Open Access Article
Open Access ArticleThe rippled β-sheet layer configuration—a novel supramolecular architecture based on predictions by Pauling and Corey†
Amaruka
Hazari‡
a,
Michael R.
Sawaya‡
b,
Niko
Vlahakis
b,
Timothy C.
Johnstone
 a,
David
Boyer
b,
Jose
Rodriguez
b,
David
Eisenberg
b and
Jevgenij A.
Raskatov
a,
David
Boyer
b,
Jose
Rodriguez
b,
David
Eisenberg
b and
Jevgenij A.
Raskatov
 *a
*a
aDept. of Chemistry and Biochemistry, UCSC, 1156 High Street, Santa Cruz, CA 95064, USA. E-mail: jraskato@ucsc.edu
bDept. of Chemistry and Biochemistry, UCLA, 607 Charles E. Young Drive East Box 951569, Los Angeles, CA 90095-1569, USA
First published on 15th July 2022
Abstract
The rippled β-sheet is a peptidic structural motif related to but distinct from the pleated β-sheet. Both motifs were predicted in the 1950s by Pauling and Corey. The pleated β-sheet was since observed in countless proteins and peptides and is considered common textbook knowledge. Conversely, the rippled β-sheet only gained a meaningful experimental foundation in the past decade, and the first crystal structural study of rippled β-sheets was published as recently as this year. Noteworthy, the crystallized assembly stopped at the rippled β-dimer stage. It did not form the extended, periodic rippled β-sheet layer topography hypothesized by Pauling and Corey, thus calling the validity of their prediction into question. NMR work conducted since moreover shows that certain model peptides rather form pleated and not rippled β-sheets in solution. To determine whether the periodic rippled β-sheet layer configuration is viable, the field urgently needs crystal structures. Here we report on crystal structures of two racemic and one quasi-racemic aggregating peptide systems, all of which yield periodic rippled antiparallel β-sheet layers that are in excellent agreement with the predictions by Pauling and Corey. Our study establishes the rippled β-sheet layer configuration as a motif with general features and opens the road to structure-based design of unique supramolecular architectures.
Introduction
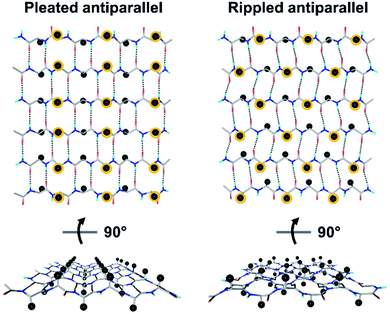
Tailored proteins and peptides hold immense potential for materials development and biomedical application. Chiral (i.e., D-amino acid) substitutions may be employed to generate self-assembling peptide architectures with unique properties,1–6 bioactive compounds with distinct activities,7–13 as well as systems with enhanced crystallization behavior.14–18 There is great interest in developing new peptidic systems that contain D-amino acid substitutions, as this would allow systematic access to a vast structural space with unique molecular properties.The rippled β-sheet is a largely neglected structural motif, hypothesized by Pauling and Corey in 1953.19 It is closely related to but distinct from the pleated β-sheet proposed by the same authors two years prior.20 More specifically, the pleated β-sheet is homochiral, i.e., enantiopure, whereas in the rippled β-sheet, every second peptide strand is of opposing chirality, i.e., racemic (Fig. 1). The mirroring of every second strand in the pleated β-sheet produces a distinct backbone topography in the rippled β-sheet. It leads to profound differences in the relative orientation of peptide sidechains between any two adjacent β-strands in the layer, which are eclipsed in the pleated but staggered in the rippled β-sheet. As a result, peptide sidechains experience much less steric crowding in the rippled β-sheet.
Research conducted in the 1970s combined experiment and theory, finding that the achiral polyglycine peptide forms chiral conformers and associates into rippled β-sheets known as polyglycine I.21–23 This work was reviewed recently by Lotz, who performed much of this early pioneering research.24 The ensuing four decades were characterized by only minimal research activity in the field.25–27 From 2010 on, the laboratories of Schneider, Nilsson and Raskatov independently published a number of key studies.1–5,11,28–31 This work was reviewed in an article written jointly by the three investigators.32
In 2022, we published the rippled β-sheet dimer structure of (L,L,L)-triphenylalanine (“FFF”) and (D,D,D)-triphenylalanine (fff) (“FFF:fff”).33 However, instead of forming the sought layers, FFF:fff packed into a herringbone structure, raising doubt as to whether the periodic rippled β-sheet layer configuration predicted by Pauling and Corey was viable. Very recent solution state NMR work further challenged the field by showing that certain model peptides preferentially form pleated and not rippled β-sheets.34,35 The development of a firm crystallographic foundation is, therefore, of utmost importance.
Herein we report on three crystal structures of periodic rippled β-sheets, formed from three distinct peptide systems. Our findings answer a longstanding structural question for the field and pave the road to the systematic structure-based design of a new class of supramolecular polymers.
Results and discussion
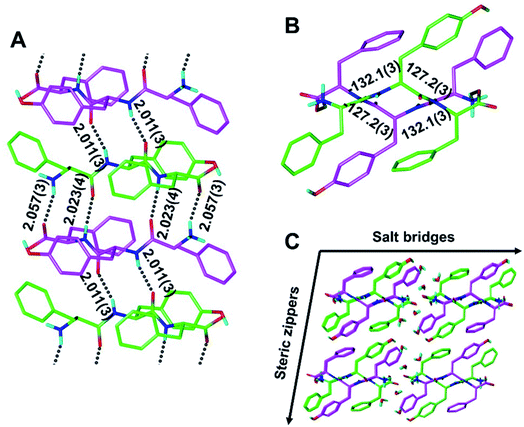
Although our understanding of what makes a peptide ripple-genic remains very limited, some rudimentary design principles are beginning to emerge. As such, our recent X-ray crystallography work identified phenylalanine as a ripple-genic residue.33 Although the FFF:fff model successfully formed cross-β dimers, the system stopped short of forming periodic β-sheet layers, possibly due to a quirk in the packing of the phenylalanine side chains, favoring end-to-edge rather than edge-to-edge interactions. To explore a greater sequence space, we designed tripeptides with different aromatic amino acids.The desired outcome was achieved by substituting the central phenylalanine residues within FFF and fff by tyrosine (FYF and fyf, respectively). The racemic mixture of FYF and its mirror-image counterpart, fyf, crystallized into small needles from a water/hexafluoroisopropanol (HFIP) mixture, which were revealed by X-ray crystallography to be composed of periodic antiparallel rippled β-sheet layers (Fig. 2). The asymmetric unit of the FYF:fyf crystal is a single tripeptide (Fig. S1†). The individual tripeptides stack in the H-bonding dimension, forming extended antiparallel rippled β-sheet layers, in which mirror-image peptide strands are arranged in strictly alternating fashion. This novel layer architecture, which we term [FYF:fyf]n, is in excellent qualitative agreement with the prediction made by Pauling and Corey (Fig. S2†). Each L-tripeptide is sandwiched between two D-tripeptides, and each D-tripeptide is sandwiched between two L-tripeptides in periodic fashion within the individual β-sheet layers, with H-bond distances ranging from 2.011(3) Å to 2.057(3) Å (Fig. 2A). Each tripeptide has four H-bonds to one of its two direct neighbors in the layer (i.e., “tight dimer”), and two H-bonds to the other (i.e., “loose dimer”). The tight FYF:fyf rippled β-sheet dimer closely resembles the FFF:fff rippled β-sheet dimer.33 The resemblance suggests that the initial step of mirror-image peptide self-assembly may be the formation of the tight dimer, which then nucleates sequence-dependent higher order assembly. The torsional angles for the central residue are measured as φ = −132.1(3)° and ψ = 127.2(3)° for the L-tripeptide, and φ = 132.1(3)° and ψ = −127.2(3)° for the inversion-related D-tripeptide (Fig. 2B). The lack of twist is in agreement with theory.29 The individual rippled antiparallel [FYF:fyf]n β-sheet columns associate periodically to form a three-dimensional crystallographic lattice (Fig. 2C). Two distinct lateral association modes are observed, one of which is governed by salt bridges between the two peptide enantiomers, and the other which is driven by hydrophobic steric zippers between the mirror-image peptides. Given the analogy to amyloids,36 we define the pairs of rippled β-sheets that are mated via steric zippers and extend along the length of the needle crystal as rippled β-sheet fibrils.
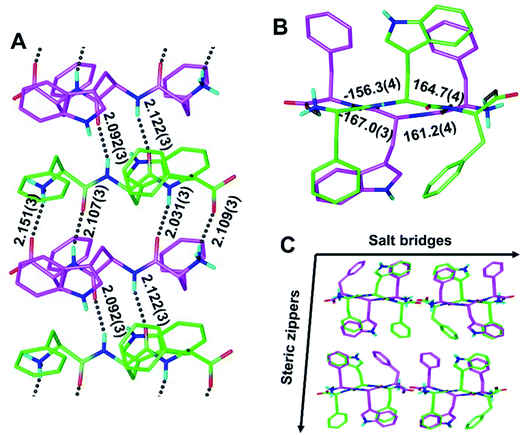
To determine whether rippled β-sheet fibrils would tolerate a bulkier sidechain, the central residue was changed to Trp (“W”). The racemic mixture of FWF and fwf was crystallized, yielding needles containing periodic [FWF:fwf]n antiparallel rippled β-sheet layers (Fig. 3). H-bond distances between peptide strands within the [FWF:fwf]n columns are slightly longer than with [FYF:fyf]n and cover a somewhat wider range from 2.031(3) Å to 2.151(3) Å (Fig. 3A). Once again, an alternation between tight and loose dimers is noted. Unlike the [FYF:fyf]n structure, the asymmetric unit of [FWF:fwf]n contains two crystallographically independent enantiomeric peptides that deviate slightly from perfect inversion symmetry (Fig. S3†). The torsional angles for the central residue are measured as φ = −156.3(4)° and ψ = 164.7(4)° for the L-tripeptide, and φ = 161.2(4)° and ψ = −167.0(3)° for the D-tripeptide (Fig. 3B; note that for each fibril there is a mirror-image fibril in the lattice, cf.Fig. 3C). The packing of individual rippled β-sheet layers in the [FWF:fwf]n lattice is similar to that of [FYF:fyf]n (Fig. 3Cvs.Fig. 2C), with pairs of [FWF:fwf]n layers mated via steric zippers to form rippled β-sheet fibrils. In close analogy to the [FYF:fyf]n system, the rippled β-sheet fibrils of [FWF:fwf]n are associated laterally through salt bridges.
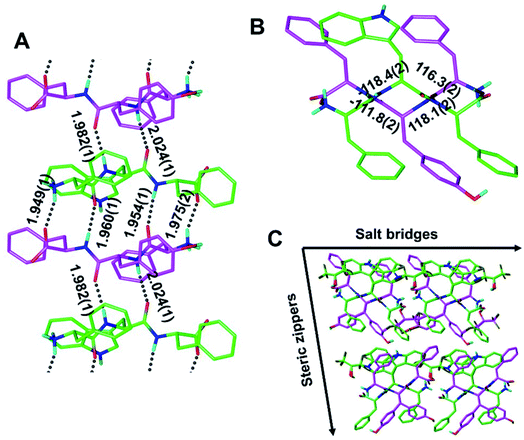
Our findings inspired us to explore the possibility of making a quasi-racemic rippled β-sheet fibril as proof of concept for an [A:B]n array, a potentially useful design principle for peptidic materials.6 An equimolar mixture of the FWF L-tripeptide and the fyf D-tripeptide was crystallized, and yielded needles containing periodic [FWF:fyf]n rippled antiparallel β-sheet layers, in which the L- and the D-tripeptides were found to strictly alternate (Fig. 4 and S4†). The backbone configuration is very similar to that found with [FYF:fyf]n and [FWF:fwf]n. The H-bond distances within the rippled β-sheet layer range from 1.949(1) Å to 2.024(1) Å. An alternation of tight and loose interfaces is once again noted, with four and two H-bonds, respectively (Fig. 4A). The torsional angles for the central residue are measured as φ = −118.4(2)° and ψ = 116.3(2)° for the FWF L-tripeptide, and φ = 118.1(2)° and ψ = −111.8(2)° for the fyf D-tripeptide (Fig. 4B). Like the systems discussed above, the individual [FWF:fyf]n columns are mated via steric zipper interfaces into rippled β-sheet fibrils that are packed laterally through salt bridges (Fig. 4C; see also Fig. S5†). Two L-tryptophan rotamers and two D-tyrosine rotamers are present (Fig. 4C; only one set of rotamers is shown in Fig. 4A; see Fig. S6† for the other rotamer set). Comparison of the Ramachandran (phi/psi) angles associated with the central residues was also of interest, revealing that the individual [FWF:fwf]n rippled β-sheet layers are substantially flatter than both [FYF:fyf]n and [FWF:fyf]n (cf.Fig. 2B, 3B and 4B). A possible explanation is that in the case of [FWF:fwf]n, the structure adopts an extended conformation to enable aromatic stacking of F1 and w2 side chains on neighboring strands. The other two rippled β-sheet systems reveal no such stacking of aromatic rings. Substantial Ramachandran angle variance in pleated β-sheets was previously noted by Eisenberg and co-workers, who found that the degree of β-strand extension and the associated pleating/flattening of the β-sheet is correlated with fibril stability.37
The preference for pleated vs. rippled is likely governed by a complex interplay of interactions including H-bonding, salt bridges, π-stacking and hydrophobic nesting that may occur both within individual β-sheets or layers and between layers.32 Wallach's rule was recently invoked to attempt to explain the tendency for racemic peptides to form rippled β-sheets in the solid state.35 It is important to note, however, that racemic peptide mixtures may produce racemic crystals without forming a rippled β-sheet. For example, the enantiomers in the racemic mixture of the GSTSTA peptide repeat from the ice-nucleation protein InaZ, self-sorted locally into pleated β-sheet layers that mated via mirror-image steric zippers in the racemic crystal (Fig. S7†), rather than forming rippled sheets.38 This shows that there are limits to using Wallach's rule to explain rippled β-sheet formation in the solid state. While the peptides sequences reported here are composed of aromatic amino acids, aromatic residues are not required for rippled β-sheet formation as both the polyglycine I system and the EKELV sequence in RV1738 adopt rippled configurations.16,24
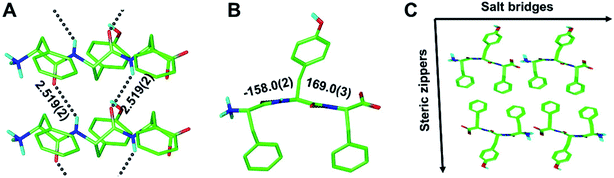
As part of this study, we also obtained crystals of the enantiopure FYF tripeptide. Whereas those crystals proved to be too small for conventional X-ray diffraction methods, micro electron diffraction (micro-ED) permitted successful structure solution and refinement (Fig. 5 and S8†). Intriguingly, [FYF:FYF]n forms parallel (pleated) β-sheets, distinct from the three rippled β-sheet systems discussed above that are all antiparallel. The H-bonds connecting the FYF monomers in the fibril are measured as 2.519(2) Å (Fig. 5A). The torsional angles for the central residue of FYF were measured as φ = −158.0(2)° and ψ = 169.0(3)° (Fig. 5B), both substantially wider than the corresponding Ramachandran angles associated with FYF in the rippled antiparallel [FYF:fyf]n and [FWF:fyf]n β-sheets. This difference is likely a consequence of the distinct architecture and associated packing forces. Similar to the rippled systems, the pleated parallel [FYF:FYF]n columns are mated via steric zippers into fibrils that associate laterally through salt bridges (Fig. 5C).
An interesting parallel is noted with a recent study of racemic Aβ40.31 Whereas enantiopure Aβ40 is known to form parallel in-register pleated β-sheets, its racemic counterpart forms antiparallel rippled β-sheets, suggesting there may be an intrinsic preference for the rippled β-sheet to adopt the antiparallel orientation. Similar observations were made for the racemic aggregated FFF:fff system,33 the racemic aggregated KLVFFAE:klvffae system,3 and for polyglycine I.21–23 However, the preference is not absolute as the MAX1/DMAX hairpin system studied by the Schneider lab did form parallel rippled β-sheets.1 Inter-sheet side-chain interactions may be another factor that could be explored to fine-tune rippled β-sheet architectures in the future.1,39
Conclusions
Crystal structures of three distinct periodic rippled β-sheet layers were presented, two of which are racemic (i.e., [FYF:fyf]n and [FWF:fwf]n), and one that is quasi-racemic (i.e., [FWF:fyf]n). The layer coordinates are in good overall agreement with the predictions of Pauling and Corey. The individual rippled β-sheet columns are mated via steric zippers into rippled β-sheet fibrils, which associate laterally via salt bridges. All three rippled β-sheet fibrils adopt the antiparallel arrangement, distinct from the pleated, homochiral [FYF:FYF]n that forms parallel β-sheet fibrils. The rippled β-sheet fibril is a novel supramolecular architecture, which may allow rational design of unique functional peptide materials.Data availability
Crystallographic files for the four structures reported in the paper have been deposited.Author contributions
A. H., M. R. S., and J. A. R. designed research; A. H., M. R. S., N. V., T. C. J. and D. B. performed research; A. H., M. R. S., N. V., T. C. J., D. B., J. R., D. E. and J. A. R. analyzed data; A. H., M. R. S. and J. A. R. wrote the paper.Conflicts of interest
There is no conflict of interest to report.Acknowledgements
This work was supported by the NIH award R01AG074954 and a generous gift of the Seaver Institute to J. A. R. We thank Duilio Cascio and the staff at the Northeastern Collaborative Access Team, which is funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Eiger 16M detector on the 24-ID-E beam line is funded by a NIH-ORIP HEI grant (S10OD021527). The Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The single-crystal X-ray diffractometer housed in UCSC X-ray diffraction facility was funded by NSF MRI grant 2018501.References
- K. Nagy-Smith, P. J. Beltramo, E. Moore, R. Tycko, E. M. Furst and J. P. Schneider, ACS Cent. Sci., 2017, 3, 586–597 CrossRef CAS PubMed.
- K. J. Nagy, M. C. Giano, A. Jin, D. J. Pochan and J. P. Schneider, J. Am. Chem. Soc., 2011, 133, 14975–14977 CrossRef CAS PubMed.
- J. M. Urban, J. Ho, G. Piester, R. Fu and B. L. Nilsson, Molecules, 2019, 24, 1983 CrossRef CAS PubMed.
- R. J. Swanekamp, J. J. Welch and B. L. Nilsson, Chem. Commun., 2014, 50, 10133–10136 RSC.
- R. J. Swanekamp, J. T. M. DiMaio, C. J. Bowerman and B. L. Nilsson, J. Am. Chem. Soc., 2012, 134, 5556–5559 CrossRef CAS PubMed.
- K. M. Wong, A. S. Robang, A. H. Lint, Y. Wang, X. Dong, X. Xiao, D. T. Seroski, R. Liu, Q. Shao, G. A. Hudalla, C. K. Hall and A. K. Paravastu, J. Phys. Chem. B, 2021, 125, 13599–13609 CrossRef CAS PubMed.
- A. R. Foley, G. P. Roseman, K. Chan, A. Smart, T. S. Finn, K. Yang, R. S. Lokey, G. L. Millhauser and J. A. Raskatov, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 28625–28631 CrossRef CAS PubMed.
- S. Dutta, T. S. Finn, A. J. Kuhn, B. Abrams and J. Raskatov, ChemBioChem, 2019, 20, 1023–1026 CrossRef CAS PubMed.
- A. R. Foley, T. S. Finn, T. Kung, A. Hatami, H.-W. Lee, M. Jia, M. Rolandi and J. A. Raskatov, ACS Chem. Neurosci., 2019, 10, 3880–3887 CrossRef CAS PubMed.
- C. J. A. Warner, S. Dutta, A. R. Foley, E. Chen, D. S. Kliger and J. A. Raskatov, Chirality, 2017, 29, 5–9 CrossRef CAS PubMed.
- S. Dutta, A. R. Foley, C. J. A. Warner, X. Zhang, M. Rolandi, B. Abrams and J. A. Raskatov, Angew. Chem., Int. Ed., 2017, 56, 11506–11510 CrossRef CAS PubMed.
- A. M. Garcia, C. Giorgiutti, Y. El Khoury, V. Bauer, C. Spiegelhalter, E. Leize-Wagner, P. Hellwig, N. Potier and V. Torbeev, Chem.–Eur. J., 2020, 26, 9889–9899 CrossRef CAS PubMed.
- M. Avital-Shmilovici, J. Whittaker, M. A. Weiss and S. B. H. Kent, J. Biol. Chem., 2014, 289, 23683–23692 CrossRef CAS PubMed.
- T. O. Yeates and S. B. Kent, Annu. Rev. Biophys., 2012, 41, 41–61 CrossRef CAS PubMed.
- M. Avital-Shmilovici, K. Mandal, Z. P. Gates, N. B. Phillips, M. A. Weiss and S. B. H. Kent, J. Am. Chem. Soc., 2013, 135, 3173–3185 CrossRef CAS PubMed.
- R. D. Bunker, K. Mandal, G. Bashiri, J. J. Chaston, B. L. Pentelute, J. S. Lott, S. B. H. Kent and E. N. Baker, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4310–4315 CrossRef CAS PubMed.
- B. L. Pentelute, K. Mandal, Z. P. Gates, M. R. Sawaya, T. O. Yeates and S. B. H. Kent, Chem. Commun., 2010, 46, 8174–8176 RSC.
- D. E. Mortenson, K. A. Satyshur, I. A. Guzei, K. T. Forest and S. H. Gellman, J. Am. Chem. Soc., 2012, 134, 2473–2476 CrossRef CAS PubMed.
- L. Pauling and R. B. Corey, Proc. Natl. Acad. Sci. U. S. A., 1953, 39, 253–256 CrossRef CAS PubMed.
- L. Pauling and R. B. Corey, Proc. Natl. Acad. Sci. U. S. A., 1951, 37, 729–740 CrossRef CAS PubMed.
- B. Lotz, J. Mol. Biol., 1974, 87, 169–180 CrossRef CAS PubMed.
- F. Colonna-Cesari, S. Premilat and B. Lotz, J. Mol. Biol., 1974, 87, 181–191 CrossRef CAS PubMed.
- W. H. Moore and S. Krimm, Biopolymers, 1976, 15, 2439–2464 CrossRef CAS PubMed.
- B. Lotz, ChemBioChem, 2022, 23, e2021006 CrossRef PubMed.
- D. M. Chung and J. S. Nowick, J. Am. Chem. Soc., 2004, 126, 3062–3063 CrossRef CAS PubMed.
- J.-H. Fuhrhop, M. Krull and G. Büldt, Angew. Chem., Int. Ed., 1987, 26, 699–700 CrossRef.
- I. Weissbuch, R. A. Illos, G. Bolbach and M. Lahav, Acc. Chem. Res., 2009, 42, 1128–1140 CrossRef CAS PubMed.
- J. A. Raskatov, ChemBioChem, 2020, 21, 2945–2949 CrossRef CAS PubMed.
- J. A. Raskatov, Biopolymers, 2021, 112, e23391 CrossRef CAS PubMed.
- S. Dutta, A. Rodriguez Foley, A. Kuhn, B. Abrams, H.-W. Lee and J. A. Raskatov, Pept. Sci., 2019, 111, e24139 CAS.
- J. A. Raskatov, A. R. Foley, J. M. Louis, W. M. Yau and R. Tycko, J. Am. Chem. Soc., 2021, 143, 13299–13313 CrossRef CAS PubMed.
- J. A. Raskatov, J. P. Schneider and B. L. Nilsson, Acc. Chem. Res., 2021, 54, 2488–2501 CrossRef CAS PubMed.
- A. J. Kuhn, B. Ehlke, T. C. Johnstone, S. R. J. Oliver and J. A. Raskatov, Chem. Sci., 2022, 13, 671–680 RSC.
- X. Liu and S. H. Gellman, ChemBioChem, 2021, 22, 2772–2776 CrossRef CAS PubMed.
- X. Li, S. E. Rios and J. S. Nowick, Chem. Sci., 2022, 13, 7739–7746 RSC.
- M. R. Sawaya, S. Sambashivan, R. Nelson, M. I. Ivanova, S. A. Sievers, M. I. Apostol, M. J. Thompson, M. Balbirnie, J. J. W. Wiltzius, H. T. McFarlane, A. O. Madsen, C. Riekel and D. Eisenberg, Nature, 2007, 447, 453–457 CrossRef CAS PubMed.
- K. A. Murray, D. Evans, M. P. Hughes, M. R. Sawaya, C. J. Hu, K. N. Houk and D. Eisenberg, ACS Nano, 2022, 16, 2154–2163 CrossRef CAS PubMed.
- C.-T. Zee, C. Glynn, M. Gallagher-Jones, J. Miao, C. G. Santiago, D. Cascio, T. Gonen, M. R. Sawaya and J. A. Rodriguez, IUCrJ, 2019, 6, 197–205 CrossRef CAS PubMed.
- V. Torbeev, M. Grogg, J. Ruiz, R. Boehringer, A. Schirer, P. Hellwig, G. Jeschke and D. Hilvert, J. Pept. Sci., 2016, 22, 290–304 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC [PDB 8DDH, 8DDF, 8DDG and CCDC 2168013]. For ESI and crystallographic data in CIF or other electronic format see https://doi.org/10.1039/d2sc02531k |
| ‡ Denotes equal contribution. |
| This journal is © The Royal Society of Chemistry 2022 |