Computational thermostability engineering of a nitrile hydratase using synergetic energy and correlated configuration for redesigning enzymes (SECURE) strategy†
Jinling
Xu
a,
Haisheng
Zhou
b,
Jiaqi
Xu
 *b,
Ziyuan
Wang
a,
Zhonglang
Yu
a,
Zhe
Wang
a,
Hongyu
Zhang
ab,
Haoran
Yu
ab,
Jianping
Wu
*b,
Ziyuan
Wang
a,
Zhonglang
Yu
a,
Zhe
Wang
a,
Hongyu
Zhang
ab,
Haoran
Yu
ab,
Jianping
Wu
 ab and
Lirong
Yang
ab and
Lirong
Yang
 *ab
*ab
aInstitute of Bioengineering, College of Chemical and Biological Engineering, Zhejiang University, Hangzhou, 310027, China. E-mail: lryang@zju.edu.cn
bZJU-Hangzhou Global Scientific and Technological Innovation Centre, NO. 733 Jianshe 3rd Road, Xiaoshan District, Hangzhou, Zhejiang 311200, China. E-mail: biojqxu@zju.edu.cn
First published on 14th September 2023
Abstract
Nitrile hydratase (NHase), an excellent biocatalyst, has been widely used for the production of amides, but the exothermic hydration reaction leads to its rapid inactivation, hindering its industrial applications, which requires the thermostability of NHase to be enhanced. In this study, employing NHase from Bordetella petrii DSM 12804 (NHAB) as the object, a computational strategy using synergetic energy and correlated configuration for redesigning enzymes (SECURE) was proposed to prune the reasonable mutant library and assemble effective single mutations, thus maximizing the thermostability of NHAB. Among the mutants, the best variant, A6M/B4M, combined six mutations in its α-subunit (S30T, A71D, A74D, A78R, S81T, and A133P) and four mutations in its β-subunit (L25F, G27Y, N59P, and A173N), showing an increase in Tm by 13.2 °C, an 866.0-fold prolonged half-life at 50 °C, and an 11.2% increase in activity. Then, the catalytic efficiency dramatically increased to 249.5 g L−1 acrylamide in 5 batches compared with that of 166.5 g L−1 by the wild type in 3 batches. Finally, the synergistic effect on the mutant represented by an overall change in structure and newly formed intermolecular interactions through dynamic simulations accounted for the enhanced thermostability. Thus, SECURE was demonstrated to be practical for the redesign of multimeric NHase, which also provided guidance for computational thermostability engineering of other industrial biocatalysts.
Introduction
Presently, the application of biocatalysts in the plastic, chemical and pharmaceutical industries1–3 has significantly increased. However, native enzymes derived from mesophilic bacteria are not robust enough to withstand the harsh conditions of long-term industrial catalysis, such as high pressure, high temperature and organic solvents.4 Thus, it is necessary to reshape existing natural enzymes or mine novel enzymes from extremophiles,5 providing biocatalysts with excellent thermostability for industrial application.NHase (EC 4.2.1.84), which can catalyze the hydration of nitriles into amides, plays a crucial role in enzymatic processes in the chemical industry.6 For example, the industrial enzymatic synthesis of acrylamide has reached several hundred thousand tons per year7 and already replaced chemical synthesis owing to its environmental friendliness and easy product recovery with high purity.8 However, the poor thermostability of NHase and the exothermic hydration reaction (68.99 kJ heat per mol acrylamide generated)9,10 result in the rapid inactivation of the unstable NHase, which requires additional cooling equipment to control the reaction temperature at below 25 °C and leads to large energy consumption costs in industrial processes.11 Thus, researchers have attempted to redesign NHase to improve its thermostability and reduce the cost of biocatalysis.
Strategies, such as protein fragment swapping,12,13 modification of β-subunit terminus, subunit fusion expression,14 site-directed mutagenesis,15 design of salt-bridge cyclization peptide tags,16 introduction of molecular chaperones17 and genome editing,18 have been applied to modify the thermostability of NHase, resulting in a 1.1–3.9-fold enhancement. Among them, most studies focused on a few specific NHases with highly accurate crystal structure,19,20 such as ptNHase (NHase from Pseudonocardia thermophila JCM3095) and ppNHase (NHase from Pseudomonas putida). The thermostability of the redesigned ptNHase was enhanced by 3.5-fold21 or showed an increase in Tm by 3.2 °C,22 while ppNHase showed a 1.4–3.5-fold enhancement.23 Besides, the reported structural modification mostly targeted the β-subunit, which is deemed to play a crucial role in maintaining the thermostability of NHase.24 The mechanism responsible for the enhanced stability is a decrease in flexibility,25 leading to an activity-stability trade-off, which may occur in thermally adapted homologous enzymes.26 This phenomenon also occurred in NHase, resulting in the inability to maximize the positive effects.27
Moreover, computational design has become an inevitable trend for the thermostability engineering of proteins.28–30 Various computational tools31–33 based on protein structure and sequence analysis have been developed for the prediction of stabilized mutations. However, the hotspot mutations given by computational tools often contain invalid mutations,34,35 which still need to be further pruned. Besides, the multi-point mutations in enzyme sequences are likely to be more destabilizing or even lead to misfolding and aggregation of proteins,36–38 indicating that the effective combination of single-point mutations is significant and worth investigating.39
In this study, a computational strategy (Synergistic Energy and Correlated Configuration for Redesigning Enzymes, SECURE) was proposed to prune the potential mutations by bioinformatics analysis and effectively assemble the mutations according to their type. To explore the feasibility of this new strategy, an NHase from Bordetella petrii DSM 12804 (NHAB) was selected as the object to be redesigned for industrial use. The redesigned mutant A6M/B4M showed an increase in Tm by 13.2 °C, a prolonged half-life at 50 °C by 866.0-fold, and an 11.2% increase in specific activity, laying the foundation for the industrial application of A6M/B4M. The SECURE strategy was proven to have a synergistic effect on thermostability and catalytic activity, demonstrating its effectiveness in the engineering of NHase for industrial applications.
Results and discussion
Computational redesign of NHAB for enhanced thermostability using SECURE strategy
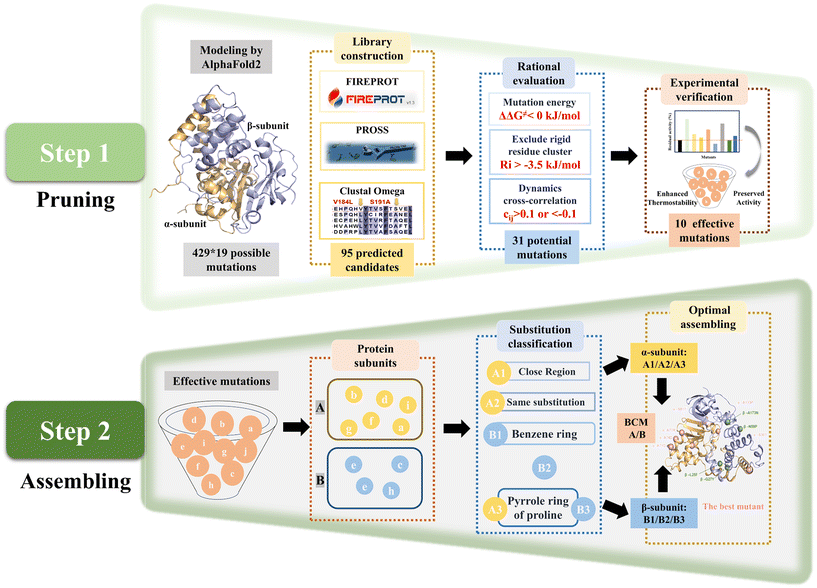
The SECURE strategy was proposed for modifying heterogeneous subunit proteins to enhance their thermostability, which involved two steps (Fig. 1). The pruning of the reasonable mutations was step 1. Firstly, a virtual library with potential stable single-point mutations was obtained by multiple strategies, and then further evaluated based on energy change, structural configuration and dynamic correlation. The unreasonable mutations that are useless to thermostability or can impair catalytic activity were eliminated to reduce the workload of the wet experiments. Based on this, a small and tailored library of single-point mutants was constructed for characterization. In step 2, the effective mutants obtained by step 1 were further assembled to achieve synergistic effects. Considering the workload of stepwise combination and the negative effects on superposed mutations, the single mutations were firstly subclassified by different subunits, and then clustered according to different mutation types to obtain combinatorial mutations. Subsequently, the combined mutants were further evaluated and assembled to obtain the final optimal mutant.NHAB, which exhibited the highest dry cell weight (DCW) crude activity of 570.3 U mg−1 toward acrylonitrile using free cells, but the worst thermostability among the high-activity strains (Table S1 and Fig. S1 and S2†), was selected as the target from 19 different NHases expressed by E. coli BL21 (DE3) to verify the feasibility of the SECURE strategy. A reliable three-dimensional structure (Fig. S3†) of NHAB was predicted by AlphaFold2.40
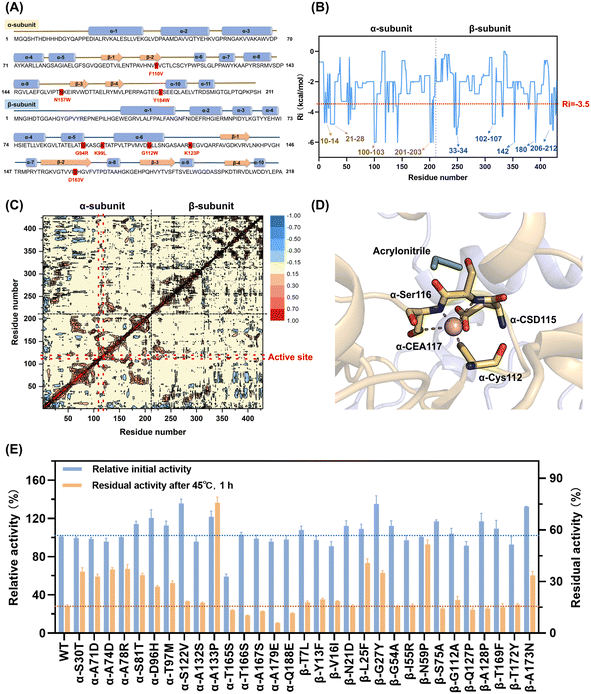
To precisely prune the effective mutant library, further rational evaluation according to the mutation energy change, configuration in structure and dynamic cross-correlation matrix (DCCM) were applied to eliminate deleterious mutations.
The mutations with free energy ΔΔG given by FIREPROT of greater than zero were regarded as unfavorable to the stability of the protein. These mutations were excluded from the library (Table S2†). Moreover, according to the configuration of NHAB, 8 mutations that would bring about steric hindrance to destroy the secondary structure and break the intermolecular interactions such as salt-bridges were marked to be filtered out (Fig. 2A). Additionally, the rigidity index (Ri) presenting a more rigid residue cluster with a lower value was calculated using CNA47 to filter out mutations in the rigid regions. Thus, 13 similar mutations with Ri values lower than −3.5 kcal mol−1 that occurred in the rigid regions were excluded (Table S2†). Finally, the DCCM analysis48 was used to explore the fluctuation correlation in NHAB and the pairwise cross-correlation coefficients (Cij) representing the degree of correlation were calculated and colored (Fig. 2C). Regions outside the active site but showing higher absolute values of Cij to the active sites were reported to be appropriate targets for stabilizing the mutations.49 Thus, the Cij between the mutation sites and the active sites (α-Cys112, α-Cys115, α-Ser116, and α-Cys117) (Fig. 2D) was used to expurgate the mutations whose Cij was between −0.1 and 0.1 (Fig. S7A†). After the rational pruning, a small and reliable mutant library including 31 single mutations was screened for further experimental verification and the Cij of selected mutation sites was also represented (Fig. S7B†).
The catalytic activity and thermostability of both the wild-type and single-point mutants were determined (Fig. 2E). Most of the mutants retained the original catalytic activity except for α-T165S, indicating the effectiveness of the rational evaluation. The residual activities after incubation at 45 °C for 1 h were assayed to characterize the thermostability, and 10 effective mutants exhibited at least a 2-fold increase. Specifically, 5 mutants from the α-subunit, including α-S30T, α-A71D, α-A74D, α-A78R and α-S81T, maintained a residual activity of 35.6%, 32.8%, 36.9%, 37.2% and 33.6%, respectively, while that maintained by the wild type was only 15.5%. Unexpectedly, both the residual activity and initial activity of A133P was 4.8-fold and 1.2-fold that of the wild type, respectively. The β-G27Y mutant also exhibited excellent thermostability with 34.9% residual activity and 135.1% relative initial activity. Besides, the other three mutants from the β-subunit, i.e., β-L25F, β-N59P and β-A173N, showed residual activity of 40.8%, 51.5% and 33.5%, which were 2.6-fold, 3.3-fold and 2.1-fold that of the wild type, respectively. Notably, in these 10 mutants, not only did their thermostability improve, also their catalytic activity did not decrease. Accordingly, the trade-off between activity and thermostability seemed to be broken in these mutants. However, to obtain the optimal mutant, these single-point mutations needed to be combined.
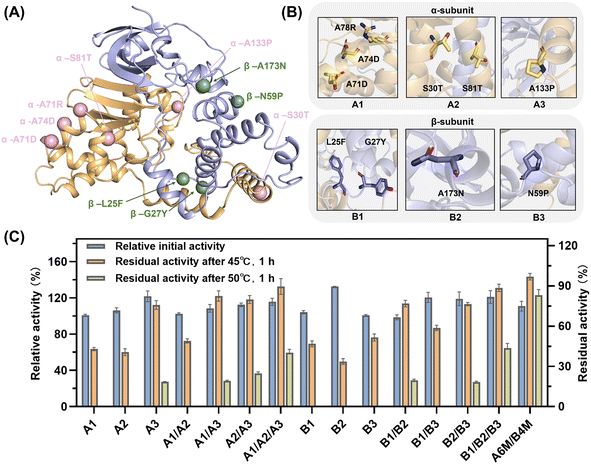
NHase is a polymer composed of two heterologous subunits. The α-subunit, where the active center is located, is usually considered to be important for catalytic activity,50 whereas molecular dynamics simulations show that the β-subunit acts mainly on the thermostability of NHase.24 Thus, the mutations distributed on the same subunit were grouped into a broad category initially (Fig. 3A). Within the same structural domain, the mutations were subdivided into different groups according to diverse amino acid substitutions. In the α-subunit, A71D, A74D and A78R were not only located on the same α-helix (Fig. 3B), but also replaced the non-polar amino acid alanine with a charged polar amino acid. Therefore, the A1 mutant was a combination of A71D, A74D and A78R. Similarly, S30T and S81T substituted with longer chain amino acids in their α-helix were assembled as the A2 mutant. Subsequently, A133P introducing a pyrrole ring was classified as a separate category, i.e., A3. For the classification of the mutations on the β-subunit, L25F and G27Y replaced by aromatic amino acids in the same loop (Fig. 3B) were combined into the B1 mutant. Besides, N59P and A173N were divided into two categories owing to the different properties of their substituted amino acids. Intra- and inter-subunit combinations were also performed. The assembled mutants exerted a superimposed effect on thermostability (Fig. 3C). In the case of the A1 mutant, it exhibited a 5.6% increase in residual activity compared to the best single mutation A78R (37.6%), which also maintained the initial enzyme activity of the wild type. The residual activities of A2, containing two threonine substitutions, and B1, consisting of L25F and G27Y, showed a similar slight increase. Intra-subunit assembly also yielded a significant enhancement. Especially, the A1/A2/A3 and B1/B2/B3 mutants, which combined all three groups, exhibited a 5.7-fold increase in residual activity at 45 °C. Moreover, when the incubation temperature was increased to 50 °C, the inactivation of the enzyme was apparently significant. The superposed mutants containing α-A133P or β-N59P still showed considerable residual activities, while the others were completely deactivated, demonstrating the crucial importance of proline substitution. A1/A2/A3 and B1/B2/B3 showed residual activity of 40.1% and 43.5%, while the inter-subunit mutant A6M/B4M (A1/A2/A3/B1/B2/B3) showed residual activity of 83.0% and 10.8% increase in catalytic activity, showing the synergy between the two subunits.
Characterization and catalytic performance of the best mutant A6M/B4M for amide production
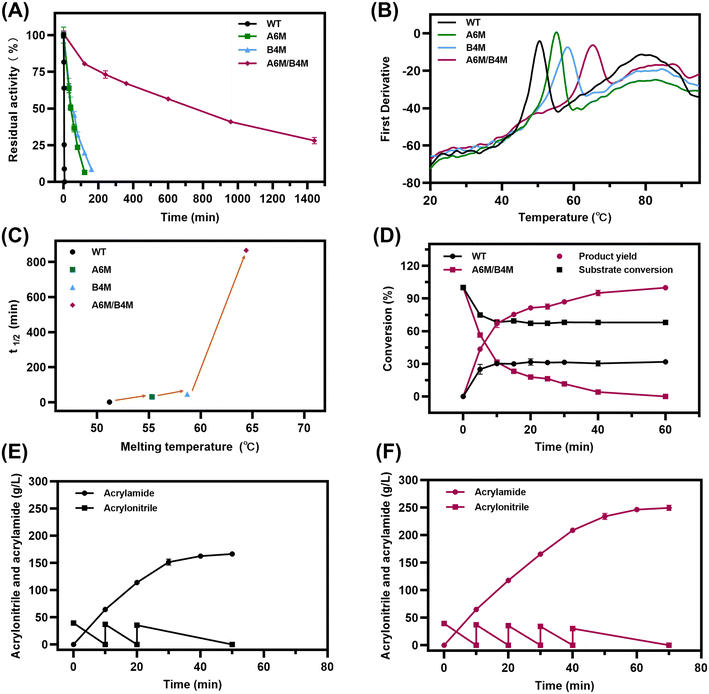
To further explore the synergy between the mutations and the practical application of the redesigned NHAB, the mutants and wild type were purified to further investigate their catalytic activity and thermostability. The expressions of crude extract and purified enzyme are presented in Fig. S8 and the specific activity of the best mutant A6M/B4M was 1697 U mg−1, which was 1.12-fold that of the wild type (Table S3†). Besides, the half-life of inactivation at 50 °C was characterized by measuring the residual activity after a period of incubation in a water bath. The wild-type NHAB was rapidly inactivated at 50 °C. Specifically, only 25.2% residual activity was detected after 3 min, and the activity was completely lost after 5 min, accounting for the extremely short half-life of 1 min. Comparatively, A1/A2/A3 and B1/B2/B3 still had the residual activity of 44.6% and 66.5% after 20 min, respectively. Surprisingly, A6M/B4M retained the residual activity of 85.4% after 2 h (Fig. 4A). A6M/B4M exhibited the best thermostability with the longest half-life of 866 min (14.4 h) at 50 °C, which was 866.0-fold longer than that of the wild type. A1/A2/A3and B1/B2/B3 also had a 30.0-fold and 46.0-fold improved half-life, respectively. Besides, the protein melting temperature Tm was also determined to characterize the thermodynamic stability of NHAB and its mutants by nano-DSF. The thermal denaturation curves of these proteins were monitored (Fig. 4B) and A6M/B4M was validated to be the most thermostable mutant with a Tm of 64.4 °C, which is 13.2 °C higher than that of the wild type (51.2 °C). Also, the Tm values of A1/A2/A3 and B1/B2/B3 were 55.3 °C and 58.7 °C, respectively. Therefore, both the Tm values and the inactivation results at 50 °C demonstrated that mutations on the α- and β-subunit synergistically played key roles in improving the thermostability of NHAB (Fig. 4C).A6M/B4M with prominently enhanced thermostability and catalytic activity exhibited great application potential. Thus, the catalytic reactions were carried out at 45 °C to test practical value of this mutant. It only took 60 min for A6M/B4M to completely transform 0.85 M acrylonitrile to acrylamide. By contrast, the reactions catalyzed by the wild type were incomplete, with a product yield of 30.7% and the reaction terminated within 10 min due to the inactivation of the enzyme (Fig. 4D). Furthermore, the industrial production process of acrylamide was simulated by fed-batch reaction. The catalytic performance of the A6M/B4M mutant was significantly better than that of the wild type (Fig. 4E and F), reaching a product concentration of 249.5 g L−1 in 5 batches, while the wild type was unable to continue the reaction after only 3 batches, ending with the concentration of 166.5 g L−1 within 50 min. Moreover, the product tolerance of A6M/B4M and WT were determined based on their residual activity after incubation in 300 g L−1 acrylamide solution for 1 h. It was observed that the wild type lost all its activity, whereas that retained by A6M/B4M was 38.4% (Table S4†). This is sufficient evidence that the product tolerance of the mutant was also improved by the stability modification, thus enabling NHase to continue the reaction at a high product concentration. The performance of A6M/B4M obtained through the SECURE strategy was remarkably improved for industrial application.
Molecular structure analysis of mutations
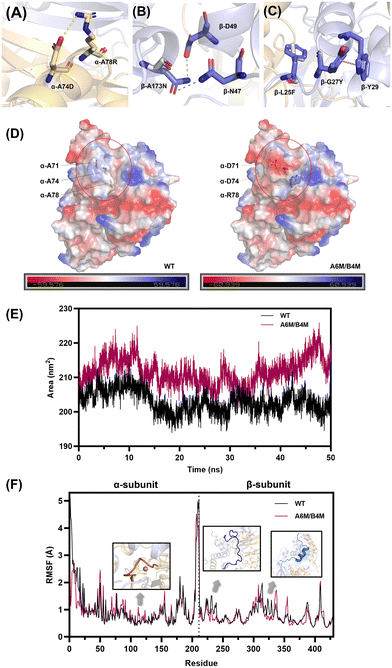
To reveal the mechanisms for the improvement of the thermostability, the intermolecular interaction and overall structural analysis of the A6M/B4M mutant and wild type were investigated by MD simulations to account for the stability of whole protein.Electrostatic interactions comprising hydrogen bonds52 and salt bridges53 were verified to significantly contribute to the protein stability. The replacement of A71D, A74D, and A78R located in the α-helix caused changes in the charge distribution on the α-subunit surface, accompanied with the formation of a new salt bridge, i.e., Asp74-Arg78 (Fig. 5A). The occupancy (O–N distance cut-off of 3.2 Å) of this salt bridge in A6M/B4M reached 74.3% with an average distance of 3.1 Å, as analyzed by the MD trajectories (Fig. S9†). A173 located in an α-helix of the β-subunit was substituted by asparagine. The amide group on the side chain shortened the distance between 173N and nearby amino acids, leading to the formation of new hydrogen bonds with D49 and N47 in the β-subunit (Fig. 5B). The probability of the existence of the two newly formed hydrogen bonds was up to 94.8% and 38.6% in the MD simulation (Fig. S9†).
Moreover, L25 and G27 located in a long loop region at the N-terminus of the β-subunit were substituted by the aromatic amino acids phenylalanine and tyrosine, leading to the formation of new aromatic interactions between nearby residues (Fig. 5C). Y27 formed a new pi-pi interaction with W29 in the α-helix and the stacked geometry was regarded as a significant contributor to stabilizing the enzyme molecule.54
Optimization of the surface charge is considered to be an effective strategy for the improvement of protein stability.55 Considering the introduction of three charged amino acids, the polarity of the α-subunit surface was significantly enhanced (Fig. 5D). Besides, the analysis of the solvent accessible surface area (SASA), a method for measuring the surface area of a protein accessible with the solvent, showed that there was an obvious increase in the SASA of A6M/B4M (Fig. 5E), indicating an increase in interaction with the hydration shell. The surface charge engineering of lipase56 proved that the hydration of the protein surface is a key factor in improving the stability of the enzyme. Moreover, the correlation between SASA and protein–water hydrogen bonds was reported to be positive57 with a correlation coefficient of over 0.9, accounting for the stability enhancement of A6M/B4M.
The root mean square fluctuation (RMSF) values by residue in the wild type and A6M/B4M were compared to illustrate the flexibility of the region (Fig. 5F). There was no significant change in the α-subunit except for the unstable N-terminal. A slight increase in the RMSF values of the activity-related region (residue 111–118) made up for the possible loss of enzyme activity caused by the enhanced stability. In the case of the β-subunit, the introduction of L25F and G27Y in A6M/B4M allowed the N-terminal (Met1-Glu28) of the β-subunit (residue 212–239) to become less flexible with lower RMSF values than the wild type. Ala101-Arg122 of the β-subunit located in the interface of the two subunits (residue 312–333) also showed a reduction in RMSF values, indicating a more rigid structure regulated by the mutations distributed in both the α- and β-subunit.
Experimental
Strains, plasmids and chemical reagents
The NHase gene from Bordetella petrii DSM 12804 and other NHase genes were cloned to the pET-30a (+) plasmid heterologously expressed in E. coli BL21 (DE3). Acrylonitrile and acrylamide were purchased from Sinopharm Chemical Reagents. Co., Ltd. and Aladdin Reagent (Shanghai) Co., Ltd.Construction of single-point mutant library
The three-dimensional structure of NHAB was obtained through AlphaFold2 (ref. 40) by uploading the protein sequence as default. The structure with the first score was selected for redesign. Web-server FIREPROT v1.3 (ref. 33) (https://loschmidt.chemi.muni.cz/fireprotweb1) and PROSS41 (https://pross.weizmann.ac.il/step/pross-terms/) were carried out for stabilizing the mutation prediction. By uploading the PDB file of NHAB and setting the input parameters (Table S5†), the relevant data output of FIREPROT were given in a few hours. Besides, the PDB file and amino acid of NHAB were input to PROSS for redesign. Two subunits were calculated separately through parameter setting (Table S6†). Consensus alignment was operated by Clustal Omega42 (https://www.ebi.ac.uk/Tools/msa/clustalo/) using the protein sequence, a new program using seeded guide trees and HMM profile-profile techniques to generate alignments between three or more sequences. Site-directed mutagenesis by a whole plasmid PCR protocol was used to construct the mutant library. The primers were designed using the CE Design V1.04 software and listed in Table S7.† The 50 μL reaction mixture contained 25 μL PrimeSTAR Max Premix (2×), 1 μL of each primer and 0.5 μL template DNA. The PCR reactions were performed using the following thermal cycling parameters: 1 cycle, 95 °C, 2 min; 35 cycles, 95 °C, 10 s, 57 °C, 20 s, 72 °C, 30 s; 72 °C, 10 min; 4 °C, 10 min.Culture and expression
The recombinant E. coli cells were first cultured for 6–8 h at 37 °C in 5 mL Luria-Bertani (LB) medium supplemented with 50 μg mL−1 kanamycin, and then transformed in the flask (50 mL LB) at 37 °C until the OD600 reached 0.8. cells were induced at 18 °C for 16 h by the addition of cobalt chloride (CoCl2, 0.4 mM) and isopropyl-β-D-thiogalactopyranoside (IPTG, 0.5 mM). The cultured bacteria were centrifuged at 4000 rpm for 10 min. The supernatant was discarded, while the precipitation was washed twice with deionized water.Purification and SDS-PAGE of NHAB and mutants
The collected cells were resuspended in Ni-50-native buffer (50 mM sodium phosphate, containing 300 mM NaCl, 50 mM imidazole, pH 7.5) and disrupted by ultrasonication in an ice bath, followed by centrifugation at 12000 rpm for 30 min to discard the cell debris. The supernatant was passed through a 0.45 μm membrane filter and loaded on an Ni-NTA column (Thermo Scientific, USA) pre-equilibrated with Ni-50-native buffer, and the proteins were eluted by an increasing gradient of imidazole (from 50 to 500 mM). The purities of the collected fractions were analyzed by SDS-PAGE. The fractions containing the pure target protein were gathered and desalted by ultrafiltration. The purified proteins were concentrated and stored in 20% (v/v) glycerol at −80 °C for further use.The expression and purification of the enzyme were analyzed by prefabricated protein gel (SurePAGE™, Bis-Tris, 10 × 8, 8–16%, GenScript USA Inc.). The gel was stained with Coomassie Brilliant Blue G-250. The protein concentration was determined using the Bradford Protein Assay Kit (Quick Start™, Bio-Rad, USA).
Enzymatic assay
The NHase activity was assayed at 28 °C in a 500 μL reaction system, as follows: 425 μL acrylonitrile water solution (50 g L−1) and 50 μL free cell or pure enzyme. After 5 min, 25 μL HCl water solution (4 mol L−1) was added to terminate the reaction. The cell-precipitation in the reaction mixture was removed by centrifugation for 3 min at 12000 rpm, and the supernatant was mixed with the same volume of acetamide water solution (20 g L−1) for gas chromatography (GC) analysis.The GC (GC950) was equipped with a Porapak Q column (OD of 3 mm × ID of 2 mm × 2 m) with the following conditions: column temperature, 210 °C; injection and detection temperature, 250 °C; and nitrogen flow rate, 60 mL min−1. The retention time of each substance is shown in Fig. S11.† One unit of NHase activity (U) was defined as the amount of enzyme required to catalyze the formation of 1 μmol acrylamide per minute.
Thermal denaturation analysis
For thermostability assessment, enzyme solutions were soaked in a water bath at 45 °C for 1 h, and the samples were taken to determine the residual enzyme activity. The residual activity of NHAB and the mutants was determined by preincubating the enzyme solutions at 45 °C or 50 °C in a water bath for different durations. The half-life was defined as the time when the residual activity retained 50% of the original activity at the measured temperature. The deactivation kinetics of NHAB was fitted to the equation ln(E/E0) = −kd × t and the half-life was calculated using the equation t1/2 = ln(2)/kd.The melting temperature (Tm) was defined as the temperature when half of the protein was unfolded. Nano-DSF (Prometheus NT.48) was used to measure the changes in intrinsic fluorescence when the protein unfolded during thermal treatment. The concentration of the enzyme solutions was controlled at 1 mg mL−1.
Product tolerance determination
The collected cells were immersed in 300 g L−1 acrylamide solution at 28 °C for 1 h, and then washed with phosphate buffer (0.25 M, pH 7.5) to remove the residual acrylamide. The cells collected by centrifugation were diluted and used for the enzyme activity assay. The activity of the cells without acrylamide immersion was defined as 100%.Catalytic reaction and fed-batch hydration
The catalytic reaction was carried out at 45 °C, and the 50 mL reaction system consisted of 0.85 M acrylonitrile substrate and 5 mL enzyme solution. Every 5 min, 100 μL reaction sample was taken to mix with 900 μL 2% HCl water solution. Then, the cell-precipitation was removed by centrifugation, and the concentration of acrylamide in the supernatant was measured by GC analysis.The fed-batch hydration was started with wet cells with a total enzyme activity of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U at 20 °C. Then, 8 g acrylonitrile substrate was added to a 200 mL reaction system in a batch. The next batch of substrate was added after detecting the complete reaction of the substrate by gas chromatography.
000 U at 20 °C. Then, 8 g acrylonitrile substrate was added to a 200 mL reaction system in a batch. The next batch of substrate was added after detecting the complete reaction of the substrate by gas chromatography.
Molecular dynamic simulations and interaction analysis
Based on the 3D structure of the wild type predicted by AlphaFold2, the structure of mutants was obtained through DS (Discovery Studio 2.5). Molecular dynamics (MD) simulations were carried out using Gromacs with the Amber-99 force field. A box was set up and added with water, and then neutralized by adding Na+. Energy minimization, 50-ps of NVT, 50-ps of NPT, and 50-ns classical MD simulations were performed on both the wild type and A6M/B4M variant.The analysis of the root-mean square deviation (RMSD), gyration radius (Rg) and solvent accessible surface area (SASA) was performed using Gromacs. Also, the last 25-ns trajectories were used for analysis of the root-mean square fluctuation (RMSF) calculated for each residue of protein. Also, the polarity of the surface was calculated by PyMOL 2.4.0 using the partial charge sum function.
The interaction analysis including salt bridges and hydrogen bonds was carried out with the Visual Molecular Dynamics (VMD 1.9.3) program. A salt bridge is considered to be formed if the distance between any of the oxygen atoms of acidic residues and the nitrogen atoms of basic residues are within the cut-off distance of 4 Å in at least one frame. The distance of the hydrogen bonds in the MD simulations trajectories was also calculated.
The last-20 ns trajectory from MD simulation was saved and converted to a dcd file type using VMD for further DCCMs analysis. The pdb and dcd files were input to Bio3D,58,59 and the Cα atoms were selected for calculating the correlation coefficients (Cij).48 Finally, the dynamics correlation matrices were visualized using Origin 2023b.
Conclusions
In this study, the two-step SECURE strategy involving the pruning and assembly of mutations that improve thermostability was proposed. The reliability of SECURE was verified in the redesign of NHAB to dramatically improve its thermostability. Ten beneficial mutations were screened from the pruned mutant library, consisting of S30T, A71D, A74D, A78R, S81T and A133P in the α-subunit and L25F, G27Y, N59P and A173N in the β-subunit. The best mutant A6M/B4M exhibited an increase in Tm by 13.2 °C and 866.0-fold extended half-life at 50 °C, together with a co-improvement in catalytic activity. The great potential for industrial application of this mutant was verified in the fed-batch biocatalytic process for the production of acrylamide. The intermolecular interaction analysis was carried out to clarify the internal mechanism of each mutation. The newly formed salt bridges, hydrogen bonds and aromatic interactions were found to contribute to the stability of A6M/B4M. Besides, the overall structural changes such as the surface electrostatic charge distribution, SASA and RMSF analysis of the dynamic simulation accounted for the synergistic effect of multiple-point mutations. In conclusion, the SECURE strategy proposed herein was demonstrated for the first time to be user-friendly and successfully applied in the redesign of a multi-subunit NHase, which no longer limits the thermostability engineering of NHase to the β-subunit, and the overall redesign of the multimeric protein maximized the thermostability accompanied by an increase in catalytic activity through effective assembly. Meanwhile, the SECURE strategy provides technology for the rational design of high-performance industrial biocatalysts.Author contributions
JLX, HSZ and LRY designed the concept of this study. JLX carried out the experimental procedures. JLX, HSZ and JQX analyzed the results and drafted the manuscript. JQX, ZYW and ZLY helped revise the manuscript. ZW performed the molecular dynamics simulations. HYZ, HRY, and JPW provided experimental help. All authors read and approved the final manuscript.Conflicts of interest
The authors declare that they have no competing financial interests.Acknowledgements
This work was funded by National Key R&D Program of China (No. 2021YFC2102003).Notes and references
- R. P. Magalhães, J. M. Cunha and S. F. Sousa, Perspectives on the role of enzymatic biocatalysis for the Degradation of Plastic PET, Int. J. Mol. Sci., 2021, 22(20), 11257 CrossRef PubMed.
- J. M. Clomburg, A. M. Crumbley and R. Gonzalez, Industrial biomanufacturing: The future of chemical production, Science, 2017, 38(355), 1–10 Search PubMed.
- S. Simić, E. Zukić, L. Schmermund, K. Faber, C. K. Winkler and W. Kroutil, Shortening synthetic routes to small molecule active pharmaceutical ingredients employing biocatalytic methods, Chem. Rev., 2022, 122(1), 1052–1126 CrossRef PubMed.
- K. M. Polizzi, A. S. Bommarius, J. M. Broering and J. F. Chaparro-Riggers, Stability of biocatalysts, Curr. Opin. Chem. Biol., 2007, 11, 220–225 CrossRef CAS PubMed.
- C. Silva, M. Martins, S. Jing, J. Fu and A. Cavaco-Paulo, Practical insights on enzyme stabilization, Crit. Rev. Biotechnol., 2018, 38(3), 335–350 CrossRef CAS PubMed.
- R. Sahu, A. K. Meghavarnam and S. Janakiraman, Nitrile hydratase-mediated conversion of acrylonitrile by Rhodococcus rhodochrous (RS-6), J. Chem. Technol. Biotechnol., 2021, 96(4), 1080–1085 CrossRef CAS.
- R. A. Sheldon, D. Brady and M. L. Bode, The Hitchhiker's guide to biocatalysis: recent advances in the use of enzymes in organic synthesis, Chem. Sci., 2020, 11(10), 2587–2605 RSC.
- M. Kobayashi, T. Nagasawa and H. Yamada, Enzymatic synthesis of acrylamide: a success story not yet over, Trends Biotechnol., 1992, 10, 402–408 CrossRef CAS PubMed.
- J. Shearer, H. L. Jackson, D. Schweitzer, D. K. Rittenberg, T. M. Leavy, W. Kaminsky, R. C. Scarrow and J. A. Kovacs, The first example of a nitrile hydratase model complex that reversibly binds nitriles, J. Am. Chem. Soc., 2002, 124(38), 11417–11428 CrossRef CAS PubMed.
- S. Jiao, F. Li, H. Yu and Z. Shen, Advances in acrylamide bioproduction catalyzed with Rhodococcus cells harboring nitrile hydratase, Appl. Microbiol. Biotechnol., 2020, 104(3), 1001–1012 CrossRef CAS PubMed.
- T. Nagasawa, H. Shimizu and H. Yamada, The superiority of the third-generation catalyst, Rhodococcus rhodochrous J1 nitrile hydratase, for industrial production of acrylamide, Appl. Microbiol. Biotechnol., 1993, 40(2), 189–195 CAS.
- Y. Cui, W. Cui, Z. Liu, L. Zhou, M. Kobayashi and Z. Zhou, Improvement of stability of nitrile hydratase via protein fragment swapping, Biochem. Biophys. Res. Commun., 2014, 450(1), 401–408 CrossRef CAS PubMed.
- X. Pei, J. Wang, Y. Wu, X. Zhen, M. Tang, Q. Wang and A. Wang, Evidence for the participation of an extra α-helix at β-subunit surface in the thermal stability of Co-type nitrile hydratase, Appl. Microbiol. Biotechnol., 2018, 102(18), 7891–7900 CrossRef CAS PubMed.
- Y. Xia, W. Cui, Z. Cheng, L. Peplowski, Z. Liu, M. Kobayashi and Z. Zhou, Improving the thermostability and catalytic efficiency of the subunit-fused nitrile hydratase by semi-rational engineering, ChemCatChem, 2018, 10(6), 1370–1375 CrossRef CAS.
- Z. Zhou, D. Ma and Z. Cheng, Engineering of the thermophilic nitrile hydratase from Pseudonocardia thermophila JCM3095 for large-scale nicotinamide production based on sequence-activity relationships, Int. J. Biol. Macromol., 2021, 191, 775–782 CrossRef CAS PubMed.
- L. Tang, J. Yang, J. Chen, J. Zhang, H. Yu and Z. Shen, Design of salt-bridge cyclization peptide tags for stability and activity enhancement of enzymes, Process Biochem., 2019, 81, 39–47 CrossRef CAS.
- Y. Chen, S. Jiao, M. Wang, J. Chen and H. Yu, A novel molecular chaperone GroEL2 from Rhodococcus ruber and its fusion chimera with nitrile hydratase for co-enhanced activity and stability, Chem. Eng. Sci., 2018, 192, 235–243 CrossRef CAS.
- Y. Liang, S. Jiao, M. Wang, H. Yu and Z. Shen, A CRISPR/Cas9-based genome editing system for Rhodococcus ruber TH, Metab. Eng., 2020, 57, 13–22 CrossRef CAS PubMed.
- A. Miyanaga, S. Fushinobu, K. Ito and T. Wakagi, Crystal structure of cobalt-containing nitrile hydratase, Biochem. Biophys. Res. Commun., 2001, 288, 1169–1174 CrossRef CAS PubMed.
- H. R. Brodkin, W. R. P. Novak, A. C. Milne, J. A. D. Aquino, N. M. Karabacak, I. G. Goldberg, J. N. Agar, M. S. Payne, G. A. Petsko, M. J. Ondrechen and D. Ringe, Evidence of the participation of remote residues in the catalytic activity of co-Type nitrile hydratase from Pseudomonas putida, Biochemistry, 2011, 50, 4923–4935 CrossRef CAS PubMed.
- L. Zhang, S. Zhao, C. Chang, J. Wang, C. Yang and Z. Cheng, N-terminal loops at the tetramer interface of nitrile hydratase act as “hooks” determining resistance to high amide concentrations, Int. J. Biol. Macromol., 2023, 245, 125531 CrossRef CAS PubMed.
- Z. Cheng, Y. Lan, J. Guo, D. Ma, S. Jiang, Q. Lai, Z. Zhou and L. Peplowski, Computational design of nitrile hydratase from Pseudonocardia thermophila JCM3095 for improved thermostability, Molecules, 2020, 25 Search PubMed.
- Y. Liu, W. Cui, Z. Liu, Y. Cui, Y. Xia, M. Kobayashi and Z. Zhou, Enhancement of thermo-stability and product tolerance of Pseudomonas putida nitrile hydratase by fusing with self-assembling peptide, J. Biosci. Bioeng., 2014, 118(3), 249–252 CrossRef CAS PubMed.
- J. D. Shen, X. Cai, Y. W. Ni, L. Q. Jin, Z. Q. Liu and Y. G. Zheng, Structural insights into the thermostability mechanism of a nitrile hydratase from Caldalkalibacillus thermarum by comparative molecular dynamics simulation, Proteins: Struct., Funct., Bioinf., 2021, 89(8), 978–987 CrossRef CAS PubMed.
- S. R. Miller, An appraisal of the enzyme stability-activity trade-off, Evolution, 2017, 71(7), 1876–1887 CrossRef CAS PubMed.
- K. S. Siddiqui, Defying the activity-stability trade-off in enzymes: taking advantage of entropy to enhance activity and thermostability, Crit. Rev. Biotechnol., 2017, 37(3), 309–322 CrossRef PubMed.
- C. Xu, L. Tang, Y. Liang, S. Jiao, H. Yu and H. Luo, Novel chaperones RrGroEL and RrGroES for activity and stability enhancement of nitrilase in Escherichia coli and Rhodococcus ruber, Molecules, 2020, 25(4), 1002 CrossRef CAS PubMed.
- Z. Yu, H. Yu, J. Xu, Z. Wang, Z. Wang, T. Kang, K. Chen, Z. Pu, J. Wu, L. Yang and G. Xu, Enhancing thermostability of lipase from Pseudomonas alcaligenes for producing L-menthol by the CREATE strategy, Catal. Sci. Technol., 2022, 12, 2531–2541 RSC.
- H. Chi, Y. Wang, B. Xia, Y. Zhou, Z. Lu, F. Lu and P. Zhu, Enhanced thermostability and molecular insights for L-Asparaginase from Bacillus licheniformis via structure- and computation-based rational design, J. Agric. Food Chem., 2022, 70, 14499–14509 CrossRef CAS PubMed.
- X. Zhang, W. Li, L. Pan, L. Yang, H. Li, F. Ji, Y. Zhang, H. Tang and D. Yang, Improving the thermostability of alginate lyase FlAlyA with high expression by computer-aided rational design for industrial preparation of alginate oligosaccharides, Front. Bioeng. Biotechnol., 2022, 10, 1011273 CrossRef PubMed.
- Y. Dehouck, J. M. Kwasigroch, D. Gilis and M. Rooman, PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality, BMC Bioinf., 2011, 12, 151 CrossRef PubMed.
- H. J. Wijma, R. J. Floor, P. A. Jekel, D. Baker, S. J. Marrink and D. B. Janssen, Computationally designed libraries for rapid enzyme stabilization, Protein Eng., Des. Sel., 2014, 27(2), 49–58 CrossRef CAS PubMed.
- D. Bednar, K. Beerens, E. Sebestova, J. Bendl, S. Khare, R. Chaloupkova, Z. Prokop, J. Brezovsky, D. Baker and J. Damborsky, FireProt: energy- and evolution-based computational design of thermostable multiple-point mutants, PLoS Comput. Biol., 2015, 11(11), e1004556 CrossRef PubMed.
- S. Li, J. Xie, S. Qiu, S. Xu, F. Cheng, Y. Wang and Y. Zheng, Semirational engineering of an aldo–keto reductase KmAKR for overcoming trade-offs between catalytic activity and thermostability, Biotechnol. Bioeng., 2021, 118(11), 4441–4452 CrossRef CAS PubMed.
- M. Klaewkla, R. Pichyangkura, T. Charoenwongpaiboon, K. Wangpaiboon and S. Chunsrivirot, Computational design of oligosaccharide producing levansucrase from Bacillus licheniformis RN-01 to improve its thermostability for production of levan-type fructo oligosaccharides from sucrose, Int. J. Biol. Macromol., 2020, 160, 252–263 CrossRef CAS PubMed.
- N. Tokuriki and D. S. Tawfik, Stability effects of mutations and protein evolvability, Curr. Opin. Struct. Biol., 2009, 19(5), 596–604 CrossRef CAS PubMed.
- J. Xu, H. Zhou, H. Yu, T. Deng, Z. Wang, H. Zhang, J. Wu and L. Yang, Computational design of highly stable and soluble alcohol dehydrogenase for NADPH regeneration, Bioresour. Bioprocess., 2021, 8, 12 CrossRef.
- P. A. Romero and F. H. Arnold, Exploring protein fitness landscapes by directed evolution, Nat. Rev. Mol. Cell Biol., 2009, 10(12), 866–876 CrossRef CAS PubMed.
- Y. Cui, Y. Chen, X. Liu, S. Dong, Y. E. Tian, Y. Qiao, R. Mitra, J. Han, C. Li, X. Han, W. Liu, Q. Chen, W. Wei, X. Wang, W. Du, S. Tang, H. Xiang, H. Liu, Y. Liang, K. N. Houk and B. Wu, Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy, ACS Catal., 2021, 11(3), 1340–1350 CrossRef CAS.
- K. Tunyasuvunakool, J. Adler, Z. Wu, T. Green, M. Zielinski, A. Žídek, A. Bridgland, A. Cowie, C. Meyer, A. Laydon, S. Velankar, G. J. Kleywegt, A. Bateman, R. Evans, A. Pritzel, M. Figurnov, O. Ronneberger, R. Bates, S. A. A. Kohl, A. Potapenko, A. J. Ballard, B. Romera-Paredes, S. Nikolov, R. Jain, E. Clancy, D. Reiman, S. Petersen, A. W. Senior, K. Kavukcuoglu, E. Birney, P. Kohli, J. Jumper and D. Hassabis, Highly accurate protein structure prediction for the human proteome, Nature, 2021, 596(7873), 590–596 CrossRef CAS PubMed.
- A. Goldenzweig, M. Goldsmith, S. E. Hill, O. Gertman, P. Laurino, Y. Ashani, O. Dym, T. Unger, S. Albeck, J. Prilusky, R. L. Lieberman, A. Aharoni, I. Silman, J. L. Sussman, D. S. Tawfik and S. J. Fleishman, Automated structure- and sequence-based design of proteins for high bacterial expression and stability, Mol. Cell, 2016, 63(2), 337–346 CrossRef CAS PubMed.
- F. Sievers and D. G. Higgins, Clustal Omega for making accurate alignments of many protein sequences, Protein Sci., 2018, 27(1), 135–145 CrossRef CAS PubMed.
- Z. Cheng, S. Jiang and Z. Zhou, Substrate access tunnel engineering for improving the catalytic activity of a thermophilic nitrile hydratase toward pyridine and pyrazine nitriles, Biochem. Biophys. Res. Commun., 2021, 575, 8–13 CrossRef CAS PubMed.
- S. Martinez, M. L. Kuhn, J. T. Russell, R. C. Holz and T. E. Elgren, Acrylamide production using encapsulated nitrile hydratase from Pseudonocardia thermophila in a sol-gel matrix, J. Mol. Catal. B: Enzym., 2014, 100, 19–24 CrossRef CAS.
- Y. Lan, X. Zhang, Z. Liu, L. Zhou, R. Shen, X. Zhong, W. Cui and Z. Zhou, Overexpression and characterization of two types of nitrile hydratases from Rhodococcus rhodochrous J1, PLoS One, 2017, 12(6), 14 Search PubMed.
- Z. Yang, X. Pei, G. Xu, J. Wu and L. Yang, Efficient production of 2,6-Difluorobenzamide by recombinant Escherichia coli expressing the Aurantimonas manganoxydans nitrile hydratase, Appl. Biochem. Biotechnol., 2019, 187(2), 439–448 CrossRef CAS PubMed.
- D. M. Krüger, P. C. Rathi, C. Pfleger and H. Gohlke, CNA web server: rigidity theory-based thermal unfolding simulations of proteins for linking structure, (thermo-)stability, and function, Nucleic Acids Res., 2013, 41(W1), W340–W348 CrossRef PubMed.
- H. Yu and P. A. Dalby, Coupled molecular dynamics mediate long- and short-range epistasis between mutations that affect stability and aggregation kinetics, Proc. Natl. Acad. Sci., 2018, 115(47), E11043–E11052 CAS.
- H. Yu and P. A. Dalby, Exploiting correlated molecular-dynamics networks to counteract enzyme activity–stability trade-off, Proc. Natl. Acad. Sci., 2018, 115(52), E12192–E12200 CrossRef CAS PubMed.
- M. T. Nelp, A. V. Astashkin, L. A. Breci, R. M. Mccarty and V. Bandarian, The alpha subunit of nitrile hydratase is sufficient for catalytic activity and post-translational modification, Biochemistry, 2014, 53(24), 3990–3994 CrossRef CAS PubMed.
- R. S. Prajapati, M. Das, S. Sreeramulu, M. Sirajuddin, S. Srinivasan, V. Krishnamurthy, R. Ranjani, C. Ramakrishnan and R. Varadarajan, Thermodynamic effects of proline introduction on protein stability, Proteins: Struct., Funct., Bioinf., 2007, 66(2), 480–491 CrossRef CAS PubMed.
- K. Saraboji, M. M. Gromiha and M. N. Ponnuswamy, Importance of main-chain hydrophobic free energy to the stability of thermophilic proteins, Int. J. Biol. Macromol., 2005, 35(3), 211–220 CrossRef CAS PubMed.
- L. Boucher, S. Somani, C. Negron, W. Ma, S. Jacobs, W. Chan, T. Malia, G. Obmolova, A. Teplyakov, G. L. Gilliland and J. Luo, Surface salt bridges contribute to the extreme thermal stability of anFN3-like domain from a thermophilic bacterium, Proteins: Struct., Funct., Bioinf., 2022, 90(1), 270–281 CrossRef CAS PubMed.
- M. L. Waters, Aromatic interactions in peptides: Impact on structure and function, Biopolymers, 2004, 76(5), 435–445 CrossRef CAS PubMed.
- S. S. Strickler, A. V. Gribenko, A. V. Gribenko, T. R. Keiffer, J. Tomlinson, T. Reihle, V. V. Loladze and G. I. Makhatadze, Protein stability and surface electrostatics: a charged relationship, Biochemistry, 2006, 45(9), 2761–2766 CrossRef CAS PubMed.
- H. Cui, L. Zhang, L. Eltoukhy, Q. Jiang, S. K. Korkunç, K. Jaeger, U. Schwaneberg and M. D. Davari, Enzyme hydration determines resistance in organic cosolvents, ACS Catal., 2020, 10(24), 14847–14856 CrossRef CAS.
- P. Pal, S. Chakraborty and B. Jana, Number of hydrogen bonds per unit solvent accessible surface area: a descriptor of functional states of proteins, J. Phys. Chem. B, 2022, 126(51), 10822–10833 CrossRef CAS PubMed.
- L. Skjærven, X. Q. Yao, G. Scarabelli and B. J. Grant, Integrating protein structural dynamics and evolutionary analysis with Bio3D, BMC Bioinf., 2014, 15(1), 399 CrossRef PubMed.
- B. J. Grant, A. P. Rodrigues, K. M. Elsawy, J. A. Mccammon and L. S. Caves, Bio3d: an R package for the comparative analysis of protein structures, Bioinformatics, 2006, 22(21), 2695–2696 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cy01102j |
| This journal is © The Royal Society of Chemistry 2023 |