 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Early detection of SARS-CoV-2 with functionalized gold and molecularly imprinted polymeric nanoparticles: a mini review
Pankaj
Singla
*a,
Harpreet
Kaur
b,
Saweta
Garg
a,
Navalpreet
Kaur
 c,
Francesco
Canfarotta
d,
Rakesh Kumar
Mahajan
c,
Francesco
Canfarotta
d,
Rakesh Kumar
Mahajan
 b and
Marloes
Peeters
b and
Marloes
Peeters
 *a
*a
aSchool of Engineering, Merz Court, Newcastle University, Claremont Road, Newcastle Upon Tyne-NE1 7RU, UK. E-mail: Pankaj.singla@ncl.ac.uk; marloes.peeters@newcastle.ac.uk
bDepartment of Chemistry, UGC-Centre for Advanced Studies-I, Guru Nanak Dev University, Amritsar-143005, India
cDepartment of Chemistry, Khalsa College Amritsar, Amritsar-143002, India
dMIP Discovery, The Exchange Building, Colworth Park, Sharnbrook, Bedford MK44 1LQ, UK
First published on 15th September 2023
Abstract
The novel coronavirus COVID-19 was first reported in Wuhan, China, in December 2019 and rapidly spread to the rest of the world, with the WHO declaring a global pandemic in March 2020. Rapid mutations of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have challenged its diagnosis and treatments. Reverse transcription-polymerase chain reaction (RT-PCR) tests are widely used for the diagnosis of COVID-19; however, they present several drawbacks including high cost, long turnaround time, and need for sophisticated lab infrastructure and trained technical personnel. Lateral flow tests based on antigen sensing are an interesting alternative since they offer rapid (15–30 min) and low-cost analysis, although their low sensitivity has led to several adopted tests being withdrawn from the market. Henceforth, the development of detection methods which are fast, robust, reliable, cost-effective, easy to use and portable is indispensable to prevent community transmission of COVID-19. We have reviewed two different emerging colloidal-based methodologies, (a) functionalized gold nanoparticles (functionalized AuNPs) and (b) molecularly imprinted polymers (MIPs), for fast, highly specific, and reliable identification of SARS-CoV-2. Different modifications of AuNPs with antibodies, antigens and nucleoproteins and their various assays including colorimetric, electrochemical, localized surface plasmon resonance (LSPR) and lateral flow immunoassays are discussed. In contrast, with MIP-based sensors, various antigen proteins and virus particles can be imprinted within the polymeric nanoplatform and hence can be detected with various readout techniques. The operating characteristics of these two emerging diagnostic platforms were critically reviewed and compared against each other.
Design, System, ApplicationRecent developments have highlighted that there remains an urgent need for accurate and low-cost point-of-care SARS-CoV-2 tests to break the chain of community virus transmission. Polymerase chain tests are not suitable for this purpose since the technique is costly and time-consuming. Furthermore, lateral flow tests lack sensitivity and are susceptible to environmental changes (e.g. pH, temperature) due to the use of antibodies as receptors. We will discuss two emerging technologies, including the use of gold and molecularly imprinted polymer nanoparticles (nanoMIPs) as highly selective synthetic recognition elements for SARS-CoV-2, combined with straightforward and rapid analysis methods. The review will focus on the design requirements for a portable test that uses nanoparticles as recognition elements and critically compare the systems (i) to traditional antibody-based assays and (ii) against each other. We will also discuss what prevents these systems from being commercialized given that lateral flow tests remain the preferred method of choice for tests in a home setting. The manuscript looks towards the future on how emerging technologies, based on versatile nanoparticles that can be modified towards other targets, can prevent future outbreaks of emerging pathogens. |
1.0 Introduction
In December 2019 (Wuhan, China), a virus called SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) (COVID-19) was reported and shortly declared as a pandemic due to its high transmission rates.1–3 There have been more than 545 million confirmed global cases as of June 2022, leading to a reported number of 6 million deaths with numbers still increasing.4 Several vaccines have been developed globally and compulsory vaccination programs were implemented, which have been completed in most of the developed countries including some of the developing countries (12.13 billion vaccine doses have been administered globally). However, in most low-income countries, vaccination programs were run on a relatively smaller scale and could cover only a fraction of the population (∼5%) due to lack of knowledge, insufficient medical campaigns, and poor medical facilities.5,6 Furthermore, emerging variants due to the fast mutation rate of the virus remain a constant threat. Currently, reverse transcription-polymerase chain reaction (RT-PCR) is considered the gold standard technique for the detection of SARS-CoV-2. This process relies on the identification of viral RNA through nucleic acid reverse transcription and amplification.7,8 However, this technique requires specialized equipment, reagents, facilities, and long turnaround time, during which community transmission occurs.9,10 However, developing countries and deprived areas lack advanced facilities and, consequently, there is rapid spread of COVID-19. In addition to this, there are several inexpensive rapid antigen lateral flow test kits on the market which are cheaper, easy to use and provide results in around 15–30 min. However, several rapid antigen test kits have been withdrawn from the market as they fail to meet the required sensitivity of the WHO norms (>80% for both symptomatic and asymptomatic cases). Consequently, the SARS-CoV-2 pandemic highlighted the need for more suitable and effective point-of-care tools for the detection and prevention of outbreaks.11 SARS-CoV-2 is composed of four types of structural proteins, viz. spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins [Fig. 1].12 The virus encompasses different protein spikes covering its outer membrane surface called S proteins. These spikes (SARS-CoV-2) have encountered mutations at multiple sites; moreover, more than one mutation type can be seen on the same mutation site. D614(7859), L5(109), L54(105), P1263(61), P681(51), S477(47), T859(30), S221(28), V483(28), and A845(24) are the top 10 mutation sites based on total number of occurrences.13 It was found from the literature that D614G is the only mutation site that all current mutations have in common.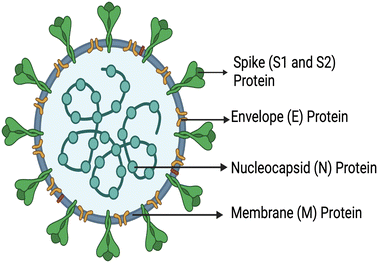 | ||
| Fig. 1 Schematic representation of the structural proteins of SARS-CoV-2, created with https://www.BioRender.com. | ||
Early work focused on these proteins because they enable the virus to enter host cells. There are two different approaches for the diagnosis of COVID-19: the first involves the detection of viral proteins, and the second serological method involves the measurement of elevated immunoglobulin M (IgM) and immunoglobulin G (IgG) levels produced by the body in response to SARS-CoV-2 infection. Nanotechnology paves the way to early, convenient, low set-up, cost-effective virus detection. The detection of COVID-19 or its antigens could be determined optically, electrochemically, and by employing mass spectrometer/hybrid transduction platforms and other different analytical techniques. Considering the advantages of optical and electrochemical sensing methods, scientists have developed several nanoscale integrated sensing platforms specific for the SARS-CoV-2 as well as its antigens/proteins and antibodies.14 There has been a growing demand for portable sensors capable of exhibiting high specificity, sensitivity and accuracy. To develop portable sensors, specifically point-of-care sensors, for the detection of SARS-CoV-2, it is crucial to identify or design a suitable molecular probe, also known as a molecular recognition element. A molecular probe should possess high affinity and selectivity and specifically recognize and bind the components of SARS-CoV-2 virus such as proteins or RNA genome. For example, spike (S1 and S2 proteins), nucleocapsid (N) and membrane (M) proteins can be targeted with specific (monoclonal) antibodies/aptamers (oligonucleotides) or MIPs. The source of the antibody can be patients with SARS-CoV-2; antibodies against these proteins were detected or can be developed via recombinant DNA technology utilizing immunogen; for example, SARS-CoV-2 antibody can be generated by sequencing peripheral blood lymphocytes of a patient exposed to SARS-CoV-2.15 Furthermore, the part of proteins (S1, S2, N and M proteins) or RNA of the virus can be recognized using antigens, peptides, antisense nucleotides, or aptamers.
Molecular recognition elements for SARS-CoV-2 can also be fabricated with molecularly imprinting technology/method; one can select the S, M and E protein or epitopes of the aforementioned proteins or the epitope of naming proteins using a range of functional monomers. There are thousands of monomers available that can be utilized for the fabrication of MIPs. Utilizing computational modeling and advanced techniques, suitable and optimal functional monomers can be selected. This approach aids in the identification of the monomers crucial for generating exceptionally selective and specific MIPs.16 For example, computational software including SiteMap for predicting protein binding sites and MM-GBSA for calculating binding free energies of monomers at each site can be utilized.
In addition to that for signal transduction, different types of monomer such as fluorescent (such as fluorescein-o-methacrylate) and electroactive monomers (such as ferrocene-o-methacrylate) can be used for fluorescence and electrochemical detection, respectively.
Furthermore, an efficient sensor requires a signal transducer, which facilitates the transmission of signals during or after the binding event and nanomaterials can serve as effective signal transducers. Examples of such nanomaterials include magnetic nanoparticles (including iron oxide nanoparticles), metal nanoparticles such as AuNPs, quantum dots, and carbon nanostructures such as single-walled carbon nanotubes, graphene, cellulose-based materials, polystyrene microbeads17–22 and many more are being used. These nanomaterials ranging in size from 1 to 100 nm possess unique properties such as conductance, thermal and chemical stability, distinctive optical characteristics, magnetic properties, increased strength, and high surface area to functionalize molecular probes.
These properties provide enhanced performance of the sensor in terms of response time, signal-to-noise (S/N) ratio, selectivity, and limits of detection. Electrochemical techniques are simple, inexpensive, and comparatively sensitive and are therefore routinely employed for the detection of bacteria, proteins, antibodies, and viruses.23 In the context of SARS-CoV-2 electrochemical sensors, AuNPs have been effectively employed to enhance both electron transfer kinetics and signal amplification. The utilization of AuNPs in these sensors accelerates the electron transfer process, thereby improving the efficiency of electrochemical detection and enhancing the overall performance of the sensor. Moreover, AuNPs can be used in colorimetric assays and are compatible with most analytical techniques.
Of these materials, functionalized AuNPs and molecularly imprinted polymeric nanoparticles (nanoMIPs) have been extensively studied for the detection of COVID-19. The colorimetric detection method employing nanoparticles is an appealing method since it offers detection by the naked eye. On the other hand, nanoMIPs are also gaining attention because they are highly robust, inexpensive and thermally stable and there is no direct need for biological counterparts for the detection. There has been no review to date focused on the comparison of functionalized AuNPs and MIPs; however, there is a pressing need to address and compare these emerging technologies and research work based on functionalized AuNPs and MIPs.
Therefore, in this mini review, the use of AuNPs functionalized with antibodies, antigens and nucleotides, aptamers and MIPs/nanoMIPs for the detection of COVID-19 is systematically compared to the gold standard.
2.0 Materials used for SARS-CoV-2 detection
2.1 Functionalized gold nanoparticles (AuNPs)
AuNPs exhibit different chemical and physical properties that make them excellent materials for the development of biosensors.24 Moreover, AuNPs can be synthesized via straightforward one-step reactions and display good biocompatibility, photostability, high electron transfer rate and catalytic activity.25 Generally, AuNPs have been widely used in sensors, particularly since colorimetric assays based on AuNPs offer simplicity, cost-effectiveness, naked-eye response, high specificity and ease of use.26 Henceforth, these have been employed for the development of efficient detection methodologies for nucleic acids, proteins and various chemicals and biomolecules. Sensing systems incorporated with AuNPs facilitate enhanced electronic transmission performance of sensors due to their electrically conductive properties.27–30| System | Detection or capture molecule/ligands | Target | Analytical method | Real sample testing/live/pseudo-virus | LOD | Ref. |
|---|---|---|---|---|---|---|
| Au–Pt NPs | Polyclonal | Spike S1 recombinant protein | Colorimetric | No | 11 ng mL−1 | 33 |
| AuNPs | SARS-CoV-2 spike monoclonal antibody | Spike S1 recombinant protein | Colorimetric and electrochemical | No | 48 ng mL−1 (colorimetric) and 1 pg mL−1 (electrochemical) | 23 |
| AuNPs on antibody-coated fluorine-doped tin oxide | SARS-CoV-2 spike S1Ab | Spike S1 protein | CV and DPV | — | 6 pg mL−1 | 34 |
| AuNPs | Anti-spike antibody | COVID-19 viral antigen and virus | Colorimetry, surface-enhanced Raman spectroscopy | Yes | ∼1 pg mL−1 (colorimetry) | 35 |
| ∼4 pg mL−1 and ∼18 virus particles/mL (SERS) | ||||||
| AuNPs | Polyclonal Abs against S1, membrane proteins and envelope proteins | Spike S1 proteins, membrane proteins and envelope proteins | Colorimetry | Yes | Not reported | 36 |
| AuNPs | Polyclonal Ab | Nucleocapsid (N) proteins | Localized surface plasmon resonance (LSPR) | No | 150 ng mL−1 | 37 |
Detection of viral antigens is more promising because antibody responses do not appear early when compared with a virus, thereby delaying the diagnosis and treatment.31,32
Fu et al. utilized polyclonal antibodies functionalized with Au–Pt alloy nanoparticles for the colorimetric detection of S1 spike proteins of SARS-CoV-2.33 Karakuş et al. performed a rapid, selective, and dual-responsive electrochemical and colorimetric detection of SARS-CoV-2 with monoclonal functionalized AuNPs. Monoclonal antibodies were conjugated with 11-mercaptoundecanoic acid (MUA)-modified AuNPs by applying EDC-NHS (N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride and N-hydroxysuccinimide) chemistry depicted in Fig. 2. The prepared system detected SARS-CoV-2 spike antigens in saliva samples with high specificity and did not show any cross-reactivity with other antigens such as Streptococcus pneumoniae, influenza A and MERS-CoV.23 In the presence of the SARS-CoV-2 spike antigen, rapid and irreversible aggregation of AuNPs was observed which was attributed to antigen–antibody interactions. The interaction could also be inferred from the change in color of the solution from red to purple. This was validated spectroscopically using UV-visible spectrometry and red shift was observed with a detection limit of 48 ng mL−1. Robert and colleagues developed an electrode consisting of a fluorine-doped tin oxide (FTO)/AuNP complex coupled with SARS-CoV-2 spike S1 antibody (SARS-CoV-2 Ab) as an immunosensor for SARS-CoV-2 spike S1 antigen as shown in Fig. 3.34 The immunosensor was optimized for response time, temperature, pH and antibody concentration. Moreover, the platform displayed excellent sensitivity towards SARS-CoV-2 spike S1 antigen with a limit of detection down to 0.63 fM in standard buffer sample and down to 120 fM (6 pg mL−1) in spiked saliva samples whilst displaying negligible cross-reactivity with other viral antigens.
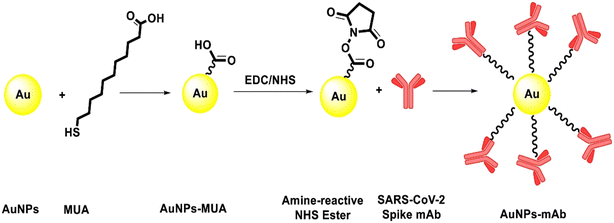 | ||
| Fig. 2 Schematic presentation of the conjugation of the mAb on the surface of MUA-modified AuNPs. Reprinted from ref. 23. E. Karakuş et al., Anal. Chim. Acta, 2021, 1182, 338939–338949. Copyright (2021) Elsevier. | ||
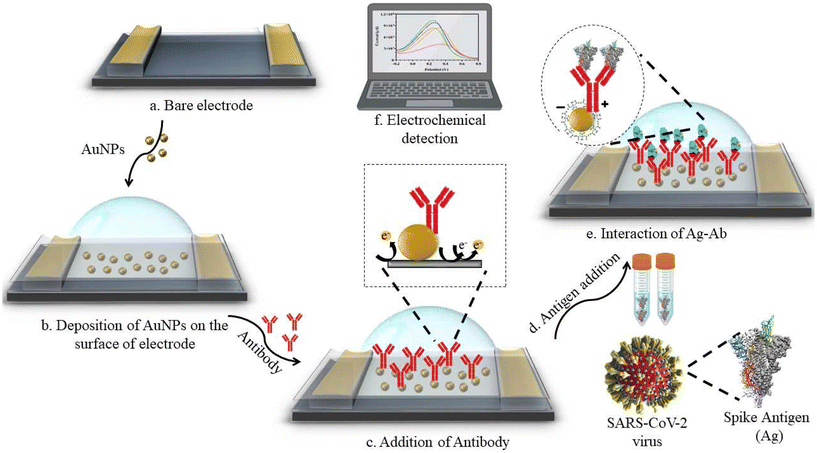 | ||
| Fig. 3 Design of electrochemical sensing of SARS-CoV-2 spike S1 antigen using an electrode system consisting of Ag/AgCl as a reference electrode and platinum as a counter electrode. Reprinted from ref. 34. A. Robert et al., Anal. Chim. Acta, 2021, 1188, 339207–339219. Copyright (2021) Elsevier. | ||
Pramanik et al. reported an assay based on colorimetric and surface-enhanced Raman spectroscopy (SERS) for the determination of pseudo-SARS-CoV-2 (genetically modified virus which is not infectious) with anti-spike antibody-functionalized PEG-coated AuNPs.35 In the presence of the antigen, AuNPs undergo aggregation due to the antigen–antibody interactions, which leads to a color change visible to the naked eye from pink to blue within 5 min. The response (LOD) for the developed sensor was 1 ng mL−1 for COVID-19 antigen and 1000 virus particles per mL for SARS-CoV-2 spike protein pseudotyped baculovirus. Ventura et al. reported highly sensitive COVID-19 detection in nasal and throat swabs by means of a colorimetric assay employing AuNPs to which three different antibodies for spike, envelope and membrane protein were attached.36 A red shift was observed in the optical profile of the antibody-decorated AuNPs in the presence of viral particles. Additionally, the optical density was compared to the threshold cycle (Ct) of a real-time PCR, and it was observed that the proposed method could bind the virus with a detection limit comparable to that of the real-time PCR test. Another group led by Behrouzi et al. developed a sensitive and specific method for the detection of viral nucleocapsid protein using polyclonal antibody-coated AuNPs.37 The method could sense the virus in 5 min with a LOD of 150 ng mL−1.
| System | Proteins | Antibody/analyte | Analytical method | Real sample testing | Ref. |
|---|---|---|---|---|---|
| AuNPs | SARS-CoV-2 spike protein | Spike protein antibody | Opto-microfluidic sensing | No | 40 |
| AuNPs | Recombinant antigen (MK201027) of SARS-CoV-2 spike protein | IgG and IgM antibodies | Lateral flow immunoassay | Yes | 41 |
| AuNPs | SARS-CoV-2 S and N epitope | IgG antibodies | Colorimetric serological assay | Yes | 42 |
| PEDOT–AuNP modified electrodes | Truncated nucleoprotein (Naa160-406aa) | IgG and IgM antibodies | Electrochemical detection | Yes | 43 |
| AuNPs | Nucleocapsid recombinant antigen ncov-ps-Ag8 | IgG and IgM antibodies | Immuno-chromatography assay | Yes | 44 |
| AuNPs | Nucleoprotein | IgM antibodies | Lateral flow immunoassay | Yes | 45 |
| AuNPs | Membrane (M), spike (S) 1 and 2, nucleocapsid (N) and envelope (E) | IgA, IgM and IgG antibodies | RNA PCR | Yes | 46 |
A lateral flow immunoassay capable of detecting IgM and IgG simultaneously has been developed by Li et al.41 The methodology could be employed for the rapid screening of both symptomatic and asymptomatic SARS-CoV-2 carriers with appreciable specificity (90.63%) and sensitivity (88.66%).
Lew et al. developed another colorimetric serological assay to conduct large-scale testing of SARS-CoV-2 IgG in human plasma.42 Four different epitopes located on the S and N proteins of the virus were conjugated to AuNPs to recognize target antibodies. The methodology was also employed to isolate and investigate the plasma samples of 35 patients and results showed 100% specificity and 83% sensitivity. Within 30 min of exposure of the sensor to the antibody, a specific optical response was observed, triggered by nanoparticle aggregation upon binding between the antibodies and AuNPs functionalized with epitopes. The limit of detection of this nanosensor was 3.2 nM with the conjugation of two antigenic epitopes (S14P5 and S21P2). Lorenzen et al. developed an electrochemical sensor for the fast and reliable detection of antibodies against COVID-19 in serum sample.43 The research group modified the electrode with electro-synthesized PEDOT and AuNPs and immobilized truncated nucleoprotein (Naa160-406aa). The sensor developed was able to detect the antibodies in about half an hour with high specificity. Liu et al. reported a colloidal gold immunochromatography assay to detect COVID-19 IgG and IgM antibodies using COVID-19 nucleocapsid recombinant antigen ncov-ps-Ag8 conjugated AuNPs within 15 min in human blood samples.44 Moreover, the developed technology was able to detect COVID-19 antibodies with high sensitivity (95.85%) and specificity (97.47%). For the rapid, specific and inexpensive on-site diagnosis of COVID, Huang et al. developed a AuNP-based lateral flow assay to detect IgM antibodies.45 The prepared AuNP-LF strips consisted of a coating of nucleoprotein on the analytical membrane and conjugation of IgM antibodies to the AuNPs. The efficacy of the assay was analyzed by testing serum samples of patients and the results were compared with that of RT-PCR, displaying a sensitivity of 100% and specificity of 93.3%. Shaw and coworkers developed another nanotechnology-based portable system for the detection of SARS-CoV-2 IgA, IgM and IgG with 88% sensitivity and 75% specificity.46
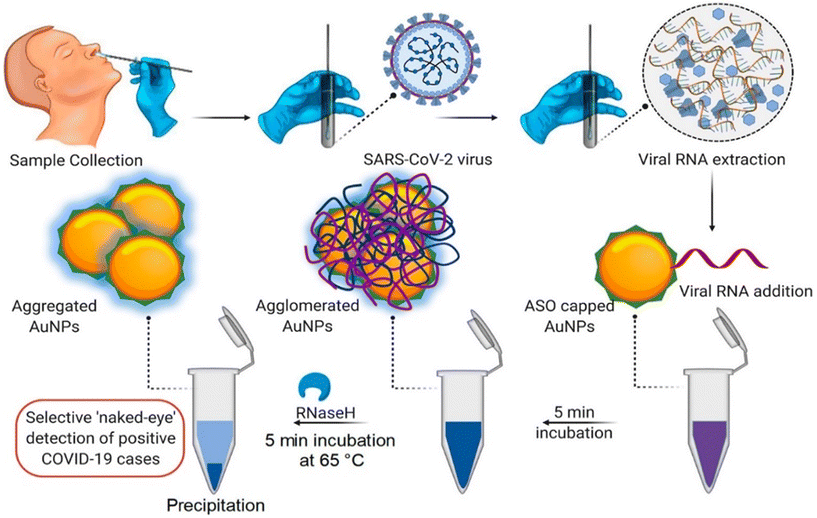 | ||
| Fig. 4 Schematic representation for the naked-eye detection of SARS-CoV-2 RNA mediated by the suitably designed ASO-capped AuNPs. Reprinted from ref. 47. P. Moitra et al., ACS Nano, 2020, 14, 7617–7627. Creative Commons public use license. | ||
Qiu et al. fabricated a dual-functional plasmonic biosensor for the clinical diagnosis of SARS-CoV-2.48 The biosensor combines the plasmonic photothermal effect and localized SPR sensing transduction of 2D gold nanoislands functionalized with complementary DNA receptors. The biosensor displayed high sensitivity towards SARS-CoV-2 with a detection limit of 0.22 pM. Feng et al. developed another immunofluorescence assay for the detection of IgM and IgG.49 The sensitivity and specificity of the immunochromatographic assay was observed to be 98.72% and 100% for IgG and 98.68% and 93.10% for IgM, respectively, when tested on serum samples from 28 clinical positive and 77 negative patients. Zhu et al. fabricated a nanoparticle-based biosensor assay coupled with RT loop-mediated isothermal amplification for COVID-19 diagnosis.50 The system was found to be highly sensitive and displayed no cross-reactivity with non-SARS-CoV-2 templates (virus, fungi, synthetic nucleic acid sequences and bacteria). The efficiency of the assay was evaluated using oropharynx swab samples from COVID-19 positive (33/33) and negative (96/96) patients, demonstrating 100% specificity and sensitivity.
Díaza et al. developed a colorimetric method for the detection of various SARS-CoV-2 proteins employing AuNPs.51 The sensor system employed different sizes of nanoparticles, sequences of oligonucleotides and buffer composition. In the presence of SARS-CoV-2, the oligonucleotides unfold, and their cholesterol moiety exposed to the aqueous medium gives a color change due to precipitation and aggregation of AuNPs [Fig. 5]. The system could detect the virus sequence in 15 min with high efficiency and selectivity.
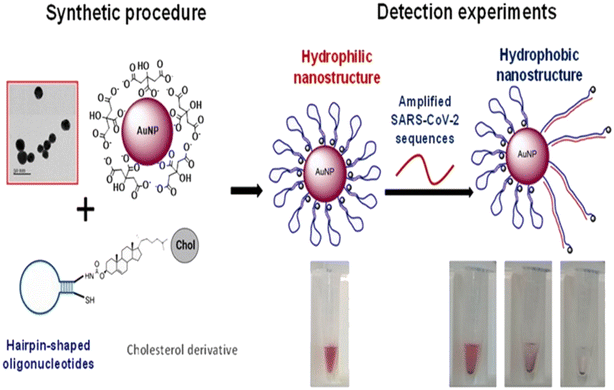 | ||
| Fig. 5 Schematic representation of the sensor. AuNPs are modified with oligonucleotides containing a cholesterol moiety. The nanostructures are exposed to SARS-CoV-2 sequences, which induce the rearrangement of the hairpin structure of the oligonucleotides to an open structure. Thus, the cholesterol is exposed to the aqueous solution, causing the aggregation of the nanoparticles, detectable by the naked eye. Reprinted from ref. 51. C. R. Díaza et al., Talanta, 2022, 243, 123393–123403. Creative Commons CC-BY license. | ||
CRISPR (clustered regularly interspaced short palindromic repeats) technology has been extensively used for the fabrication of highly specific and sensitive molecular assays including the recognition of viral RNA of SARS-CoV-2.52 Based on this technology, Cao et al. reported the development of a colorimetric assay employing trans-cleavage activity of the CRISPR/Cas system to facilitate sequence-dependent aggregation of AuNPs as displayed in Fig. 6.53 The assay provided a specific and sensitive detection of the viral RNA of SARS-CoV-2 which could also be observed with the naked eye with a response time of <1 min.
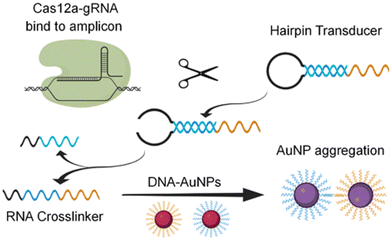 | ||
| Fig. 6 Schematic representation of the principle of Cas-mediated AuNP aggregation and corresponding change in color from red to purple. Reproduced from ref. 53. Y. Cao et al., Chem. Comm., 2021, 57, 6871–6876. Copyright (2021) Royal Society of Chemistry. | ||
Another CRISPR–Cas-based biosensor equipped with a smartphone readout was developed for the ultrasensitive and selective determination of the virus by Ma et al.54 The disaggregation of AuNPs induced by the degradation of single-stranded DNA led to observable color changes, which could be easily read on a smartphone device with a Color Picker app. Alafeef et al. developed a biosensor for the viral nucleocapsid phosphoprotein (N-gene) using highly specific antisense oligonucleotides (ssDNA) capped on AuNPs.55 The authors optimized the conjugation of ssDNA concentration with AuNPs. Upon the addition of SARS-CoV-2 RNA to a graphene–AuNP-based platform, a red shift was observed through hyperspectral imaging [Fig. 7].
