resulting in glucose detection with an improved LOD of 320 nM.30 Using a similar approach, Borah et al. detected formaldehyde and benzaldehyde in the linear range of 10–150 nM and 150–750 nM, respectively.31 | ||
| Fig. 3 Organic dye sensors. (A) Tollen's reagent oxidizes aldehydes and produces a visible silver precipitate. (B) Reaction of ninhydrin with amino acids. (C) Initially yellow LH chemosensor loses Cu2+ and becomes colorless in the presence of ATP, but not ADP or AMP.32 | ||
Ninhydrin, another well-known representative of the color-changing reagents, is widely used for detection of amines including amino acids (Fig. 3(B)). Recently, Alimohammadi et al. developed a ninhydrin-based array of metal-doped carbon dots that can simultaneously distinguish all 20 amino acids in the format of a ‘chemical tongue’33 with a detection limit of 1.0 μM for lysine and 80 μM for arginine.33 Mondal and Manivannan synthesized a colorimetric sensor based on Cu2+ chelate (‘LH’ in Fig. 3(C)). Cysteine (LOD = 60 nM) or ATP (LOD = 130 nM) displaced LH from the complex with Cu2+, which resulted in the loss of a yellow color.32 An enzyme-free reaction, while being affordable, demonstrates moderate sensitivity due to the lack of signal amplification capabilities.
2.4. Enzyme and DNAzyme catalysed reactions
Horse radish peroxidase (HRP) is a well-known colorogenic label used in numerous applications.34 HRP oxidizes a broad range of colorless organic substrates, thereby turning them into colored products. The advantages of such visually detectable signals are low reagent cost and additional signal amplification through multiple substrate turnover by HRP.Martell et al. converted HRP into a true molecular sensor by splitting it into two fragments and fusing each fragment with a protein of interest (P1 and P2 in Fig. 4(A)).35 The enzymatic activity of the split HRP was restored when the P1 and P2 bind each other. This can be used for detecting protein–protein interactions.35 Due to a broad specificity of HRP, it is a versatile signal reporter. For example, HRP-catalysed oxidation of Amplex Red into resorufin enables a fluorescence output signal.36 Oxidation of a phenol moiety of a phenol–biotin conjugate generates radicals to tag proteins of interest with biotin for subsequent detection by pull-down experiments.37 DAB (3,3′-diaminobenzidine) is oxidized by HRP into a polymer that attracts OsO4 as a ‘contrast’ agent to enable an electron microscopy study of HRP-containing cells.38 It was recently reported that ascorbate peroxidase has advantages over HRP in biotechnological applications due to the lack of glycans and cysteines in its structure and greater stability.39 Another enzyme with a colorimetric output is alkaline phosphatase (ALP), which is typically used as a label.40 However, to date, ALP-based molecular sensors (e.g., split versions of ALP) have not been developed. HRP is also smaller in size than ALP making it a tool of choice among colorimetric enzymes. Recently, Bae et al. presented an excellent review describing split enzyme systems that can be used for designing molecular sensors.41
 | ||
| Fig. 4 Principles of peroxidase-based sensors. (A) Split horse radish peroxidase (HRP).35 (B) Peroxidase-like DNA machine for detection of folded RNA analytes.42,43 Tight binding of the RNA analyte is ensured by long Arm 1 and Arm 3, while high selectivity is guaranteed by short Arm 1 complementary to the SNP site. (C) G4-based aptameric structural switch sensor for the detection of non-nucleic acid analytes.44–46 (D) Apollon aptamer-based DNAzyme sensor for detection of oligonucleotides.47 | ||
The success of HRP in analytical applications inspired a search for non-protein materials with peroxidize-like activity.48,49 Examples of such materials include nanocrystals of platinum-group metals,49,50 whose peroxidase-like activity is ∼104-fold greater than that of HRP.51
A well-known molecule that enhances the peroxidizing properties of a heme moiety is a DNA G-quadruplex (G4) introduced by Sen and colleagues.52 G4 can be divided into two parts and used as a sensor for nucleic acids.53,54 Recently, Gorbenko et al.42 integrated a split G4 sensor into a DNA nanostructure called the DNA machine for the detection of folded RNA by using three RNA-binding arms (Fig. 4(B)). The sensor enabled naked-eye detection of a bacterial RNA amplicon obtained from 100 bacterial genome equivalents. The same approach was reported to be efficient in detecting dsDNA amplicons obtained by stem-loop primer-assisted isothermal amplification (SPA), but not by PCR or LAMP.43
A structure switching G4 system fused with aptamers can be applied for detecting a variety of non-nucleic acid analytes (Fig. 4(C)).44–46,55 For example, Ahmadi et al. designed G4-based structural switches for sensing quinine with a LOD as low as 600 nM.44 However, the authors noted challenges in the design of such sensors. For example, their attempts to produce a sensor for ATP or Staphylococcus enterotoxin B were unsuccessful.44 Tang et al. reported a G4-based sensor for detecting antibiotic tetracycline in food with a LOD of 3.1 nM via altering the enzymatic activity of the G4–hemin complex.45 Xiang et al. developed a multistage procedure for the label-free colorimetric sensing of phenylalanine at 190 nM level.46 This LOD was possible only after the cascade signal amplification. Similarly, Gerasimova et al. succeeded in designing a 4G cascade for amplification-free detection of mycobacteria 16S rRNA with a detection limit of ∼12.5 ng of total bacterial RNA in a 3 μL sample or about 106 bacteria cells.55
Recently, Volek et al. have selected a DNA sequence named Apollon that can specifically dephosphorylate a colorless 4-nitrophenyl phosphate to produce yellow 4-nitrophenol (Fig. 4(D)).47 The Apollon structure can be converted into a structural switch for sensing of specific nucleic acids with a prohibitively high LOD of 20 μM.47 A sensor variant targeting RNase A exhibited a more practically relevant LOD of 10 nM.47 This work represents one of the few alternatives to the peroxidase reaction for producing colored outputs.
2.6. Fluorescent sensors
Fluorescent sensors are widely used due to their availability and higher sensitivity than colorimetric sensors due to low background when measured using an instrument. Recently, Tsuchiya et al. synthesized a series of pH sensitive fluorescent dyes (an example of a sulfo-containing piperazine-based cyanine dye is shown in Fig. 5(A)) that can be used for photoacoustic imaging of cancer cells.56 Fluorescent dyes are well known as sensors of temperature,57 or ions.58 Such compounds often have low toxicity and can be used in living cells.59 Organic dyes have been extensively applied for bioimaging and sensing,60–62 usually with a fluorescence microscope or a fluorometer. | ||
| Fig. 5 Examples of organic dyes that can be used as fluorescent sensors. (A) The functional architecture of a pH-responsive cyanine dye for cancer cell photoacoustic imaging developed by Tsuchiya et al.56 The maximum absorption spectral shifts from ∼700 nm to ∼800 nm when pH changes from 7 to 3.5 in the tumour environment. (B) A self-immolative sensor Ac-DEVD-PABCNaph for detecting caspase-3 (Cas3) activity.63 Cas 3 cleavage followed by a self-immolative elimination of a naphthalimide derivative causes a red shift, in the emission maximum to allow the ratio of fluorescence intensities (475 nm/535 nm) be used for quantification of Cas 3 activity. | ||
Self-immolative sensors undergo irreversible disassembly upon analyte recognition accompanied by chromophore release or chemiluminescence.64 For example, Wynne synthesised a ‘self-immolative’ sensor for detecting caspase-3 activity, as shown in Fig. 5(B), with a LOD of ∼4.96 ng mL−1 (0.15 nM).63 A boronate-based self-immolative sensor was developed for specific detection of peroxynitrite with a dynamic range of around 4 orders of magnitude and a LOD of 2.5 μM.65 The sensor did not respond to the presence of H2O2 or HClO, which is important for the specific detection of peroxynitrite inside cells. In some cases, self-immolative sensors are capable of signal amplification because of their cascade-like chemical reactions. One limitation of such sensors is the potential toxicity of the cleavage products.64
A fluorescence output can also be detected with the naked eye at micromolar or high sub-micromolar concentrations. For example, Yuan et al. detected Hg2+ at 1 μM (0.2 ppm) using quantum dots.66 In another study, Wei et al. detected Δ9-tetrahydrocannabinol (THC) with a LOD of 0.6 μM using an aptamer-based sensor in the instrument-free format.67 These examples highlight the potential of fluorescent sensors to function effectively without a fluorometer.
Dyes that change fluorescence intensity upon binding to nucleic acids can be used as molecular sensors.68 Well-known examples include ethidium bromide, SYBR Green, SYBR Gold, and Gel Red. These dyes bind to nucleic acids non-specifically and, therefore, are generally used for visualizing DNA and RNA in a gel. For example, SYBR Green is widely used for device-free visualization of the LAMP products. To achieve sequence-specific nucleic acid recognition, a fluorescent light-up aptamer with affinity to an intrinsically low-fluorescent dye can be used in a format of binary (split) aptameric sensors.69 In this approach, two DNA or RNA strands bind to the abutting positions of the nucleic acid analyte and form a dye-binding site (Fig. 6(A)).70 Once bound, the dye exhibits enhanced fluorescence to an extent that naked eye-based detection is possible. In this case, the sample was irradiated with a flashlight or a simple portable light source. For example, Reed et al. demonstrated feasibility of visualising the presence of 100 nM mycobacterial DNA with a split dapoxyl aptamer probe71 using a fluorimeter upon transilluminator-assisted excitation of fluorescence and capturing the images with a smartphone camera.72
 | ||
| Fig. 6 Examples of nucleic acid sensors producing a fluorescent response. (A) Binary (split) light-up aptameric sensor: two RNA or DNA strands bind a nucleic acid analyte to form a binding site for a fluorogenic dye.69,70 (B) MB probe-based OWL-2 sensor for highly specific detection of nucleic acids.73 OWL-2 binds nucleic acid analyte T2, R, P and T4 arms followed by binding a universal MB probe (UMB) thus producing fluorescence. The SNV stie is complementary to the DNA scaffold-detached P strand to achieve gretest selectivity.73 (C) DNAzyme sensor for sensing RNase A: RNAse A activates inactive Aurora DNAzyme by cleaving its inhibitory rUAAA fragment. Active Aurora DNAzyme dephosphorylates 4-methylumbelliferyl phosphate (4-MUP) and turns it fluorescent.72 | ||
Molecular beacon (MB) probes, fluorophore and quencher labelled DNA hairpins,74 are versatile sensors for the analysis of nucleic acids75 and proteins.76 For example, recently Mueller et al. reported an MB probe-based sensor that forms a so called OWL nanostructure with nucleic acid analytes (Fig. 6(B)).73 The advantage of the sensor is in its ability to bind folded RNA and DNA and differentiate single nucleotide variations (SNVs) with great accuracy under ambient conditions.73 MB probes can also be used for fluorometer-free visual detection of analytes at sub-micromolar concentrations.77 To increase sensitivity of stoichiometric binding of analytes to the MB probes, organic fluorescent dyes can be replaced with nanoparticles. For example, recently Su et al. demonstrated detection of miR down to 1 aM using a MB probe-like sensor equipped with a lanthanide fluorophore that can be excited in the infrared range and GNP as a quencher.78
Recently, Volek et al. reported a DNA probe with dephosphorylating catalytic activity, named Aurora (Fig. 6(C)).79 The Aurora sequence was converted into a sensor for RNases by adding an extra rUAAA fragment that both inhibited Aurora's catalytic activity and served as a recognition site for RNase A. This innovative design achieved a LOD of 100 pM for the detection of RNase A.79 This low LOD was possible due to multiple substate turnover by RNase A.
Even though used mostly with fluorometers, fluorescent sensors can also be adopted for a naked-eye fluorometer free-format. The LOD of such a detection format is comparable with that of colorimetric sensors. However, a light source for excitation is unavoidable for fluorescent sensors, which complicates the detection assay. The sensitivity can be improved through catalytic signal amplification by protein enzymes80 or by catalytic nucleic acids.81 For example, RNA-cleaving DNAzyme sensors can cleave fluorophore- and quencher-labelled substrates to produce a fluorescence signal.82 The method can detect double-stranded DNA fragments.83 The LOD for fluorimeter-based detection can reach picomolar84 and subpicomolar ranges.85 Even though these sensors can also be used in a fluorometer-free format, the LOD for such output is yet to be determined.
2.7. Chemiluminescent sensors
Chemiluminescence is a phenomenon of light emission resulting from chemical transformation. Unlike fluorescence signalling, such a format does not require light sources for excitation, which also contributes to the reduced background and, consequently, to higher sensitivity as well as greater affordability.86The chemiluminescent signal can be produced via reactions catalysed by bacterial luciferase, beetle luciferase, and a system of HRP coupled to the luminol or by a variety of nanomaterials87 according to the reactions shown in Fig. 7(A)–(C). Bacterial luciferase is a more complex protein than beetle luciferase, but it requires no ATP to emit light. Split luciferase re-forms the active luciferase analyte upon binding, similar to split HRP (Fig. 4(A)).88 One of the recently developed biosensors, based on split luciferase, can be used for measuring the level of cyclic di-GMP, an intracellular signalling molecule in bacteria.89
 | ||
| Fig. 7 Examples of reactions used for chemiluminescent signalling. (A) Oxidation of the flavin mononucleotide by a beetle luciferase. (B) The basic reaction of luciferin by a beetle luciferase. (C) Decomposition of luminol to aminophthalate by HRP. (D) Oxidation of the CDP-star luminophore by Supernova DNAzyme.90 | ||
Alternatively, luminescence can be achieved without any protein enzymes. For example, luminol oxidation (Fig. 7(C))91 can be catalysed by a G4-hemin system (Fig. 4(B) and (C)).92 Such systems find applications in in vivo imaging and in vitro measurement of reactive oxygen species. Performance of luminol biosensors can be amplified with the use of nanoparticles, and, in some cases, such sensors achieve impressive femtomolar LOD93 in the case of instrument-based detection. However, to the best of our knowledge, no examples using naked eye visualization have been published. Besides luminol, other substrates such as acridinium ester, peroxyoxalate, 1,2-dioxetan, as well as various nanoparticles have also been used for chemiluminescence signalling.94 Recently, Chen et al. reported a G4 catalysed luminol oxidation sensor for the cell phone-assisted detection of nucleic acids.95 Svehlova et al.90 introduced a DNAzyme capable of catalysing dephosphorylation of luminophore CDP-Star (Fig. 7(D)) and designed a nucleic acid sensor akin to the Apolon DNAzyme sensor (Fig. 4(D)). The sensor demonstrated a LOD of ∼100 nM for a synthetic oligonucleotide target.
Chemiluminescence is devoid of photobleaching and phototoxicity and is suitable for in vivo studies. Unlike fluorescence, it does not require a light source for excitation. This also increases sensitivity due to the reduced background irradiation. As a result, chemiluminescence provides a dynamic range of around four orders of magnitude. The naked eye detection was confirmed only for luciferase-based assays, such as the forensic blood detection test.96 The disadvantage is the inability to multiplex and the lack of versatility of sensor design.
2.8. Other visually detectable formats
Aggregation and sedimentation of nanoparticles can be used as a visually detectable readout.97,98 For example, Hu et al.99 detected the product of nucleic acid amplification in a flocculation sedimentation assay. The DNA amplicon was captured by magnetic beads followed by flocculation only in the presence of the DNA product.Turbidity has been used to detect products of LAMP.100 The method is based on precipitation of Mg2P2O7, a by-product of DNA-polymerization according to the following equations:
| (DNA)n−1 + dNTP → (DNA)n + P2O74− | (1) |
| P2O74− + 2Mg2+ → Mg2P2O7↓ | (2) |
It is possible to achieve real-time detection using a turbidimeter.101 The precipitate can be observed by the naked eye and does not require labeling of nucleic acid primers or probes. However, the method detects a bulk amount of DNA with low specificity and is not applicable for the detection of SNVs. As such, it is prone to false-positives due to non-specific amplification.
The analyte concentration can be detected visually using a distance-based assay,102 which originally was reported by Zuk et al. in 1985.103 In this method, the analyte concentration can be estimated by visual assessment of the length of the colored area. For example, Daurai and Gogoi developed a paper distance-based assay utilizing the starch-triiodide method to diagnose acute pancreatitis via α-amylase quantification.104 The blue color of the starch–triiodide complex fades away and becomes colorless when α-amylase breaks the starch chain at the glycosidic bond. The distance covered by the change in the color is directly proportional to the concentration of α-amylase in a sample. Xue et al. applied a similar approach for quantification of uracil–DNA glycosylase.105
The possibility of an instrument-free readout is the greatest advantage of the sensors producing visible outputs. This makes such sensing systems cheaper and easier to use than corresponding instrument-dependent systems. With the exception of distance-based assays, the visual format is limited by qualitative detection: analyte quantification still relies on measuring devices. Another limitation is a relatively low sensitivity, which is set up by the sensitivity of a human eye and spectrophotometers detecting only ∼submicromolar concentrations of a typical small organic dye. This LOD can be brought down to a sub-nanomolar range by implementing nanoparticles, enzymes, or signal amplifications cascades. The dynamic range of visual detection methods is generally narrow, further complicating quantitative assessments.
3. Detection by touch
Covering the area of about 2 m2, skin is our largest sense organ. Pressure, temperature, and pain can be sensed by skin receptors. Interestingly, skin is sensitive to electricity and, therefore, can be used for the detection of electric output signals. Tactile sensors, devices that produce signal upon touch, are used in our daily practice (e.g., buttons) and are under extensive development primarily in robotics, computer hardware, and security systems.106 However, there are no commercially available molecular sensors that produce an output detectable by touch. Such a sensing mode promises to become instrument-free and accessible to blind and colour-blind people.Lin et al. studied the change in the mechanical properties of the polyacrylamide gel linked to oligonucleotides upon crosslinking of complementary sequences.107 Oligonucleotide-modified hydrogels can be used for drug delivery, tissue engineering, sensing, and cancer therapy.108
A molecule sensor with tactile output was reported by Fedotova et al. for sensing ATP and nucleic acids.109 The sensor designed for detecting specific nucleic acids is made of two DNA strands, which form a G4 structure upon binding an analyte. In the presence of hemin, H2O2, and acrylamide, the G4 structure initiates a polyacrylamide formation, which can be detected both visually and by touch as a gel. The samples that lack the analyte and fail to form G4 remained liquid (Fig. 8(A)). Importantly, the sensor was highly selective: it did not produce an output in the presence of a single base mismatched DNA (sample 4, Fig. 8(B)). This was predetermined by the split sensor design, in which strand PxD-B has a short analyte-binding arm sensitive to the SNV even at room temperature.110 Instead of using perishable H2O2 as an oxidizer, it is possible to generate H2O2in situ by UV light irradiation of TiO2/H2O. It was shown that such a mode of H2O2 generation is compatible with the tactile detection because it changes its aggregation state from liquid to gel, which could be sensed by touch.111 This method also provides an option of visual detection since the polymerization event can be visually detected if, for example, the tubes with pre-stained samples turned upside down (Fig. 8(B)).
 | ||
| Fig. 8 Polymerization-based visual and tactile detection of a DNA analyte using peroxidase-like deoxyribozyme (PxD).109 (A) Sensor design: two DNA strands PxD-A and PxD-B hybridize to a DNA analyte and form a G4 structure, which binds hemin and catalyses polymerization of acrylamide solution. (B) Tactile and visual signal outputs. Polyacrylamide gel is observed in the presence of the fully matched DNA analyte (3), but not in its absence (2) or the presence of a mismatched analyte (4). Sample 5 did not contain DNA strands (negative control); sample 1 contained the full G4 sequence as a positive control. Low panel: the gel fragments can be detected by touch after their transfer on a filter paper. Reproduced from ref. 109 with permission from Royal Society of Chemistry, copyright 2017. | ||
Considering the high accessibility, tangible detection is worth attention in molecular sensing. A disadvantage of the test at its current stage is a relatively high limit of detection of ∼10 μM, which is suitable for detecting ATP, K+, and Na+, but insufficient for DNA, proteins, and environmentally significant ions, such as Hg+ or Pb2+. Furthermore, the sensor lacks a dynamic range, providing only a qualitative yes/no output.
4. Detection by smell
Via a sense of smell, living organisms can feel the presence of chemical substances in the air. The olfactory system consists of hundreds of receptors that recognize odorants with low specificity, producing a response ‘fingerprint’ known as a smell. The idea of such a recognition platform dubbed a ‘chemical nose’ has been adopted in chemosensors.112 Ethyl mercaptan is added to natural gas as an odorant, giving it a distinct, strong smell that allows people to detect a leak of the odorless gas. However, there are only a few reports of molecular sensors that rely on human sense of smell for signal detection.Mohapatra and Phillips described compound 1 (Fig. 9(A)) that reacts with micromolar concentrations of hydrogen peroxide to trigger the release of both a fluorescent compound 2 and ethyl mercaptan as an odorant reporter. Compound 1 detects β-D-galactosidase with a LOD of 21 nM and a dynamic range extending up to 500 nM.113 Since compound 1 is sensitive to the presence of H2O2, it can be used, for example, for detecting glycose oxidase activity which generates hydrogen peroxide as a side product of a process of glucose oxidation.
 | ||
| Fig. 9 Chemical reactions producing odorants suitable for analyte detection by smell. (A) Detection of H2O2 by both smell and fluorescence.113 (B) Detection of tryptophanase (Tpase) activity by smell.114 (C) Detection of lipase activity by smell.108 | ||
Xu et al. proposed to use reactions catalyzed by an enzyme tryptophanase (Tpase) for generating either methyl mercaptan or indole (Fig. 9(B)), both of which can be detected by the human nose with threshold concentrations of 10 pM and 1 pM, respectively.114 When linked to antibodies, this system was able to detect specific proteins in an ELISA-like assay format with a detection threshold of 85 nM. The system can also be used in a coupled enzymatic assay for detecting ATP down to 320 nM.115 This technology was converted to an inkjet-printed bioactive paper sensor that reports ATP through odor generation. The methyl mercaptan assay allowed ATP detection by the human nose at 1.36 μM in sealed tubes and 9.4 μM in swabs.115
Duncan et al. used a lipase-catalyzed hydrolysis of compound 3 (Fig. 9(C)) to release phenylethyl alcohol having rose scent.116 The system was adopted for detecting bacteria down to 100 cfu mL−1 in a 15 min assay.116 The assay was not bacterial species-specific.
Miller et al. developed a yeast cell culture that produces isoamyl alcohol scent as a readout in response to the presence of galactose or hormone estradiol. The detection threshold found for estradiol with a panel of human volunteers (n = 49) was 39 nM.117
The sensors with smell as an output may provide lower LODs than assays utilizing visual instrument-free formats. They can be useful for blind and color-blind people or used in the dark. Due to the lack of systematic research, the advantages and disadvantages of such sensors over sensors with visual outputs are yet to be determined.
5. Bubble release with pressure-based detection
In the process of writing this review, we discovered an arguably most versatile and highly promising in practice output – production of gas bubbles. This output can be produced by a variety of reactions and sensed by a variety of human senses. An excellent review on this subject was published recently by Shi et al.118In 2015, Zhu et al. proposed using gas release as a signal readout measured using a portable pressure meter.119 An example of such a sensor can be oxygen release upon H2O2 disproportionation catalysed by Pt nanoparticles (Scheme 2(a)). When combined with an antibody-based ELISA-like assay for prostate specific antigen (PSA) the analyte could be detected down to 0.687 pM, which surpasses the sensitivity of the standard ELISA with a colorimetric readout.119
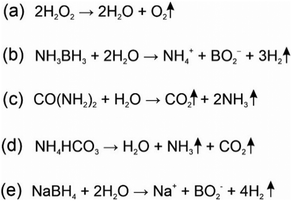 | ||
| Scheme 2 Gas releasing reactions used for biosensing. (a) H2O2 disproportionation catalyzed by catalase or Pt nanopaticles;118–121 (b) decomposition of ammonia borane by CuO/Co3O4122 or CuO–NiO/C;123 (c) catalysed by urease;124 (d) catalyzed by GNP irradiated by NIR;125,126 and (e) can be catalyzed by H+.127 | ||
Nesterova and colleagues reported a G4-based split sensor (part of the sensor is shown in Fig. 4(B)) to detect DNA analytes using catalase activity of G4 according to eqn (a) in Scheme 2.120 In this case, bubbles of oxygen generated in response to the presence of the DNA analyte could be detected visually with a LOD of ∼ 200 nM. The approach enables naked eye quantitative analysis down to 500 nM DNA. Similarly, Shi et al. used O2 production to detect specific microRNA within the range of 10 fM to 10 pM with a LOD of 7.6 fM using a portable test system. In this case, the sensor output was efficiently amplified by the peroxidase activity by Pt nanoparticles.121
Ding et al.122 used the reaction of ammonia borane decomposition liberating H2 gas (Scheme 2(b)) catalysed by CuO/Co3O4 heterojunction nanofibers for the detection of cancer cells with a LOD of 50 cells per mL. In this approach, CuO/Co3O4 heterojunction nanofibers were labelled with folic acid, which had affinity to folic acid receptors on the cell surface. Wang et al.123 used CuO–NiO/C nanofibers to decompose ammonia borane resulting in both the release of H2 and reduction of rhodamine 6G. The latter caused fluorescence quenching. In this case, the LOD achieved with a pressure meter was 50 cells per mL.
Li et al.128 proposed a sandwich ELISA method, in which HRP generated N2 as a byproduct of luminol decomposition (Fig. 7(C)). The gas was released in the capillaries containing a dye-filled line allowing the dye to move along the capillaries upon the gas release. The system enabled detection of human chorionic gonadotropin (as in a pregnancy test) with a practically significant LOD of ∼1.4 ng mL−1.128 The test accuracy was insured by elegant implementation of the internal control. This is an example of an instrument-free portable quantitative test of the future.
To increase the sensitivity of bubble-based analyte detection, the assay can take advantage of a chemical reaction allowing for the release of more than one type of gas. For example, Yuan et al.124 proposed a method to detect urea via its decomposition into CO2 and NH3 (Scheme 2(c)) catalysed by urease conjugated to gold nanoflowers. The quantity of the released gases measured using a portable pressure meter enabled urea detection with a clinically significant LOD of 0.08 mM.
Pressure-based detection can also be coupled to temperature changes during a chemical reaction. For example, Tian et al.125 created hollow CuS nanoparticles that can be visualized by localized surface plasmon resonance (LSPR) via photothermal effects and through NH4HCO3 decomposition to NH3 and CO2 (Scheme 2(d)). In this approach, hyaluronic acid was used to attract cell surface receptors to CuS nanoparticles. Breast cancer was detected with LOD 9 and 4 cells per mL by temperature or pressure, respectively.125 A similar approach was adapted to Ag–AgCl nanocubes that were able to detect receptors to folic acids on the cell surface with ammonium bicarbonate decomposition and temperature release caused by TMB oxidation transformed via NIR.126 The LOD was comparable to those of the previous study: 6 and 5 cells per mL for pressure-based and thermal detection, respectively. Decomposition of NaBH4 is another promising reaction for producing large quantities of gas output (Scheme 2(e)). Based on this reaction, Shi et al. developed an assay for detection of glucose, acetylcholine and ATP.127
Though the gas release itself can work well as an easy-to-read signal, the gaseous output can be translated into the actuating function beyond signalling, e.g., mechanical work, that can be sensed by an impressive variety of means including ink bars in volumetric bar chart chips or capillary tubes,123 which may open up new avenues for application of such sensors due to their ability to translate quantitatively and detect analytes. We believe that the bubble outputs can be further refined in point-of-care test designs since they can be combined to device-free readouts achieving practically significant LODs.
6. Detection by taste and hearing
Taste, also known as gustation, is the sensory perception of food that occurs when a substance in the mouth reacts chemically with receptor cells in the taste buds. Given this mechanism, molecular sensors designed to detect taste can be effectively utilized to monitor harmful substances or pathogens in food or in the air. These sensors could trigger a bitter or sour taste when they detect minor concentrations of potentially harmful compounds, thereby discouraging consumption and promoting food safety.During preparation of this article, we have not found any test systems that employ taste as an output. Considering that the smell is sometimes felt by taste buds, the tests for sensing described earlier in the smell section can be possibly adapted to the taste purposes (e.g., E. coli sensing).116 A critical consideration for such taste-based systems is ensuring the biocompatibility and non-toxicity of all products resulting from the aforementioned reactions.
To date, no molecular sensors have been developed that produce sound as an output. Creating hearing-based molecular sensors presents unique challenges, primarily due to the difficulty of generating sound signals directly from chemical reactions used by molecular sensors. However, though such challenges lie outside of this review scope, potentially sensors could be coupled with specific devices to convert the produced signals into audible sounds. The current state of the art relies on the electrical source of audio signals that come from any other type of input. For example, a hospital heart monitor alerts with a signal reporting severe alteration in the heartbeat and a smoke detector alerts fire. Hearing and smelling hybridized probe detectors can be used as background monitors for constant screening in situations that constrain electrical transmitters. For example, such detectors can be integrated into air conditioning systems to identify specific compounds or pathogens in the air. Alternatively, they could be used by blind people who cannot perceive optical signals.
7. Conclusion
Humans predominantly rely on visual information, which often overshadows the contributions of other sensory inputs. Most common molecular sensors explored so far rely on detection of signals by human vision. Such sensors include those that produce colour changes, changes in transparency or formation of precipitates or luminescence that can be visualized without instrumentation. The LOD of such sensors is typically limited by 100 nM unless catalytic signal amplification steps are implemented. The signals are difficult to quantify without specialized instrumentation, even though semiconduction and quantification using a distance-based assay102–104 are possible.Molecular sensors with tactile outputs, though being low sensitive, can be used by visually impaired people and can be considered as a more accessible option to visual sensors. The liquid-to-gel transition, particularly when reversible, presents a promising solution for applications in bionic skin and tactile detection.129,130 Additionally, the olfactory outputs represent another avenue that could facilitate communication between humans and molecular robots.120
Sensors that generate chemical outputs (e.g., those related to smell and taste) have a potential application beyond diagnostics. These sensors can be integrated into the human body as components of a signalling network. In this context, the substances produced by these sensors could be designed to target specific receptors or modulate the functions of natural or artificial organs. For instance, they could serve as linkers for glucose sensing in artificial pancreas systems. In such scenarios, it is essential that the sensing reactions, along with all resultant products and by-products, meet biocompatibility requirements to ensure safety.
Out of all outputs, we consider gas bubbles as outputs produced by sensors as the most promising (see the recent review by Shi et al.).118 Such outputs can be visualized or converted to tactile outputs or sounds. Most importantly, sensor outputs can be quantified using a hand-held portable pressure meter. Achieving a picomolar LOD is possible using catalytic amplification, considering that the chemistry of gas production is versatile and compatible with reactions catalysed by HRP and other accessible enzymes. The multiplexing capability of such a detection format has already been demonstrated.128
Among the challenges that will be faced in the future development of molecular sensors, the authors highlight a significant lack of sensitivity, particularly in less common formats such as tactile and olfactory sensors. Furthermore, there is notable inability to assess the selectivity of many of these methods when applied to real samples. Some studies mentioned in this minireview do not declare successful detection in real or even mock samples. This limitation may be caused by various factors, including difficulties in accessing materials or the inherent low selectivity of the developed systems.
The authors encourage a paradigm shift within the scientific community urging exploration of molecular sensors with outputs other than visual including tangible and gas release. This shift could facilitate the introduction of sensors across a wider variety of contexts and environments, thereby enhancing their applicability and effectiveness in real-world scenarios.
Even though intentionally excluded from this article, we do consider promising the development of communication interfaces between molecular sensors and electronic computers. This can be achieved by using electrochemical sensors and molecular logic gates as reported earlier.131,132 This option will enable using machine learning and AI resources for biosensing.
Author contributions
The manuscript was written through contributions of all the authors. All the authors gave an approval to the final version of the manuscript. Authors used Elicit and ChatGPT AI-tools for data aggregation and language improvement.Data availability
This is a review article, and no new data were generated or analyzed during the preparation of this manuscript. All data sources referenced in the article are publicly available and cited appropriately.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
D. M. K. is grateful to Yulia V. Gerasimova for the careful reading of the manuscript. M. R. acknowledges the support from the St. Petersburg State University (project 116742818) and the Academic Writing Lab at ITMO University for their assistance with proofreading.Notes and references
- P. Mehrotra, J. Oral Biol. Craniofacial Res., 2016, 6, 153–159 CrossRef PubMed.
- C. S. Kosack, A.-L. Page and P. R. Klatser, Bull. W. H. O., 2017, 95, 639 CrossRef PubMed.
- B. K. Ashley and U. Hassan, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2021, 13, e1701 Search PubMed.
- L. Zhang, C. Gu, H. Ma, L. Zhu, J. Wen, H. Xu, H. Liu and L. Li, Anal. Bioanal. Chem., 2019, 411, 21–36 CrossRef CAS PubMed.
- A. Hulanicki, S. Glab and F. Ingman, Pure Appl. Chem., 1991, 63, 1247–1250 CrossRef.
- B. Wang and Y. Lu, Nano-Micro Lett., 2024, 18, 155 CrossRef PubMed.
- D. M. Kolpashchikov, Acc. Chem. Res., 2019, 52, 1949–1956 CrossRef CAS PubMed.
- D. D. Nedorezova, A. F. Fakhardo, T. A. Molden and D. M. Kolpashchikov, ChemBioChem, 2020, 21, 607–611 CrossRef CAS PubMed.
- H. Li, H. Xu, Y. Li and X. Li TrAC, Trends Anal. Chem., 2024, 174, 117700 CrossRef CAS.
- S. Lu, J. Yang, Y. Gu, D. He, H. Wu, W. Sun, D. Xu, C. Li and C. Guo, ACS Sens., 2024, 9, 1134–1148 CrossRef CAS PubMed.
- A. Tittl, A. John-Herpin, A. Leitis, E. R. Arvelo and H. Altug, Angew. Chem., Int. Ed., 2019, 58, 14810–14822 CrossRef CAS PubMed.
- G.-J. Luo, P.-H. Wu, M.-L. Lin, F.-F. Chou and J. J.-M. Lin, J. Chem. Educ., 2023, 100, 4298–4306 CrossRef CAS.
- A. Steinegger, O. S. Wolfbeis and S. M. Borisov, Chem. Rev., 2020, 120, 12357–12489 CrossRef CAS PubMed.
- L. Yang, Q.-Y. Yuan, C.-W. Lou, J.-H. Lin and T.-T. Li, Text. Res. J., 2024, 00405175231225831 Search PubMed.
- K.-K. Kunimoto, H. Sugiura, T. Kato, H. Senda, A. Kuwae and K. Hanai, Spectrochim. Acta, Part A, 2001, 57, 265–271 CrossRef CAS PubMed.
- H. Takezawa, S. Akiba, T. Murase and M. Fujita, J. Am. Chem. Soc., 2015, 137, 7043–7046 CrossRef CAS PubMed.
- L. Zhang, Y. Liang, Z. Xiao, Y. Meng, J. Huang, X. Lin, J. Li, H.-C. Zhou and Y. Fang, Sci. China: Chem., 2024, 67, 1–7 CrossRef PubMed.
- N. A. Tanner, Y. Zhang and T. C. J. Evans, Biotechniques, 2015, 58, 59–68 CrossRef CAS PubMed.
- Y. Zhang, E. A. Hunt, E. Tamanaha, I. R. Corrêa Jr and N. A. Tanner, Commun. Biol., 2022, 5, 999 CrossRef CAS PubMed.
- H. G. Loehr and F. Voegtle, Acc. Chem. Res., 1985, 18, 65–72 CrossRef CAS.
- Z. Kowser, U. Rayhan, T. Akther, C. Redshaw and T. Yamato, Mater. Chem. Front., 2021, 5, 2173–2200 RSC.
- J.-D. Lian, H.-Y. Hu, Y.-H. Lin, P. Raghunath, M.-C. Lin and W.-S. Chung, J. Org. Chem., 2023, 88, 14292–14302 CrossRef CAS PubMed.
- M. S. Tabatabaei, R. Islam and M. Ahmed, Anal. Chim. Acta, 2021, 1143, 250–266 CrossRef CAS PubMed.
- P. D. Howes, R. Chandrawati and M. M. Stevens, Science, 2014, 346, 1247390 CrossRef PubMed.
- A. N. Baker, G. W. Hawker-Bond, P. G. Georgiou, S. Dedola, R. A. Field and M. I. Gibson, Chem. Soc. Rev., 2022, 51, 7238–7259 RSC.
- L. Guo, Y. Xu, A. R. Ferhan, G. Chen and D.-H. Kim, J. Am. Chem. Soc., 2013, 135, 12338–12345 CrossRef CAS PubMed.
- P. G. McDonough, Fertil. Steril., 2003, 80, 1534–1535 CrossRef PubMed.
- Y. Wang and L. Tang, Biosens. Bioelectron., 2015, 67, 18–24 CrossRef CAS PubMed.
- F. Ozefe and A. A. Yildiz, Analyst, 2020, 145, 5816–5825 RSC.
- H. Xiang, S. Huang, D. Zhu, L. Yu, R. Liu, Y. Guo and L. Xu, Chem. – Eur. J., 2023, 29, e202300454 CrossRef CAS PubMed.
- N. Borah, D. Gogoi, N. N. Ghosh and C. Tamuly, Food Chem., 2023, 399, 133975 CrossRef CAS PubMed.
- A. Mondal and V. Manivannan, Spectrochim. Acta, Part A, 2024, 322, 124734 CrossRef CAS PubMed.
- M. Alimohammadi, H. Sharifi, J. Tashkhourian, M. Shamsipur and B. Hemmateenejad, Lab Chip, 2023, 23, 3837–3849 RSC.
- V. G. Grigorenko, I. P. Andreeva, M. Y. Rubtsova and A. M. Egorov, Biochem., 2015, 80, 408–416 CAS.
- J. D. Martell, M. Yamagata, T. J. Deerinck, S. Phan, C. G. Kwa, M. H. Ellisman, J. R. Sanes and A. Y. Ting, Nat. Biotechnol., 2016, 34, 774–780 CrossRef CAS PubMed.
- D. Dębski, R. Smulik, J. Zielonka, B. Michałowski, M. Jakubowska, K. Dębowska, J. Adamus, A. Marcinek, B. Kalyanaraman and A. Sikora, Free Radical Biol. Med., 2016, 95, 323–332 CrossRef PubMed.
- N. D. Udeshi, K. Pedram, T. Svinkina, S. Fereshetian, S. A. Myers, O. Aygun, K. Krug, K. Clauser, D. Ryan, T. Ast, V. K. Mootha, A. Y. Ting and S. A. Carr, Nat. Methods, 2017, 14, 1167–1170 CrossRef CAS PubMed.
- Z. Liposits, T. Görcs, F. Gallyas, B. Kosaras and G. Sétáló, Neurosci. Lett., 1982, 31, 7–11 CrossRef CAS PubMed.
- S. Škulj, M. Kožić, A. Barišić, A. Vega, X. Biarnés, I. Piantanida, I. Barisic and B. Bertoša, Comput. Struct. Biotechnol. J., 2024, 23, 742–751 CrossRef PubMed.
- L. Liu, Y. Chang, J. Lou, S. Zhang and X. Yi, Molecules, 2023, 28, 6565 CrossRef CAS PubMed.
- J. Bae, J. Kim, J. Choi, H. Lee and M. Koh, ChemBioChem, 2024, 25, e202400123 CrossRef CAS PubMed.
- D. A. Gorbenko, L. A. Shkodenko, M. S. Rubel, A. V. Slita, E. V. Nikitina, E. A. Martens and D. M. Kolpashchikov, Chem. Commun., 2022, 58, 5395–5398 RSC.
- Y. I. Maltzeva, D. A. Gorbenko, E. V. Nikitina, M. S. Rubel and D. M. Kolpashchikov, Int. J. Mol. Sci., 2023, 24, 7812 CrossRef CAS PubMed.
- Y. Ahmadi, R. Soldo, K. Rathammer, L. Eibler and I. Barišić, Anal. Chem., 2021, 93, 5161–5169 CrossRef CAS PubMed.
- Y. Tang, X. Huang, X. Wang, C. Wang, H. Tao and Y. Wu, Food Chem., 2022, 366, 130560 CrossRef CAS PubMed.
- J. Xiang, J. Zhang, S. Li, R. Yuan and Y. Xiang, Anal. Chim. Acta, 2022, 1230, 340393 CrossRef CAS PubMed.
- M. Volek, J. Kurfürst, M. Kožíšek, P. Srb, V. Veverka and E. A. Curtis, Nucleic Acids Res., 2024, 52, 9062–9075 CrossRef CAS PubMed.
- Z. Chi, Q. Wang and J. Gu, Analyst, 2023, 148, 487–506 RSC.
- Z. Wei, Z. Xi, S. Vlasov, J. Ayala and X. Xia, Chem. Commun., 2020, 56, 14962–14975 RSC.
- Y. Liu and Z.-G. Wang, ACS Nano, 2023, 17, 13000–13016 CrossRef CAS PubMed.
- Z. Xi, K. Wei, Q. Wang, M. J. Kim, S. Sun, V. Fung and X. Xia, J. Am. Chem. Soc., 2021, 143, 2660–2664 CrossRef CAS PubMed.
- P. Travascio, Y. Li and D. Sen, Chem. Biol., 1998, 5, 505–517 CrossRef CAS PubMed.
- D. M. Kolpashchikov, J. Am. Chem. Soc., 2008, 130, 2934–2935 CrossRef CAS PubMed.
- R. P. Connelly, C. Verduzco, S. Farnell, T. Yishay and Y. V. Gerasimova, ACS Chem. Biol., 2019, 14, 2701–2712 CrossRef CAS PubMed.
- Y. V. Gerasimova, E. M. Cornett, E. Edwards, X. Su, K. H. Rohde and D. M. Kolpashchikov, ChemBioChem, 2013, 14, 2087–2090 CrossRef CAS PubMed.
- K. Tsuchiya, H. Takakura, K. Nakajima, N. Ieda, T. Kaneko, T. Hirasawa, M. Kobayashi, Y. Yamaoka, M. Ishihara, T. Taketsugu and M. Ogawa, J. Photochem. Photobiol., A, 2024, 453, 115634 CrossRef CAS.
- N. G. Zhegalova, A. Aydt, S. T. Wang and M. Y. Berezin, Reporters, Markers, Dyes, Nanoparticles, and Molecular Probes for Biomedical Applications V, SPIE, 2013, vol. 8596, pp. 84–92 Search PubMed.
- D. Wu, A. C. Sedgwick, T. Gunnlaugsson, E. U. Akkaya, J. Yoon and T. D. James, Chem. Soc. Rev., 2017, 46, 7105–7123 RSC.
- E. A. Specht, E. Braselmann and A. E. Palmer, Annu. Rev. Physiol., 2017, 79, 93–117 CrossRef CAS PubMed.
- M. Dai, Y. J. Yang, S. Sarkar and K. H. Ahn, Chem. Soc. Rev., 2023, 52, 6344–6358 RSC.
- G. Jiang, H. Liu, H. Liu, G. Ke, T. Ren, B. Xiong, X. Zhang and L. Yuan, Angew. Chem., Int. Ed., 2024, 136, e202315217 CrossRef.
- A. Martin and P. Rivera-Fuentes, Nat. Chem., 2024, 16, 28–35 CrossRef CAS PubMed.
- C. Wynne and R. B. P. Elmes, Front. Chem., 2024, 12, 1418378 CrossRef CAS PubMed.
- J. Yan, S. Lee, A. Zhang and J. Yoon, Chem. Soc. Rev., 2018, 47, 6900–6916 RSC.
- J. Kim, J. Park, H. Lee, Y. Choi and Y. Kim, Chem. Commun., 2014, 50, 9353–9356 RSC.
- C. Yuan, K. Zhang, Z. Zhang and S. Wang, Anal. Chem., 2012, 84, 9792–9801 CrossRef CAS PubMed.
- Y. Wei, L. Yang, Y. Ye, L. Liao, H. Dai, Z. Wei, Y. Lin and C. Zheng, Chem. Commun., 2024, 60, 5205–5208 RSC.
- A. S. Klymchenko, Acc. Chem. Res., 2017, 50, 366–375 CrossRef CAS PubMed.
- D. M. Kolpashchikov, J. Am. Chem. Soc., 2005, 127, 12442–12443 CrossRef CAS PubMed.
- D. M. Kolpashchikov and A. A. Spelkov, Angew. Chem., Int. Ed., 2021, 133, 5040–5051 CrossRef.
- N. Kikuchi, A. Reed, Y. V. Gerasimova and D. M. Kolpashchikov, Anal. Chem., 2019, 91, 2667–2671 CrossRef CAS PubMed.
- M. A. Reed and Y. V. Gerasimova, Front. Chem., 2022, 10, 951279 CrossRef CAS PubMed.
- B. L. Mueller, M. J. Liberman and D. M. Kolpashchikov, Nanoscale, 2023, 15, 5735–5742 RSC.
- S. Tyagi and F. R. Kramer, Nat. Biotechnol., 1996, 14, 303–308 CrossRef CAS PubMed.
- D. M. Kolpashchikov, Scientifica, 2012, 2012, 928783 CrossRef PubMed.
- H. Shi, X. He, X. Yang, K. Wang, Q. Wang, Q. Guo and X. Huo, Sci. China: Chem., 2010, 53, 704–719 CrossRef CAS PubMed.
- A. C. Bardales, Q. Vo and D. M. Kolpashchikov, J. Chem. Educ., 2024, 101, 4360–4369 CrossRef CAS.
- Y. Sun, X. Qu, P. Qiu and C. Mao, Talanta, 2023, 260, 124602 CrossRef CAS PubMed.
- M. Volek, J. Kurfürst, M. Drexler, M. Svoboda, P. Srb, V. Veverka and E. A. Curtis, Nucleic Acids Res., 2024, 52, 9049–9061 CrossRef CAS PubMed.
- Y. V. Gerasimova and D. M. Kolpashchikov, Chem. Soc. Rev., 2014, 43, 6405–6438 RSC.
- Z. Huang, X. Wang, Z. Wu and J. Jiang, Chem. – Asian J., 2022, 17, e202101414 CrossRef CAS PubMed.
- D. D. Nedorezova, M. S. Rubel and A. A. Rubel, Biochem., 2024, 89, S249–S261 CAS.
- T. A. Lyalina, E. A. Goncharova, N. Y. Prokofeva, E. S. Voroshilina and D. M. Kolpashchikov, Analyst, 2019, 144, 416–420 RSC.
- A. A. El-Deeb, S. S. Zablotskaya, M. S. Rubel, M. A. Y. Nour, L. I. Kozlovskaya, A. A. Shtro, A. B. Komissarov and D. M. Kolpashchikov, ChemMedChem, 2022, 17, e202200382 CrossRef CAS PubMed.
- Z. Hussein, M. A. Y. Nour, A. V. Kozlova, D. M. Kolpashchikov, A. B. Komissarov and El. A. A. Deeb, Anal. Chem., 2023, 95, 18667–18672 CrossRef CAS PubMed.
- Y. Yan, P. Shi, W. Song and S. Bi, Theranostics, 2019, 9, 4047 CrossRef CAS PubMed.
- R. Wang, N. Yue and A. Fan, Analyst, 2020, 145, 7488–7510 RSC.
- T. Azad, A. Tashakor and S. Hosseinkhani, Anal. Bioanal. Chem., 2014, 406, 5541–5560 CrossRef CAS PubMed.
- A. Petchiappan, S. Y. Naik and D. Chatterji, Biophys. Rev., 2020, 12, 719–730 CrossRef CAS PubMed.
- K. Svehlova, O. Lukšan, M. Jakubec and E. A. Curtis, Angew. Chem., Int. Ed., 2022, 134, e202109347 CrossRef.
- S. Deepa, R. Venkatesan, S. Jayalakshmi, M. Priya and S.-C. Kim, J. Environ. Chem. Eng., 2023, 11, 109853 CrossRef CAS.
- D. Li, W. Cheng, Y. Li, Y. Xu, X. Li, Y. Yin, H. Ju and S. Ding, Anal. Chem., 2016, 88, 7500–7506 CrossRef CAS PubMed.
- Y. Zhang, J. Liu, T. Liu, H. Li, Q. Xue, R. Li, L. Wang, Q. Yue and S. Wang, Biosens. Bioelectron., 2016, 77, 111–115 CrossRef CAS PubMed.
- K. Imai, Detection-Oriented Derivatization Techniques in Liquid Chromatography, CRC Press, 2021, pp. 359–379 Search PubMed.
- H. Chen, Y. Feng, F. Liu, C. Tan, N. Xu, Y. Jiang and Y. Tan, Biosens. Bioelectron., 2024, 247, 115929 CrossRef CAS PubMed.
- F. Barni, S. W. Lewis, A. Berti, G. M. Miskelly and G. Lago, Talanta, 2007, 72, 896–913 CrossRef CAS PubMed.
- Y. Li, Z. Cui, L. Huang, D. Zhang, Y. Shen, J. Cheng and J. Wang, Aggregate, 2024, 5, e559 CrossRef CAS.
- K. Shirato, Microbiol. Immunol., 2019, 63, 407–412 CrossRef CAS PubMed.
- J. Hu, Y. Wang, H. Ding, C. Jiang, Y. Geng, X. Sun, J. Jing, H. Gao, Z. Wang and C. Dong, Int. J. Food Microbiol., 2020, 331, 108691 CrossRef CAS PubMed.
- Y. Mori, M. Kitao, N. Tomita and T. Notomi, J. Biochem. Biophys. Methods, 2004, 59, 145–157 CrossRef CAS PubMed.
- X. Zhang, S. B. Lowe and J. J. Gooding, Biosens. Bioelectron., 2014, 61, 491–499 CrossRef CAS PubMed.
- T. Tian, J. Li, Y. Song, L. Zhou, Z. Zhu and C. J. Yang, Lab Chip, 2016, 16, 1139–1151 RSC.
- R. F. Zuk, V. K. Ginsberg, T. Houts, J. Rabbie, H. Merrick, E. F. Ullman, M. M. Fischer, C. C. Sizto, S. N. Stiso and D. J. Litman, Clin. Chem., 1985, 31, 1144–1150 CrossRef CAS.
- B. Daurai and M. Gogoi, RSC Adv., 2024, 14, 24365–24372 RSC.
- W. Xue, Y. Wu, X. Li, Q. Zhang, Y. Wu, Y. Chang and M. Liu, Biosens. Bioelectron., 2024, 264, 116687 CrossRef CAS PubMed.
- K. S. Kumar, P.-Y. Chen and H. Ren, Research., 2019, 2019, 3018568 CAS.
- D. C. Lin, B. Yurke and N. A. Langrana, J. Biomech. Eng., 2004, 126, 104–110 CrossRef PubMed.
- S. Khajouei, H. Ravan and A. Ebrahimi, Adv. Colloid Interface Sci., 2020, 275, 102060 CrossRef CAS PubMed.
- T. A. Fedotova and D. M. Kolpashchikov, Chem. Commun., 2017, 53, 12622–12625 RSC.
- M. Stancescu, T. A. Fedotova, J. Hooyberghs, A. Balaeff and D. M. Kolpashchikov, J. Am. Chem. Soc., 2016, 138, 13465–13468 CrossRef CAS PubMed.
- Y. V. Lanchuk, S. A. Ulasevich, T. A. Fedotova, D. M. Kolpashchikov and E. V. Skorb, RSC Adv., 2018, 8, 37735–37739 RSC.
- R. Haddad, A. Medhanie, Y. Roth, D. Harel and N. Sobel, PLoS Comput. Biol., 2010, 6, e1000740 CrossRef PubMed.
- H. Mohapatra and S. T. Phillips, Angew. Chem., Int. Ed., 2012, 51, 11145–11148 CrossRef CAS PubMed.
- Y. Xu, Z. Zhang, M. M. Ali, J. Sauder, X. Deng, K. Giang, S. D. Aguirre, R. Pelton, Y. Li and C. D. M. Filipe, Angew. Chem., Int. Ed., 2014, 53, 2620–2622 CrossRef CAS PubMed.
- Z. Zhang, J. Wang, R. Ng, Y. Li, Z. Wu, V. Leung, S. Imbrogno, R. Pelton, J. D. Brennan and C. D. M. Filipe, Analyst, 2014, 139, 4775–4778 RSC.
- B. Duncan, N. D. B. Le, C. Alexander, A. Gupta, G. Yesilbag Tonga, M. Yazdani, R. F. Landis, L.-S. Wang, B. Yan and S. Burmaoglu, ACS Nano, 2017, 11, 5339–5343 CrossRef CAS PubMed.
- R. A. Miller, S. Lee, E. J. Fridmanski, E. Barron, J. Pence, M. Lieberman and H. V. Goodson, ACS Sens., 2020, 5, 3025–3030 CrossRef CAS PubMed.
- L. Shi, C. Yang and Y. Jin, Trends Anal. Chem., 2024, 180, 11789 CrossRef.
- Z. Zhu, Z. Guan, D. Liu, S. Jia, J. Li, Z. Lei, S. Lin, T. Ji, Z. Tian and C. J. Yang, Angew. Chem., Int. Ed., 2015, 127, 10594–10599 CrossRef.
- E. E. Iwaniuk, T. Adebayo, S. Coleman, C. G. Villaros and I. V. Nesterova, Nucleic Acids Res., 2023, 51, 1600–1607 CrossRef CAS PubMed.
- L. Shi, J. Lei, B. Zhang, B. Li, C. J. Yang and Y. Jin, ACS Appl. Mater. Interfaces, 2018, 10, 12526–12533 CrossRef CAS PubMed.
- E. Ding, J. Hai, T. Li, J. Wu, F. Chen, Y. Wen, B. Wang and X. Lu, Anal. Chem., 2017, 89, 8140–8147 CrossRef CAS PubMed.
- Z. Wang, J. Hai, T. Li, E. Ding, J. He and B. Wang, ACS Sustainable Chem. Eng., 2018, 6, 9921–9929 CrossRef CAS.
- Y. Yuan, Y. He, D. Pei, L. Tong, S. Hu, L. Liu, X. Yi and J. Wang, Microchem. J., 2022, 179, 107450 CrossRef CAS.
- M. Tian, J. Wang, W. Xiang, Z. Zheng, Z. Luo, S. Jing, Y. Zheng, S. He, H. Wei and C.-Y. Yu, Chem. Eng. J., 2024, 486, 150186 CrossRef CAS.
- S. Liu, S. Lu, S. Sun, J. Hai, G. Meng and B. Wang, Anal. Chem., 2021, 93, 14307–14316 CrossRef CAS PubMed.
- L. Shi, Q. Tang, B. Yang, B. Li, C. Yang and Y. Jin, Microchim. Acta, 2023, 190, 69 CrossRef CAS PubMed.
- Y. Li, J. Xuan, T. Xia, X. Han, Y. Song, Z. Cao, X. Jiang, Y. Guo, P. Wang and L. Qin, Anal. Chem., 2015, 87, 3771–3777 CrossRef CAS PubMed.
- Y. Wang, X. Jiang, X. Li, K. Ding, X. Liu, B. Huang, J. Ding, K. Qu, W. Sun, Z. Xue and W. Xu, Mater. Horiz., 2023, 10, 4033–4058 RSC.
- D. Zhao, Y. Zhu, W. Cheng, W. Chen, Y. Wu and H. Yu, Adv. Mater., 2021, 33, 2000619 CrossRef CAS PubMed.
- S. Mailloux, Y. V. Gerasimova, N. Guz, D. M. Kolpashchikov and E. Katz, Angew. Chem., Int. Ed., 2015, 54, 6562–6566 CrossRef CAS PubMed.
- N. Guz, T. A. Fedotova, B. E. Fratto, O. Schlesinger, L. Alfonta, D. M. Kolpashchikov and E. Katz, ChemPhysChem, 2016, 17, 2247–2255 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |



