Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Selective detection of peroxynitrite using an isatin receptor and a naphthalimide fluorophore†
Yueci
Wu‡
 a,
Hai-Hao
Han‡
a,
Hai-Hao
Han‡
 def,
Liu
He
c,
Li
Li
b,
Yi
Zang
def,
Liu
He
c,
Li
Li
b,
Yi
Zang
 d,
Jia
Li
d,
Jia
Li
 *def,
Xiao-Peng
He
*def,
Xiao-Peng
He
 *c,
Yaping
Ding
*c,
Yaping
Ding
 *b,
Weiguo
Cao
*b,
Weiguo
Cao
 *b and
Tony D.
James
*b and
Tony D.
James
 *ag
*ag
aDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
bDepartment of Chemistry, Shanghai University, Shanghai 200444, China. E-mail: wgcao@shu.edu.cn
cKey Laboratory for Advanced Materials and Joint International Research Laboratory of Precision Chemistry and Molecular Engineering, Feringa Nobel Prize Scientist Joint Research Center, Frontiers Center for Materiobiology and Dynamic Chemistry, School of Chemistry and Molecular Engineering, East China University of Science and Technology, 130 Meilong Rd., Shanghai 200237, China. E-mail: xphe@ecust.edu.cn
dState Key Laboratory of Drug Research, Molecular Imaging Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China. E-mail: jli@simm.ac.cn
eUniversity of Chinese Academy of Sciences, No. 19A Yuquan Road, Beijing, 100049, China
fShandong Laboratory of Yantai Drug Discovery, Bohai Rim Advanced Research Institute for Drug Discovery, Yantai, Shandong 264117, China
gSchool of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang 453007, China
First published on 6th April 2023
Abstract
Peroxynitrite is a reactive oxygen and nitrogen species that participates in various biological reactions. Therefore, it is important to readily detect and track peroxynitrite in biological systems. Here, a novel turn-on probe encapsulated in PEG DSPE-PEG/HN-I was used to fluorescently detect ONOO− rapidly. The encapsulation of HN-I using DSPE-PEG2000 optimizes the sensing performances of the naphthalimide probe and avoids ACQ. Using DSPE-PEG/HN-I to detect changes in the levels of exogenous ONOO− in HepG2 cells and endogenous ONOO− induced by LPS in RAW 267.4 cells was demonstrated.
Peroxynitrite (ONOO−) is a short-lived bioactive agent, belonging to the category of reactive oxygen and nitrogen species (ROS/RNS). Peroxynitrite is an oxidant that generates free radicals, and acts as a Lewis base in living systems.1 Upon exposure to peroxynitrite, biochemical cycles can be promoted while cellular function and viability may be adversely affected, depending on the concentrations of peroxynitrite.2 Peroxynitrite at low concentrations can contribute to programmed cell death while higher concentrations of peroxynitrite promote a disruption of cellular energy production resulting in necrotic cell death.3–5 Peroxynitrite is involved in mediating numerous reactions, including the promotion of mitochondrial dysfunction, the regulation of cellular signalling pathways, the breaking of cellular redox status, and the induction of pain response under inflammatory conditions.6–10 While, Hooper and Padalko have reported that peroxynitrite can mediate the immune response to virus infection.11,12 However, due to a lack of rapid measurement tools suitable to detect peroxynitrite and quantify its concentrations in vivo, most of the biological reactions peroxynitrite participates in have only been investigated in vitro.2 As such, in order to further explore the role of peroxynitrite in disease pathogenesis, it is necessary to investigate and develop effective methods for peroxynitrite detection.
Fluorescent probes can detect analytes based on changes in spectroscopic properties promoted by targeted reactions.13–15 In general, a fluorescent probe consists of three units: a fluorophore that produces spectroscopic signals; a receptor that is able to react with a specific analyte; a linker that is suitable for connecting the fluorophore with the receptor.13,16,17 According to the reaction between the receptor and the targeted analyte, a fluorescent probe can exhibit specified fluorescence responsiveness to its targeted analyte.18 In addition, turn-on fluorescent probes can exhibit enhanced monitoring capabilities against dark backgrounds, which results in reduced background interference.18
Aryl boronate groups and α-ketoamide groups have been widely applied in the design of peroxynitrite targeting probes due to their good sensitivity and rapid response.19 However, these receptors can also react with other ROS species, such as hydrogen peroxide and hypochlorite.20–24 Significantly, isatin exhibits a sensitive, selective and rapid response to peroxynitrite.25–28
Naphthalimide fluorophores are on the whole cell permeable and exhibit good photostabilities.29 Due to the electron-withdrawing ability of its imide core, naphthalimide exhibits strong intramolecular charge transfer (ICT) in its solution state when the C-4 site of naphthalimide is modified by electron donors.30–32 These properties make naphthalimide an excellent fluorophore candidate for the construction of chemiluminescent probes for analyte detection in biological systems.30,33–37 However, one of the main drawbacks of naphthalimide-based systems is low solubility. Poor solubility and the planar structure make naphthalimide undergo π–π stacking easily, which results in aggregation-caused quenching (ACQ).29,38,39
In most cases, structural modification is used to solve solubility problems and overcome the ACQ of naphthalimide-based fluorophores.40–42 However, this approach requires additional time-consuming synthesis. Therefore, we decided to explore a much simpler approach and use PEG encapsulation to improve solubility.43,44 Polyethylene glycol (PEG) is widely used in the field of drug delivery as an excipient to improve molecular solubility.45–47 In addition, DSPE-PEG2000 (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-poly (ethylene glycol)) can reduce aggregation, improve the stability of nanoparticles and increase the circulation times of molecules in vivo.47–49
Here, we selected 4-hydroxy-1,8-naphthalimide (HN) as the fluorophore and a 1-methylindoline-2,3-dione moiety as the receptor to construct a novel fluorescent probe HN-I for the detection of ONOO−. According to the specific redox reaction between ONOO− and 1-methylindoline-2,3-dione, the fluorophore HN is released (Scheme 1).32,50 In order to ensure the improvement of water solubility and the reduction of ACQ effect, we used DSPE-PEG2000 to encapsulate the fluorescent probe HN-I (Scheme 1).
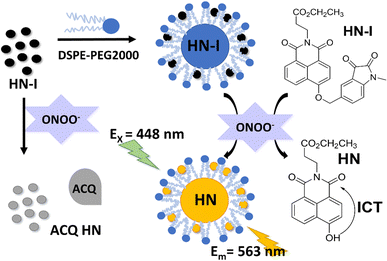 | ||
| Scheme 1 Schematic for use of DSPE-PEG/HN-I in the detection of peroxynitrite reaction mechanism of HN-I. | ||
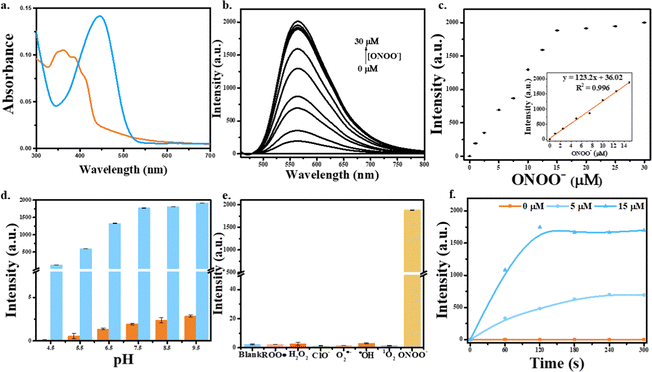
We were inspired by the group of Bruemmer who found that isatin-based groups could detect ONOO− with high selectivities without any interference from other ROS/RNS.25 As such, we prepared HN where the C-5 position of a 1-methylindoline-2,3-dione moiety was linked to a naphthalimide (Fig. S1, ESI†). Upon the addition of ONOO−, the fluorescence of HN-I can be turned on (Fig. S2 and S3, ESI†). In addition, by using DSPE-PEG2000, the aqueous solubility of DSPE-PEG/HN-I can be improved. As we mentioned above, the improvement in solubility of HN-I when using DSPE-PEG2000 decreases the π–π stacking of HN-I, which reduces ACQ and contributes to the recovery of the fluorescence (Fig. S4 and S5, ESI†). In other words, the sensing performances of DSPE-PEG/HN-I were optimized by the encapsulation of DSPE-PEG2000 (Fig. S4 and S5, ESI† and Fig. 1a).
First, the absorption spectra of DSPE-PEG/HN-I were explored (Fig. 1a). Initially, the ICT effect of the probe was inhibited so that the probe was nonfluorescent. However, on the addition of ONOO−, the naphthalimide fluorophore of DSPE-PEG/HN was released, and the recovery of the ICT effect resulted in fluorescence enhancement. As such, when the solution was excited at 448 nm, the fluorescence emission of DSPE-PEG/HN at 563 nm was enhanced. The fluorescence intensity of DSPE-PEG/HN-I increased with increasing concentrations of ONOO− from 0 μM to 30 μM (Fig. 1b). For ONOO− from 0–15 μM, the fluorescence response was linear (Fig. 1c). The limit of the detection of DSPE-PEG/HN-I for ONOO− was calculated to be 22 nM (Fig. 1c). The reaction rate between DSPE-PEG/HN-I and ONOO− was rapid. From Fig. 1f, the fluorescence intensities reached the highest level at around 240 s. In addition, the selectivity of DSPE-PEG/HN-I was evaluated. Based on the graph of Fig. 1e, DSPE-PEG/HN-I exhibited no fluorescent responses upon the addition of other ROS including ROO˙, H2O2, O2˙−, ˙OH, 1O2 and ClO−. The pH sensitivity of DSPE-PEG/HN-I was then evaluated from 4.5 to 9.5 (Fig. 1d).9,51,52 Over a pH range from 6.5 to 9.5, DSPE-PEG/HN-I exhibited low pH sensitivity, which confirms that DSPE-PEG/HN-I can be used for monitoring ONOO− in biological systems.
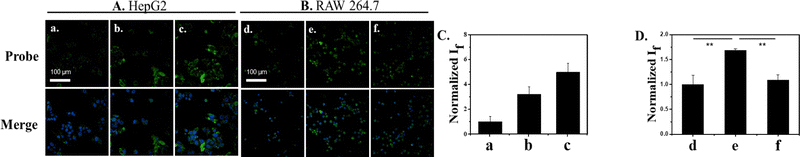
Based on excellent sensing performances of DSPE-PEG/HN-I in solution, we evaluated DSPE-PEG/HN-I for imaging ONOO− in a human liver cancer cell line (HepG2). Before cell fluorescence imaging tests, the cell viability of DSPE-PEG/HN-I was investigated in live HepG2 cells by a cell counting kit-8 (CCK-8) assay. The results suggested that DSPE-PEG/HN-I showed almost no cytotoxicity (cell viability ≈ 100% treated with a 40/40 μM DSPE-PEG/HN-I) (Fig. S7, ESI†). DSPE-PEG/HN-I was evaluated with and without SIN-1 (a typical ONOO− exogenous donor). As expected, without SIN-1, there was no fluorescence observed (Fig. 2a). However, with a concentration increase of SIN-1, the fluorescence intensity enhanced 5-fold (Fig. 2a–c). Then, DSPE-PEG/HN-I was also shown to detect exogenous ONOO− in SIN-1-treated human cervical cancer cell line (HeLa). However, after pretreatment of cells with N-acetylcysteine (NAC, an ONOO− scavenger), it attenuated the increase in the fluorescence of DSPE-PEG/HN-I induced by SIN-1 (Fig. S8, ESI†). Furthermore, the possibility of using DSPE-PEG/HN-I for endogenous ONOO− detection was also investigated in a macrophage cell line (RAW 264.7). RAW 264.7 cells were incubated with lipopolysaccharide (LPS) which can promote inflammatory response and upregulate the concentration of ONOO−.53–56 As shown in Fig. 2D, the fluorescence intensity of DSPE-PEG/HN-I exhibited a 1.5-fold increase for LPS-loaded RAW 264.7 cells. After RAW 264.7 cells were incubated with both LPS and NAC, weak fluorescence was observed. All these results indicated that DSPE-PEG/HN-I can be applied for the detection of both exogenous and endogenous ONOO−.
In summary, a novel turn-on probe was designed for the highly selective detection of ONOO−. To avoid the ACQ effect, a disadvantage of naphthalimide fluorophores, DSPE-PEG2000 was used to encapsulate HN-I to improve the sensing performances. We determined that DSPE-PEG/HN-I can detect ONOO− rapidly in solution. In addition, DSPE-PEG/HN-I can be used to image exogenous and endogenous ONOO−. These results confirm the potential of the DSPE-PEG/HN-I for the monitoring of ONOO− in biological systems.
Y. W wishes to thank China Scholarship Council, the University of Bath and Shanghai University. TDJ wishes to thank the Royal Society for a Wolfson Research Merit Award and the Open Research Fund of the School of Chemistry and Chemical Engineering, Henan Normal University for support (2020ZD01). X.-P. H. thanks the National Natural Science Foundation of China (No. 21788102, 91853201 and 9185920077). J. L. and Y. Z. thank the National Natural Science Foundation of China (No. 82130099, 82151219, 31871414 and 81971265) and the Shanghai Municipal Science and Technology Major Project (No. 22ZR1415200). H.-H. H. thanks the National Natural Science Foundation of China (No. 22107029) and Project funded by the China Postdoctoral Science Foundation (No. 2020M681196).
Conflicts of interest
There are no conflicts to declare.Notes and references
- G. Ferrer-Sueta, N. Campolo, M. Trujillo, S. Bartesaghi, S. Carballa, N. Romero, B. Alvarez and R. Radi, Chem. Rev., 2018, 118(3), 1338–1408 CrossRef CAS PubMed.
- C. Szabo, H. Ischiropoulos and R. Radi, Nat. Rev. Drug Discovery, 2007, 6, 662–680 CrossRef CAS PubMed.
- C. Szabo, Toxicol. Lett., 2003, 140, 105–112 CrossRef PubMed.
- P. Jagtap and C. Szabo, Nat. Rev. Drug Discovery, 2005, 4, 421–440 CrossRef CAS PubMed.
- L. Virag, E. Szabo, P. Gergely and C. Szabo, Toxicol. Lett., 2003, 140, 113–124 CrossRef PubMed.
- N. Nin, A. Cassina, J. Boggia, E. Alfonso, H. Botti, G. Peluffo, A. Trostchansky, C. Batthyany, R. Radi, H. Rubbo and F. J. Hurtado, Intensive Care Med., 2004, 30, 2271–2278 CrossRef PubMed.
- A. L. Levonen, R. P. Patel, P. Brookes, Y. M. Go, H. Jo, S. Parthasarathy, P. G. Anderson and V. M. Darley-Usmar, Antioxid. Redox Signaling, 2001, 3, 215–229 CrossRef CAS PubMed.
- D. Salvemini, T. M. Doyle and S. Cuzzocrea, Biochem. Soc. Trans., 2006, 34, 965–970 CrossRef CAS PubMed.
- R. Radi, J. Biol. Chem., 2013, 288, 26464–26472 CrossRef CAS PubMed.
- P. Pacher, J. Beckman and L. Liaudet, Physiol. Rev., 2007, 87, 315–424 CrossRef CAS PubMed.
- D. C. Hooper, R. B. Kean, G. S. Scott, S. V. Spitsin, T. Mikheeva, K. Morimoto, M. Bette, A. M. Rohrenbeck, B. Dietzschold and E. Weihe, J. Immunol., 2001, 167, 3470–3477 CrossRef CAS PubMed.
- E. Padalko, T. Ohnishi, K. Matsushita, H. Sun, K. Fox-Talbot, C. Bao, W. M. Baldwin and C. J. Lowenstein, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 11731–11736 CrossRef CAS PubMed.
- X. H. Li, X. H. Gao, W. Shi and H. M. Ma, Chem. Rev., 2014, 114, 590–659 CrossRef CAS PubMed.
- H. H. Han, H. Tian, Y. Zang, A. C. Sedgwick, J. Li, J. L. Sessler, X. P. He and T. D. James, Chem. Soc. Rev., 2021, 50, 9391–9429 RSC.
- W. T. Dou, H. H. Han, A. C. Sedgwick, G. B. Zhu, Y. Zang, X. R. Yang, J. Yoon, T. D. James, J. Li and X. P. He, Sci. Bull., 2022, 67, 853–878 CrossRef CAS PubMed.
- A. P. de Silva, H. Q. N. Gunaratne, T. Gunnlaugsson, A. J. M. Huxley, C. P. McCoy, J. T. Rademacher and T. E. Rice, Chem. Rev., 1997, 97, 1515–1566 CrossRef CAS PubMed.
- W. Shi and H. M. Ma, Chem. Commun., 2012, 48, 8732–8744 RSC.
- T. Ueno and T. Nagano, Nat. Methods, 2011, 8, 642–645 CrossRef CAS PubMed.
- W. L. Cui, M. H. Wang, Y. H. Yang, J. Y. Wang, X. Z. Zhu, H. T. Zhang and X. X. Ji, Coord. Chem. Rev., 2023, 474, 214848 CrossRef CAS.
- J. W. Chen, T. C. Wu, W. Liang, J. J. Ciou and C. H. Lai, Drug Delivery Transl. Res., 2022 DOI:10.1007/s13346-022-01248-w.
- D. H. Tian, J. R. Liu, S. Y. Wang, S. Yan, Z. H. Chai, F. Dai, S. X. Zhang and B. Zhou, Sens. Actuators, B, 2022, 368, 132149 CrossRef CAS.
- L. Zhen, J. S. Lan, S. A. Zhang, L. Liu, R. F. Zeng, Y. Chen and Y. Ding, Anal. Methods, 2022, 14, 2147–2152 RSC.
- X. L. Xie, X. E. Yang, T. H. Wu, Y. Li, M. M. Li, Q. Tan, X. Wang and B. Tang, Anal. Chem., 2016, 88, 8019–8025 CrossRef CAS PubMed.
- W. Shu, Y. L. Wu, S. P. Zang, S. Su, H. Kang, J. Jing and X. L. Zhang, Sens. Actuators, B, 2020, 303, 127284 CrossRef CAS.
- K. J. Bruemmer, S. Merrikhihaghi, C. T. Lollar, S. N. S. Morris, J. H. Bauer and A. R. Lippert, Chem. Commun., 2014, 50, 12311–12314 RSC.
- X. Y. Lu, H. H. Su, J. Zhang, N. N. Wang, H. Wang, J. Y. Liu and W. L. Zhao, Spectrochim. Acta, Part A, 2022, 267, 120537 CrossRef PubMed.
- J. H. Xiong, W. W. Wang, C. X. Wang, C. Zhong, R. Q. Ruan, Z. Q. Mao and Z. H. Liu, ACS Sens., 2020, 5, 3237–3245 CrossRef CAS PubMed.
- W. W. Wang, J. H. Xiong, X. J. Song, Z. Wang, F. Zhang and Z. Q. Mao, Anal. Chem., 2020, 92, 13305–13312 CrossRef CAS PubMed.
- S. Mukherjee and P. Thilagar, Chem. – Eur. J., 2014, 20, 8012–8023 CrossRef CAS PubMed.
- S. A. Choi, C. S. Park, O. S. Kwon, H. K. Giong, J. S. Lee, T. H. Ha and C. S. Lee, Sci. Rep., 2016, 6(1), 26203 CrossRef CAS PubMed.
- S. U. Hettiarachchi, B. Prasai and R. L. McCarley, J. Am. Chem. Soc., 2014, 136, 7575–7578 CrossRef CAS PubMed.
- X. Lv, G. B. Ge, L. Feng, J. Troberg, L. H. Hu, J. Hou, H. L. Cheng, P. Wang, Z. M. Liu, M. Finel, J. N. Cui and L. Yang, Bios. Bioelectron., 2015, 72, 261–267 CrossRef CAS PubMed.
- R. K. Jackson, Y. Shi, X. D. Yao and S. C. Burdette, Dalton Trans., 2010, 39, 4155–4161 RSC.
- A. P. de Silva, A. Goligher, H. Q. N. Gunaratne and T. E. Rice, ARKIVOC, 2003, 7, 229–243 Search PubMed.
- A. P. de Silva and T. E. Rice, Chem. Commun., 1999, 163–164 RSC.
- L. Ingrassia, F. Lefranc, R. Kiss and T. Mijatovic, Curr. Med. Chem., 2009, 16, 1192–1213 CrossRef CAS PubMed.
- J. Liu, S. L. Zhong, L. L. Zhang, M. W. Yi, X. J. Liu, T. Bing, N. Zhang and D. H. Shangguan, Chem. Commun., 2021, 57, 6558–6561 RSC.
- J. Gierschner, L. Luer, B. Milian-Medina, D. Oelkrug and H. J. Egelhaaf, J. Phys. Chem. Lett., 2013, 4, 2686–2697 CrossRef CAS.
- D. L. Reger, J. D. Elgin, R. F. Semeniuc, P. J. Pellechia and M. D. Smith, Chem. Commun., 2005, 4068–4070 RSC.
- C. G. Xu, T. Wu, L. Z. Duan and Y. M. Zhou, Chem. Commun., 2021, 57, 11366–11369 RSC.
- S. Sharma, S. Srinivas, S. Rakshit and S. Sengupta, Org. Biomol. Chem., 2022, 20, 9422–9430 RSC.
- J. Y. Zheng, J. Cao, Y. J. Tu, C. P. Huang, M. M. Liu and M. J. Zhang, J. Mol. Struct., 2022, 1250, 131868 CrossRef CAS.
- H. H. Han, A. C. Sedgwick, Y. Shang, N. Li, T. T. Liu, B. H. Li, K. Q. Yu, Y. Zang, J. T. Brewster, M. L. Odyniec, M. Weber, S. D. Bull, J. Li, J. L. Sessler, T. D. James, X. P. He and H. Tian, Chem. Sci., 2020, 11, 1107–1113 RSC.
- L. Li, Z. M. Lin, X. C. Lu, C. Chen, A. Q. Xie, Y. P. Tang and Z. Q. Zhang, RSC Adv., 2022, 12, 33358–33364 RSC.
- K. Knop, R. Hoogenboom, D. Fischer and U. S. Schubert, Angew. Chem., Int. Ed., 2010, 49, 6288–6308 CrossRef CAS PubMed.
- R. G. Strickley, Pharm. Res., 2004, 21, 201–230 CrossRef CAS PubMed.
- J. Che, C. I. Okeke, Z. B. Hu and J. Xu, Curr. Pharm. Des., 2015, 21, 1598–1605 CrossRef CAS PubMed.
- S. Y. Geng, B. Yang, G. W. Wang, G. Qin, S. Wada and J. Y. Wang, Nanotechnology, 2014, 25, 275103 CrossRef PubMed.
- N. O. Knudsen, S. Ronholt, R. D. Salte, L. Jorgensen, T. Thormann, L. H. Basse, J. Hansen, S. Frokjaer and C. Foged, Eur. J. Pharm. Biopharm., 2012, 81, 532–539 CrossRef CAS PubMed.
- J. Cao, W. W. An, A. G. Reeves and A. R. Lippert, Chem. Sci., 2018, 9, 2552–2558 RSC.
- A. Denicola, J. M. Souza and R. Radi, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 3566–3571 CrossRef CAS PubMed.
- J. P. Crow, C. Spruell, J. Chen, C. Gunn, H. Ischiropoulos, M. Tsai, C. D. Smith, R. Radi, W. H. Koppenol and J. S. Beckman, Free Radical Biol. Med., 1994, 16, 331–338 CrossRef CAS PubMed.
- A. Kumar, S. H. Chen, M. B. Kadiiska, J. S. Hong, J. Zielonka, B. Kalyanaraman and R. P. Mason, Free Radical Biol. Med., 2014, 73, 51–59 CrossRef CAS PubMed.
- H.-H. Han, H.-M. Wang, P. Jangili, M. Li, L. Wu, Y. Zang, A. C. Sedgwick, J. Li, X.-P. He, T. D. James and J. S. Kim, Chem. Soc. Rev., 2023, 52, 879–920 RSC.
- M. Weber, H.-H. Han, B.-H. Li, M. L. Odyniec, C. E. F. Jarman, Y. Zang, S. D. Bull, A. B. Mackenzie, A. C. Sedgwick, J. Li, X.-P. He and T. D. James, Chem. Sci., 2020, 11, 8567–8571 RSC.
- X.-L. Hu, H.-Q. Gan, F.-D. Meng, H.-H. Han, D.-T. Shi, S. Zhang, L. Zou, X.-P. He and T. D. James, Front. Chem. Sci. Eng., 2022, 16, 1425–1437 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cc06425a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |