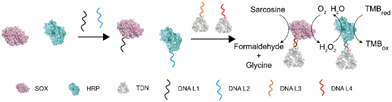
The enhancement of enzyme cascading via tetrahedral DNA framework modification†
Haipei
Zhao
a,
Mingqiang
Li
 a,
Shasha
Lu
c,
Nan
Cao
a,
Xiaolei
Zuo
a,
Shasha
Lu
c,
Nan
Cao
a,
Xiaolei
Zuo
 ab,
Shaopeng
Wang
*b and
Min
Li
*b
ab,
Shaopeng
Wang
*b and
Min
Li
*b
aSchool of Chemistry and Chemical Engineering, and Zhang Jiang Institute for Advanced Study, Shanghai Jiao Tong University, Shanghai 200240, China
bInstitute of Molecular Medicine, Shanghai Key Laboratory for Nucleic Acid Chemistry and Nanomedicine, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 200127, China. E-mail: wspsinap@gmail.com; mlisinap@163.com
cSchool of Materials Science and Engineering, Suzhou University of Science and Technology, Suzhou 215009, China
First published on 16th January 2023
Abstract
Enzyme clustering is widely used in many organisms to increase the catalytic efficiency of cascade reactions. Inspired by nature, organizing enzymes within a cascade reaction also draws much attention in both basic research and industrial processes. An important step for organizing enzymes precisely in vitro is enzyme modification. However, modifying enzymes without sacrificing their activity remains challenging until now. For example, labeling enzymes with DNA, one of the well-established enzyme modification methods, has been shown to significantly reduce the enzymatic activity. Herein we report an enzyme conjugation method that can rescue the reduction of enzymatic activity caused by DNA labeling. We demonstrate that immobilizing DNA-modified enzymes on the vertex of TDNs (tetrahedral DNA nanostructures) enhances the enzymatic activity compared with their unmodified counterparts. Using this strategy, we have further developed an ultra-sensitive and high-throughput electrochemical biosensor for sarcosine detection, which holds great promise for prostate cancer screening.
Introduction
Enzymatic reactions play critical roles in many cellular processes and are essential for cell survival. In many circumstances, multiple enzymes are involved in one reaction to produce the final products.1 To improve the catalytic efficiency of such cascade reactions, related enzymes are usually assembled into a functional co-cluster in cells.2 Clustering enzymes together can promote the formation of protein tunnels that connect the active sites of enzymes. Under such circumstances, the intermediary metabolic product can be passed to the downstream enzymes directly, which accelerates the processing of intermediates. This phenomenon is called substrate channeling and has been shown to enhance the cascade enzymatic activity significantly.3,4 In addition to maximizing the catalytic efficiency, functional co-clusters also minimize the interference between different reactions by restricting the diffusion of intermediate metabolites.3 Clustering related enzymes together for increasing the cascade enzymatic activity is also one of the primary considerations in many industrial processes such as the synthesis of pharmaceutical, cosmetic, and nutritional compounds.5 Thus, considerable efforts have been devoted to developing optimal strategies for constructing enzyme complexes in recent years. For example, various synthetic scaffolds have been used to organize unmodified enzymes into confined spaces to increase the activity, stability, and reusability of enzymes.6–9 The arrangement of different enzymes can be achieved more precisely by immobilizing modified enzymes on pre-designed platforms.10–20 Some platforms such as DNA nanostructures provide opportunities to manipulate the distance and the stoichiometry of different enzymes, which is ideal for deciphering the influence of those parameters on the enzymatic activity.21–24 More importantly, DNA nanostructures also allow the possibility of studying cascade enzymatic reactions in 3D. For example, a recent study utilized a 3D DNA wireframe octahedron to organize glucose oxidase and horseradish peroxidase and deciphered the influence of the spatial organization on the cascade enzymatic activity.25Despite the remarkable progress that has been made in recent years, there are still limitations to cluster enzymes in vitro. One of the big challenges that remain unsolved in this field is how to accurately immobilize enzymes on the scaffold without influencing their activity. As noted above, enzyme modification in combination with DNA nanostructures provides the possibility to immobilize enzymes with nanoscale precision. However, it has been demonstrated that linking DNA to enzymes may impair their activity.26 One possible explanation is that the single-stranded DNA may interact with the enzyme due to its flexibility and influence the substrate accessibility of the active site.27,28 Thus, decreasing the flexibility of the single-stranded DNA is a possible way of diminishing its impairment on the activity of the conjugated enzymes.
Tetrahedral DNA nanostructures (TDNs) are ideal platforms for assembling different functional biomolecules and have been used widely in biomedical areas including bioimaging, biodetection, and drug delivery.29 Due to the structural rigidity, TDNs can also influence the flexibility of the DNA strand assembled on the vertex through base pairing. Here, we used TDNs as scaffolds for enzyme immobilization and accessed their influence on the enzymatic activity. Consistent with previous findings, we found that linking with the single-stranded DNA (ssDNA) decreases the activity of enzymes. Surprisingly, our results revealed that coupling DNA-conjugated enzymes with TDNs significantly enhanced the enzyme activity. A 5.5-fold increase in the Michaelis constant kcat of the TDN enzymes was observed with the Michaelis–Menten model compared to that of single-stranded DNA conjugated enzymes. We have also demonstrated that TDN-based cascade enzyme immobilization is suitable for the analysis of cancer markers with high sensitivity.
Results and discussion
Scheme of the TDN-directed enzyme cascade
To investigate the influence of DNA modification and TDN conjugation on the activity of enzymes, we chose sarcosine oxidase (SOX) and horseradish peroxidase (HRP). There are two reasons for this choice: (1) sarcosine is the substrate of SOX and has been identified as a biomarker of prostate cancer which can be detected in body fluids including urine,30,31 thus developing simple and sensitive detection methods holds promise for an earlier prostate cancer diagnosis;32 and (2) the product of SOX can be used as a substrate of HRP. These two enzymes can form a cascade reaction and have been widely used to detect sarcosine (Fig. 1).Characterization of TDN-immobilized enzyme cascade
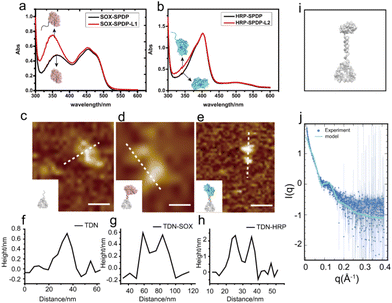
To construct the TDN-immobilized enzyme cascade, we first labeled SOX and HRP with the single-stranded DNA (DNA L1 and DNA L2 respectively) using the SPDP crosslinker (Fig. 1 and Fig. S1†). The structures of the enzymes are shown in Fig. S2.† We performed absorbance measurements to assess the protein–DNA coupling efficiency. A significant increase in the absorbance at 343 nm was observed after DNA conjugation (Fig. 2a and b), indicating the successful labeling of enzymes with DNA (Fig. S1†). Next, TDNs were prepared as described previously (Fig. S3†).33 The successful assembly of TDNs was confirmed by both native polyacrylamide gel electrophoresis (Fig. S4 and S5†) and atomic force microscopy (AFM) imaging (Fig. 2c, f and Fig. S6†). DNA-conjugated enzymes were then immobilized on the vertex of TDNs containing a pre-designed pendant strand (DNA L3 and DNA L4 respectively) through base pairing (Fig. 1). AFM imaging was performed to verify the assembly of TDN–SOX (Fig. 2d, g and Fig. S7†) and TDN–HRP (Fig. 2e, h and Fig. S8†). The PAGE results confirmed the formation of TDN enzymes (Fig. S9 and S10†). The average inter-distance between TDNs and enzymes was measured to be 8 nm, which is consistent with the predicted length of the linker DNA (Fig. 2g and h). The structure of TDN enzymes was rendered by PYMOL (Fig. 2i), and the size of the TDN–SOX was predicted to be 17.7 nm. Next, we used synchrotron-based SAXS to characterize the in situ structure of the TDN–enzymes conjugation (Fig. 2j). By fitting the SAXS data, we found that the size of TDN–SOX was 16.1 nm, which is consistent with the size predicted by PYMOL. Taken together, these data demonstrated that the TDN–enzyme conjugations were assembled successfully as designed and were suitable for further analysis.TDNs conjugation enhances the cascade enzymatic activity
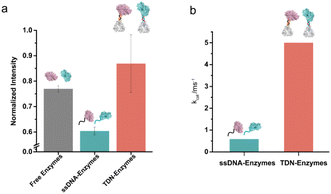
Next, we examined the influence of the TDNs conjugation on the activity of enzymes using the well-established SOX–HRP cascade reaction. As shown in Fig. 1, SOX catalyzes the oxidative demethylation of sarcosine and produces hydrogen peroxide (H2O2), which is the substrate of horseradish peroxidase (HRP). The activity of HRP can be visualized by the conversion of its colorless TMB (3,3′,5,5′-tetramethylbenzidine) starting substrate into its green oxidation state. We compared the cascade catalytic efficiency of free enzymes, ssDNA–enzymes, and TDN–enzymes by measuring the absorbance change of each reaction at 370 nm in the absence and presence of 100 μM sarcosine. Consistent with previous studies, we found that the enzymatic activity of DNA labeled enzymes was 80% of the activity of unmodified enzymes (Fig. 3a). The activity of TDN-conjugated enzymes was 1.44-fold higher than that of ssDNA–enzymes, indicating that TDN conjugation enhanced the activity of enzymes. More surprisingly, the activity of TDN-conjugated enzymes was even higher than that of unmodified enzymes (Fig. 3a and Fig. S11†). We further fitted our experimental data with the Michaelis–Menten model and found that the Michaelis constants kcat of the TDN–enzymes and the ssDNA–enzymes were 0.91 ± 0.022 ms−1 and 5.0 ± 1.2 ms−1, respectively (Fig. 3b). This indicated that a 5.5-fold enhancement of the enzymatic activity could be achieved by immobilizing ssDNA–enzymes on TDNs.Sensitive sarcosine detection with TDN–enzymes
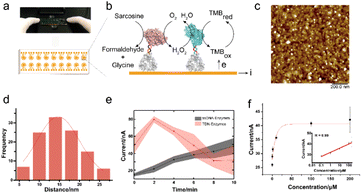
To further test the performance of the TDN-conjugated enzyme cascade and explore its potential diagnostic application, we immobilized TDN-conjugated enzymes on the interface of a 16-channel electrochemical chip to electrochemically detect sarcosine (Fig. 4a and b). We first characterized the morphology of the enzyme-immobilized electrode by AFM (Fig. 4c). After immobilization, TDN-conjugated enzymes were uniformly distributed on the interface. The inter-enzyme distance of TDN–enzymes was ∼20 nm (Fig. 4d), which is within the critical coupling length (CCL) for efficient reactions.34 Next, we performed the sarcosine detection and found that the cascade catalytic efficiency of TDN–enzymes was significantly higher than that of ssDNA–enzymes (Fig. 4e). Catalytic kinetics analysis demonstrated that there was a 5-fold increase in the speed of the TDN–enzyme-mediated cascade reaction compared to that of ssDNA–enzymes. Lastly, we employed the modular platform established here to detect sarcosine in complex biological samples. The electrochemical interrogation of complex biological samples with spike-in sarcosine exhibited a linear dose–response curve. The detection limit of the TDN–enzyme-immobilized electrochemical chip was estimated to be 100 nM, which makes it suitable for clinical use.Experimental
Materials
Horseradish peroxidase (HRP, P6782), dimethyl sulfoxide (DMSO), tris(2-carboxyethyl)phosphine hydrochloride (TCEP) and D-glucose N-succinimidyl 3-(2-pyridyldithio) propionate (SPDP) were purchased from Sigma-Aldrich, USA. 3,3′,5,5′-Tetramethylbenzidine (TMB, 34021) was purchased from ThermoFisher Scientific, Inc., USA. Sarcosine oxidase (SOX, 60105) was purchased from Realbio (Shanghai, China). All DNA sequences used in this work were synthesized and modified by Sangon Biotech. Co. Ltd (Shanghai, China). All other chemicals were purchased from Sinopharm Chemical Reagent Co. Ltd (Shanghai, China). All solutions used in this work were prepared with Milli-Q water (18 MΩ cm−1).Assembly of tetrahedral DNA nanostructure
Tetrahedron DNA nanostructures (TDNs) were synthesized by mixing equimolar quantities (1 μM) of DNA strands in TM buffer (20 mM Tris, 50 mM MgCl2, pH 8.0). The mixture was then heated to 95 °C for 10 min and rapidly cooled to 4 °C within 30 s using a T100 Touch thermal cycler (BioRad). The final concentration of the TDN was 1 μM.Characterization of TDNs
All TDNs were characterized by native polyacrylamide gel electrophoresis (PAGE). Briefly, DNA solutions were resuspended in gel sample buffer (6× sample buffer: TEK buffer, pH 8.0, 50% glycerol, 0.25% bromphenol blue) and were analysed by 8% native PAGE. The electrophoresis was performed in 1× TBE buffer (12.5 mM Mg2+; pH 8.0) with a constant voltage of 80 V for about 120 min. The gel was then visualized on a chemiluminescence imaging system (G:Box Chemi-XL) after Gel Red staining.Preparation of DNA–enzyme conjugations
SPDP was used to crosslink enzymes with DNA strands. First, 100 μl of the 100 μM SOX solution was reacted with a 5-fold excess of SPDP in 1× DPBS (pH 8.0) for two hours (a 100-fold excess of SPDP was used to react with HRP for 4 h). Excess SPDP was removed with Millipore 30 kDa cutoff filters. Next, a 10-fold excess of thiol-modified DNA was incubated with SPDP-modified enzymes. The efficiency of DNA–enzyme conjugation was evaluated by monitoring the release of pyridine-2-thione (343 nm, extinction coefficient: 8080 M−1 cm−1) as shown in Fig. S1.† Excess DNA was then removed with Millipore 30 kDa cutoff filters. The enzymes and DNA-modified enzymes were quantified with a PerkinElmer lambda 850 spectrophotometer (USA). SOX was linked with the single-stranded DNA linker 1 (L1). HRP was linked with the single-stranded DNA linker2 (L2). All the reactions above were performed at room temperature.Preparation of TDN–enzyme conjugations
To immobilize DNA-labeled enzymes on the vertex of TDNs, ssDNA–enzymes were incubated with equimolar quantities of TDNs in TM buffer at 37 °C for two hours and then the solution mixture was cooled to 4 °C. The solution was stored at 4 °C until further use. The successful assembly of TDN–enzyme conjugations was confirmed by PAGE, as shown in Fig. S4 and S5.† TDN–enzyme conjugations were then used in enzyme cascading in solution or at the interface.Enzyme concentration quantification
Concentrations of free enzymes, ssDNA–enzymes, and TDN–enzymes were quantified using a UV-vis spectrophotometer and calculated according to the Lambert–Beer law. Specifically, the molar concentrations of HRP were measured spectrophotometrically at 403 nm using ε403 = 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 as the molar absorption coefficient. The molar concentrations of SOX were measured spectrophotometrically at 452 nm using ε452 = 12
000 M−1 cm−1 as the molar absorption coefficient. The molar concentrations of SOX were measured spectrophotometrically at 452 nm using ε452 = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 M−1 cm−1 as the molar absorption coefficient.
700 M−1 cm−1 as the molar absorption coefficient.
AFM characterization
Enzymes, TDNs, and TDN–enzyme conjugations were characterized by AFM (Multimode Nanoscope VIII, Bruker). 20 μL of APTES (0.5%) was added to the surface of freshly cleaved mica for 3 min. Excess APTES was removed by washing with Milli-Q water. 10 μL of the sample was deposited on the surface of APTES-treated mica and incubated for 3 min to allow the sample to be absorbed onto the substrate. After removing the excess sample by washing it with Milli-Q water, the sample was scanned in TM buffer in the ScanAsyst mode using a Multimode Nanoscope VII (Bruker) instrument.Enzyme cascade assay in solution
0.25 μM SOX and HRP were used for enzyme cascade assays. The enzymatic activity of the SOX–HRP cascade was quantified in 0.04 g L−1 TMB (phosphate buffer, pH 7.0) with varying sarcosine concentrations by measuring the absorbance change at 370 nm. A SpectraMax iD5 96 well plate reader (Molecular Devices, USA) was used to monitor the absorbance change.Electrochemical measurements
All electrochemical measurements were performed on 16-channel electrochemical chips with a Model CHI 1040C electrochemical workstation (CH Instruments, Inc., Austin, TX). Cyclic voltammetry (CV) was carried out at a scan rate of 50 mV s−1. The amperometric detection was conducted at 100 mV. The electroreduction current was measured for 1 min after the catalytic reaction reached the steady state. Electrochemical detection of the SOX–HRP cascade reaction was performed as described previously. Briefly, the 16-channel electrochemical chips were washed with 75% ethanol followed by Milli-Q water. Next, 7 μL of the mixture of TDN–enzyme conjugations (TDN1–SOX and TDN2–HRP, with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was dropped onto the chips and incubated overnight at room temperature. The excess enzymes were rinsed with 10 mM PBS (12 mM phosphate buffer, 137 mM NaCl and 2.7 mM KCl) and then the modified chips were used in the electrochemical experiment.
1) was dropped onto the chips and incubated overnight at room temperature. The excess enzymes were rinsed with 10 mM PBS (12 mM phosphate buffer, 137 mM NaCl and 2.7 mM KCl) and then the modified chips were used in the electrochemical experiment.
Fetal bovine serum (FBS) was filtered using a 10 kDa filter before being mixed with different concentrations of sarcosine. Sarcosine detection in complex biological fluids was mimicked by performing the assay in solutions containing 10% FBS. Briefly, 7 μL of an equally mixed TDN1–SOX and TDN2–HRP mixture (0.5 μM) or 7 μL of an equally mixed ssDNA–SOX and ssDNA–HRP (0.5 μM) mixture was added on the surface of 16-channel electrochemical chips and incubated overnight at room temperature. ssDNA–SOX and ssDNA–HRP were immobilized on 16-channel electrochemical chips with DNA L3′ and DNA L4′ respectively. Then excess enzymes were removed by washing with 10 mM PBS. The enzymatic activity of the cascade reaction at the interface was measured in 0.144 g L−1 TMB (pH 7.0) containing 10% FBS.
Conclusions
In this work, we have reported a strategy to cluster enzymes in vitro without sacrificing their activity. Consistent with previous studies, we found that labeling enzymes with single-stranded DNA impaired their activity. We demonstrated that this impairment can be rescued by immobilizing ssDNA–enzymes on the vertex of TDNs. A 5.5-fold enhancement of the cascade enzyme activity was observed after the assembly of ssDNA–enzymes on TDNs. More importantly, the activity of TDN–enzymes was found to be even higher than that of unmodified enzymes. Taking advantage of the high controllability of TDN assembly, we developed an ultra-sensitive and high-throughput platform for the detection of sarcosine by immobilizing TDN–enzymes on the interface of a multi-channel electrochemical chip. The enhancement of the detection sensitivity was achieved for two reasons. First, the TDN conjugation increased the activity of enzymes as compared with unmodified enzymes. Second, organizing TDN–enzymes on the interface of the electrochemical chip brought the enzyme pairs within the critical coupling length (∼20 nm), which is beneficial for cascade reactions. The detection limit of sarcosine in complex biological samples was 100 nM. Thus, the method established here holds great potential for prostate cancer screening.Author contributions
Haipei Zhao: writing original draft, conceptualization, data curation, and formal analysis. Mingqiang Li, Shasha Lu and Nan Cao: methodology, visualization. Xiaolei Zuo: funding acquisition, and supervision. Shaopeng Wang and Min Li: methodology, funding acquisition, and project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research and Development Program of China (No. 2021YFF1200300 and 2020YFA0909000), the National Natural Science Foundation of China (No. 22025404 and 21904086) and the Innovative Research Team of High-Level Local Universities in Shanghai (No. SHSMU-ZLCX20212602).References
- M. L. Contente and F. Paradisi, Nat. Catal., 2018, 1, 452–459 CrossRef CAS.
- S. An, R. Kumar, E. D. Sheets and S. J. Benkovic, Science, 2008, 320, 103–106 CrossRef CAS PubMed.
- I. Wheeldon, S. D. Minteer, S. Banta, S. C. Barton, P. Atanassov and M. Sigman, Nat. Chem., 2016, 8, 299–309 CrossRef CAS PubMed.
- V. Pareek, Z. Sha, J. He, N. S. Wingreen and S. J. Benkovic, Mol. Cell, 2021, 81, 3775–3785 CrossRef CAS PubMed.
- E. T. Hwang and S. Lee, ACS Catal., 2019, 9, 4402–4425 CrossRef CAS.
- C. Myhrvold, J. K. Polka and P. A. Silver, ACS Synth. Biol., 2016, 5, 1396–1403 CrossRef CAS PubMed.
- Y. Ouyang, P. Zhang and I. Willner, Sci. Adv., 2022, 8, eabn3534 CrossRef CAS PubMed.
- J. S. Qin, S. Yuan, C. Lollar, J. Pang, A. Alsalme and H. C. Zhou, Chem. Commun., 2018, 54, 4231–4249 RSC.
- W.-H. Chen, M. Vázquez-González, A. Zoabi, R. Abu-Reziq and I. Willner, Nat. Catal., 2018, 1, 689–695 CrossRef CAS.
- H. Lee, W. C. DeLoache and J. E. Dueber, Metab. Eng., 2012, 14, 242–251 CrossRef CAS PubMed.
- O. I. Wilner, Y. Weizmann, R. Gill, O. Lioubashevski, R. Freeman and I. Willner, Nat. Nanotechnol., 2009, 4, 249–254 CrossRef CAS PubMed.
- J. E. Dueber, G. C. Wu, G. R. Malmirchegini, T. S. Moon, C. J. Petzold, A. V. Ullal, K. L. Prather and J. D. Keasling, Nat. Biotechnol., 2009, 27, 753–759 CrossRef CAS PubMed.
- E. A. Berckman and W. Chen, Chem. Commun., 2020, 56, 11426–11428 RSC.
- Z. Ge, J. Fu, M. Liu, S. Jiang, A. Andreoni, X. Zuo, Y. Liu, H. Yan and C. Fan, ACS Appl. Mater. Interfaces, 2019, 11, 13881–13887 CrossRef CAS PubMed.
- D. Wang, Y. Chai, Y. Yuan and R. Yuan, ACS Appl. Mater. Interfaces, 2020, 12, 2871–2877 CrossRef CAS PubMed.
- D. Wang, Y. Chai, Y. Yuan and R. Yuan, Anal. Chem., 2019, 91, 3561–3566 CrossRef CAS PubMed.
- M. Liu, Z. Li, Y. Li, J. Chen and Q. Yuan, Chin. Chem. Lett., 2019, 30, 1009–1012 CrossRef CAS.
- Y. Liu, R. Lv, S. Sun, D. Tan, F. Dong, Y. A. Golubev, X. Nie, O. B. Kotova, J. Liu and K. Wang, Chin. Chem. Lett., 2022, 33, 807–811 CrossRef CAS.
- F. Zhang and W. B. Zhang, Chin. J. Chem., 2020, 38, 864–878 CrossRef CAS.
- F. Yang, X. Zuo, C. Fan and X.-E. Zhang, Natl. Sci. Rev., 2018, 5, 740–755 CrossRef CAS.
- J. Fu, Y. R. Yang, A. Johnson-Buck, M. Liu, Y. Liu, N. G. Walter, N. W. Woodbury and H. Yan, Nat. Nanotechnol., 2014, 9, 531–536 CrossRef CAS PubMed.
- M. Li, F. Yin, L. Song, X. Mao, F. Li, C. Fan, X. Zuo and Q. Xia, Chem. Rev., 2021, 121, 10469–10558 CrossRef CAS PubMed.
- M. Li, H. Ding, M. Lin, F. Yin, L. Song, X. Mao, F. Li, Z. Ge, L. Wang, X. Zuo, Y. Ma and C. Fan, J. Am. Chem. Soc., 2019, 141, 18910–18915 CrossRef CAS PubMed.
- X. Yang, J. Li, H. Pei, Y. Zhao, X. Zuo, C. Fan and Q. Huang, Anal. Chem., 2014, 86, 3227–3231 CrossRef CAS PubMed.
- J. S. Kahn, Y. Xiong, J. Huang and O. Gang, JACS Au, 2022, 2, 357–366 CrossRef CAS PubMed.
- J. Fu, M. Liu, Y. Liu, N. W. Woodbury and H. Yan, J. Am. Chem. Soc., 2012, 134, 5516–5519 CrossRef CAS PubMed.
- R. Kosinski, J. M. Perez, E.-C. Schöneweiß, Y. B. Ruiz-Blanco, I. Ponzo, K. Bravo-Rodriguez, M. Erkelenz, S. Schlücker, G. Uhlenbrock, E. Sanchez-Garcia and B. Saccà, Sci. Adv., 2022, 8, eabk0425 CrossRef CAS PubMed.
- L. C. Bock, L. C. Griffin, J. A. Latham, E. H. Vermaas and J. J. Toole, Nature, 1992, 355, 564–566 CrossRef CAS PubMed.
- Q. Hu, H. Li, L. Wang, H. Gu and C. Fan, Chem. Rev., 2019, 119, 6459–6506 CrossRef CAS PubMed.
- A. Sreekumar, L. M. Poisson, T. M. Rajendiran, A. P. Khan, Q. Cao, J. Yu, B. Laxman, R. Mehra, R. J. Lonigro, Y. Li, M. K. Nyati, A. Ahsan, S. Kalyana-Sundaram, B. Han, X. Cao, J. Byun, G. S. Omenn, D. Ghosh, S. Pennathur, D. C. Alexander, A. Berger, J. R. Shuster, J. T. Wei, S. Varambally, C. Beecher and A. M. Chinnaiyan, Nature, 2009, 457, 910–914 CrossRef CAS PubMed.
- B. Cavaliere, B. Macchione, M. Monteleone, A. Naccarato, G. Sindona and A. Tagarelli, Anal. Bioanal. Chem., 2011, 400, 2903–2912 CrossRef CAS PubMed.
- N. Cernei, Z. Heger, J. Gumulec, O. Zitka, M. Masarik, P. Babula, T. Eckschlager, M. Stiborova, R. Kizek and V. Adam, Int. J. Mol. Sci., 2013, 14, 13893–13908 CrossRef PubMed.
- M. Lin, J. Wang, G. Zhou, J. Wang, N. Wu, J. Lu, J. Gao, X. Chen, J. Shi, X. Zuo and C. Fan, Angew. Chem., Int. Ed., 2015, 54, 2151–2155 CrossRef CAS PubMed.
- P. Song, J. Shen, D. Ye, B. Dong, F. Wang, H. Pei, J. Wang, J. Shi, L. Wang, W. Xue, Y. Huang, G. Huang, X. Zuo and C. Fan, Nat. Commun., 2020, 11, 838 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an02097a |
| This journal is © The Royal Society of Chemistry 2023 |