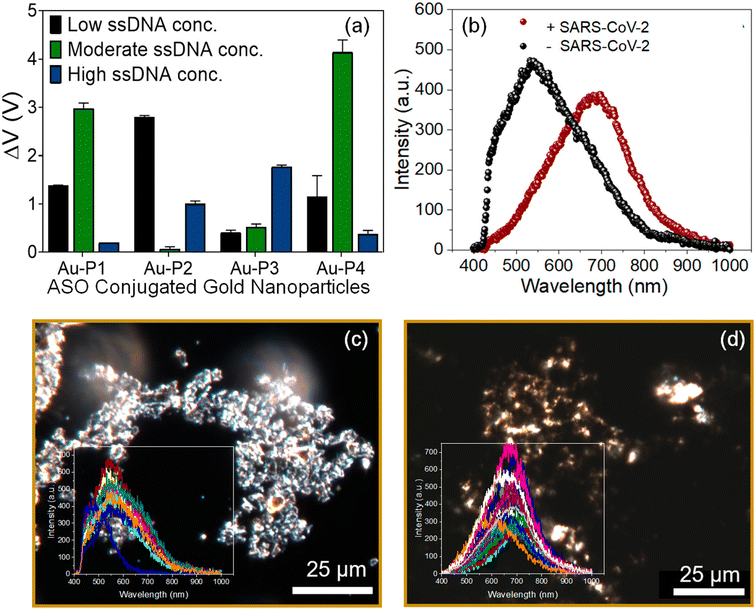 | ||
| Fig. 7 (a) Optimization of the antisense ssDNA probes conjugated with AuNPs at different ratios to achieve maximum sensitivity and optimal signal output. (b) Average of the spectra obtained from the hyperspectral imaging from the graphene–Au–Pmix in the absence and presence of SARS-CoV-2 viral RNA. Enhanced dark-field microscopic-hyperspectral image (EDFM-HSI) of the graphene–Au–Pmix (c) in the absence of SARS-CoV-2 RNA and (d) in the presence of SARS-CoV-2 RNA with spectra collected from several pixels shown in the inset. Reprinted with permission from ref. 55. M. Alafeef et al., ACS Nano, 2020, 14, 17028–17045. Copyright (2021) American Chemical Society. | ||
The same research group developed another colorimetric method for rapid and naked eye detection of the virus using a unique approach integrating nucleic acid amplification and plasmonic sensing of the virus with a response time of less than an hour.56 The method employs plasmonic AuNPs capped with antisense oligonucleotides as a colorimetric sensor to determine the nucleic acid specifically resulting in the aggregation of the AuNPs. Furthermore, the result can be monitored using a handheld optical reader to quantify the response. The dual detection of both Abs and antigens is highly of interest; therefore, Sadique et al. developed a novel category of electrochemical immunosensors for dual detection of SARS-CoV-2 antibodies and antigens by fabricating graphene oxide–gold nanocomposites.57 DPV was employed to quantify the detection of SARS-CoV-2 antigens/antibodies [Fig. 8]. The LOD for the antigen immunosensor immunosensor was 3.99 ag mL−1, while for the antibody this was 1 fg mL−1 and showed good consistency among patient serum and swab samples (Table 3).
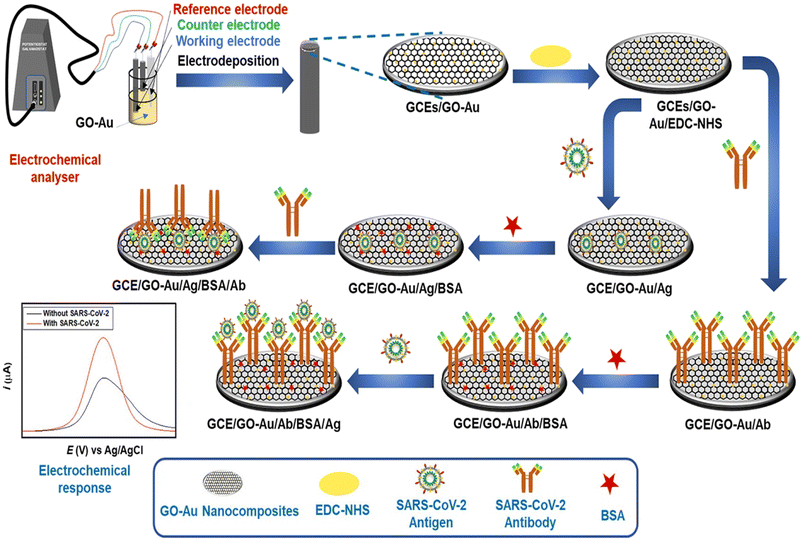 | ||
| Fig. 8 Schematic of steps involved in the fabrication of SARS-CoV-2 antigen and SARS-CoV-2 antibody immunosensors. Reprinted from ref. 57. M. A. Sadique et al., ACS Appl. Bio Mater., 2022, 5, 2421–2430. Copyright (2021) Creative Commons public use license. | ||
| System | Detection or capture molecule/ligands | Target | Analytical method | Real sample testing/live/pseudo-virus | LOD | Ref. |
|---|---|---|---|---|---|---|
| AuNPs | Thiol-modified antisense oligonucleotides | N-gene (nucleocapsid phosphoprotein) | Colorimetric | Yes | 0.18 ng μL−1 | 47 |
| Gold nanoislands (AuNIs) | DNA receptors | Spike proteins, membrane proteins and envelope proteins | Plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) | Yes | 0.22 pM | 48 |
| AuNPs | Nucleocapsid protein | IgM and IgG antibodies | Immunofluorescence assay | Yes | Not reported | 49 |
| AuNPs | F1ab (opening reading frame 1a/b) and np (nucleoprotein) genes | — | RT loop-mediated isothermal amplification | Yes | Not reported | 50 |
| AuNPs | Oligonucleotides | E and R (RNA-dependent RNA polymerase) genes, the insertion region of S protein | Colorimetric | Yes | 10 nM (E), 20 nM (R), 500 pM each (E + R) | 51 |
| AuNPs | Cas12a-gRNA ribonucleoprotein | N and E gene | Colorimetric | Yes | Not reported | 53 |
| AuNPs | Single-stranded DNA | N gene | CRISPR-Cas12a powered visual biosensor | Yes | 1 copy per μL for SARS-CoV-2 pseudoviruses | 54 |
| 6.9 copies per μL | ||||||
| Plasmonic AuNPs | Antisense oligonucleotides | Nucleic acid | Colorimetric | Yes | 10 copies per μL | 55 |
| Graphene oxide–gold nanocomposites | Antigen/antibody | Antigen/antibody | Electrochemical immunosensors | Yes | 3.99 ag mL−1 (antigen), 1 fg mL−1 (antibody) | 56 |
Aithal and colleagues utilized aptamer-functionalized AuNPs for rapid detection test for SARS-CoV-2, which can detect 3540 genome copies per μL of inactivated SARS-CoV-2.60 Researchers used two aptamers which have been developed previously with SELEX technology and can target the receptor-binding domain (RBD) of the SARS-CoV-2 spike proteins. The test uses plasmon absorbance spectra to detect 16 nM and higher concentrations of spike protein in phosphate-buffered saline. The sensing signal is enhanced by adding a coagulant, MgCl2 salt solution, to induce nanoparticle agglomeration, which depends on the amount of aptamer–protein binding. Adeel et al. developed a flexible aptamer-based electrochemical sensor for the rapid, label-free detection of SARS-CoV-2 S protein using a platform made of a porous and flexible carbon cloth coated with AuNPs.61 The sensor displayed good selectivity, repeatability, and was tested in diluted human saliva spiked with different SARS-CoV-2 SP concentrations, providing lower limits of detection (LODs) of 0.11 ng mL−1 and 37.8 ng mL−1, respectively. AuNPs were used to coat the flexible carbon cloth platform to increase its conductivity and electrochemical performance [Fig. 9]. The thiol-functionalized DNA aptamer was assembled with the AuNPs via thiol–Au bonds for the selective recognition of the SARS-CoV-2 spike protein.
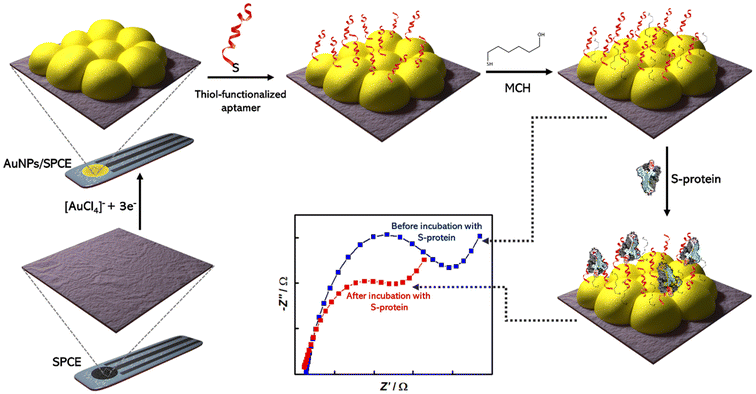 | ||
| Fig. 9 Schematic representation of fabrication of aptasensor for SARS-CoV-2 S-protein detection using screen printed carbon electrode (SPCE); Reprinted from ref. 62; Abrego-Martinez et al., Biosen. Bioelectron., 2022, 113595. Copyright (2022) Elsevier. | ||
Guan et al. fabricated a SERS nanoprobe by mixing AuNPs with Raman reporter Nile blue A (NBA).63 These nanoprobes were then functionalized with spike protein aptamers and magnetic beads and redispersed in PBS-T buffer solution. The probes modified with aptamers cocktail, recognize the spike protein to form sandwich complexes, which are then detected by a handheld Raman spectrometer within 5 min with a LOD of 18 fM for the pseudovirus. Daniel et al. developed a low-cost, handheld device for rapid detection of SARS-CoV-2 RNA using aptamer-functionalized AuNPs synthesized by a novel method.64 The device can detect SARS-CoV-2 RNA in less than 5 min by utilizing the selectivity of the aptamer to SARS-CoV-2 RNA.
Abrego-Martinez et al. demonstrated an aptamer-functionalized AuNP-based electrochemical biosensor for the detection of RBD in S protein of SARS-CoV-2.62 The biosensor demonstrated excellent sensing performance, with a LOD of 1.30 pM (66 pg mL−1) for SARS-CoV-2 S protein and was successfully applied for the detection of a SARS-CoV-2 pseudovirus. Gu et al. fabricated a new sensor based on spherical cocktail aptamer–AuNP (SCAP) for ultrasensitive detection of SARS-CoV-2 S protein based on surface-enhanced Raman scattering (SERS).65 The aptamers are used to specifically bind to the SARS-CoV-2 S protein, while the AuNPs enhance the Raman signal through SERS. The developed detection platform can detect the SARS-CoV-2 S protein with a LOD as low as 0.7 fg mL−1. Sun and co-workers reported the development of a new neutralizing agent called spherical neutralizing aptamer–AuNP (SNAP) to inhibit SARS-CoV-2 infection and suppress mutational escape.66 SNAP exhibits exceptional binding affinity against the RBD of SARS-CoV-2 S protein and potent neutralization against authentic SARS-CoV-2 and mutant pseudoviruses.
Incorporating aptamer-functionalized AuNPs for SARS-CoV-2 detection exemplifies a pioneering approach at the forefront of biosensing technology, harnessing the prowess of aptamers' precision, stability, and design versatility. This innovative strategy not only showcases enhanced sensitivity and specificity but also heralds a new era of rapid, reliable, and adaptable diagnostic solutions for addressing the challenges posed by infectious diseases like COVID-19.
2.2 Molecularly imprinted polymeric nanoparticles
Molecularly imprinted polymeric nanoparticles (nanoMIPs) are artificial receptors, which possess tailor-made recognition sites that are complementary to the size, spatial orientation, and chemical functionality functional groups of the template molecule.67,68 These artificial receptors display high selectivity and binding affinity towards a particular analyte (template) and can compete with antibodies in terms of affinity.69 Moreover, these nanoparticles are highly robust and exhibit high chemical and thermal stability. Henceforth, these nanoMIPs have attracted significant attention in the development of biosensors for the detection of bacterial endotoxins and viruses.70,71 Fabrication of nanoMIPs can be accomplished via various approaches, namely nanoprecipitation, mini-emulsion and solid-phase polymerization. In the nanoprecipitation method, a solution containing a mixture of monomers and template is polymerized with a suitable initiator and, after removal of the solvent (via evaporation or ultrasonication), nanoMIPs are obtained via precipitation.72,73 Mini-emulsion polymerization is widely used to prepare nanoMIPs opposite to microemulsion where formation of emulsion is spontaneous. Moreover, high shear force needs to be employed to break up the emulsion into nanoscale particles to obtain mini-emulsions. In both nanoprecipitation synthesis and mini-emulsion polymerization, the template is removed using different methods including centrifugation, dialysis and extraction.74 In solid-phase synthesis, the template is covalently bound to a solid phase such as glass beads which is used as an affinity medium to separate and collect high-affinity nanoMIPs.75 In this approach, unreacted monomers and low-affinity nanoMIPs are removed using low-temperature elution, and then the high-affinity nanoMIPs are purified using elevated-temperature elution (60 °C) as shown in Fig. 10. The advantage of this approach lies in the covalent immobilization of the template molecule which ensures that the resulting high-affinity nanoMIPs are template-free. Furthermore, the covalent immobilization of the template molecule allows for a specific orientation, which results in the production of a more homogeneous binding site affinity, much like monoclonal antibodies.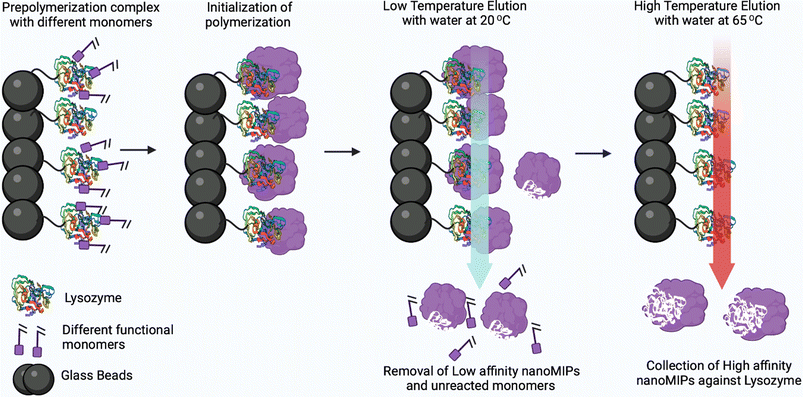 | ||
| Fig. 10 Solid-phase synthesis to produce nanoMIPs against protein lysozyme (instead of lysozyme, other proteins or peptides can be imprinted). Reprinted from ref. 75. P. Singla et al., Anal. Bioanal. Chem., 2023, 415, 4467–4478; copyright (2023) Springer Nature. | ||
Given their synthetic nature, MIPs possess excellent thermal and chemical stability. Moreover, MIP-based platforms can be deployed at different pH ranges, where most of the lateral flow rapid antigen tests fail to record accurate results.
For synthesizing MIPs against SARS-CoV-2, one can use a range of monomers. For instance, N-isopropylacrylamide and (hydroxyethyl)methacrylate can be used for hydrogen bonding and N-tert-butylacrylamide and 2-ethylhexylacrylate for hydrophobic interactions. Moreover, cationic monomers such as N-(3-aminopropyl)methacrylamide, 2-aminoethylmethacrylamide hydrochloride, 2-aminoethyl methacrylate, 3-guanidino-propylmethacrylate, methacrylic acid (MAA) and acrylic acid (AAc) are typically the monomers of choice to facilitate ionic interactions. It has been reported that a mix of monomers that enable hydrogen bond formation or ionic and hydrophobic interactions, particularly for protein targets, is crucial to produce MIPs with high affinity and selectivity. The most commonly used crosslinkers for organic polymerisation are ethylene glycol dimethacrylate (EGDMA) and trimethylolpropane trimethacrylate (TRIM), whilst for aqueous polymerisation typically N,N-methylenebis(acrylamide) is used. To synthesize nanoMIPs, the entire protein can be imprinted; however, due to the fragile nature of proteins, their native confirmation might be altered during the polymerization process. Furthermore, proteins are often expensive and/or not available in sufficient amounts. To overcome these issues, short surface-exposed fragments of protein (i.e. antigenic determinants of a protein that binds to an antibody, known as epitopes) can be used as a template.76
Scientists at Rice University developed different methodologies for the detection of viruses; one for the detection of viral particles in the air and another consisting of a mobile phone reader with a chip capable of detecting virus in a blood sample.77 A thin-film electronic device capable of detecting virus in air was developed under the RAPID (real-time amperometry platform using molecular imprinting for the selective detection of SARS-CoV-2) project, in addition to a plug-in tool to diagnose small amounts of virus in less than an hour. Another research group at Harvard and MIT are attempting to develop a face mask which lights up when it detects the presence of SARS-CoV-2 virus.78,79 Using molecularly imprinted technology, scientists from Sixth Wave company were the first to develop MIPs for the rapid detection of SARS-CoV-2. A United States patent was filed on Accelerated Molecularly Imprinted Polymers (AMIPs™) technology and they incorporated this technology into a face mask. This mask will show a color change upon exposure to the virus exhaled in the breath.36 Moreover, this company achieved the colorimetric detection of the delta variant of the SARS-CoV-2 virus causing COVID-19 infection using the same AMIPs™ technology.80,81
MIP Discovery (formerly MIP Diagnostics), a UK-based company, has developed a sensor for SARS-COV-2 using nanoMIPs synthesized via the proprietary solid phase approach. McClements and colleagues have used these nanoMIPs for the detection of the SARS-CoV-2 in clinical samples as well as the RBD (receptor binding domain) of spike protein using the heat transfer method.82 The system was thermally stable and offered superior sensitivity towards antibodies with a LOD found to be in the range of fg mL−1. In addition to this, Bhalla et al. developed a nanoplasmonic biosensor using the same nanoMIPs employed in the previous study for the detection of different variants of SARS-CoV-2.83 The technology enabled the detection of spike proteins of the SARS-CoV-2 with a LOD of 9.71 fM (1.3 pg mL−1), 7.32 fM (1.04 pg mL−1) and 8.81 pM (1.2 ng mL−1) for the alpha, beta and gamma variants of the proteins, respectively. Cennamo et al. developed a low-cost optical–chemical MIP sensor.84 The sensor system displayed a change in the refractive index of the MIP nanofilm upon binding with the viral particles, which was investigated using a simple plasmonic D-shaped POF platform. The sensor employed a plasmonic plastic optical fiber sensor coupled with a synthetic MIP nanolayer for the detection of the spike protein of SARS-CoV-2. The research group firstly established the effectiveness of the MIP receptor to bind the spike protein and then measured clinical nasal swabs. The test results obtained using MIPs were compared with the RT-PCR results and were found to be more sensitive than RT-PCR with relatively fast response time. Raziq et al. reported on a MIP-based electrochemical sensor for the detection of SARS-CoV-2 antigen (ncovNP) as displayed in Fig. 11.85 The ncovNP sensor displayed a detection and quantification limit of 15 fM and 50 fM (0.7–2.2 pg mL−1), respectively, in lysis buffer. Additionally, the sensor could differentiate the target nucleoprotein from interfering proteins such as, S1, BSA, CD48 and E2 HCV.
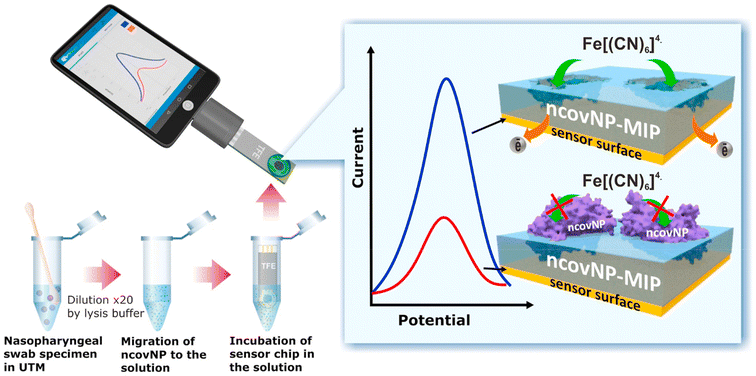 | ||
| Fig. 11 COVID-19 diagnosis by the ncovNP electrochemical sensor analyzing the samples obtained from nasopharyngeal swabs of COVID patients. Reprinted from ref. 85. A. Raziq et al., Biosens. Bioelectron., 2021, 178, 113029–113036. Copyright (2021) Elsevier. | ||
Parisi et al. synthesized plastic antibodies based on MIPs using an inverse microemulsion method against the SARS-CoV-2 spike protein, employing the hydrophilic monomers acrylic acid and acrylamide. The synthesized nanoMIPs could recognize and bind to novel coronavirus SARS-CoV-2 RBD with high selectivity to inhibit the activity of spike protein. Moreover, these nanoMIPs inhibited the virus replication in Vero cell culture and thus are deemed suitable for the prevention and treatment of SARS-CoV-2 infection.86 Sharif and colleagues electropolymerized water-soluble N-hydroxymethylacrylamide (NHMA) as a functional monomer and cross-linked this with N,N-methylenebisacrylamide using SARS-CoV-2 pseudoparticles (PPs) as a template to imprint. The obtained LOD was 88.0 fg mL−1, suggesting its suitability for detection of SARS-CoV-2 with minimal sample preparation (Table 4).87
| System | Imprinted template | Target | Analytical method | Real sample testing/live/pseudovirus | LOD | Ref. |
|---|---|---|---|---|---|---|
| nanoMIPs | Spike protein epitope | Spike protein | Heat transfer method | Yes | ∼9.9 fg mL−1 (alpha) and ∼6.1 fg mL−1 (delta) variant | 81 |
| nanoMIPs | Spike protein epitope | Spike protein | Nanoplasmonic biosensor | Yes | 1.3 pg mL−1, 1.04 pg mL−1 and 1.2 ng mL−1 for the alpha, beta and gamma variants | 82 |
| MIPs | Spike protein (S1 subunit, His-Tag) | S1 protein | Optical–chemical | Yes | 3.8 mg mL−1 | 83 |
| MIPs | Spike protein | Spike protein S1 antigen | Electrochemical | Yes | 0.7 pg mL−1 | 84 |
| MIPs | SARS-CoV-2 (2019-nCoV) spike protein (RBD) | Spike protein | Multiple techniques | Yes | 1 ng | 85 |
| e-MIPs | SARS-CoV-2 pseudotyped particles (PPs) | SARS-CoV-2 PPs | Electrochemical | Yes | 88.0 fg mL−1 | 87 |
3.0 Comparison of functionalized AuNP- and nanoMIP-based platforms
For the development of highly sensitive, rapid, and reliable diagnostic tools for SARS-CoV-2 virus detection, both functionalized AuNPs and nanoMIPs are promising approaches. AuNPs functionalized with molecular recognition probes (antibodies, antigens, nucleotides, and aptamers) possess unique optical and physical properties, whilst MIPs exhibit high selectivity, sensitivity, robustness, reusability and batch-to-batch consistency. The major advantages of the nanoMIP platform are that there is no direct need to conjugate antibodies, proteins/antigens, nucleotides, and aptamers specific for SARS-COV-2. In contrast, AuNP-based sensor systems rely on the cross-linking of these nanoparticles with the aforementioned molecular recognition elements/biological counterparts. Immunosensors utilizing antibody-functionalized AuNPs and antigen-functionalized AuNPs are undoubtedly the most employed binders for detection of SARS-CoV-2 (and in diagnostics in general). They are relatively expensive and have limited shelf life and stability. Besides this, MIPs are cost-effective and can withstand high temperature and pH conditions without compromising the binding affinity and selectivity. Another crucial aspect to consider is the fabrication duration of individual sensors employing nanoparticles. Typically, immunosensors that utilize nanoparticles functionalized with antibodies involve more extensive procedures, demanding several hours to effectively immobilize biomolecules. The phase of obstructing nonspecific bindings using blocking solutions like BSA consumes a substantial period, approximately around 30 min. Aptamer functionalized AuNPs are also a viable alternative option to fabricate aptamer sensors for SARS-CoV-2 as it has excellent stability and cost-effectiveness as compared to antibodies, but the production time to design and fabricate aptamers can take weeks to months. Conversely, sensors based on MIPs might exhibit a shorter production time, particularly when employing the solid-phase synthesis method. The removal of the template through the dialysis technique or via immersing the electrode in extraction solutions (in the case of electropolymerization) might constitute one of the most time-intensive stages, lasting a few hours. Nonetheless, by utilizing solid-phase synthesis, the separation of high-affinity nanoMIPs can be achieved within a few minutes. There are varied functionalities of AuNPs modified for detecting antigens and antibodies. The nucleotide-functionalized AuNPs exhibit dual detection mode for both antigens and antibodies. The LOD with antibody-functionalized AuNPs is as low as 1 fg mL−1, whilst with MIPs it is 6.1 fg mL−1, both of which are suitable for lateral flow immunoassay in portable devices.4.0 Conclusions
This review summarized and compared two of the most important and widely researched nanoparticle systems for detection of SARS-CoV-2. The detection of SARS-CoV-2 is currently a global health concern and diagnostics play a crucial role in controlling the spread of infectious diseases. The diagnostic tools based on functionalized AuNPs as well as MIPs offer many advantages such as high selectivity, robustness, rapid response time, high specificity and in some cases, reusability. Furthermore, we have discussed the concept of molecular design for developing portable sensors capable of detecting the SARS-CoV-2 virus. This approach has the potential to be extended for detecting other pathogenic viruses, offering a valuable tool in preventing and addressing future pandemics. All these methodologies can identify the virus with high selectivity and sensitivity with a response time of less than an hour and, in a few cases, even minutes. However, functionalized AuNPs rely on other molecular recognition elements such as antibodies (especially mAbs), antigens, nucleotides, aptamers, and others. However, their very nature also brings some disadvantages: relatively high manufacturing costs together with difficult functionalization chemistry and relatively short shelf life at room temperature as well as time-consuming and sometimes complex production processes involving animals. Intriguing alternatives to these biological counterparts as molecular recognition are MIPs. Current imprinting techniques suffer from a heterogeneous “polyclonal” distribution of binding sites, poor performance in water, low capacity and slow mass transfer. The use of solid-phase approaches overcomes these issues, as the immobilized template enables an oriented immobilization, thus reducing the “polyclonality” of the imprinted sites. The synthetic nature of nanoMIPs allows them to withstand bacterial degradation. They therefore have a longer shelf life, even in the absence of refrigeration. This makes them viable for use in more remote geographical areas where healthcare infrastructure is more limited. Hence, nanoMIPs represent ideal candidates for replacing antibodies in lateral flow tests to be deployed in equatorial countries across the globe. However, they do encounter some challenges and involve ensuring reliable fabrication and reproducibility, achieving selective binding in complicated mixtures, and keeping their performance steady over longer time frames. Additionally, factors like making nanoMIPs work seamlessly with devices, efficiently scaling up production, and managing costs need to be considered. To overcome these challenges, future progress might involve fine-tuning the methods used to develop them to gain better control over their size and properties. Enhancing their design to make their binding strength and kinetics even better, and creating surfaces that fit perfectly with devices, are also potential avenues. Investigating new ways to cross-link these materials could also make them more durable. Using computer modelling could help predict and optimize their behaviour. It is very important to carefully check if these materials work well with living things. Finding good methods to deal with tricky samples and having experts from different areas like materials, chemistry, biology, and engineering collaborate is a crucial way to directly deal with these challenges. This comprehensive approach is absolutely essential to make sure that nanoMIPs become seamlessly integrated into a wide range of applications.Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
Dr. Pankaj Singla would like to acknowledge the European Union's Horizon 2020 research and innovation program for a Marie Sklodowska-Curie Postdoctoral fellowship (grant agreement number 893371, TEMPER).References
- C. Huang, et al., Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China, Lancet, 2020, 395, 497–506 CrossRef CAS PubMed.
- https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---22-march-2022 .
- B. Gates, Responding to Covid-19—a once-in-a-century pandemic?, N. Engl. J. Med., 2020, 382, 1677–1679 CrossRef CAS PubMed.
- Wikipedia, The New York Times, JHU CSSE COVID-19 Data and Our World in Data.
- https://ourworldindata.org/covid-vaccinations .
- G. Forni and A. Mantovani, COVID-19 vaccines: where we stand and challenges ahead, Cell Death Differ., 2021, 28, 626–639 CrossRef CAS PubMed.
- E. Hunter, et al., First experience of COVID-19 screening of health-care workers in England, Lancet, 2020, 395, 77–78 CrossRef PubMed.
- N. Cennamo, et al., Proof of Concept for a Quick and Highly Sensitive On-Site Detection of SARS-CoV-2 by Plasmonic Optical Fibers and Molecularly Imprinted Polymers, Sensors, 2021, 21, 1681–1698 CrossRef CAS PubMed.
- Y. Li, et al., Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19, J. Med. Virol., 2020, 92, 903–908 CrossRef CAS PubMed.
- M. Teymouri, et al., Recent advances and challenges of RT-PCR tests for the diagnosis of COVID-19, Pathol., Res. Pract., 2021, 221, 153443 CrossRef CAS.
- A. Parihar, et al., Point-of-care biosensor-based diagnosis of COVID-19 holds promise to combat current and future pandemics, ACS Appl. Bio Mater., 2020, 3, 7326–7343 CrossRef CAS PubMed.
- R. Gorkhali, et al., Structure and function of major SARS-CoV-2 and SARS-CoV proteins, Bioinf. Biol. Insights, 2021, 15, 11779322211025876 Search PubMed.
- L. Guruprasad, Human SARS CoV-2 spike protein mutations, Proteins: Struct., Funct., Genet., 2021, 89, 569–576 CrossRef CAS PubMed.
- S. Iravani, Nano-and biosensors for the detection of SARS-CoV-2: challenges and opportunities, Mater. Adv., 2020, 1, 3092–3103 RSC.
- A. Pramanik, et al., The rapid diagnosis and effective inhibition of coronavirus using spike antibody attached gold nanoparticles, Nanoscale Adv., 2021, 3, 1588–1596 RSC.
- A. Szymczak, et al., Antibodies specific to SARS-CoV-2 proteins N, S and E in COVID-19 patients in the normal population and in historical samples, J. Gen. Virol., 2021, 102, 001692 CrossRef CAS PubMed.
- M. V. Sullivan, et al., Toward rational design of selective molecularly imprinted polymers (MIPs) for proteins: computational and experimental studies of acrylamide based polymers for myoglobin, J. Phys. Chem. B, 2019, 123, 5432–5443 CrossRef CAS.
- V. Kesarwani, et al., Column Agglutination Assay Using Polystyrene Microbeads for Rapid Detection of Antibodies against SARS-CoV-2, ACS Appl. Mater. Interfaces, 2022, 14, 2501–2509 CrossRef CAS.
- A. F. Nahhas and T. J. Webster, The promising use of nano-molecular imprinted templates for improved SARS-CoV-2 detection, drug delivery and research, J. Nanobiotechnol., 2021, 19, 1–4 CrossRef.
- F. Curti, et al., Folding-Based Electrochemical Aptasensor for the Single-Step Detection of the SARS-CoV-2 Spike Protein, ACS Appl. Mater. Interfaces, 2022, 14, 19204–19211 CrossRef CAS PubMed.
- K. Wu, et al., One-step, wash-free, nanoparticle clustering-based magnetic particle spectroscopy bioassay method for detection of SARS-CoV-2 spike and nucleocapsid proteins in the liquid phase, ACS Appl. Mater. Interfaces, 2021, 13, 44136–44146 CrossRef CAS PubMed.
- E. Piccinini, et al., Surface engineering of graphene through heterobifunctional supramolecular-covalent scaffolds for rapid COVID-19 biomarker detection, ACS Appl. Mater. Interfaces, 2021, 13, 43696–43707 CrossRef CAS PubMed.
- E. Karakuş, E. Erdemir, N. Demirbilek and L. Liv, Colorimetric and electrochemical detection of SARS-CoV-2 spike antigen with a gold nanoparticle-based biosensor, Anal. Chim. Acta, 2021, 1182, 338939 CrossRef PubMed.
- C. Yu and J. Irudayaraj, Multiplex biosensor using gold nanorods, Anal. Chem., 2007, 79, 572–579 CrossRef CAS PubMed.
- D. Zhang, M. C. Huarng and E. C. Alocilja, A multiplex nanoparticle-based biobarcoded DNA sensor for the simultaneous detection of multiple pathogens, Biosens. Bioelectron., 2010, 26, 1736–1742 CrossRef CAS PubMed.
- R. Ahirwar and P. Nahar, Development of a label-free gold nanoparticle-based colorimetric aptasensor for detection of human estrogen receptor alpha, Anal. Bioanal. Chem., 2016, 408, 327–332 CrossRef CAS PubMed.
- C. Chang, et al., Gold nanoparticle-based colorimetric strategies for chemical and biological sensing applications, Nanomaterials, 2019, 9, 861–885 CrossRef CAS PubMed.
- M. H. Jazayeri, et al., Colorimetric detection based on gold nano particles (GNPs): an easy, fast, inexpensive, lowcost and short time method in detection of analytes (protein, DNA, and ion), Sens. Bio-Sens. Res., 2018, 20, 1–8 CrossRef.
- H. Aldewachi, et al., Gold nanoparticle-based colorimetric biosensors, Nanoscale, 2018, 10, 18–33 RSC.
- J. P. Oliveira, et al., Impact of conjugation strategies for targeting of antibodies in gold nanoparticles for ultrasensitive detection of 17b-estradiol, Sci. Rep., 2019, 9, 13859–13867 CrossRef PubMed.
- B. Diao, et al., Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection, Clin. Microbiol. Infect., 2021, 27, 289–2933 Search PubMed.
- B. Di, et al., Monoclonal antibody-based antigen capture enzyme-linked immunosorbent assay reveals high sensitivity of the nucleocapsid protein in acute-phase sera of severe acute respiratory syndrome patients, Clin. Diagn. Lab. Immunol., 2020, 12, 135–140 Search PubMed.
- Z. Fu, et al., Porous Au@Pt nanoparticles with superior peroxidase-like activity for colorimetric detection of spike protein of SARS-CoV-2, J. Colloid Interface Sci., 2021, 604, 113–121 CrossRef CAS PubMed.
- A. Robert, S. Mahari, D. Shahdeo and S. Gandhi, Label-free detection of SARS-CoV-2 Spike S1 Antigen triggered by electroactive gold nanoparticles on Antibody coated Fluorine-Doped Tin Oxide (FTO) Electrode, Anal. Chim. Acta, 2021, 1188, 339207 CrossRef PubMed.
- A. Pramanik, et al., The rapid diagnosis and effective inhibition of coronavirus using spike antibody attached gold nanoparticles, Nanoscale Adv., 2021, 3, 1588–1598 RSC.
- B. D. Ventura, et al., Colorimetric Test for Fast Detection of SARS-CoV-2 in nasal and Throat Swabs, ACS Sens., 2020, 5, 3043–3048 CrossRef CAS PubMed.
- K. Behrouzi and L. Lin, Gold nanoparticle based plasmonic sensing for the detection of SARS-CoV-2 nucleocapsid proteins, Biosens. Bioelectron., 2022, 195, 113669 CrossRef CAS PubMed.
- A. C. Hoste, et al., Two serological approaches for detection of antibodies to SARS-CoV-2 in different scenarios: a screening tool and a point-of-care test, Diagn. Microbiol. Infect. Dis., 2020, 98, 115167 CrossRef CAS PubMed.
- S. N. Amrun, et al., Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity, EBioMedicine, 2020, 58, 102911 CrossRef PubMed.
- R. Funari, K. Y. Chu and A. Q. Shen, Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip, Biosens. Bioelectron., 2020, 169, 112578 CrossRef CAS PubMed.
- Z. Li, et al., Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis, J. Med. Virol., 2020, 92, 1518–1524 CrossRef CAS PubMed.
- T. T. S. Lew, et al., Epitope-Functionalized Gold Nanoparticles for Rapid and Selective Detection of SARS-CoV-2 IgG Antibodies, ACS Nano, 2021, 15, 12286–12297 CrossRef CAS PubMed.
- A. L. Lorenzen, et al., PEDOT-AuNPs-based impedimetric immunosensor for the detection of SARS-CoV-2 antibodies, Electrochim. Acta, 2022, 404, 139757 CrossRef CAS PubMed.
- C. Liu, et al., A facile assay for rapid detection of COVID-19 antibodies, RSC Adv., 2020, 10, 28041–28048 RSC.
- C. Huang, et al., Rapid Detection of IgM Antibodies against the SARS-CoV-2 Virus via Colloidal Gold Nanoparticle-Based Lateral-Flow Assay, ACS Omega, 2020, 21, 12550–12556 CrossRef PubMed.
- A. M. Shaw, et al., Real-world evaluation of a novel technology for quantitative simultaneous antibody detection against multiple SARS-CoV-2 antigens in a cohort of patients presenting with COVID-19 syndrome, Analyst, 2020, 145, 5638–5646 RSC.
- P. Moitra, et al., Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles, ACS Nano, 2020, 14, 7617–7627 CrossRef CAS PubMed.
- G. Qiu, et al., Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS PubMed.
- M. Feng, et al., Development of a sensitive immunochromatographic method using lanthanide fluorescent microsphere for rapid serodiagnosis of COVID-19, ACS Sens., 2020, 5, 2331–2337 CrossRef CAS PubMed.
- X. Zhu, et al., Reverse transcription loop-mediated isothermal amplification combined with nanoparticles-based biosensor for diagnosis of COVID-19, Biosens. Bioelectron., 2020, 166, 112437 CrossRef CAS PubMed.
- C. R. Díaza, et al., Development of colorimetric sensors based on gold nanoparticles for SARS-CoV-2 RdRp, E and S genes detection, Talanta, 2022, 243, 123393 CrossRef PubMed.
- S. K. Kailasa, et al., An overview of molecular biology and nanotechnology based analytical methods for the detection of SARS-CoV-2: Promising bio-tools for the rapid diagnosis of COVID-19, Analyst, 2021, 146, 1489–1513 RSC.
- Y. Cao, et al., CRISPR/Cas12a-mediated gold nanoparticle aggregation for colorimetric detection of SARS-CoV-2, Chem. Commun., 2021, 57, 6871–6876 RSC.
- L. Ma, et al., A smartphone-based visual biosensor for CRISPR-Cas powered SARS-CoV-2 diagnostics, Biosens. Bioelectron., 2022, 195, 113646 CrossRef CAS PubMed.
- M. Alafeef, et al., Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip, ACS Nano, 2020, 14, 17028–17045 CrossRef CAS PubMed.
- M. Alafeef, et al., RNA-extraction-free nano-amplified colorimetric test for point-of-care clinical diagnosis of COVID-19, Nat. Protoc., 2021, 16, 3141–3162 CrossRef CAS PubMed.
- M. A. Sadique, Highly Sensitive Electrochemical Immunosensor Platforms for Dual Detection of SARS-CoV-2 Antigen and Antibody based on Gold Nanoparticle Functionalized Graphene Oxide Nanocomposites, ACS Appl. Bio Mater., 2022, 5, 2421–2430 CrossRef CAS PubMed.
- A. Mahmoudi, et al., Harnessing aptamers against COVID-19: a therapeutic strategy, Drug Discovery Today, 2023, 28, 103663 CrossRef CAS PubMed.
- G. Marrazza, Aptamer sensors, Biosensors, 2017, 7, 5 CrossRef PubMed.
- S. Aithal, et al., SARS-CoV-2 detection with aptamer-functionalized gold nanoparticles, Talanta, 2022, 236, 122841 CrossRef CAS PubMed.
- M. Adeel, et al., Label-free electrochemical aptasensor for the detection of SARS-CoV-2 spike protein based on carbon cloth sputtered gold nanoparticles, Biosen. Bioelectron., 2022, 12, 100256 CrossRef CAS PubMed.
- J. C. Abrego-Martinez, et al., Aptamer-based electrochemical biosensor for rapid detection of SARS-CoV-2: Nanoscale electrode-aptamer-SARS-CoV-2 imaging by photo-induced force microscopy, Biosens. Bioelectron., 2022, 195, 113595 CrossRef CAS PubMed.
- P. C. Guan, et al., Rapid Point-of-Care Assay by SERS Detection of SARS-CoV-2 Virus and Its Variants, Anal. Chem., 2022, 94, 17795–17802 CrossRef CAS PubMed.
- S. K. Daniel, et al., Handheld, low-cost, aptamer-based sensing device for rapid SARS-CoV-2 RNA detection using novelly synthesized gold nanoparticles, IEEE Sens. J., 2022, 22, 18437–18445 CAS.
- M. M. Gu, et al., Ultrasensitive detection of SARS-CoV-2 S protein with aptamers biosensor based on surface-enhanced Raman scattering, J. Chem. Phys., 2023, 158, 024203 CrossRef CAS PubMed.
- M. Sun, et al., Spherical neutralizing aptamer inhibits SARS-CoV-2 infection and suppresses mutational escape, J. Am. Chem. Soc., 2021, 143, 21541–21548 CrossRef CAS PubMed.
- L. Ch, S. Xu and J. Li, Recent advances in molecular imprinting technology: current status, challenges and highlighted applications, Chem. Soc. Rev., 2011, 40, 2922–2942 RSC.
- J. W. Lowdon, et al., MIPs for commercial application in low-cost sensors and assays–An overview of the current status quo, Sens. Actuators, B, 2020, 325, 128973 CrossRef CAS PubMed.
- Z. Altintas, et al., Detection of Waterborne Viruses Using High Affinity Molecularly Imprinted Polymers, Anal. Chem., 2015, 87, 6801–6807 CrossRef CAS PubMed.
- R. Liu, et al., Preparation of sialic acid-imprinted fluorescent conjugated nanoparticles and their application for targeted cancer cell imaging, ACS Appl. Bio Mater., 2017, 9, 3006–3015 CrossRef CAS PubMed.
- Z. Altintas, et al., Ultrasensitive detection of endotoxins using computationally designed nanoMIPs, Anal. Chim. Acta, 2016, 935, 239–248 CrossRef CAS PubMed.
- R. Liu, et al., Preparation of sialic acid-imprinted fluorescent conjugated nanoparticles and their application for targeted cancer cell imaging, ACS Appl. Mater. Interfaces, 2017, 9, 3006–3015 CrossRef CAS PubMed.
- H. Zhang, et al., Molecularly imprinted nanoparticles for biomedical applications, Adv. Mater., 2020, 32, 1806328 CrossRef CAS PubMed.
- I. Capek, et al., On inverse miniemulsion polymerization of conventional water-soluble monomers, Adv. Colloid Interface Sci., 2010, 156, 35–61 CrossRef CAS PubMed.
- P. Singla, et al., Electrochemical and thermal detection of allergenic substance lysozyme with molecularly imprinted nanoparticles, Anal. Bioanal. Chem., 2023, 415, 4467–4478 CrossRef CAS PubMed.
- F. Canfarotta, et al., A novel thermal detection method based on molecularly imprinted nanoparticles as recognition elements, Nanoscale, 2018, 10(4), 2081–2089 RSC.
- https://www.ricethresher.org/article/2021/03/rice-researchers-develop-two-new-sensors-to-detect-covid-19 .
- https://wyss.harvard.edu/technology/wfdcf-face-masks-a-wearable-covid-19-diagnostic/ .
- https://www.businessinsider.in/science/news/harvard-and-mit-researchers-aredeveloping-a-face-mask-that-lights-up-when-it-detects-the coronavirus/articleshow/75723321.cms .
- https://www.canhealth.com/2020/06/17/companies-develop-covid-19-detecting-face-mask/ .
- https://sixthwave.com/sixth-wave-amips-detects-delta-variant-of-sars-cov-2/ .
- J. McClements, et al., Molecularly Imprinted Polymer Nanoparticles Enable Rapid, Reliable, and Robust Point-of-Care Thermal Detection of SARS-CoV-2, ACS Sens., 2022, 7, 1122–1131 CrossRef CAS PubMed.
- N. Bhalla, et al., Nanoplasmonic biosensor for rapid detection of multiple viral variants in human serum, Sens. Actuators, B, 2022, 365, 131906–131914 CrossRef CAS PubMed.
- N. Cennamo, et al., Proof of Concept for a Quick and Highly Sensitive On-Site Detection of SARS-CoV-2 by Plasmonic Optical Fibers and Molecularly Imprinted Polymers, Sensors, 2021, 21, 1681–1698 CrossRef CAS PubMed.
- A. Raziq, et al., Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen, Biosens. Bioelectron., 2021, 178, 113029 CrossRef CAS PubMed.
- O. I. Parisi, et al., Design and development of plastic antibodies against SARS-CoV-2 RBD based on molecularly imprinted polymers that inhibit in vitro virus infection, Nanoscale, 2021, 13, 16885–16899 RSC.
- H. E. Sharif, et al., Evaluation of electropolymerized molecularly imprinted polymers (E-MIPs) on disposable electrodes for detection of SARS-CoV-2 in saliva, Anal. Chim. Acta, 2022, 1206, 339777 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |







