Open Access Article
Open Access ArticleA rapid investigation of near-infrared (NIR) fluorescent switch-on probes for detection and in cellulo tracking of G-quadruplex and double-stranded DNA
Hazeena Shinziya,
Revathi S Menon and
Avijit Kumar Das *
*
Department of Chemistry, Christ University, Hosur Road, Bangalore, Karnataka 560029, India. E-mail: avijitkumar.das@christuniversity.in
First published on 25th September 2024
Abstract
This review provides a comprehensive overview of the recent advancements in Near Infrared (NIR) fluorescence switch-on probes designed for the detection and in cellulo tracking of G-quadruplex and double-stranded DNA (dsDNA). G-quadruplexes, non-canonical DNA structures, play pivotal roles in regulating various biological processes, making them critical targets for therapeutic and diagnostic applications. The unique properties of NIR fluorescence probes, such as deep tissue penetration, minimal photodamage, and low autofluorescence background, offer significant advantages for bioimaging. We critically analyze the design strategies, photophysical properties, and binding mechanisms of various NIR fluorescence switch-on probes. Additionally, we discuss their efficacy and specificity in identifying G-quadruplexes and dsDNA within cellular environments. Key challenges and future directions for improving the sensitivity, selectivity, and biocompatibility of these probes are also highlighted. This review aims to underscore the potential of NIR fluorescence probes in advancing our understanding of DNA dynamics and their applications in biomedical research.
1. Introduction
DNA plays a vital role in numerous cellular processes and serves as the repository of all genetic information.1 Various small molecules can bind to DNA and influence its function.2 DNA binding can occur through intercalation,3 minor groove binding,4 or a combination of both.5 Effective transcription inhibitors that bind to DNA are small compounds with potential therapeutic uses as antiviral, anticancer, and antiparasitic medicines. Since DNA is a universal cellular component, its detection both in vivo and in vitro is crucial.6 For this use, ligands that bind to DNA are great choices. Consequently, the development of small molecules that specifically interact with DNA has numerous applications and remains a vibrant area of research.7 It is imperative to do research on the photochemical characteristics of dyes in combination with different biopolymers, including proteins and nucleic acids. Often these properties play a vital role in the application of dyes and related compounds in biology and medicine, especially in developing spectral-fluorescent methods for analyzing and studying biomolecules. These methods have become essential for researchers in biomedicine and clinical practice. The benefits of spectral-fluorescent analysis highlight the necessity to find new, promising dye probes, which requires an in-depth examination of the excited state properties of their molecules in complexes with biopolymers.8 Near-infrared (NIR) fluorescence switch-on probes are an innovative class of molecular tools designed for the detection and in cellulo tracking of specific DNA structures, particularly G-quadruplexes (G4s) and double-stranded DNA (dsDNA). These probes offer several advantages over traditional detection methods, including high sensitivity, selectivity, and real-time monitoring capabilities.9 G-quadruplexes are non-canonical DNA structures formed by the stacking of guanine (G) rich sequences, stabilized by Hoogsteen hydrogen bonding.10 They are involved in various biological processes, including gene regulation, telomere maintenance, and DNA replication. Similarly, double-stranded DNA is the most common form of DNA in cells and serves as the template for gene expression and replication.11 The design of NIR fluorescence switch-on probes typically involves a fluorophore-quencher pair linked by a DNA sequence that can specifically interact with the target DNA structure. In the absence of the target, the fluorophore is quenched by the proximity of the quencher, resulting in low fluorescence intensity. However, upon binding to the target DNA structure, conformational changes occur, leading to the separation of the fluorophore from the quencher and the restoration of fluorescence emission, thus enabling the detection of the target.12–18To date, numerous turn-on fluorescence probes have been developed to target DNA.19 Some well-known probes include EtBr,13 Hoechst dyes,17 and DAPI.18 Numerous of these probes have the capacity to change the DNA's structure after binding, and they frequently have significant toxicity and poor cell permeability. Furthermore, their emission and absorption spectra may coincide with those of biological constituents. To address these limitations, a newer class of far-red or near-infrared (NIR) fluorescent probes has been created. These probes are advantageous because biological molecules exhibit low absorbance and emission in the NIR wavelength range of 700–900 nm. Using NIR probes helps avoid interference from the absorbance and autofluorescence of cellular components. Because biological compounds have low absorbance and emission in the 700–900 nm NIR wavelength region, these probes are useful. Consequently, NIR probes have garnered significant attention in recent years.20 Therefore, in this current review, we have assembled various types of NIR probes for selective binding of Double-Stranded DNA, G Quadruplex DNA and both the DNA at the same time.
2. The structural features of G quadruplex and double-stranded DNA
Two lengthy polynucleotide chains, each made up of four different kinds of nucleotide subunits, make up a DNA molecule. A DNA strand, often known as a chain, is any one of these chains. The two chains are held together by hydrogen bonds that form between the base parts of the nucleotides. A five-carbon sugar, one or more phosphate groups, and a base that contains nitrogen make up a nucleotide. When it comes to the nucleotides in DNA, the base can be either adenine (A), cytosine (C), guanine (G), or thymine (T), and the sugar is deoxyribose linked to a single phosphate group (hence the term deoxyribonucleic acid). The sugars and phosphates form a “backbone” of alternating sugar-phosphate-sugar-phosphate because they are covalently bonded to the nucleotides in a chain. Each polynucleotide chain in DNA is comparable to a necklace (the backbone) strung with four different types of beads (the four bases A, C, G, and T) because only the base differs in each of the four types of subunits. These same symbols (A, C, G, and T) are also commonly used to denote the four different nucleotides—that is, the bases with their attached sugar and phosphate groups.1There exist a variety of non-canonical nucleic acid structures, such as G-quadruplexes, triplexes, hairpin loops, internal loops, junctions, and higher-ordered structures. G-quadruplex structures have garnered significant attention due to their potential roles in both in vitro and in vivo settings. A G-quadruplex is a four-stranded DNA structure with stacked guanine tetrads, G-quartets, which are held together via eight Hoogsteen hydrogen bonds (Fig. 1).10
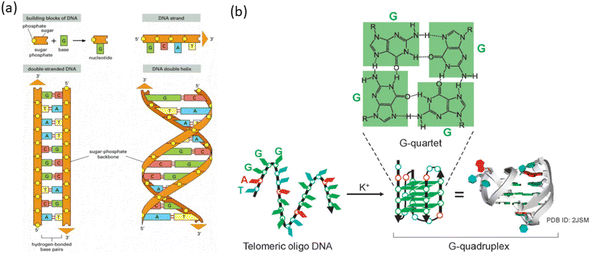 | ||
| Fig. 1 (a) DNA and its building blocks.21 (b) Diagram showing how telomeric DNA folds into a G-quadruplex structure when potassium ions are present. (This figure has been adapted from ref. 21 with permission from American Society for Cell Biology, copyright 2021). | ||
3. Fluorescent probes for G quadruplex and double-stranded DNA: design strategies
Near-infrared (NIR) probes designed to interact with duplex and quadruplex DNA often share common structural features and key functional groups that enhance their binding and fluorescence properties. These probes typically have aromatic systems, such as phenyl, naphthyl or various heterocyclic rings, which stack with DNA bases through π–π interactions, stabilizing the probe-DNA complex.22 They often possess planar or semi-planar structures that enable intercalation between DNA bases or interaction with DNA grooves, and rigid or semi-rigid backbones that facilitate fitting into the DNA structure, whether in the major groove of duplex DNA or the G-quadruplex. Key functional groups include nitrogen and oxygen donors, such as amines and carbonyls, which form hydrogen bonds with DNA bases, and charge-interacting groups, like sulfonic acids or ammonium groups, that engage electrostatically with the DNA's negatively charged phosphate backbone.23 Different NIR fluorophores like cyanine dyes, thiazole orange, benzothiazole, thiacarbocyanine are often substituted with various functional groups to enhance binding through non-covalent interactions with DNA and emit fluorescence in the near-infrared range. As shown in cartoon depictions of Scheme 1 of these mechanisms might include intercalation of probes between DNA base pairs, groove binding into the major or minor grooves of the DNA double helix, and binding to the grooves or loops of G-quadruplexes.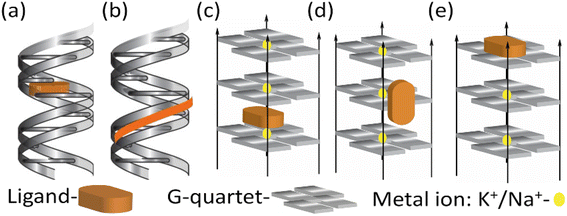 | ||
| Scheme 1 Cartoon diagram of (a) intercalation and (b) groove binding with duplex DNA. (c) Intercalation (d) groove binding and (e) end stacking with quadruplex DNA. | ||
4. NIR fluorescent probes for detection of double-stranded DNA
Pugachev et al. reported photomodulated NIR-fluorescence molecules (1a–c) which showed binding to DNA molecules by molecular docking study.24 All these compounds are characterized by reversible isomerization under activating radiation (usually UV) between the colorless spirocyclic (Sp) and brightly colored merocyanine (Mc) forms (Fig. 2a). The results of molecular docking analysis revealed that the minor groove of DNA is more energetically favorable for the formation of spiropyran complexes compared to other interaction types (Fig. 2b–d).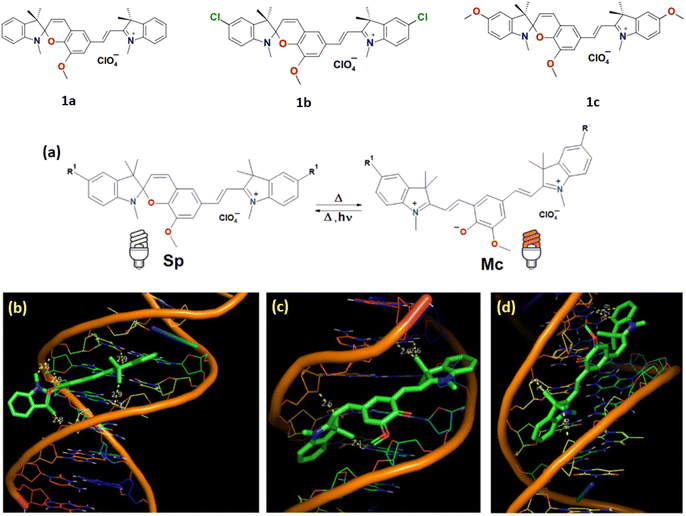 | ||
| Fig. 2 (a) Equilibrium between Sp and Mc isomers of SPs 4 in acetonitrile solution. Molecular docking results with DNA in minor groove modes for 1a: (b) Sp isomer; (c) Mc isomer with TTC configuration; (d) Mc isomer with TTT configuration (highest affinity, ΔGest = −10.33 kcal mol−1, at 25 °C). (This figure has been adapted from ref. 24 with permission from Royal Society of Chemistry, copyright 2023). | ||
In 2015, Govindaraju et al. reported a DNA minor groove detection using an NIR-fluorescence switch-on probe 2 based on quinone cyanine–dithiazole (QCy–DT) within the framework of one-donor-two-acceptor (D2A).25 This design capitalizes on the unique internal charge transfer (ICT) process for the specific recognition of the minor groove in AT-rich DNA. Notably, absorption spectra of probe 2 showed a progressive red shift in the absorption maxima from 463 to 479 nm and 530 to 564 nm upon the stepwise introduction of AT-rich DNA, accompanied by an increase in hyperchromicity. And probe 2 showed a ∼200-fold NIR emission enhancement with the blue shifted (Δλem = 27 nm) emission maxima at 650 nm in the presence of AT-rich DNA compared to basal fluorescence of the probe alone distinguishing the probe selectivity towards AT-rich DNA as compared to GC-rich and single-stranded DNAs. Ligand 2 showed the maximum binding affinity (Ka = 2.9 × 106 M−1) for (A-T), which is 2-fold higher than that for Drew–AT (Ka = 1.5 × 106 M−1). Probe 2 displayed selective nuclear staining of P. falciparum, effectively inhibiting its growth and propagation. This inhibitory effect, possibly attributed to the targeting of cellular DNA, suggests probe 2 as a potential therapeutic agent against specific parasitic infections, akin to other compounds that bind to the minor groove. Live cell imaging investigations validated the permeability of mammalian cells by confirming low toxicity and selective staining capacity for nuclear DNA without the need for RNase treatment (Fig. 3).
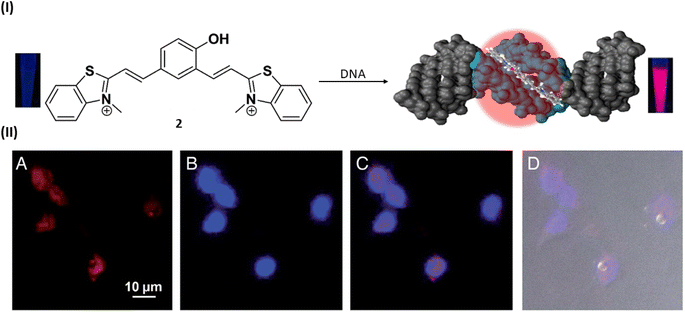 | ||
| Fig. 3 (I) Molecular structure of NIR probe 2 and DNA minor groove recognition of QCy–DT through switch-on NIR-fluorescence response. (II) Cellular uptake properties of QCy–DT in live MCF-7. (A–D) Fluorescence microscope images of live MCF-7 cells incubated with 2. (A) Probe 2, (B) Hoechst, (C) overlay image of (A) and (B), (D) differential interference contrast (DIC, bright field image) with overlay of (A) and (B) (this figure has been adapted from ref. 25 with permission from Oxford University Press, copyright 2015). | ||
In 2021, Basu et al. utilized same NIR probe 2 for demonstrating the DNA-bound structure and the DNA binding mechanism of quinone cyanine dithiazole.26 The nuclear magnetic resonance (NMR) structure of probe 2 showed slight groove binding without observable distinct ligand-DNA interactions because of an endothermic and entropy-driven binding process, as well as for the minimal disturbance of the DNA structure. Generally, the cyanine-based probe 2 undergoes cis–trans isomerization facilitated by overlapping methine bridges, resulting in 16 potentials slowly interconverting cis/trans isomers. Utilizing a combination of NMR, DFT and free energy calculations, authors demonstrated that the DNA-free and DNA-bound environments of probe 2 favor distinct isomers and the slow kinetics arise from cis–trans isomerization results preferably minor groove binding of DNA with the unstable cis/trans isomer of probe 2(Fig. 4).
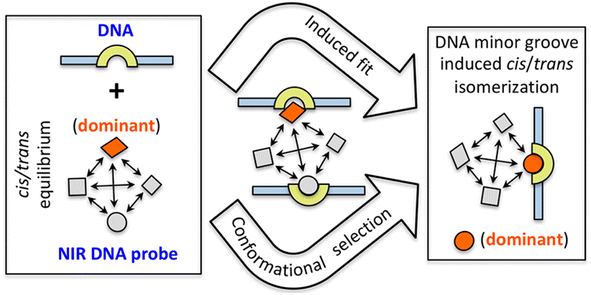 | ||
| Fig. 4 Diagram illustrating the two possible routes that probe 2 can take to reach the DNA-bound form (this figure has been adapted from ref. 26 with permission from American Chemical Society, copyright 2021). | ||
Zhang et al. included fluorescence resonance energy transfer (FRET) mechanism in near-infrared fluorescent oligonucleotide probes 3–5 for protein binding to oligo deoxyribonucleotide duplex and this technique has the potential to be employed for direct detection of protein–DNA interactions.27 Efficient fluorescence resonance energy transfer (FRET) was noted in both 3–4 and 3–5 fluorochrome pairs, demonstrating sensitivity to the spatial arrangement of the dyes. The increased FRET efficiency observed in the 3–4 pair was attributed to a more substantial overlap between the emission spectrum of the oligonucleotide-linked 3 and the excitation spectrum of 4. The potential application of NIR FRET effects lies in the detection of protein–DNA interactions. The feasibility of this detection relies on the efficiency of FRET and the specific positions of fluorochromes within the binding sites of oligonucleotides.
In 2016, sequence-dependent fluorescence intensity has been measured by Somoza et al. by developing similar type of compounds like 6 (Cy3) and 7 (Cy5) bound to the 5′ end of all 1024 possible double-stranded DNA 5 mers.28 Fluorescence variations were also noted when bound to the 5′ end of fixed-sequence double-stranded DNA with a variable sequence 3′ overhang next to the dyes 6 and 7. It was observed that both dyes' fluorescence intensity responded to each of the five bases or base pairs. The sequence dependence was found to be more pronounced for double-stranded DNA compared to single-stranded DNA (Fig. 5). Additionally, the dyes 6 and 7 exhibited sensitivity to both the adjacent double-stranded DNA sequence and the 3′-single-stranded DNA overhang. Higher fluorescence was observed in purine-rich sequences which resulted valuable insights for estimating measurement error in experiments involving fluorescent-labeled DNA and optimizing fluorescent signals by considering the nucleobase environment of the labeling cyanine dye (Fig. 5). Fluorescence enhancement was observed due to the restriction of the cis–trans isomerization through stacking interactions of the purines relative to the pyrimidines.
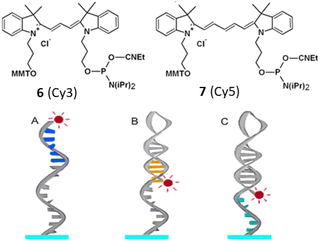 | ||
| Fig. 5 Modes of interaction between (red) dye on DNA. (A) 5′ dye in ssDNA with nearby nucleobases (blue). (B) The orange 5′ dye in dsDNA has base-paired nucleobases. (C) Next to a terminal dye on dsDNA is a 5′ dye containing the green nucleobases of ssDNA (This figure has been adapted from ref. 28 with permission from American Chemical Society, copyright 2016). | ||
Similarly, two meso-substituted cationic thiacarbocyanine dyes 8 and 9 developed by Pronkin et al. for the detection of ds-DNA in aqueous solution creating a noncovalent complex with DNA accompanied by a notable fluorescence enhancement.29 The nature of the dye–DNA interaction was examined using a variety of techniques, such as molecular docking, complexation of the dyes with ssDNA, melting of DNA in the dye–DNA complex, and displacement of dye by Na+ ions from the complex. These findings showed that the dyes 8 and 9 formed bonds with DNA, occupying both the grooves of the DNA molecule and intercalating between base pairs. The binding constants and detection limits of 8 and 9 with ds-DNA have been calculated as 1.22 × 104, 1.27 ×104 L mol−1 and 0.2, 0.022 μmol L−1 respectively (Fig. 6).
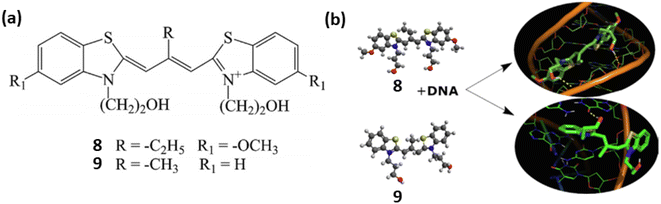 | ||
| Fig. 6 (a) Chemical structures of compounds 8 and 9. (b) Probable binding mode of ligands 8 and 9 with ds DNA (This figure has been adapted from ref. 29 with permission from Elsevier, copyright 2021). | ||
Along with the use of only cyanine dye for DNA binding study, a polymer conjugate like polyethylenimine (PEI) coupled with NIR indocyanine dye (10) was developed by Masotti et al. for the binding with DNA and its delivery can be observed in vivo through noninvasive optical imaging methods.30 Dye 10 exhibited an excellent chemical stability and favorable optical characteristics, absorbing light at 665 nm, emitting at 780 nm with a significant large stokes shift of 115 nm.30 To investigate the localization of DNA, calf thymus DNA was labeled with the near-infrared dye 10 and the labeled DNA was complexed with an unlabeled PEI solution. This approach allowed to track fluorescence in the NIR region of the 10-DNA complex and provided insights, particularly in the initial stages, into the localization of DNA. The results revealed a similar accumulation of DNA in the liver as observed with the IR820-PEI/DNA complex (Fig. 7). Notably, high cationic charge density potential polyamine polymer led to form an electrostatic interactions with DNA, which has been employed as a versatile vector for several biomedical applications like gene therapy.
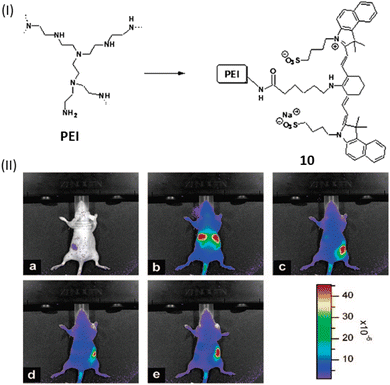 | ||
| Fig. 7 (I) Chemical structures of compound 10. (II) Pre-injection optical pictures of a naked mouse receiving a 2 mg mL−1 tail vein injection of 10-DNA (a), and after 4 min (b), 1 h (c), 2 h (d), and 3 h (e) from the injection. The animal's fluorescence emission signal efficiency is indicated by the color bar on the right side (This figure has been adapted from ref. 30 with permission from American Chemical Society, copyright 2016). | ||
Similarly, Chen et al. reported an activatable NIR fluorescence nanoprobe 11 containing of NIR quantum dots (QDs) and Al(III)-gatifloxacin (Al-GFLX) complexes for the sensitive detection of double-stranded DNA (dsDNA) both in aqueous solution and in living cells.31 By photoinduced electron transfer mechanism (PET) the first strong NIR fluorescence of QDs in QD-Al-GFLX was quenched by Al-GFLX complex. The significant binding affinity between the Al-GFLX complex and double-stranded DNA (dsDNA) may cause 11 to dissociate upon contact, hence impairing the PET process. This led to a notable increase in NIR fluorescence at 710 nm by 7-folds (Fig. 8). The complex 11 demonstrated sensitivity and specificity in detecting dsDNA in aqueous solutions, achieving a detection limit of 6.83 ng mL−1.
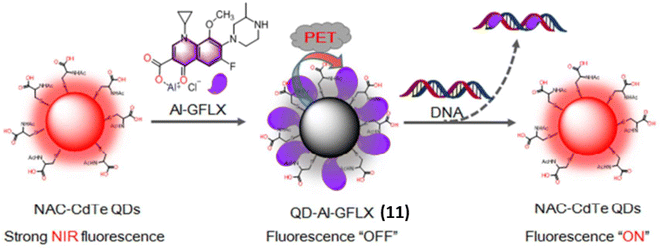 | ||
| Fig. 8 The general design of QD-Al-GFLX nanoprobe 11 with tunable NIR fluorescence and and DNA detection. QD-Al-GFLX showed an “OFF” NIR fluorescence due to the PET process between QDs and AI-GFLX. The subsequent binding of Al-GFLX with DNA can trigger QD-Al-GFLX dissociation, inducing fluorescence “ON” (This figure has been adapted from ref. 31 with permission from American Chemical Society, copyright 2017). | ||
5. NIR fluorescent probes for detection of G quadruplex DNA
Wurthner et al. introduced a near infrared amphiphilic squaraine dye 12 which is water soluble and binds with the parallel topology of G4-DNA.32 Dye 12 selectively detects the parallel over non-parallel and non G4 topologies. The probe 12 demonstrated strong selectivity of G4 over duplex, and the dicyanovinyl squaraine dye was able to provide exceptional selectivity over non-G4 structures, but it has certain limitations including moderate solublity and low binding constants, thus the moeity of this dye is modified by water soluble group that lead to the formation of water-soluble amphiphilic dye. When G4-DNA was added to dicyanovinyl squaraine dye 12 with water-soluble amphiphilic dye complex, resulted a bathochemically shifted band at ≈700 nm. The binding of G4-DNA with water soluble amphiphillic squaraine dye is through end stacking, which results NIR light up fluorescence response having quantum yield above ϕf = 0.7 and the binding constant was found to be 107–108 M−1, which shows 12 is an excellent binder to G4-DNA (Fig. 9).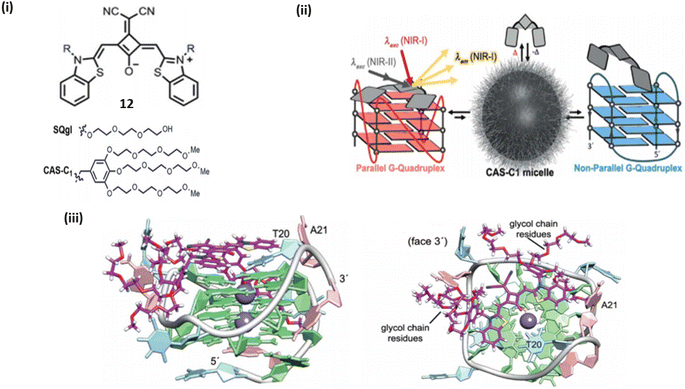 | ||
| Fig. 9 (i) Dicyanovinyl squarine dye and its derivative. (ii) Schematic representation of parallel and non-parallel G4 DNA with Dicyanovinyl squarine dye. (iii) Lateral view of G4 and water soluble amphiphilic squaraine dye complex. (ii) Top view of G4 and water soluble amphiphilic squaraine dye complex (This figure has been adapted from ref. 32 with permission from Royal Society of Chemistry, copyright 2018). | ||
In 2019, thiazole orange based styryl derivatives styryl derivatives were developed by Liu et al. and those probes specifically recognize G4-DNA showing significant biological functions.33 The derivatives of thiazole orange 13a–13d are used as a fluorescent probe for G4 DNA. In the derivatives of thiazol orange styryl group were introduced to its ortho-position in the quinoline ring. These derivatives of thiazole orange are highly selective towards G4.
The modification of thiazol orange to its 4-(diethylamine) styryl derivative 13a enhances its selectivity towards G4 DNA, while the 4-(methylpiperzine-1-yl) styryl derivative 13b exhibits a stronger affinity for antiparallel G4 DNA. Significantly all these derivatives of thiazol orange can penetrate live cells effectively, making them excellent tools for genome recognition within live cells. Notably, 13b produces intense fluorescence upon binding with nucleoli. Binding of G4 DNA with 13a results in a significant red shift in absorbance peaks, indicating high selectivity towards G4 DNA over single and double-stranded DNA. Similarly, 13b shows a substantial red shift in absorbance peaks specifically for G4 DNA. Circular Dichroism spectroscopy reveals that binding with 13a does not induce any conformational changes in G4 DNA whereas the binding of 13b with antiparallel G4 DNA leads to conformational alterations.
Similarly, Zhang et al. reported thiazol orange based probe 14 by combining with 9- vinyljulolidine for binding with G4 DNA through end-stacking.34 Upon significant binding of 14 with G-quadruplexes (G4s), particularly those arranged in an antiparallel manner, a significant increase in fluorescence intensity was observed, reaching a maximum enhancement of 2742-fold. This enhancement was accompanied by a considerable Stokes shift of 198 nm, with the emission peak reaching 694 nm in the near-infrared region. The molecule 14 exhibited remarkable sensitivity and specificity towards G4 structures, particularly favoring antiparallel G4s over other DNA configurations such as single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA). Binding of the ligand 14 with the antiparallel DNA does not cause conformational change but bring down the structural stability. In parallel DNA also it does not cause conformational change. The intensity of fluorescence increases as it has high selectivity and sensitivity towards G4 DNA compared to single and double stranded DNA. The limits of detection (LODs) for human antiparallel telomere G-quadruplexes (G4s) like Hum24 and 22AG in the presence of Na+ were determined to be remarkably low, reaching levels as low as 164 pM and 231 pM, respectively. It has been suggested by molecular docking studies that the 9- vinyljulolidine side chain present in the ortho-position of 1-methylquinolinium can improve the interaction between 14 and the G4s groove and loop regions. Additionally, experiments involving cellular localization and imaging demonstrate that 14 has the capability to penetrate live cells and localize within the mitochondria (Fig. 10).
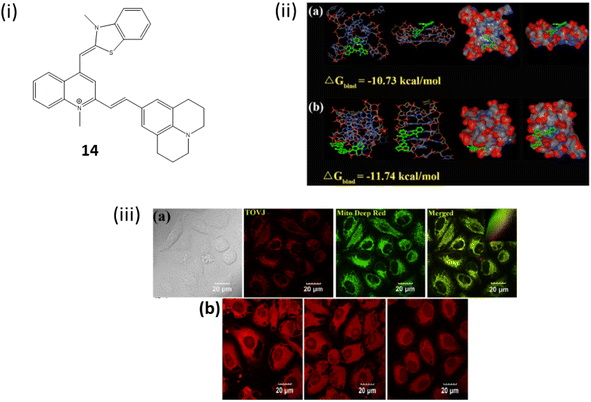 | ||
| Fig. 10 (i) Structure of 9-vinyljulolidineto Thiazol Orange 14. (ii) (a) The docking study's molecular model of the 14-parallel 22AG complex. (b) Molecular model of the 14 antiparallel 22AG complex from the docking study (iii) (a) Confocal fluorescence images of HeLa cells stained with 14 (1 μM) and Mito Deep Red (1 μM). Inset: intensity scatter plots of red and green channels. (b) Confocal pictures of the fixed HeLa cells stained with 14 (1 μM) with or without the DNase I or RNase A treatment (This figure has been adapted from ref. 34 with permission from American Chemical Society, copyright 2021). | ||
A conceptually new light-up nucleic acid fluorescent coumarin–naphthalene diimide dyad (15) was developed by Freccero et al. by linking a coumarin to a naphthalene diimide (NDI) for NIR G-quadruplex sensing.35 Ligand 15 demonstrates a distinct emission wavelength of 498 nm in solution and emits additional red/NIR light upon binding to G-quadruplex DNA. The specific light-up reaction, centered at 666 nm, shows remarkable specificity for quadruplex DNA compared to duplex DNA or RNA quadruplexes. Upon attaching to G4 DNA via its NDI moiety, ligand 15 experiences a displacement of its coumarin unit, consequently inhibiting the D–A interaction and the associated quenching by electron transfer. The binding constant for 15 with the parallel G4-DNA was found to be 6.9 ± 2.9 × 106 M−1(Fig. 11).
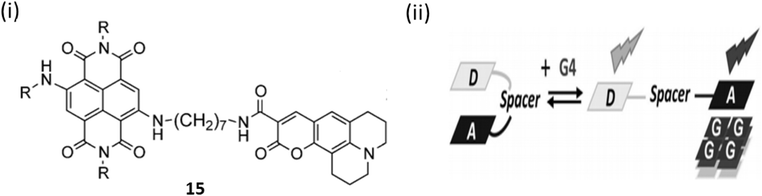 | ||
| Fig. 11 (i) Structure of Coumarin– naphthalene diimide (15). (ii) Method of sensing of G4 DNA by 15 (This figure has been adapted from ref. 35 with permission from Wiley-VCH, copyright 2015). | ||
A NIR probe 16 was engineered by Hu et al. combining G4 ligand and a mitochondrion-targeting AIE luminogen for binding with G4s via a distinctive mode.36 Cell-based experiments suggests that 16 can track mitochondrial DNA G4 formation in live cells. On exciting at 538 nm, NIR ligand 16 emitted weakly at 660 nm. And it was noted that with the addition of 1 μM of mitochondrial DNA (mt DNA) there was a 28-fold increase in the fluorescence enhancement causing an intense hypochromicity accompanied by a red shift. A strong induced CD (ICD) signal at 300–400 nm, indicating interaction with the grooves of mt DNA like mt6363, was observed upon incubation with 16, while the absence of an ICD signal in the triphenylamine-based indolium salt absorption region suggested stacking on the terminal G-tetrad, and CD melting studies showed that 16 significantly stabilized mt6363's structure, forming a tight complex. Furthermore, 16 was treated with live A549 cells a red fluorescent foci was observed (Fig. 12b). And with the subsequent treatment with the PDS (a non-fluorescent G4 ligand) it abolished the red foci in live A549 cells (Fig. 12c) and this suggests 16 may bind to G4 structures in mitochondria.
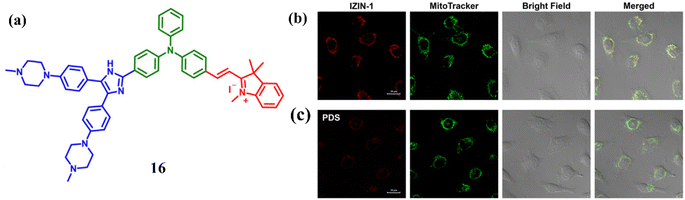 | ||
| Fig. 12 (a) Structure of the engineered molecule IZIN-1 (16). (b) Pictures of living A549 cells stained with 0.1 mM of MitoTrackerTM Green (green) and 4 mM of IZIN-1 (red channel, stimulated at 543 nm) channel, excited for 30 minutes at 488 nm. (c) Pictures of living A549 cells treated with a standard G4 ligand PDS (20 mM) after being labeled for 30 minutes with 4 mM of IZIN-1 and 0.1 mM of MitoTrackerTM Green (This figure has been adapted from ref. 36 with permission from Elsevier, copyright 2021). | ||
Kumari et al. synthesized a cationic styryl dye which consist of dimethylamine donor and pyridinium acceptor group 17 and examined their interactions with double stranded and quadruplex DNAs.37 The titration of ligand 17a with ds-DNA resulted in a 4.6-fold fluorescent enhancement accompanied by a hypochromic shift. Ligand 17b shows emission at 758 nm causing 9-fold enhancement. With G4-DNA ligand 17a showed a hypsochromic shift with 35-fold emission intensity enhancement. Ligand 17b exhibited a 43 nm blue shift with a 73-fold intensity enhancement. Ligand 17b bearing two positive charges enabled the notable emission changes and binding affinity with G4-DNA. In addition, ligand 17b possessed significant conformation stability to bind and stabilize the G4-DNA. One of the naphthyl rings stack over dG1, while the pyridinium ring stacks partially over dG13 as the dynamic developed. Besides this in ligand 17b the center-to-center distance of the naphthyl and pyridinium rings is around the same as the guanine residues (Fig. 13). This enables effective stacking between the rings of ligand 17b and the top quartet. The enhanced water solubility and prominent binding along with strong red emission provide a favourable opportunity for the creation of fluorescent probes that target biologically significant G4-DNAs.
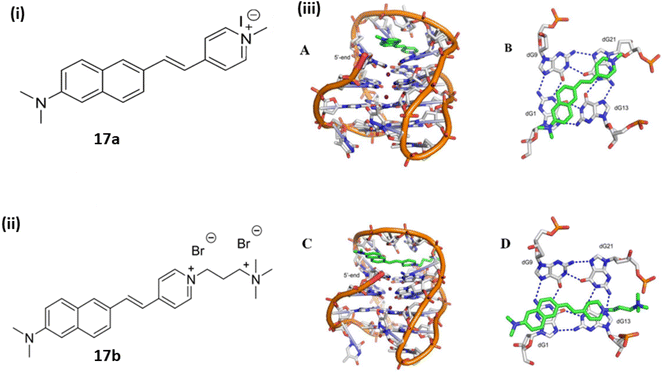 | ||
| Fig. 13 The structures of fluorophores (i) 17a (ii), 17b and (iii) Final MD snapshots of ligands 17a and 17b obtained at the conclusion of 100 ns simulations bound to telomeric G4-DNA. (A) Side view of the G4-DNA-bound 17a (B) axial perspective of 17a layered over the top quartet (C) side view of G4-DNA bound 17b (D) axial view of 17b over the top quartet (This figure has been adapted from ref. 37 with permission from Elsevier, copyright 2019). | ||
Nie et al. developed a G4 probe ThT (18) and introduced derivatives of benzothiazole ThT-DB, ThT-NA extending π-conjugation to identify and visualize G4 structures in tissue samples and living cells.38 Upon treating with c-MYC (G4s DNA), it showed a 117 nm redshift with a 12-fold enhancement in the emission intensity at 610 nm. ThT-NA exhibits excellent photophysical properties (Fig. 14a), characterized as powerful two-photon fluorescence emission, high sensitivity selectivity for G4s (1600-fold fluorescence turn-on ratio), substantial Stokes shift (>100 nm), and red emission (610 nm). When ThT-DB was mixed with double-stranded DNA, an 8-fold fluorescence enhancement was observed, indicating poor specificity towards the G4 structure. The extended linkages of probes between the electron acceptor (A) and donor (D) groups typically increase the response to double-stranded DNA, making elongation of the conjugated π-electron chain insufficient for developing a red fluorescence emission ThT analogue with reasonable specificity. Consequently, through monitoring alterations in RNA G4 folding, these features enable ThT-NA to detect the cell cycle and reveal the endogenous RNA G4 distribution in real cells (Fig. 14b).
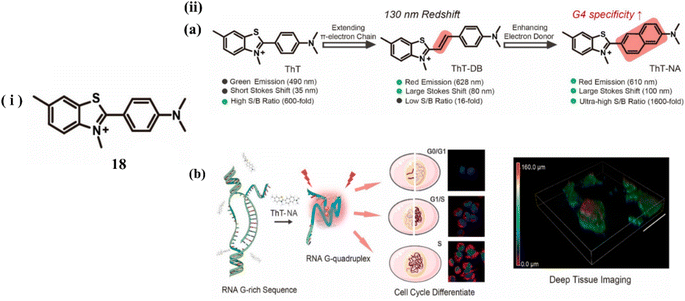 | ||
| Fig. 14 (i) Molecular structure of the probe 18. (ii) (a) Probe Characteristics and ThT Modification. (b) ThT-NA interacts with Intracellular RNA G4 (This figure has been adapted from ref. 38 with permission from American Chemical Society, copyright 2024). | ||
Along with benzothiazole based cyanine dye, Chen and Tan et al. developed a benzoselenazoloium-based hemicyanine dye, 19 for the selective detection of G-quadruplexes.39 The probe 19 is an activatable fluorescent probe whose fluorescence was activated in the presence of G-quadruplexes in buffer solution. To validate the binding of 19 with G-quadruplex Pu22, molecular docking studies are performed. Through the π–π interaction, it is discovered that 19 packed exactly on the surface of both terminal G-quartet planes of Pu22 (Fig. 15a). The dissociation constant (KD) values of 19 with G-quadruplexes were around 0.22∼ 4.33 μM. In order to evaluate the behavior of 19 with cells, HeLa cells were stained with 1 μM 19 and a strong fluorescent focus was observed (Fig. 15b). To further investigate the source of emerging fluorescence enzyme digestion experiments were carried out in DNase and RNase before staining of 19 and it was observed that the fluorescence intensity of 19 decreased. This suggests that 19 could bind to both rDNA and rRNA in cells. When a classical quadruplex ligand, BRACO19 was introduced to compete with 19 for its potential rDNA and rRNA target, the fluorescence intensity of 19 was decreased (Fig. 15c). Besides this 19 was treated with stained live cells, it was confirmed that 19 is fully functional and its cytotoxicity was determined (Fig. 15d). In addition, a mitochondria-targeted G-quadruplex model ligand RHPS4 was also used as a competitor against 19 and it was found that 19 foci in mitochondria was significantly depressed with the addition of RHPS4 (Fig. 15e).
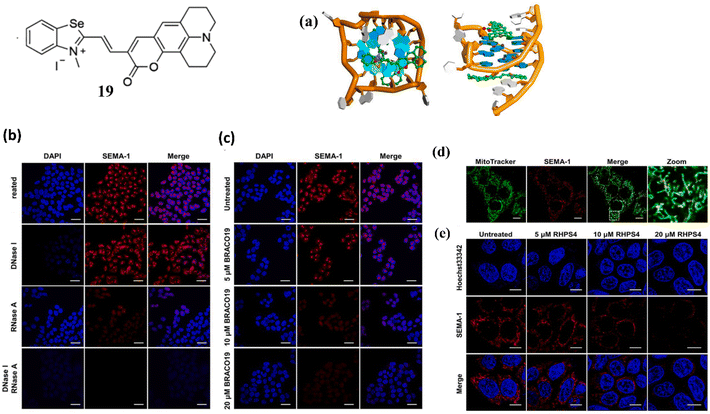 | ||
| Fig. 15 Structure of ligand 19. (a) Views of the binding mechanisms of 19 with G-quadruplex Pu22 from the top and side. (b) Fixed HeLa cells were stained with 1 μM 19 and treated by DNase and RNase. (c) Following the application of 1 μM 19 (red) dye, fixed HeLa cells were exposed to varying doses of BRACO19. (d) After being stained with 1 μM 19, RHPS4 was applied at varying doses to live HeLa cells (This figure has been adapted from ref. 39 with permission from Elsevier, copyright 2022). | ||
In addition of normal linear NIR cyanine dye, Guan et al. designed a benzothiophenyl rotor, an activatable cyanine probe 20 with a higher signal-to-ratio probe for G4 recognition and imaging.40 The ligand 20 have various properties including red emission, a rotor π structure, fluorescence quenching by H-aggregation in aqueous medium. When ligand 20 was interacted with G4s, a 98-fold fluorescent enhancement was seen with a visible color change with naked eye was seen due to the dissembling of G aggregates with the detection limit at 1.51 nM. Introducing a flexible meso-aromatic heterocyclic substituent on a polymethine chain increased the surface area for π-stacking with G4s but weakened duplex DNA intercalation compared to unsubstituted form of ligand 20. Planar “Y”-shaped structure of 20 resulted to stack on the 5′-end of the G-plane of c-myc via terminal stacking, with constrained torsion of the C–C bonds between the polymethine chain and meso-benzothiophenyl showing significant fluorescence enhancement.
Time-progressive imaging experiments demonstrated that a turn on fluorescence of 20 can be detected in the cytoplasm of HepG2 cells (Fig. 16a). In order to confirm the specific staining of intracellular G4s by DNase, RNase, certain competition experiments were performed (Fig. 16b). After the treatment with DNase, the fluorescence of 20 was completely disappeared and thus the ligand 20 provided better imaging targeting at 0.5 μM staining concentration for mitochondrial G4s (Fig. 16c). These results suggests that the meso-substituted cyanine dyes can explore the G4 targeting anticancer drugs.
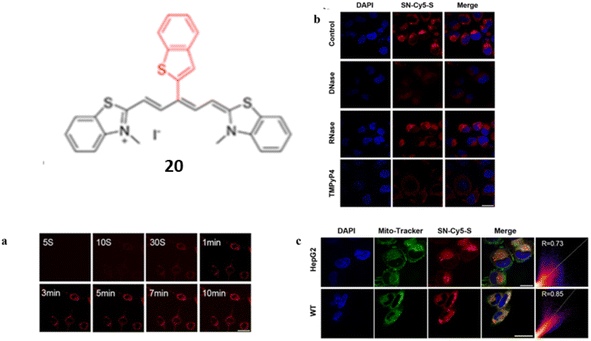 | ||
| Fig. 16 Structure of SN-Cy5-S (20). (a) Time-progressive images of living HepG2 cells at different time points after the addition of 20. (b) Confocal microscopy pictures of fixed HepG2 cells treated with DNase I, RNase A, or TMPyP4 after being stained with 20. (c) HepG2 and WT cells co-stained with DAPI (2 μg mL−1), 20 (0.5 μM), and Mito-Tracker Green (0.2 μM) in confocal fluorescence pictures (This figure has been adapted from ref. 40 with permission from American Chemical Society, copyright 2023). | ||
Muraoka et al. developed a NIR fluorescence switch-on backbone by the modification of tripodal quinone-cyanine fluorescent backbone 21 in order to visualize the guanine-quadruplex (G4) nucleic acids in cells.41 The backbone 21 was identified to have higher selectivity and large stock shifts towards G4-DNA. In one of the form of 21, fluorescence response of QCy(BnBT)3 towards various dsDNAs and G4 DNAs were recorded and significantly the fluorescence response towards dsDNAs were suppressed wherein with G4DNA there was a 500-fold enhancement. These results indicate the increased selectivity due to the modification of tripodal quinone-cyanine fluorescent ligand. The fluorescence quantum yield of QCy(BnBT)3 in the absence and presence of c-Myc G4 DNA is 2.3 × 10−2 and 3.0 × 10−4 respectively. The dissociation constant KD-app of QCy(BnBT)3 with G4 DNA were 10−7 to 10−8 M. The selective imaging of G4DNA in cells without the blocking signal from dsDNA in the nuclei is responsible for this remarkable selectivity. Furthermore, G4 imaging of 21 in fixed HeLa cells were performed (Fig. 17). The huge fluorescence response of 21 with the addition of RNA G4 sequences suggests that 21 might stain both DNA G4 and RNA G4 in cells with NIR fluorescence.
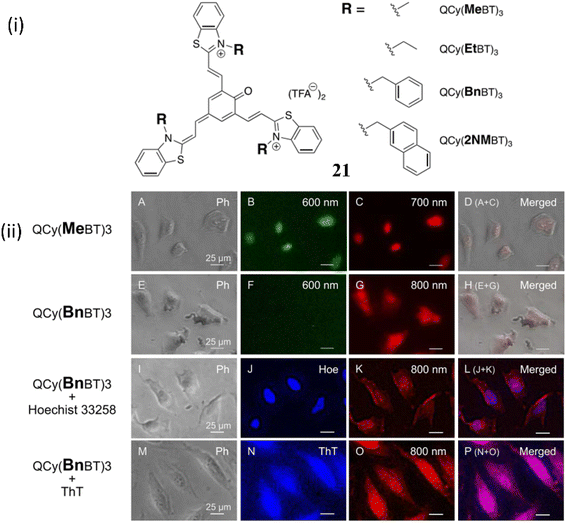 | ||
| Fig. 17 (i) Structure of D3A type tripodal quinone-cyanine backbone 21. (ii) G4 imaging with QCy(MeBT)3 or QCy(BnBT)3 in fixed HeLa cells (This figure has been adapted from ref. 41 with permission from American Chemical Society, copyright 2023). | ||
Together with the growing interest in NIR cyanine dyes based on benzothiazole and benzoselenazole moieties, quinolinium-based cyanine dyes are also highly effective for binding with G4-DNA. In 2023, Verma et al. reported red-emissive styryl quinolinium-based molecular probes 22a-c for the selective sensing of telomeric G4 DNA, with a distinct preference over DNA duplexes (Fig. 18).42 The fluorescence response of the chemicals in the presence of different nucleic acids was documented in order to assess the nucleic acid selectivity. The largest fluorescence enhancement was observed in 22a. While 22b and 22c displayed 5-fold lower fluorescence compared to 22a. Upon addition of various DNAs, human telomeric G4 DNA showed the maximum fluorescence of 660 nm with a 154-fold enhancement with the binding affinity as Ka = 0.51× 106 M−1. Molecular docking experiment suggested that the interactions of 22a in the grooves of the G-quadruplex DNA (Fig. 18ii). These findings lead to the development of a red-emissive molecular scaffold like 22a–c with selective recognition of human telomeric G-quadruplex DNA.
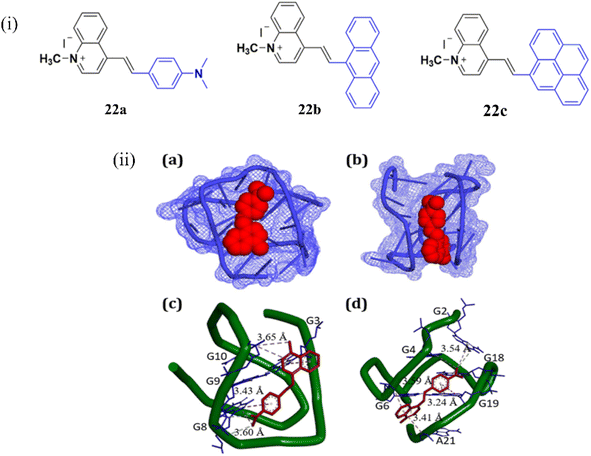 | ||
| Fig. 18 (i) Chemical structures of styryl quinolinium derivatives 22a–c. (ii) (a and c) the docking results showing the comparative binding of 21a with the human telomeric G-quadruplex DNA, (b, d) c-MYC G-quadruplex DNA sequences (This figure has been adapted from ref. 42 with permission from American Chemical Society, copyright 2022). | ||
In another type of NIR dye, Wang et al. developed a fluorescent probe YON (23), a thread-like molecule with an A–π–D–π–A structure, for the sensitive and selective monitoring for mt-DNA with an enhanced fluorescence and large stokes shift (λex = 435 nm, λem = 640 nm) based on twisted intramolecular charge transfer (TICT) mechanism.43 The molecular docking experiments suggest that 23 bounds strongly with mt-DNA through minor grooves and has created little damage to mt-DNA (Fig. 19ii). The fluorescence signal strength of 23 at 640 nm exhibited a good linear relationship with a lower detection limit of 71 ng mL−1 and a binding constant of 8.5 × 105 M−1. The cancer cell line HepG2 was chosen in order to confirm the real-time imaging capability of 23 for mt DNA in living cells which indicates a strong fluorescence signal at 600–660 nm in HepG2 cells (Fig. 19iii). In DNA digestive experiment cells were treated with DNA digestive enzymes and the fluorescence intensity drops to 2.4 times when the incubation period of DNA digestion enzymes is extended to 90 minutes. The DNA digestion enzymes broke down the closed circular double-stranded structure of mt-DNA, allowing the probe 23 to rotate freely (Fig. 19iv). When HepG2 cells were pretreated with CCCP and 23, the fluorescent intensity of 23 remained unchanged, whereas the fluorescent intensity Of CCCP was increased significantly (Fig. 19v). The cell imaging results suggests that 23 can significantly accumulate in the mitochondria due to its good amphiphilic nature.
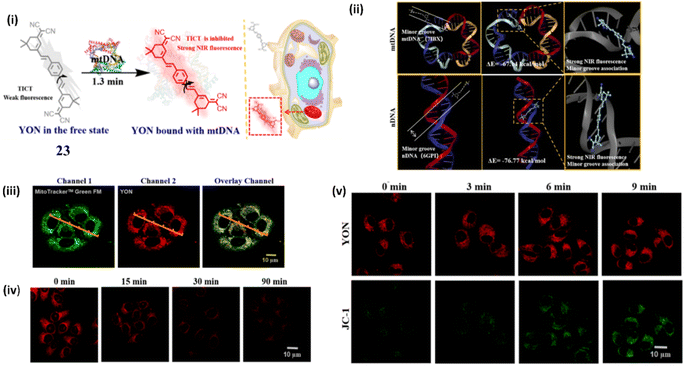 | ||
| Fig. 19 (i) Structure of 23 in the free state and its structure when bound to DNA. (ii) Molecular docking studies between 23 and DNA. (iii) Imaging in HepG2 cells. (iv) Imaging of DNA digestive enzymes in Living HepG2 cells. (v) Cell imaging of living HepG2 cells with different mitochondrial membrane potentials. (This figure has been adapted from ref. 43 with permission from American Chemical Society, copyright 2022). | ||
Richter et al. investigated a red-NIR light-up prototype core-extended naphthalene di imide based probe 24 for the binding of G-quadruplex DNA.44 This ligand 24 when bound with the G4 DNA provides a wide range of biological advantage by acting as an inhibitor for various type of pathological process (Fig. 20).
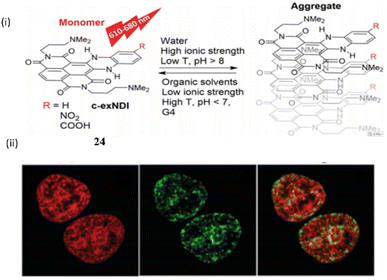 | ||
| Fig. 20 (i) The structure of Naphthalene diimide probe (c-exNDI) (24) and the aggregation properties. (ii) Cell imaging study with 24. (This figure has been adapted from ref. 44 with permission from Royal Society of Chemistry, copyright 2024). | ||
The tri- and tetra-substituted naphthalene diimide are also an excellent probe as well as an alkylating agent that specifically target the guanine rich nucleic acid in G4-DNA. Binding constants of the ligand 24 with c-myc and TBA were found to be 3.43 ± 0.3 × 107 M−1 and 1.2 ± 0.5 × 105 M−1 respectively. The core-extended naphthalene diimide selectively binds with the viral G4 of HIV virus rather than the cellular G4.
6. NIR fluorescent probes for detection of double-stranded DNA and G quadruplex DNA
Along with the development of various NIR cyanine probes for the individual binding of duplex DNA and quadruplex DNA, there are few reports on NIR probes that can bind both G4-DNA and ds-DNA. In this respect, Sakamoto et al. reported a tripodal quinone-cyanine dye 25 having one quinone donor and three acceptors N-methylbenzothiazolium moieties for binding with G4-DNA and ds-DNA with the emission enhancement at NIR region at 700 nm (λex = 570 nm) and 600 nm (λex = 470 nm) respectively.45 The findings from docking simulations and the reaction to viscosity alterations indicated that the dual-fluorescence reaction stemmed from variations in the binding mode of ligand 25, depending on the structure of the DNA. Additionally, ligand 25 can also bind to the DNA minor groove of the ds (AAATTT) region, enhancing its fluorescence. Results from fluorescence microscopy imaging experiments employing the ligand 25 implied the potential for visualizing G4 DNAs and dsDNAs within the cell nucleus using near-infrared (NIR, 700 nm) and red (600 nm) fluorescence emissions (Fig. 21).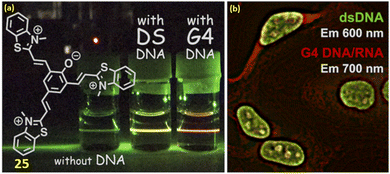 | ||
| Fig. 21 (a) Chemical structure of ligand 25. (b) Fluorescence microscopy imaging inside the cell (This figure has been adapted from ref. 45 with permission from American Chemical Society, copyright 2022). | ||
Similarly, Ihmels et al. reported a NIR probe 3-(4-(N,N-dimethylamino)phenyl)naphtho[1,2-b]quinolizinium (26) using the Suzuki–Miyaura reaction for binding with RNA, duplex DNA and G4-DNA through photometric, fluorimetric, and polarimetric titrations as well as DNA denaturation studies.46 The compound 26 binds with DNA and RNA by intercalation and G4-DNA by terminal π stacking (Fig. 22). The ligand 26 exhibits a fluorescence light-up effect upon complexation with the nucleic acids (Φfl = 0.01–0.05, λfl = 725–750 nm). Furthermore, triple-exponential fluorescence decay of 26 upon attaching to biomacromolecules inside a cell makes them easier to see within this environment and enables the selective labeling of different cellular components.
 | ||
| Fig. 22 Chemical structure of ligand 26 and the binding mode of DNA with ligand 26 (This figure has been adapted from ref. 46 with permission from Wiley-VCH, copyright 2021). | ||
7. Conclusion
The development and application of Near-Infrared (NIR) fluorescence switch-on probes for the detection and in cellulo tracking of G-quadruplex and double-stranded DNA represent a significant advancement in the field of bioimaging and molecular diagnostics. These probes offer several advantages, including deep tissue penetration, minimal photodamage, and low background fluorescence, making them highly suitable for live-cell imaging and real-time tracking of nucleic acid structures. Our review highlights the diverse strategies employed in the design of NIR fluorescence switch-on probes, emphasizing the importance of specificity and sensitivity in detecting G-quadruplex and ds-DNA. The unique structural and photophysical properties of these probes enable them to distinguish between various nucleic acid conformations, thereby providing valuable insights into the dynamic processes of DNA regulation and interaction within the cellular environment. Despite the promising results, there are still challenges to be addressed, such as improving the selectivity and reducing the cytotoxicity of these probes. Future research should focus on refining the molecular design and enhancing the biocompatibility of NIR probes to facilitate their broader application in clinical diagnostics and therapeutic monitoring. In conclusion, NIR fluorescence switch-on probes for G-quadruplex and double-stranded DNA detection and tracking offer a powerful tool for molecular and cellular biology. Their continued development will undoubtedly contribute to significant advancements in the understanding of nucleic acid biology and the diagnosis and treatment of genetic diseases.Abbreviations
| NIR | Near-infrared |
| DNA | Deoxyribonucleic acid |
| RNA | Ribonucleic acid |
| dsDNA | Double-stranded DNA |
| ssDNA | Single-stranded DNA |
| G4s | G-quadruplexes |
| EtBr | Ethidium bromide |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| Sp | Spirocyclic |
| Mc | Merocyanine |
| QCy–DT | Quinone cyanine–dithiazole |
| D2A | One-donor-two-acceptor |
| ICT | Internal charge transfer |
| PET | Photoinduced electron transfer |
| DIC | Differential interference contrast |
| NMR | Nuclear magnetic resonance |
| FRET | Fluorescence resonance energy transfer |
| PEI | Polyethylenimine |
| QDs | Quantum dots |
| LODs | Limits of detection |
| NDI | Naphthalene diimide |
| mtDNA | Mitochondrial DNA |
| TICT | Twisted intramolecular charge transfer |
| TBA | Thrombin binding aptamer |
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors would like to thank Christ University, Bengaluru for the research facilities and Centre for Research, Christ University for the seed money grant (grant approval number SMSS-2105). Avijit Kumar Das specially acknowledges State University Research Excellence (SERB-SURE) of the Science and Engineering Research Board (SERB) (File Number: SUR/2022/002461) under Department of Science and Technology, Government of India, for financial support by the research grant and research fellowship for Hazeena Shinziya.References
- A. Travers and M. Georgi, FEBS J., 2015, 12, 2279–2295 CrossRef PubMed.
- S. Bhaduri, N. Ranjan and D. P. Arya, Beilstein J. Org. Chem., 2018, 14, 1051–1086 CrossRef CAS PubMed.
- (a) H. Ihmels and D. Otto, In Supermolecular Dye Chemistry, 2005, 258, 161–204 Search PubMed; (b) S. Nafisi, A. A. Saboury, N. Keramat, J. F. Neault and H. A. Tajmir- Riahi, J. Mol. Struct., 2007, 827, 35–43 CrossRef CAS; (c) T. Mahata, A. Kanungo, S. Ganguly, E. K. Modugula, S. Choudhury, S. K. Pal, G. Basu and S. Dutta, Angew. Chem., 2016, 55, 7733–7736 CrossRef CAS PubMed.
- (a) S. Neidle, Nat. Prod. Rep., 2001, 18, 291–309 RSC; (b) A. Rahman, P. O'Sullivan and I. Rozas, MedChemComm, 2019, 10, 26–40 RSC; (c) F. Han, N. Taulier and T. V. Chalikian, Biochemistry, 2005, 44, 9785–9794 CrossRef CAS PubMed.
- (a) D. Banerjee and S. K. Pal, J. Phys. Chem., 2008, 112, 1016–1021 CrossRef CAS PubMed; (b) S. Ganguly, D. Ghosh, N. Narayanaswamy, T. Govindaraju and G. Basu, PLoS One, 2020, 15, e0239145 CrossRef CAS PubMed; (c) A. K. Das, S. I. Druzhinin, H. Ihmels, M. Müller and H. Schönherr, Chem.–Eur. J., 2019, 25, 12703–12707 CrossRef CAS PubMed; (d) A. K. Das, H. Ihmels and S. Kölsch, Photochem. Photobiol. Sci., 2019, 18, 1373 CrossRef CAS PubMed; (e) S. Vishnu, A. Nag and A. K. Das, Anal. Methods, 2024, 16, 5263–5271 RSC.
- (a) A. Rahman, P. O'Sullivan and I. Rozas, MedChemComm, 2019, 10, 26–40 RSC; (b) N. J. Wheate, C. R. Brodie, J. G. Collins, S. Kemp and J. R. Aldrich-Wright, Med. Chem., 2007, 7, 627–648 CAS.
- B. Juskowiak, Anal. Bioanal. Chem., 2011, 399, 3157–3176 CrossRef CAS PubMed.
- Y. V. Suseela, N. Narayanaswamy, S. Pratihar and T. Govindaraju, Chem. Soc. Rev., 2018, 47, 1098–1131 RSC.
- (a) H. Kobayashi, M. R. Longmire, M. Ogawa and P. L. Choyke, Chem. Soc. Rev., 2011, 40, 4626–4648 RSC; (b) D.-H. Li, C. L. Schreiber and B. D. Smith, Angew. Chem., 2020, 59, 12154–12161 CrossRef CAS PubMed.
- (a) S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402–5415 CrossRef CAS PubMed; (b) M. L. Bochman, K. K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS PubMed.
- M. A. Goldman, Drug Discovery Today, 2003, 8, 294–296 CrossRef PubMed.
- H. Ihmels and D. Otto, Top. Curr. Chem., 2005, 258, 161–204 CrossRef CAS.
- S. Nafisi, A. A Saboury, N. Keramat, J. F. Neault and H. A. T. Riahi, J. Mol. Struct., 2007, 827, 35–43 CrossRef CAS.
- T. Mahata, A. Kanungo, S. Ganguly, E. K. Modugula, S. Choudhury, S. K. Pal, G. Basu and S. Dutta, Angew. Chem., 2016, 55, 7733–7736 CrossRef CAS PubMed.
- S. Neidle, Nat. Prod. Rep., 2001, 18, 291–309 RSC.
- A. Rahman, P. O'Sullivan and I. Rozas, MedChemComm, 2019, 10, 26–40 RSC.
- F. Han, N. Taulier and T. V. Chalikian, Biochemistry, 2005, 44, 9785–9794 CrossRef CAS PubMed.
- D. Banerjee and S. K. Pal, J. Phys. Chem. B, 2008, 112, 1016–1021 CrossRef CAS PubMed.
- B. Juskowiak, Anal. Bioanal. Chem., 2011, 399, 3157–3176 CrossRef CAS PubMed.
- Y. V. Suseela, N. Narayanaswamy, S. Pratihar and T. Govindaraju, Chem. Soc. Rev., 2018, 47, 1098–1131 RSC.
- B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter, Molecular Biology of the Cell, Garland Science, New York, 4th edn, 2002 Search PubMed.
- Z. Liu, Y. Liu, Y. Sun, G. Chen and Y. Chen, RSC Adv., 2017, 7, 52581–52587 RSC.
- R. V. Balayeshwanth and J. Alzeer, Chembiochem, 2013, 1–20 Search PubMed.
- A. D. Pugachev, A. S. Kozlenko, N. I. Makarova, I. A. Rostovtseva, I. V. Ozhogin, V. S. Dmitriev, G. S. Borodkin, V. V. Tkachev, A. N. Utenyshev, M. A. Sazykina, I. S. Sazykin, T. N. Azhogina, S. K. Karchava, M. V. Klimova, A. V. Metelitsa and B. S. Lukyanov, Photochem. Photobiol. Sci., 2023, 22, 2651–2673 CrossRef CAS PubMed.
- N. Nagarjun, S. Das, P. K. Samanta, K. Banu, G. P. Sharma, N. Mondal, S. K. Dhar, S. K. Pati and T. Govindaraju, Nucleic Acids Res., 2015, 18, 8651–8663 Search PubMed.
- S. Ganguly, N. A. Murugan, D. Ghosh, N. Narayanaswamy, T. Govindaraju and G. Basu, Biochemistry, 2021, 60, 2084–2097 CrossRef CAS PubMed.
- S. Zhang, V. Metelev, D. Tabatadze, P. C. Zamecnik and A. Bogdanov, Proc. Natl. Acad. Sci. U.S.A., 2008, 105, 4156–4161 CrossRef CAS PubMed.
- N. Kretschy, M. Sack and M. M. Somoza, Bioconjugate Chem., 2016, 27, 840–848 CrossRef CAS PubMed.
- G. P. Pronkin and S. A. Tatilolov, Spectrochim. Acta Mol. Biomol. Spectrosc., 2021, 263, 120171 CrossRef PubMed.
- A. Masotti, P. Vicennati, F. Boschi, L. Calderan, A. Sbarbati and G. Ortaggi, Bioconjugate Chem., 2008, 19, 983–987 CrossRef CAS PubMed.
- S. Yizhong, N. Zhang, Y. Sun, W. W. Zhao, D. Ye, J. J. Xu and H. Y. Chen, ACS Appl. Mater. Interfaces, 2017, 30, 25107–25113 Search PubMed.
- V. Grande, C. A. Shen, M. Deiana, M. Dudek, J. Olesiak-Banska, K. Matczyszyn and F. Würthner, Chem. Sci., 2018, 44, 8375–8381 RSC.
- Z. Lingling, X. Liu, S. Lu, J. Liu, S. Zhong, Y. Wei, T. Bing, N. Zhang and D. Shangguan, ACS Appl. Bio Mater., 2020, 5, 2643–2650 Search PubMed.
- J. Ming, J. Li, Y. Chen, J. Zhao, J. Zhang, Z. Zhang, P. Du, L. Zhang and X. Lu, ACS Appl. Mater. Interfaces, 2021, 28, 32743–32752 Search PubMed.
- M. Zuffo, F. Doria, V. Spalluto, S. Ladame and M. Freccero, Chem. Eur J., 2015, 49, 17596–17600 CrossRef PubMed.
- M. H. Hu, Anal. Chim. Acta, 2021, 1169, 338600 CrossRef CAS PubMed.
- B. Kumari, A. Yadav, S. P. Pany, P. I. Pradeepkumar and S. Kanvah, J. Photochem. Photobiol., B, 2019, 190, 128–136 CrossRef CAS PubMed.
- X. Lu, X. Wu, S. Kuang, C. Lei and Z. Nie, Anal. Chem., 2022, 94, 10283–10290 CrossRef CAS PubMed.
- Z. C. Li, T. Y. Wu, S. T. Zeng, L. Fang, J. X. Mao, S. B. Chen, Z. S. Huang, X. C. Chen and J. H. Tan, Bioorg. Med. Chem., 2022, 70, 128801 CrossRef CAS PubMed.
- L. Guan, Y. Zhou, X. Li, Y. Mao, A. Li, Y. Fu, W. Liu, S. Dong, Z. Liang, Y. Zhang, Q. Zhao and L. Zhang, Anal. Chem., 2023, 95, 9288–9296 CrossRef CAS PubMed.
- Y. Muraoka, J. Muramoto, Y. Yasuhara, M. Kawatake and T. Sakamoto, Anal. Chem., 2023, 47, 17162–17165 CrossRef PubMed.
- S. Verma, R. K. Patidar, K. Tiwari, R. Tiwari, J. Baranwal, R. Velayutham and N. Ranjan, J. Phys. Chem. B, 2022, 38, 7298–7309 CrossRef PubMed.
- Y. Wang, H. Niu, K. Wang, G. Wang, J. Liu, T. D. James and H. Zhang, Anal. Chem., 2022, 94, 7510–7519 CrossRef CAS PubMed.
- F. Doria, M. Nadai, M. Zuffoa, R. Perrone, M. Freccero and S. N. Richter, Chem. Commun., 2017, 53, 2268 RSC.
- T. Sakamoto, Z. Yu and Y. Otani, Anal. Chem., 2022, 94, 4269–4276 CrossRef CAS PubMed.
- P. J. Wickhorst, S. I. Druzhinin, H. Ihmels, M. Müller, M. S. Sardo, H. Schönherr and G. Viola, ChemPhotoChem, 2021, 12, 1079–1088 CrossRef.
| This journal is © The Royal Society of Chemistry 2024 |