A ‘smart’ aptamer-functionalized continuous label-free cell catch–transport–release system†
Bozhen
Zhang‡
a,
Canran
Wang‡
a,
Yingjie
Du
a,
Rebecca
Paxton
b and
Ximin
He
 *a
*a
aDepartment of Materials Science and Engineering, University of California, Los Angeles, CA 90095, USA. E-mail: ximinhe@ucla.edu
bUniversity of Arizona College of Medicine – Phoenix, Phoenix, AZ 85004, USA
First published on 1st July 2021
Abstract
Label-free cell sorting devices are of great significance for biomedical research and clinical therapeutics. However, current platforms for label-free cell sorting cannot achieve continuity and selectivity simultaneously, resulting in complex steps and limited reliability. Here, an immunoaffinity-based cell catch–transport–release thermo-chemo-mechanical coupling hydrogel (iCatch) device is reported. It contains a temperature-responsive hydrogel that can generate spatial movement synergically with the reversible binding of affinity handle modified. The functionalized hydrogel is embedded inside a biphasic microfluidic platform to enable cell transportation between the flows. The cell sorting capability and biocompatibility of the iCatch device were validated with CCRF-CEM cells as a proof-of-concept, and CCRF-CEM-specific aptamers with thermo-responsive affinity as well as a hydrogel with temperature-dependent volume were employed accordingly. A cell catching efficiency of ∼40% and a recovery rate of ∼70% were achieved. The iCatch device provides a high-throughput (∼900 cells mm−1 s−1) platform for cell sorting and is ultimately valuable for downstream biomedical applications.
1. Introduction
Recent advances in microfluidics have further enabled cell separation to take place in simple miniaturized devices for applications such as regenerative medicine,1,2 cancer research,3,4 and clinical therapeutics,5,6 where sorting and isolating target cells from heterogeneous samples are fundamental for subsequent analysis and culture. While there have been a few cell separation approaches used in practice with microfluidic devices, label-free cell sorting has become especially attractive. Label-free cell sorting exploits different cell phenotypical properties and has become a preferential choice because it avoids the high cost of cell labelling and minimizes cell damage.7–9Typically, a physical process-based label-free cell separation relies on internal fluid dynamics forces10–12 and external forces, such as filtration,13 centrifugal,14 acoustophoretic,15 magnetophoretic,16 and dielectrophoretic forces,17,18 to separate target cells. However, these approaches often require microfluidic devices with dedicated designs and complex fabrication procedures, resulting in limited reconfigurability.7 Additionally, most of these methods have limited selectivity due to the variance in physical properties among the targeted cells.19
In contrast to physical process-based methods, immunoaffinity-based methods rely on chemical interactions between affinity ligands (antibodies or aptamers) and cells to separate cells, which have been reported to have higher selectivity.20,21 Based on affinity-ligand-functionalized materials, these methods include cell affinity chromatographs,22,23 pseudo-chromatographic methods24,25 and two-phase partitioning.26,27 However, these designs require elution or releasing steps28 (e.g., nucleases,29,30 complementary strands,31,32 and electrochemical reduction33) that are spatially and temporarily separated from the catching step. This discontinuity limits their convenience and efficiency.34,35 Currently, few reports have successfully achieved the continuous separation and collection of target cells through an immunoaffinity-based method.
Recently, continuous cell separation has received much attention due to lower cost, convenient quality control, and scalability.36,37 Notably, compared with intermittent cell separation, continuous separation can release cells in situ following the catching step and allow cells to stay in the same environment with little changes in the surrounding chemical composition, which is crucial for retaining the cell functionality. These methods are also promising candidates for rapid, simple, and high-throughput cell separation due to the continuous separation process. Thus, a continuous cell separation device with high selectivity and high throughput is demanded.
‘Smart’ stimuli-responsive materials can respond to specific signals by changes in their mechanical and chemical properties, which makes them promising materials in biomedical applications.38,39 By utilizing ‘smart’ materials synchronized with affinity handles that respond to the same thermal, chemical or mechanical stimuli, we can generate spatial movements and cargo catch–transport–release simultaneously. Thus, the transportation and releasing processes are integrated, making the process more streamlined without complex instruments and time-consuming processes. In our previous work, we reported a ‘smart’ microfluidic system consisting of aptamers and a responsive hydrogel with microscopic fins that achieved chemo-mechanical modulation for continuous biomolecule separation.40 Similar devices can be employed for controllable continuous inlet separation and spatial cell transportation. However, to expand the capacity of the platform from separating nm-sized proteins to micron-sized cells, the device should be modified for higher binding affinity via enlarging the contact surface and optimizing the flow rates in the microfluidic device.
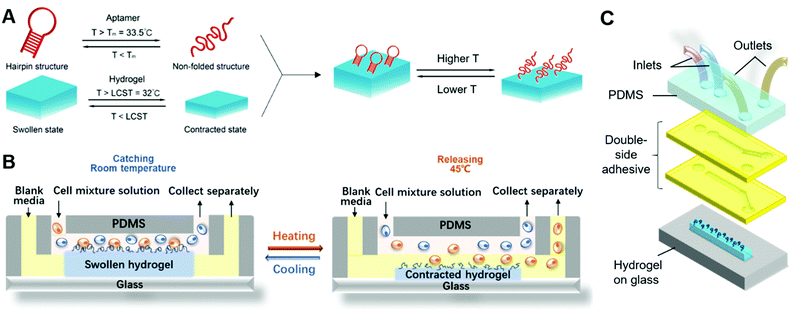
Here, we report an immunoaffinity-based cell catch–transport–release thermo-chemo-mechanical coupling hydrogel (iCatch) device that is fabricated by an equipment-free method from a ‘smart’ hydrogel and affinity handles that are synchronized to respond to temperature changes and realize a sequential and autoregulated target cell sorting and transportation between fluids. The captured cells can be transported and released in situ through the thermally triggered actuation of the ‘smart’ hydrogel-based material, resulting in sample-in-answer-out continuous cell separation. The microfluidic system contains three biocompatible components: (1) microfluidic channels that generate two parallel laminar flows for target cell transportation, (2) dynamic affinity handles that can reversibly and responsively catch and release target cells, and (3) a responsive hydrogel that exerts controllable actuation to transport target cells. In our system, the aptamers, known for high selectivity and scalable production, are employed as the dynamic affinity handles due to their capability to catch and release cells in response to temperature changes reversibly. As the stimuli-responsive hydrogel loaded with aptamers generates thermo-triggered volume changes, it will transport target cells from one stream (upper) to another (lower). The crucial factor for realizing target cell catch–transport–release is that the cell catching and releasing by the aptamer are synchronized with the volume changes of the hydrogel. To achieve this, both the functionalization and denaturation of the aptamers and the swelling and contraction of the hydrogels are designed to be regulated by temperature changes.
To demonstrate the efficiency of our system, we selected the leukemia CCRF-CEM cell line as the model target. Lymphoblastic leukemia is a hemopathy characterized by abnormal white blood cells that occurs in the blood and bone marrow.41,42 Due to its malignancy, early detection of leukemia cells is required for timely diagnosis and therapy.43 Accordingly, sgc8c aptamers were selected using cell-SELEX strategies for CCRF-CEM affinity targeting the cell-membrane protein PTK7,44,45 and the poly(N-isopropylacrylamide) (PNIPAAm) hydrogel is selected for its thermo-responsive expansion and contraction.46 Our ‘smart’ cell transportation microfluidics realize selective cell binding, controllable transportation between flows, and responsive cell release. In this manner, we developed a controllable thermo-chemo-mechanically modulated microfluidic device for label-free and continuous CCRF-CEM cell sorting with a facile fabrication process.
2. Results and discussion
2.1 Working principle and design of the iCatch device
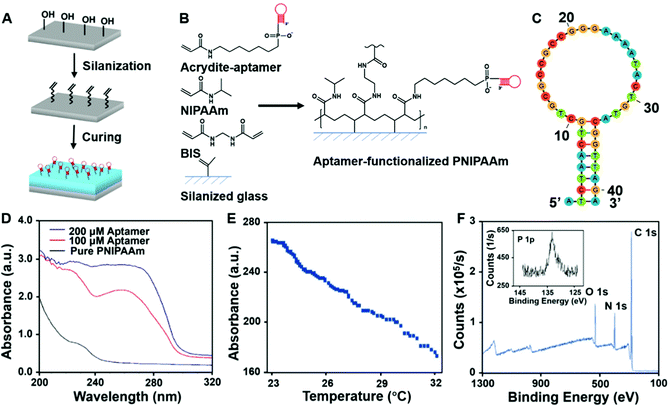
To achieve immunoaffinity-based cell catch and release, a well-designed responsive material and a matching microfluidic system are required. Our responsive material is a PNIPAAm hydrogel functionalized with sgc8c aptamers (TTT TTT ATC TAA CTG CTG CGC CGC CGG GAA AAT ACT GTA GGG TTA GAT). The PNIPAAm is a commonly used thermo-responsive hydrogel that switches from the hydrophilic swollen state to the hydrophobic contracted state when the temperature is increased above its lower critical solution temperature (LCST) of ∼32 °C.47–49 The sgc8c aptamer adopts a hairpin structure that is capable of binding to CCRF-CEM cells by recognizing specific membrane proteins (Fig. 2C) at temperatures below its melting point (Tm = 33.5 °C).50 However, when the temperature increases above Tm, the aptamer will be temporarily denatured and lose its ability to bind to cells. Based on the matching responsiveness and similar critical temperatures of the hydrogel and aptamer, these two selected components can respond to temperature changes synergistically (Fig. 1A). At RT, the functionalized hydrogel swells, and the cells attach to the nearby aptamers on the hydrogel surface. When the temperature increases to be above their common critical temperatures, the material shrinks and the cells are released due to aptamer denaturation. This synergic response makes it possible for the material to transport target cells from one site to another. Thus, in a typical catch–transport–release process, the sample is first pumped into the upper microfluidic channel and the target cells are captured by the aptamers on the swollen hydrogel at room temperature. Then, warm buffer is pumped into the lower channel, making the hydrogel contract and release the captured cells into the lower channel. The processed sample can be retrieved from the upper channel and the target cell solution is collected from the lower one.Unlike our previous work on a pH-responsive protein-catching hydrogel,40 the iCatch system is designed to catch cells rather than proteins, therefore requiring higher binding affinity due to larger cargo volume. To address this challenge, we fabricated the hydrogel as a non-structured thin film directly functionalized with aptamers instead of using aptamer-carrying epoxy microfins mounted on the hydrogel. This design endows our ‘smart’ hydrogel with increased contact area for cells as well as continuous interactions with multiple proteins on the cell membrane. Such a simplified design is more feasible for catching cells than proteins because cells are large enough to avoid being absorbed into the pores of a non-structured hydrogel. Additionally, it accelerates the response of the material due to the removal of the non-responsive epoxy. Another difference between the iCatch system and the protein-catching system is that a thermal stimulus is used instead of a pH stimulus. This stimulus is more advantageous for remote delivery and applicable to more cargos (e.g., cells) without changing their chemical environment.
For the microfluidic system, we used a straightforward two-channel design. The design is illustrated in Fig. 1C. Two channels with distinct inlets and outlets are placed above the hydrogel on a glass substrate. The mixture solution is pumped into the upper channel, while the sorted CCRF-CEM cells flow out from the lower channel. The channels are simply formed from laser-cut double-sided tapes, which makes the system highly cost-efficient and reconfigurable. Laminar flows in the channels are guaranteed by controlling the flow rates. In a typical cell sorting process (Fig. 1B), we begin with the swollen hydrogel with its top surface in the upper channel at room temperature, catching the targeted cells flowing in the mixture solution. Warm buffer is then pumped into the lower channel, heating the hydrogel to a temperature above its critical temperature. The hydrogel shrinks so that its top surface is immersed in the lower channel. The aptamers are denatured, and the cells are released into the channel due to the warm buffer. The aptamers do not contact the warm buffer in the lower channel and denature before the hydrogel shrinks, which prevents the captured cells from being released to the upper channel (detailed discussion on the responding sequence of the aptamer and hydrogel can be found in Note S1, ESI†). The sorted cells and residue solution are collected separately from two outlets following a Y-shaped junction.
In cell catching of the microfluidic system, the mild shear force provided by the microfluid also assists with the cell attachment on the hydrogel, which resembles the cell attachment of leukocytes on vascular endothelial cells during leukocyte recruitment to inflamed tissues.51,52 In this process, the shear forces result in the rolling and deformation of the recruited cells, which enhance the contact area with the targeted surface and the probability of forming multiple bindings between the cells and aptamers. All of these factors lead to sufficient interactions between the cells and the functionalized hydrogel surface. Our previous simulation study on the catching and releasing behaviour of aptamers immobilized on hydrogels in fluid further confirms our design.53
To determine whether the aptamers were successfully attached to the hydrogels, UV absorption experiments were conducted. The UV absorption spectra of the pure PNIPAAm and hydrogel synthesized with different concentrations of acrydite-functionalized aptamers are shown in Fig. 2D. The features at 260 nm, which can be attributed to the DNA only present in the spectra of functionalized hydrogels, confirm that the functionalization was successful.54
The UV absorption results indicate the chemical composition in the bulk material, while the material surface is more crucial for cell–material interactions. Therefore, XPS was further conducted to acquire information on the surface elemental composition of the material. The XPS results of the functionalized hydrogel and pure PNIPAAm are shown in Fig. 2F and Fig S1 (ESI†). Both spectra contain C 1s, N 1s, and O 1s signals, while the phosphorous signal from the phosphate group in the DNA was only detected in the functionalized hydrogel. This confirms that there were exposed aptamers on the surface of the aptamer-functionalized material.
To characterize the geographic response of the hydrogel to thermal stimuli, we assessed its temperature-induced volume change (Fig. S2, ESI†). Upon a sudden heating up (from room temperature to around 32 °C), the thickness of the gel decreased rapidly, while it slowly recovered to the original state when the temperature gradually decreased (Video S1, ESI†). The relationship between the hydrogel thickness and temperature is demonstrated in Fig. 2E, showing a maximum shrinkage of 37% at a rate of 12 μm °C−1. This fast response shows the high sensitivity of the hydrogel part of the device.
2.2 Evaluation of iCatch capability of selective cell catch–transport–release
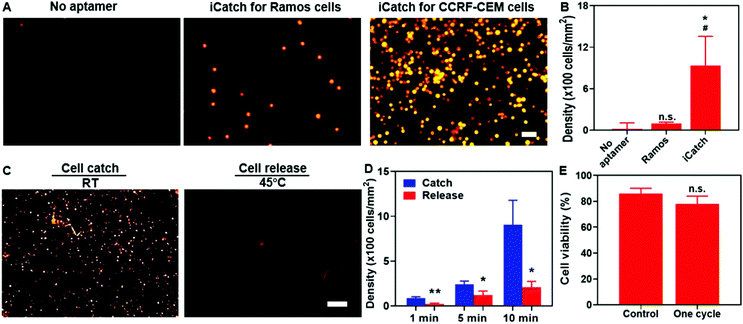
For affinity-based microfluidic cell sorting, non-specific interactions with the device components should be avoided. The PNIPAAm surface is known for cell adhesion after co-culturing for a long time.54,55 We anticipated that the porous hydrogel beside the aptamers would not trap cells in minutes during cell separation. Indeed, when we incubated cells on the surface of PNIPAAm without and with sgc8c aptamer modification for 10 min, the CCRF-CEM cell attachment was only observed in the latter group (Fig. 3A). Compared with 20 ± 12 cells per mm2 for bare PNIPAAm, the cell density on the aptamer-modified hydrogel could achieve 905 ± 273 cells per mm2, indicating the capacity of the aptamer-functionalized hydrogel for high-throughput cell catching (Fig. 3B). Apart from preventing non-specific interactions between the cells and the hydrogel, realizing certain selectivity among different cell species is required to achieve cell sorting. We confirmed the selectivity of our system by using RAMOS cells that have no affinity with sgc8c aptamers as the control group. As shown in Fig. 3A and B, after incubation, the captured Ramos cells were at a density of 98 ± 21 cells per mm2. The captured cell density for the CCRF-CEM cells was 10 times higher than that for the Ramos cells, demonstrating the cell-specific affinity and selectivity of our system.We verified that the selectivity of cell catching was attributed to sgc8c aptamers immobilized on the hydrogel by introducing scrambled aptamers with acrydite linkers for immobilization and sgc8c aptamers without acrydite linkers to the PNIPAAm hydrogel. The cell affinities of each group were defined as the density of cells remaining after incubating and washing. Our experimental group, utilizing sgc8c aptamers with acrydite linkers, exerted a significantly higher cell density than the group utilizing scrambled aptamers (60 ± 50 cells per mm2) and the group without acrydite linkers (20 ± 13 cells per mm2) (Fig. S3, ESI†). Collectively, these results demonstrated the importance of covalent aptamer attachment and cell-specific designs in catching cells with high selectivity and efficiency.
To evaluate the performance of the aptamers in thermo-responsive cell ‘catch–transport–release’, we first conducted experiments to assess the cell density changes on the aptamer-functionalized hydrogel at various time points (1, 5, and 10 min) when the temperature increased to 45 °C from room temperature (RT). After one-min incubation and 30 s wash at 45 °C, cell release was quantified. It was evident that the cell release was triggered at an increased temperature (Fig. 3C). The quantification in Fig. 3D indicates that the cell density after releasing was 207 ± 65 cells per mm2 and thus the recovery rate of our system reached 77.2%. To confirm that the cell release was not caused by the mechanical force generated by the flow, we washed the aptamer-functionalized hydrogel at room temperature (RT) first and then subsequently at 45 °C. It was observed that most cells were washed away at elevated temperatures (Fig. S4, ESI†), further confirming the thermo-responsive cell releasing capability. Additionally, both the catching of cells in the flow and the remaining cells after washing at RT indicated that the binding affinity between the aptamer and cells could be maintained in spite of the shear force generated from the flow (Fig. S4, ESI†). We also observed that only 25 ± 17 and 18 ± 9 cells per mm2 remained attached to the pure PNIPAAm hydrogel at RT and 45 °C, respectively (Fig. S5, ESI†), suggesting the limited non-specific interaction with the hydrogel in the whole process. Notably, one cycle of the catch–transport–release process was finished in 12 min. The ability for rapid and high-throughput cell sorting of our simply controlled device makes it suitable for dealing with large samples.
Maintaining biocompatibility during cell separation is also required for preventing cell damage before further analytical applications. It is essential to test whether the short-term temperature elevation and aptamer affinity influence cell viability. We characterized the cell conditions by performing the live/dead staining assay. It was observed that after flowing through the channel, 86 ± 4% of total cells were alive in the buffer. Particularly, 78 ± 6% of cells remained alive after one “catch–transport–release” cycle, suggesting the excellent biocompatibility of our system (Fig. 3E). Taken together, these results proved the capability of our aptamer–hydrogel system in selective cell ‘catch–transport–release’ in response to temperature change with good compatibility. It is worth noting that thermo-responsive aptamers could realize cell catching at RT and release at 45 °C, synchronizing with the swelling and contraction of the hydrogel, respectively.
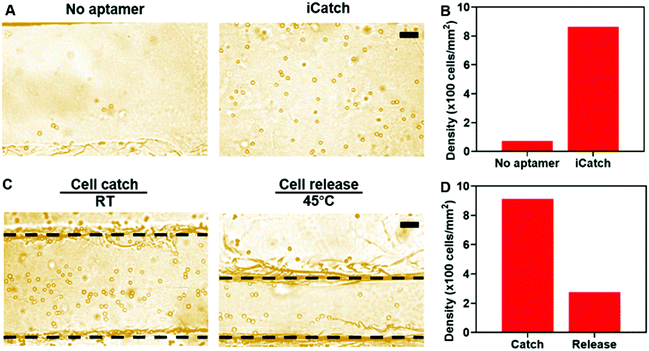
We applied the aptamer–hydrogel system to a microfluidic chip to investigate the cell sorting capability of our whole device by first pumping cells at the concentration of 5 × 106 mL−1 in the device. The non-specific interaction is a common issue that should be dealt with in cell sorting devices. As shown in Fig. 4A, this aspect was not presented in the control group, where the hydrogel inside the microfluidic device was not modified with aptamers, suggesting that the affinity is mediated by aptamers. The quantitative characterization further confirmed the specific interaction between the cells and the iCatch device. Compared with the iCatch device, where the cell density reached 863 cells per mm2, the non-aptamer modified group only caught cells at a density of 72 cells per mm2, which is consistent with our results in the hydrogel material with and without the aptamer (Fig. 4B).
Then, the efficacy of the thermo-chemo-mechanical modulated cell separation device was assessed. As shown in Fig. 4C and Video S2 (ESI†), when warm buffer was pumped into the device, the hydrogel contracted and generated spatial movement. At the same time, the captured CCRF-CEM cells were released from the hydrogel in a few minutes, demonstrating the thermo-chemo-mechanically modulated cell separation with high efficiency. After releasing, the density of cells remaining in the device was decreased to 272 cells per mm2 from 915 cells per mm2 with a 70% recovery rate approximately (Fig. 4D). Accordingly, the throughput of the iCatch device could be calculated as 888 cells mm−1 s−1. Additionally, the throughput of cell sorting could be further optimized by increasing the hydrogel surface area and aptamer modification density. We also tested the limit of detection (LOD) of the iCatch device. It was calculated from Fig. 4B that 3% of the cells could be caught in the device without the aptamer, which was set as the threshold of detection. A series of experiments were then conducted with cell concentrations ranging from 10 to 5 × 106 cells per mL. It was observed that the LOD of the iCatch device could reach 100 cells per mm2. The LOD correlates with those of many previous reports on sgc8 aptamer-mediated CCRF-CEM cell detection, indicating that our design maintained the selectivity and sensitivity of the aptamer as the key catching agent.56,57
In summary, we have developed a miniature iCatch device for cell sorting integrated with thermo-responsive aptamer catch-release and PNIPAAm hydrogel swelling and contraction to realize the target cell spatial movement in a synergistic way. It is observed that the iCatch device shows high throughput (888 cells mm−1 s−1), efficiency (39.4% in 15 s) and effectiveness (>70% recovery rate) on CCRF-CEM cell sorting.9,58 Due to the continuous manner of the cell catch–transport–release process, the cell solution which has been processed once could be pumped back into the microfluidic device and be processed repeatedly. The cycling enabled by the continuity could further increase the catching efficiency. Besides, the efficiency could be improved by altering the concentration of the aptamers and the cell solution. The recovery rate can be further enhanced by tuning the response rate, releasing time and flow rate of the warm buffer. The reversibly responsive components, the aptamer and PNIPAAm hydrogel, in the iCatch device work synergistically, enabling the continuous and cyclable label-free cell sorting and guaranteeing the high selectivity at the same time. This synergism also integrates the transport and release processes, which shortens the operation time. Also, the microfluidic design endows the device with high throughput. The facile fabrication of the iCatch device can reduce the cost for scaling-up as well. These advantages address many challenges in cell sorting, particularly when dealing with large amounts of samples. Compared to that of the high-throughput continuous physical process-based methods,59,60 such as inertial based ones, the selectivity of our method is higher owing to the immunoaffinity-based mechanism. Compared to other stimuli-responsive continuous immunoaffinity-based methods, such as electric-field-based ones,61,62 the iCatch device shows higher throughput and its fabrication is simple. Though its one-cycle catching efficiency is limited, the separation rate could be enhanced by increasing the number of cycles. Additionally, compared with triggers such as electric fields and pH changes, regulating the responsive materials with temperature is preferred due to less alternation in cell properties such as membrane electric potential.7,63 We proved that the isolated cells from the iCatch device can maintain their viability, which is essential for downstream research and clinical applications.
Although we tailored an iCatch device for CCRF-CEM cell sorting as a proof-of-concept, realizing a high catching efficacy (∼900 cells per mm2), it is easy to design aptamer sequences using cell-SELEX strategies and optimize fabrication parameters (aptamer concentration and hydrogel volume) to serve the demands of sorting various cells and reaching even higher efficiency. For instance, in future applications, the iCatch device can be adjusted to extract T-cells from peripheral blood mononuclear cells (PBMCs) for immunotherapy preparation.64 It can also be integrated into other sample-in-answer-out microfluidic platforms as an independent module. Additionally, with the advancement in SELEX strategies, the iCatch device applications could be expanded. Compared to antibodies, aptamer–cell interactions depend on cell surface properties rather than specific protein markers. They enable the distinguishment of even a small subset of cells for the early detection of disease.65,66 With the merits of ease of use, high-throughput, and scalability, we conclude that our iCatch device provides a powerful cell sorting platform that will benefit broad applications ranging from biomedical research to clinical diagnostics and therapeutics.
As a proof-of-concept study, thermal-regulated continuous and high-throughput cell separation has been achieved in this work. To further understand the microscopic process of catch–transport–release, more computational simulations could be useful. Due to the separation of catching and releasing sites, the target cells can be collected while still remaining in an environment similar to the original mixture. This makes it possible to conduct continuous rounds of cell separation of one mixture to further increase the separation efficiency and maintain the health and functionality of the cells. In the future, iCatch devices can also be connected in series to separate different cells sequentially and continuously from a practical mixture of interest such as body fluids with more complicated components.
3. Experimental
3.1 Chemicals
3-(Trimethoxysilyl)propyl methacrylate (Sigma M6514), N-isopropyl acrylamide (NIPAAM), N,N′-methylenebisacrylamide (BIS), N,N,N′,N′-tetramethylethylenediamine (TEMED) and ammonium persulfate (APS) were purchased from Sigma-Aldrich. Sgc8c aptamers and scrambled aptamers were purchased from Integrated DNA Technologies. Polydimethylsiloxane (PDMS) (Dow-Sylgard 184) was purchased from Ellsworth.3.2 Surface modification of glass
We used glass slides as the substrate for synthesizing the hydrogel. The glass substrates were modified by silanization, which was inspired by a previous reported method.67 Glass cover slides were cleaned by ultrasonication for 30 min with a soap solution (ALconox) and acetone. Then, the slides were washed with ultra-pure water and air dried. Afterwards, they were immersed in a silanization solution which was prepared by diluting 1 mL of 3-(trimethoxysilyl)propyl methacrylate in 200 mL ethanol. Prior to immersion, 6 mL of an acetic acid solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 glacial acetic acid: water) was added to the solution. The glass slides were then incubated for 1 h, until the reaction was terminated by washing them with ethanol followed by water.
10 glacial acetic acid: water) was added to the solution. The glass slides were then incubated for 1 h, until the reaction was terminated by washing them with ethanol followed by water.
3.3 Synthesis of aptamer-functionalized hydrogel
A precursor solution was prepared by dissolving 70.0 mg of NIPAAM, 1.5 mg of BIS and 5.0 μL of TEMED in 600 μL demineralized water and purging the solution for 30 min in a water/ice bath. Afterwards, 400 μL of 500 μM aptamer (or 200 μL of 500 μM aptamer and 200 μL demineralized water for hydrogel with low aptamer concentration for characterization) and 13.3 μL of 0.5 M APS were added to initiate the reaction. The solution was mixed and transferred to a Petri dish in 5 μL aliquots. Silanized glass slides and cover slides serving as spacers were put on top of the solution droplets. The solution was then left to cure for 2 hours at room temperature, and aptamer-functionalized gels attached to glass slides were obtained. After the glass slides were peeled off, the hydrogels were washed for two days in 1× TBE (Tris/Borate/EDTA, pH = 8) to remove the unreacted components.3.4 Microfluidic device fabrication
The microfluidic device was composed of 4 layers: the bottom layer of responsive hydrogel on glass, the top layer of flat PDMS, and two layers of double-sided adhesive tapes with a laser-cut pattern in between. The rectangular hydrogel (0.5 mm × 8 mm) with the aptamer was synthesized by using the method mentioned above, on top of which the patterned adhesive tapes were placed. There were rectangular channels with the same size as the hydrogel on both tapes. Besides them, there were distinct inlet and outlet holes, allowing 2 independent paths for fluids. To collect two separate solutions from these paths, the microfluidic channel was designed to branch into two outlets with a Y-shaped junction, diverting the fluids to distinct collection outlets. Subsequently, the channels and holes were capped with a PDMS sheet integrated with PDMS tubing, allowing the inlet of fluid from syringe pumps and the outlet to collection devices. The calibrated flow rates of the pumps were used to define and maintain the laminar flows of the two layers of fluids.3.5 Chemical characterization
UV absorption spectroscopy of the thermo-responsive materials with 2 different aptamer concentrations was performed with a plate reader (Multiskan SkyHigh, Thermo-Fisher, USA). Pure PNIPAAm was used as a reference. X-ray photoelectron spectroscopy of the hydrogel was performed using an XPS spectrometer (Thermo K-alpha XPS, Thermo-fisher, USA, beam size 400 μm) after it was dried.3.6 Thermo-responsive swelling test
To investigate the thermo-responsivity of the functionalized PNIPAAm hydrogel, the hydrogel swelling at different temperatures was monitored by confocal microscopy (SP5 TCS confocal microscope, Leica, Germany, 10× objective). Fluorescent spheres with a diameter of 1 μm were added to the sample to ease the visualization of the gel. The sample was immersed in hot water and the temperature was allowed to equilibrate to room temperature, while the temperature was measured with a thermocouple simultaneously. In this process, the thickness of the gel was monitored by capturing the z-stacks of a specific part of the gel every 60 s. A temperature change cycle approximately lasted 10 min. The experiment was also repeated by adding hot water to the swollen hydrogel. Fluorescence images were collected with an excitation wavelength of 488 nm and an emission range of 500–570 nm. Each z-stack contained 270 slices with 1 μm spacing and was acquired with a Leica DMI#3000.3.7 Cell culture and staining
CCRF-CEM cells (CCL-119, ATCC, USA) and Ramos cells (CRL-1596, ATCC, USA) were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1.2% Penn Strep antibiotic. Cell solutions were stored at 37 °C in a humidified incubator containing 5% CO2. Prior to the cell experiments, cells were suspended to obtain a cell concentration of 5 × 106 cells per mL. Here, a cell binding buffer was employed since normal cell media contain nucleases that are potentially harmful to the aptamers. For cell visualization, samples were labeled with Vybrant dye (DiL, Invitrogen, USA) according to the protocol provided by the manufacturer.3.8 Cell catch-and-release experiment of hydrogels
For cell catch experiments, samples were incubated with the CCRF-CEM cell solution (5 × 106 cells per mL) at room temperature for varying amounts of time (1, 5 and 10 min). Afterwards, unbound cells were removed by placing 50 μL of the cell binding buffer on top of each hydrogel and shaking the sample at 90 rpm for 30 s in an orbital shaker. The remaining attached cells were imaged with an inverted fluorescence microscope (Olympus IX71, Munday Scientific, USA) and counted manually. For cell release, samples were incubated at 45 °C for 1 min in the oven. Subsequently, the cell catch was converted to cell release by adding 50 μL of cell binding buffer at a temperature of 45 °C. The detached cells were gently removed by shaking the sample at 90 rpm for 30 s and the remaining attached cells were imaged using an inverted microscope.3.9 Live/dead staining
Staining for cell viability assessment was performed with the Live/Dead Viability/Cytotoxicity kit (Invitrogen, USA) according to the protocols provided by the manufacturer. After incubating for 30 min, the samples were washed with DPBS gently and detected using a fluorescence microscope at the wavelengths of 488 nm and 544 nm.3.10 Cell catch-and-release experiment on the iCatch device
At RT, 100 μL of cell solution (2% (vol/vol) fetal calf serum in PBS, 5 μL min−1) was pumped into the top layer at the same time as 100 μL of buffer (20 μL min−1) was pumped into the bottom layer. The cell solution eluted from the top layer was collected and re-pumped into the channel for 10 cycles, until 45 °C buffer (20 μL min−1) was pumped into the bottom layer to collect the released cells that were captured by the hydrogel surface. The catching efficiency and the cell recovery rate of the iCatch device were calculated as follows:| Efficiency% = [total number of cells caught on the device/(flow rate of cells × cell solution flowing time)] × 100% |
| Recovery rate% = [1 − (total number of cells remaining on the device after releasing)/(total number of cells on the device before releasing)] × 100% |
Throughput = two-dimensional density of captured cells × velocity (derivation of this expression can be found in Note S2, ESI†).
3.11 Video imaging and cell quantification
To quantify the cell “catch–transport–release” properties of the iCatch device, the whole process was imaged with movies taken with a camera (Olympus IX71, Munday Scientific, USA). The cells were quantified using the ImageJ program (National Institute of Health, USA).Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was supported by the NSF CAREER award 1724526.References
- P. Zhang and A. R. Abate, Adv. Mater., 2020, 32, e2005346 CrossRef PubMed.
- R. N. Judson, M. Low, C. Eisner and F. M. Rossi, Methods Mol. Biol., 2017, 1668, 93–103 CrossRef CAS PubMed.
- S. C. Hur, J. Che and D. Di Carlo, Methods Mol. Biol., 2017, 1634, 65–79 CrossRef CAS PubMed.
- N. Sun, X. Li, Z. Wang, Y. Li and R. Pei, Biosens. Bioelectron., 2018, 102, 157–163 CrossRef CAS PubMed.
- M. Labib, Z. Wang, S. U. Ahmed, R. M. Mohamadi, B. Duong, B. Green, E. H. Sargent and S. O. Kelley, Nat. Biomed. Eng., 2021, 5, 41–52 CrossRef CAS PubMed.
- T. N. G. Adams, A. Y. L. Jiang, N. S. Mendoza, C. C. Ro, D. H. Lee, A. P. Lee and L. A. Flanagan, Biosens. Bioelectron., 2020, 152, 111982 CrossRef CAS PubMed.
- R. Nasiri, A. Shamloo, S. Ahadian, L. Amirifar, J. Akbari, M. J. Goudie, K. Lee, N. Ashammakhi, M. R. Dokmeci, D. Di Carlo and A. Khademhosseini, Small, 2020, 16, e2000171 CrossRef PubMed.
- S. Zhang, Y. Chen, H. Liu, Z. Wang, H. Ling, C. Wang, J. Ni, B. Çelebi-Saltik, X. Wang, X. Meng, H.-J. Kim, A. Baidya, S. Ahadian, N. Ashammakhi, M. R. Dokmeci, J. Travas-Sejdic and A. Khademhosseini, Adv. Mater., 2020, 32, 1904752 CrossRef CAS PubMed.
- H. Tavassoli, P. Rorimpandey, Y. C. Kang, M. Carnell, C. Brownlee, J. E. Pimanda, P. P. Y. Chan and V. Chandrakanthan, Small, 2021, 17, e2006176 CrossRef PubMed.
- Z. Liu, R. Chen, Y. Li, J. Liu, P. Wang, X. Xia and L. Qin, Adv. Biosyst., 2018, 2, 1800200 CrossRef.
- X. Zhang, Z. Zhu, N. Xiang, F. Long and Z. Ni, Anal. Chem., 2018, 90, 4212–4220 CrossRef CAS PubMed.
- J. Feng, J. Mo, A. Zhang, D. Liu, L. Zhou, T. Hang, C. Yang, Q. Wu, D. Xia, R. Wen, J. Yang, Y. Feng, Y. Huang, N. Hu, G. He and X. Xie, Nanoscale, 2020, 12, 5103–5113 RSC.
- T.-C. Sung, W.-L. Huang, L.-K. Ban, H. H.-C. Lee, J.-H. Wang, H.-Y. Su, S. H. Jen, Y.-H. Chang, J.-M. Yang, A. Higuchi and Q. Ye, J. Mater. Chem. B, 2020, 8, 10577–10585 RSC.
- L. Yin, Z. Yang, Y. Wu, V. Denslin, C. C. Yu, C. A. Tee, C. T. Lim, J. Han and E. H. Lee, Biomaterials, 2020, 240, 119881 CrossRef CAS PubMed.
- P. Li, Z. Mao, Z. Peng, L. Zhou, Y. Chen, P.-H. Huang, C. I. Truica, J. J. Drabick, W. S. El-Deiry, M. Dao, S. Suresh and T. J. Huang, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4970 CrossRef CAS PubMed.
- W. Zhao, T. Zhu, R. Cheng, Y. Liu, J. He, H. Qiu, L. Wang, T. Nagy, T. D. Querec, E. R. Unger and L. Mao, Adv. Funct. Mater., 2016, 26, 3990–3998 CrossRef CAS PubMed.
- N. Sobahi and A. Han, Biosens. Bioelectron., 2020, 166, 112458 CrossRef CAS PubMed.
- A. A. Nawaz, M. Urbanska, M. Herbig, M. Nötzel, M. Kräter, P. Rosendahl, C. Herold, N. Toepfner, M. Kubánková, R. Goswami, S. Abuhattum, F. Reichel, P. Müller, A. Taubenberger, S. Girardo, A. Jacobi and J. Guck, Nat. Methods, 2020, 17, 595–599 CrossRef CAS PubMed.
- A. Lee, J. Park, M. Lim, V. Sunkara, S. Y. Kim, G. H. Kim, M.-H. Kim and Y.-K. Cho, Anal. Chem., 2014, 86, 11349–11356 CrossRef CAS PubMed.
- T. Gao, Z. Mao, W. Li and R. Pei, J. Mater. Chem. B, 2021, 9, 746–756 RSC.
- K. Bacon, A. Lavoie, B. M. Rao, M. Daniele and S. Menegatti, Acta Biomater., 2020, 112, 29–51 CrossRef CAS PubMed.
- F. Mohr, S. Przibilla, F. Leonhardt, C. Stemberger, S. Dreher, T. R. Mueller, S. P. Fraessle, G. P. Schmidt, M.-L. Kiene, H. Stadler and D. H. Busch, Sci. Rep., 2018, 8, 16731 CrossRef PubMed.
- A. Srivastava, A. K. Shakya and A. Kumar, Enzyme Microb. Technol., 2012, 51, 373–381 CrossRef CAS PubMed.
- G. Xu, Y. Tan, T. Xu, D. Yin, M. Wang, M. Shen, X. Chen, X. Shi and X. Zhu, Biomater. Sci., 2017, 5, 752–761 RSC.
- R. Saranya, R. Murugan, M. Hegde, J. Doyle and R. Babu, Affinity Membranes for Capture of Cells and Biological Substances, Springer, Cham, Switzerland, 2018 Search PubMed.
- M. Gonzalez-Gonzalez and M. Rito-Palomares, J. Mol. Recognit., 2015, 28, 142–147 CrossRef CAS PubMed.
- M. Gonzalez-Gonzalez, R. C. Willson and M. Rito-Palomares, Sep. Purif. Technol., 2016, 158, 103–107 CrossRef CAS.
- L. Wu, Y. Wang, L. Zhu, Y. Liu, T. Wang, D. Liu, Y. Song and C. Yang, ACS Appl. Bio Mater., 2020, 3, 2743–2764 CrossRef CAS.
- J. Li, C. Qi, Z. Lian, Q. Han, X. Wang, S. Cai, R. Yang and C. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 2511–2516 CrossRef CAS PubMed.
- H. Shen, J. Yang, Z. Chen, X. Chen, L. Wang, J. Hu, F. Ji, G. Xie and W. Feng, Biosens. Bioelectron., 2016, 81, 495–502 CrossRef CAS PubMed.
- N. Sun, M. Liu, J. Wang, Z. Wang, X. Li, B. Jiang and R. Pei, Small, 2016, 12, 5090–5097 CrossRef CAS PubMed.
- Y. Lin, L. Jiang, Y. Huang, Y. Yang, Y. He, C. Lu and H. Yang, Chem. Commun., 2019, 55, 5387–5390 RSC.
- T.-T. Zhai, D. Ye, Q.-W. Zhang, Z.-Q. Wu and X.-H. Xia, ACS Appl. Mater. Interfaces, 2017, 9, 34706–34714 CrossRef CAS PubMed.
- K. A. Hyun, T. Y. Lee, S. H. Lee and H. I. Jung, Biosens. Bioelectron., 2015, 67, 86–92 CrossRef CAS PubMed.
- S. Sun, S. Yang, X. Hu, C. Zheng, H. Song, L. Wang, Z. Shen and Z. S. Wu, ACS Sens., 2020, 5, 3870–3878 CrossRef CAS PubMed.
- A. C. Fisher, M. H. Kamga, C. Agarabi, K. Brorson, S. L. Lee and S. Yoon, Trends Biotechnol., 2019, 37, 253–267 CrossRef CAS PubMed.
- P. H. Chen, Y. T. Cheng, B. S. Ni and J. H. Huang, Appl. Biochem. Biotechnol., 2020, 191, 151–163 CrossRef CAS PubMed.
- X. Gong, C. Hou, Q. Zhang, Y. Li and H. Wang, ACS Appl. Mater. Interfaces, 2020, 12, 51225–51235 CrossRef CAS PubMed.
- M. Qu, X. Jiang, X. Zhou, C. Wang, Q. Wu, L. Ren, J. Zhu, S. Zhu, P. Tebon, W. Sun and A. Khademhosseini, Adv. Healthcare Mater., 2020, 9, 1901714 CrossRef CAS PubMed.
- A. Shastri, L. M. McGregor, Y. Liu, V. Harris, H. Nan, M. Mujica, Y. Vasquez, A. Bhattacharya, Y. Ma, M. Aizenberg, O. Kuksenok, A. C. Balazs, J. Aizenberg and X. He, Nat. Chem., 2015, 7, 447–454 CrossRef CAS PubMed.
- J. M. Klco and C. G. Mullighan, Nat. Rev. Cancer, 2021, 21, 122–137 CrossRef CAS PubMed.
- K. A. Foon and R. F. Todd, Blood, 1986, 68, 1–31 CrossRef CAS PubMed.
- X. Pang, C. Cui, M. Su, Y. Wang, Q. Wei and W. Tan, Nano Energy, 2018, 46, 101–109 CrossRef CAS PubMed.
- W. Niu, X. Chen, W. Tan and A. S. Veige, Angew. Chem., Int. Ed., 2016, 55, 8889–8893 CrossRef CAS PubMed.
- L. Ding, Y. Wu, Y. Duan, S. Yu, F. Yu, J. Wang, Y. Tian, Z. Gao, Z. Wan and L. He, ACS Sens., 2020, 5, 440–446 CrossRef CAS PubMed.
- Z. Chen, J. Liu, Y. Chen, X. Zheng, H. Liu and H. Li, ACS Appl. Mater. Interfaces, 2021, 13, 1353–1366 CrossRef CAS PubMed.
- L. Yang, X. Fan, J. Zhang and J. Ju, Polymers, 2020, 12, 389 CrossRef CAS PubMed.
- T. Sarwan, P. Kumar, Y. E. Choonara and V. Pillay, Front. Mater., 2020, 7, 73 CrossRef.
- K. Nagase and T. Okano, J. Mater. Chem. B, 2016, 4, 6381–6397 RSC.
- Q. Zhang, W. Wang, S. Huang, S. Yu, T. Tan, J. R. Zhang and J. J. Zhu, Chem. Sci., 2020, 11, 1948–1956 RSC.
- M. Abadier, A. B. Pramod, S. McArdle, A. Marki, Z. Fan, E. Gutierrez, A. Groisman and K. Ley, Cell Rep., 2017, 21, 3885–3899 CrossRef CAS PubMed.
- S. Okhota, I. Melnikov, Y. Avtaeva, S. Kozlov and Z. Gabbasov, Int. J. Mol. Sci., 2020, 21, 7804 CrossRef CAS PubMed.
- Y. Liu, O. Kuksenok, X. He, M. Aizenberg, J. Aizenberg and A. C. Balazs, ACS Appl. Mater. Interfaces, 2016, 8, 30475–30483 CrossRef CAS PubMed.
- Y. Wang, N. Gan, Y. Zhou, T. Li, Y. Cao and Y. Chen, Biosens. Bioelectron., 2017, 87, 508–513 CrossRef CAS PubMed.
- J. A. Reed, A. E. Lucero, M. A. Cooperstein and H. E. Canavan, J. Appl. Biomater. Biomech., 2008, 6, 81–88 CAS.
- J. Tan, Z. Lai, L. Zhong, Z. Zhang, R. Zheng, J. Su, Y. Huang, P. Huang, H. Song, N. Yang, S. Zhou and Y. Zhao, Nanoscale Res. Lett., 2018, 13, 66 CrossRef PubMed.
- X. Pang, C. Cui, M. Su, Y. Wang, Q. Wei and W. Tan, Nano Energy, 2018, 46, 101–109 CrossRef CAS PubMed.
- W. Liang, J. Liu, X. Yang, Q. Zhang, W. Yang, H. Zhang and L. Liu, Microfluid. Nanofluid., 2020, 24, 26 CrossRef.
- S. Zhu, D. Wu, Y. Han, C. Wang, N. Xiang and Z. Ni, Lab Chip, 2020, 20, 244–252 RSC.
- J. F. Edd, A. Mishra, T. D. Dubash, S. Herrera, R. Mohammad, E. K. Williams, X. Hong, B. R. Mutlu, J. R. Walsh, F. M. de Carvalho, B. Aldikacti, L. T. Nieman, S. L. Stott, R. Kapur, S. Maheswaran, D. A. Haber and M. Toner, Lab Chip, 2020, 20, 558–567 RSC.
- S. Dey, R. Vaidyanathan, L. G. Carrascosa, M. J. A. Shiddiky and M. Trau, ACS Sens., 2016, 1, 399–405 CrossRef CAS.
- S. Dey, K. M. Koo, Z. Wang, A. A. I. Sina, A. Wuethrich and M. Trau, Lab Chip, 2019, 19, 738–748 RSC.
- Y. Zhou and R. M. Raphael, Biophys. J., 2007, 92, 2451–2462 CrossRef CAS PubMed.
- K. K. Dijkstra, C. M. Cattaneo, F. Weeber, M. Chalabi, J. van de Haar, L. F. Fanchi, M. Slagter, D. L. van der Velden, S. Kaing, S. Kelderman, N. van Rooij, M. E. van Leerdam, A. Depla, E. F. Smit, K. J. Hartemink, R. de Groot, M. C. Wolkers, N. Sachs, P. Snaebjornsson, K. Monkhorst, J. Haanen, H. Clevers, T. N. Schumacher and E. E. Voest, Cell, 2018, 174, 1586–1598 CrossRef CAS PubMed.
- D. Shangguan, Y. Li, Z. Tang, Z. C. Cao, H. W. Chen, P. Mallikaratchy, K. Sefah, C. J. Yang and W. Tan, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11838 CrossRef CAS PubMed.
- J. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef CAS PubMed.
- Z. Zhang, N. Chen, S. Li, M. R. Battig and Y. Wang, J. Am. Chem. Soc., 2012, 134, 15716–15719 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Note S1 and S2, Table S1, Fig. S1–S5 and Videos S1 and S2. See DOI: 10.1039/d1tb00739d |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |