Open Access Article
Open Access ArticleSupramolecular assembly of DNA-constructed vesicles†
Simon
Rothenbühler
 a,
Ioan
Iacovache
b,
Simon M.
Langenegger
a,
Benoît
Zuber
a,
Ioan
Iacovache
b,
Simon M.
Langenegger
a,
Benoît
Zuber
 *b and
Robert
Häner
*b and
Robert
Häner
 *a
*a
aDepartment of Chemistry and Biochemistry, University of Bern, Freiestrasse 3, CH – 3012 Bern, Switzerland. E-mail: robert.haener@dcb.unibe.ch; Web: http://haener.dcb.unibe.ch
bInstitute of Anatomy, University of Bern, Baltzerstrasse 2, CH – 3012 Bern, Switzerland. E-mail: benoit.zuber@ana.unibe.ch
First published on 29th June 2020
Abstract
The self-assembly of DNA hybrids possessing tetraphenylethylene sticky ends at both sides into vesicular architectures in aqueous medium is demonstrated. Cryo-electron microscopy reveals the formation of different types of morphologies from the amphiphilic DNA-hybrids. Depending on the conditions, either an extended (sheet-like) or a compact (columnar) alignment of the DNA hybrids is observed. The different modes of DNA arrangement lead to the formation of vesicles appearing either as prolate ellipsoids (type I) or as spheres (type II). The type of packing has a significant effect on the accessibility of the DNA, as evidenced by intercalation and light-harvesting experiments. Only the vesicles exhibiting the sheet-like DNA alignment are accessible for intercalation by ethidium bromide or for the integration of chromophore-labelled DNA via a strand exchange process. The dynamic nature of type I vesicles enables their elaboration into artificial light-harvesting complexes by DNA-guided introduction of Cy3-acceptor chromophores. DNA-constructed vesicles of the kind shown here represent versatile intermediates that are amenable to further modification for tailored nanotechnology applications.
Introduction
DNA nanotechnology offers the possibility of a rational design and construction of precise architectures due to the reliability of nucleobase pairing.1–4 The programmable bottom-up construction of nano-sized structures via DNA origami provides a variety of different shapes, such as rings, tubes, or polyhedra, among others.5–7 Engineering short single-stranded overhangs, known as sticky ends, extend potential strategies for the assembly of DNA into nanostructures ranging from two-dimensional tiles, to three-dimensional DNA crystals, or nanocapsules.8–12 Merging purely artificial DNA nucleotide surrogates with natural DNA nucleotides lead to DNA conjugates and the resulting functional supramolecular assemblies have recently gained much attention in the fields of nanotechnology and materials science.13–19 Such functional supramolecular polymers feature properties beyond the classical role of DNA in biological systems, with applications in optoelectronic devices, drug delivery systems, and diagnostics to name a few.20–28 However, vesicular morphologies are predominantly reported from lipid–DNA conjugates or DNA functionalized liposomes,29–34 while the field apart from lipid-based DNA constructs remains largely unexplored.35 In previous work, we demonstrated the self-assembly of phenanthrene–DNA conjugates into vesicular structures with light-harvesting features.36 Combining this approach with the rapidly evolving area of aggregation-induced emission (AIE),37–40 we herein present DNA constructs assembled from DNA conjugates, functionalized with the AIEgen tetraphenylethylene (TPE). The resulting AIE-active supramolecular assemblies were characterized by cryo-electron microscopy (cryo-EM)41,42 at a resolution level of the width of a DNA duplex. Additionally, the accessibility of the DNA duplex within the two types of architectures was tested by DNA intercalation experiments using ethidium bromide (EthBr) and by light-harvesting experiments.Results and discussion
The chemically modified oligonucleotides utilized in this work are depicted in Fig. 1. Strands A and B were prepared via solid-phase synthesis and are modified at their 3′-ends with three phosphodiester-linked E-TPE units. The synthesis of the corresponding TPE phosphoramidite building block was adapted from published procedures.43 Due to the complementarity of DNA single strands A and B, they hybridize and form the DNA duplex A·B, which contains TPE overhangs (sticky ends) on both sides.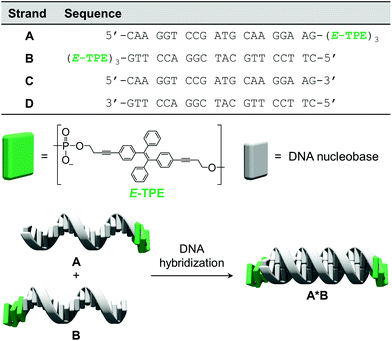 | ||
| Fig. 1 Sequences of oligomers A–D, chemical structure of the TPE modification and illustration of DNA hybrid A·B. | ||
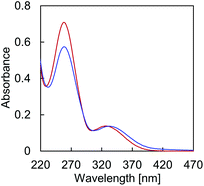
Temperature dependent absorption profiles of A·B (Fig. 2) show a combination of absorption by TPE and the DNA nucleobases in the region around 260 nm (Fig. S13, ESI†). The structureless band around 330 nm, on the other hand, is due to TPE absorption only. The hypochromic effect of the 260 nm band upon controlled cooling from 75 °C to 20 °C originates from DNA hybridization. In addition, cooling of the solution results in a bathochromic shift of the maximum at 326 nm to 333 nm, due to TPE aggregation.
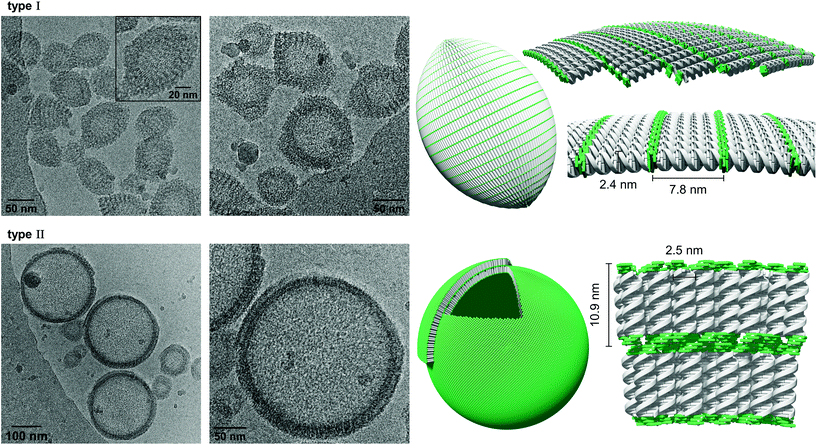
Cryo-EM enables the characterization of structures in their actual morphology, since vitrified specimens preserve the structures present in solution.41,42 Therefore, the formation of supramolecular assemblies was investigated by cryo-EM (Fig. 3), which revealed the coexistence of two distinct morphologies. Hybrid A·B either self-assembles into vesicles with an extended alignment of the DNA duplexes (type I, Fig. 3, top) or in a compact arrangement (type II, Fig. 3, bottom). More than 94% of all observed vesicles exhibit the rugby-ball-shaped morphology (type I) after assembly by a simple cooling procedure under these conditions. A notable feature of the extended architecture is the appearance of a regular pattern of discrete bands. The average distance between the bands is 7.8 ± 0.5 nm (Fig. 3 and Fig. S16, ESI†),44–46 which corresponds well with the length of a 23-mer A·B, assuming a contact distance of 3.4 Å between base pairs or between stacked TPE units. The width of the DNA duplex A·B (see Fig. 3, inset) is also observable and found to be about 2.4 nm. The two morphologies exhibit different diameters: while the size range of type I vesicles is roughly between 50 and 100 nm, the diameter of type II vesicles ranges from 200 to 350 nm. A thickness of 10.9 ± 0.5 nm was observed for the membrane of the compact vesicles (Fig. 3, bottom), which agrees well with the length of hybrid A·B. The width of the DNA duplex A·B in type II vesicles was measured to be about 2.5 nm. In vesicles with this morphology, the TPE sticky ends are positioned on both sides of the membrane. Therefore, the assembly of additional layers of the same type is facilitated via π-stacking. As a result, double layers are clearly discernible in many areas of the images. The presence of spermine is essential for the assembly of the vesicles. DNA duplexes are tightly packed in both types of vesicles and the resulting coulombic repulsion between the negatively charged phosphate backbones is neutralized by the polycation. In the absence of spermine, no vesicles were visible by cryo-EM (Fig. S17, ESI†). The self-assembly of duplex A·B into vesicles is further affirmed by atomic force microscopy (AFM) images (Fig. S23, ESI†).
As described above, assembly of the vesicles from duplex A·B was accomplished in the presence of 20 vol% ethanol. In order to investigate the effect of solvent composition on the morphology, ethanol was removed by dialysis against 10 mM sodium phosphate buffer pH 7.2, containing 0.1 mM spermine·4 HCl. As evidenced by cryo-EM images (Fig. 4), the ratio of the two types of vesicles is substantially affected by dialysis. Whereas the vast majority of vesicles belonged to type I before dialysis, more than 90% exhibited the compact type II morphology (membrane thickness: 10.6 ± 0.6 nm) after removal of ethanol. Additionally, also the average diameter had decreased to 50–150 nm. Performing the thermally controlled assembly process in the absence of ethanol yielded only small, ill-defined aggregates (Fig. S20, ESI†). Thus, ethanol is required to assemble the vesicles in the first instance, which renders this two-step procedure necessary to obtain well-defined vesicles in the absence of ethanol.
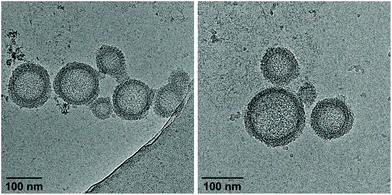 | ||
| Fig. 4 Cryo-EM images of vesicles from hybrid A·B after removal of ethanol by dialysis. Conditions: 1 μM A·B, 10 mM sodium phosphate buffer pH 7.2, 0.1 mM spermine·4 HCl. | ||
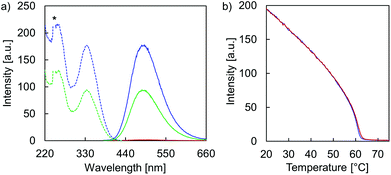
Temperature-dependent fluorescence spectra (Fig. 5a) and fluorescence quantum yields (ΦFL, Table 1) demonstrate the AIE properties of hybrid A·B. At 75 °C, emission is close to zero (ΦFL < 0.75%) after TPE excitation at 335 nm. This suggests that the two single strands A and B are completely disassembled and that the TPE units show only negligible aggregation within the single stranded oligomers. In agreement with AIE properties, TPE fluorescence emerges during the assembly process (slowly cooling the solution to 20 °C). The maximum of the structureless emission band is centered around 490 nm with a substantially increased ΦFL (31%). The excitation spectrum confirms that fluorescence originates from the AIE-active TPE units (Fig. 5a). The mechanism of the supramolecular assembly process was examined by fluorescence monitored annealing curves (Fig. 5b). The shape of the curve is non-sigmoidal, featuring a sharp increase in fluorescence with an onset temperature of 62 °C. This strongly suggests a cooperative assembly process47,48 of the vesicles, with a nucleation temperature around 62 °C.
| 75 °C | 20 °C, after assembly process | 20 °C, after dialysis | |
|---|---|---|---|
| Φ FL [%] | <0.75 | 31 ± 1 | 23 ± 2 |
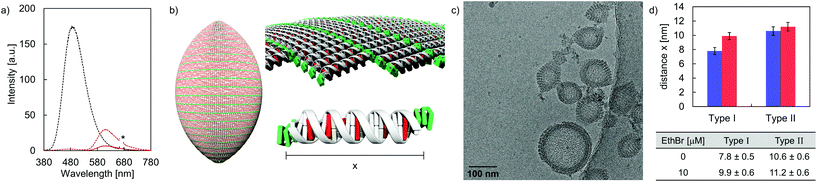
DNA intercalation experiments using EthBr were performed in order to gain information on the accessibility of DNA in the formed vesicles. Taking the neighbor exclusion principle49 into account, up to 10 EthBr can intercalate into the 20-mer DNA duplex C·D (Fig. S28, ESI†).50 Upon intercalation, the fluorescence emission signal is enhanced, compared to the intrinsic fluorescence of free EthBr in solution (Fig. S29–S32, ESI†).51,52 After the addition of EthBr (10 μM) to the rugby-ball-shaped type I vesicles, TPE emission is almost completely quenched upon TPE excitation (Fig. 6a). Instead, a new band centered around 610 nm, due to EthBr emission emerged, which implies excitation energy transfer from the TPE units to EthBr. Based on the relative integrated fluorescence intensities (EthBr excitation at 520 nm) between the DNA duplex C·D and hybrid A·B, it was calculated that about 6 EthBr intercalate per duplex A·B (Fig. 6b and Table S4, ESI†). On the other hand, only small amounts of EthBr (≤2 per DNA) may intercalate in type II vesicles, indicating that just the extended arrangement in type I vesicles enables efficient EthBr intercalation. Cryo-EM imaging supports EthBr intercalation into type I vesicles (Fig. 6c), which demonstrates a statistically significant widening of the discrete bands from originally 7.8 ± 0.5 nm to 9.9 ± 0.6 nm after the addition of EthBr (Fig. 6d). Assuming a lengthening of the helix by 3.4 Å per intercalator,53,54 approximately 6 EthBr are intercalated per hybrid A·B, which is in line with the results obtained from fluorescence spectroscopy. No statistically significant extension (from 10.6 ± 0.6 nm to 11.2 ± 0.6 nm) was observed when EthBr was added to type II vesicles (Fig. S22, ESI†).
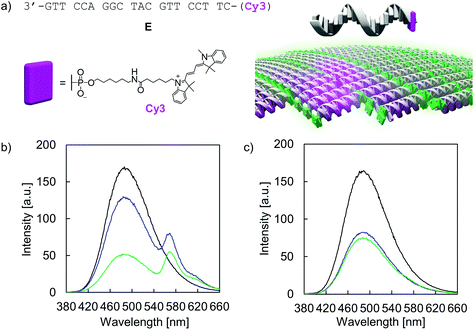
The addressability and accessibility of the vesicles was further studied by incorporation experiments of a Cy3-labelled DNA single strand E (Fig. 7a). Strand E is complementary to strand A. After incorporation of strand E into the vesicular membrane by DNA strand exchange, excitation energy transfer from the TPE donors to Cy3 acceptor is expected (λex.: 335 nm). Doping of type I vesicles with Cy3 (1 mol% per TPE unit) was accomplished by simple addition of strand E to the preformed vesicles at 20 °C and waiting for 10 min before measurement. This doping process leads to a reduction of TPE emission, along with the appearance of Cy3 emission at 570 nm (Fig. 7b). Energy transfer is presumably taking place via a Förster resonance energy transfer (FRET) mechanism. The FRET efficiency was calculated according to integrated TPE fluorescence intensities (Fig. S36 and Table S5, ESI†). Based on this, 22 ± 4 TPE units contribute to the observed excitation energy transfer to the Cy3 acceptor in this artificial light-harvesting complex. Light-harvesting properties are maintained after dialysis, indicating that strand E is not removed by dialysis (Fig. 7b). Control experiments with a non-complementary, Cy3-modified DNA single strand F show no energy transfer and thus confirming the sequence specificity of the doping process (Fig. S38 and S39, ESI†). When strand E was added to type II vesicles, only marginal energy transfer was observed (Fig. 7c). Since type II vesicles are partially multi-lamellar, this observation can be explained by edge effects, i.e. strand exchange and incorporation of strand E can only take place at the exposed areas but not in the core of the compact, columnar arrangement.
Conclusions
In conclusion, the assembly of an amphiphilic DNA possessing TPE sticky ends at both sides into two different types of vesicular constructs has been demonstrated. Cryo-EM imaging reveals a regular alignment of DNA duplexes in both types of vesicular morphologies. Vesicles of type I appear as prolate ellipsoids with a diameter of 50–100 nm. The dimensions of the vesicular membrane suggest an extended arrangement of DNA duplexes that interact via AIE-active TPE sticky ends in the presence of spermine. Type II vesicles, on the other hand, exhibit a thicker membrane, which is compatible with a model of DNA duplexes arranged in a more compact (columnar) manner. The abundance as well as the size of type II vesicles is influenced by the solvent composition (i.e. ethanol content). The type of arrangement plays a crucial role for the accessibility of the DNA duplexes. Thus, only type I vesicles exhibiting the extended DNA duplex alignment are amenable to ethidium intercalation or to the incorporation of a Cy3-labelled DNA via strand exchange. The incorporation of acceptor chromophores into type I vesicles resulted in the formation of light-harvesting vesicular constructs. Ongoing research focuses on exploring the suitability of the presented vesicles for light-harvesting applications or as DNA-addressable nanocarriers for targeted delivery.Author contributions
S. R. designed the project, synthesized the oligomers, performed the experiments, analyzed the data, and wrote the paper. I. I. performed cryo-EM experiments, analyzed the data, and contributed to the writing of the paper. S. M. L. designed the project, analyzed the data, created the artwork, and contributed to the writing of the paper. B. Z. designed and supervised the project and contributed to the writing of the paper. R. H. designed and supervised the project, analyzed the data, and wrote the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support by the Swiss National Foundation (200020_188468 to RH and 179520 to BZ) is gratefully acknowledged. Cryo-electron microscopy was performed on equipment supported by the Microscopy Imaging Center (MIC), University of Bern, Switzerland.References
- U. Feldkamp and C. M. Niemeyer, Angew. Chem., Int. Ed., 2006, 45, 1856–1876 CrossRef CAS.
- N. C. Seeman, Nature, 2003, 421, 427–431 CrossRef PubMed.
- Y.-J. Chen, B. Groves, R. A. Muscat and G. Seelig, Nat. Nanotechnol., 2015, 10, 748–760 CrossRef CAS PubMed.
- H. Ramezani and H. Dietz, Nat. Rev. Genet., 2020, 21, 5–26 CrossRef CAS PubMed.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- J. R. Burns, E. Stulz and S. Howorka, Nano Lett., 2013, 13, 2351–2356 CrossRef CAS PubMed.
- R. P. Thomsen, M. G. Malle, A. H. Okholm, S. Krishnan, S. S. R. Bohr, R. S. Sørensen, O. Ries, S. Vogel, F. C. Simmel, N. S. Hatzakis and J. Kjems, Nat. Commun., 2019, 10, 5655 CrossRef PubMed.
- N. C. Seeman, DNA Cell Biol., 1991, 10, 475–486 CrossRef CAS PubMed.
- E. Ban and C. R. Picu, Biomacromolecules, 2014, 15, 143–149 CrossRef CAS PubMed.
- M. N. Hansen, A. M. Zhang, A. Rangnekar, K. M. Bompiani, J. D. Carter, K. V. Gothelf and T. H. LaBean, J. Am. Chem. Soc., 2010, 132, 14481–14486 CrossRef CAS PubMed.
- Y. P. Ohayon, C. Hernandez, A. R. Chandrasekaran, X. Wang, H. O. Abdallah, M. A. Jong, M. G. Mohsen, R. Sha, J. J. Birktoft, P. S. Lukeman, P. M. Chaikin, S. L. Ginell, C. Mao and N. C. Seeman, ACS Nano, 2019, 13, 7957–7965 CrossRef CAS PubMed.
- D. Bhatia, S. Mehtab, R. Krishnan, S. S. Indi, A. Basu and Y. Krishnan, Angew. Chem., Int. Ed., 2009, 48, 4134–4137 CrossRef CAS PubMed.
- M. Vybornyi, Y. Vyborna and R. Häner, Chem. Soc. Rev., 2019, 48, 4347–4360 RSC.
- S. P. W. Wijnands, E. W. Meijer and M. Merkx, Bioconjugate Chem., 2019, 30, 1905–1914 CrossRef CAS PubMed.
- M. Madsen and K. V. Gothelf, Chem. Rev., 2019, 119, 6384–6458 CAS.
- C. Gong, S. Sun, Y. Zhang, L. Sun, Z. Su, A. Wu and G. Wei, Nanoscale, 2019, 11, 4147–4182 RSC.
- J.-F. Lutz, ACS Macro Lett., 2020, 9, 185–189 CrossRef CAS.
- C. J. Serpell, M. Barłóg, K. Basu, J. F. Fakhoury, H. S. Bazzi and H. F. Sleiman, Mater. Horiz., 2014, 1, 348–354 RSC.
- E. M. Estirado, M. A. A. Garcia, J. Schill and L. Brunsveld, J. Am. Chem. Soc., 2019, 141, 18030–18037 CrossRef PubMed.
- P. Ensslen, S. Gärtner, K. Glaser, A. Colsmann and H.-A. Wagenknecht, Angew. Chem., Int. Ed., 2016, 55, 1904–1908 CrossRef CAS PubMed.
- X. Wang, R. Sha, M. Kristiansen, C. Hernandez, Y. Hao, C. Mao, J. W. Canary and N. C. Seeman, Angew. Chem., Int. Ed., 2017, 56, 6445–6448 CrossRef CAS PubMed.
- H. Bui, S. A. Díaz, J. Fontana, M. Chiriboga, R. Veneziano and I. L. Medintz, Adv. Opt. Mater., 2019, 7, 1900562 CrossRef.
- M. Kownacki, S. M. Langenegger, S.-X. Liu and R. Häner, Angew. Chem., Int. Ed., 2019, 58, 751–755 CrossRef CAS PubMed.
- B. L. Li, R. Li, H. L. Zou, K. Ariga, N. B. Li and D. T. Leong, Mater. Horiz., 2020, 7, 455–469 RSC.
- D. Bousmail, L. Amrein, J. J. Fakhoury, H. H. Fakih, J. C. C. Hsu, L. Panasci and H. F. Sleiman, Chem. Sci., 2017, 8, 6218–6229 RSC.
- C.-H. Lu and I. Willner, Angew. Chem., Int. Ed., 2015, 54, 12212–12235 CrossRef CAS PubMed.
- Z.-Y. Wang, C.-P. Zhang and C.-Y. Zhang, Chem. Commun., 2020, 56, 2119–2122 RSC.
- B. Shin, W.-K. Kim, S. Yoon and J. Lee, Sens. Actuators, B, 2020, 305, 127471 CrossRef.
- S. R. Bandara, T. G. Molley, H. Kim, P. A. Bharath, K. A. Kilian and C. Leal, Mater. Horiz., 2020, 7, 125–134 RSC.
- Y. H. Roh, J. B. Lee, P. Kiatwuthinon, M. R. Hartman, J. J. Cha, S. H. Um, D. A. Muller and D. Luo, Small, 2011, 7, 74–78 CrossRef CAS PubMed.
- A. Patwa, A. Gissot, I. Bestel and P. Barthélémy, Chem. Soc. Rev., 2011, 40, 5844–5854 RSC.
- N. Dave and J. Liu, ACS Nano, 2011, 5, 1304–1312 CrossRef CAS PubMed.
- P. M. G. Löffler, O. Ries, A. Rabe, A. H. Okholm, R. P. Thomsen, J. Kjems and S. Vogel, Angew. Chem., Int. Ed., 2017, 56, 13228–13231 CrossRef PubMed.
- S. Matoori and J.-C. Leroux, Mater. Horiz., 2020, 7, 1297–1309 RSC.
- Y. Kamiya, Y. Yamada, T. Muro, K. Matsuura and H. Asanuma, ChemMedChem, 2017, 12, 2016–2021 CrossRef CAS PubMed.
- C. D. Bösch, J. Jevric, N. Bürki, M. Probst, S. M. Langenegger and R. Häner, Bioconjugate Chem., 2018, 29, 1505–1509 CrossRef PubMed.
- J. Luo, Z. Xie, J. W. Y. Lam, L. Cheng, H. Chen, C. Qiu, H. S. Kwok, X. Zhan, Y. Liu, D. Zhu and B. Z. Tang, Chem. Commun., 2001, 1740–1741 RSC.
- J. Mei, N. L. C. Leung, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Chem. Rev., 2015, 115, 11718–11940 CrossRef CAS PubMed.
- Y. Chen, J. W. Y. Lam, R. T. K. Kwok, B. Liu and B. Z. Tang, Mater. Horiz., 2019, 6, 428–433 RSC.
- Y. Zhang, Y. Wang, J. Wang and X.-J. Liang, Mater. Horiz., 2018, 5, 799–812 RSC.
- J. Dubochet, M. Adrian, J.-J. Chang, J.-C. Homo, J. Lepault, A. W. McDowall and P. Schultz, Q. Rev. Biophys., 1988, 21, 129–228 CrossRef CAS PubMed.
- M. Adrian, B. ten Heggeler-Bordier, W. Wahli, A. Z. Stasiak, A. Stasiak and J. Dubochet, EMBO J., 1990, 9, 4551–4554 CrossRef CAS PubMed.
- S. Li, S. M. Langenegger and R. Häner, Chem. Commun., 2013, 49, 5835–5837 RSC.
- C. T. Rueden, J. Schindelin, M. C. Hiner, B. E. DeZonia, A. E. Walter, E. T. Arena and K. W. Eliceiri, BMC Bioinf., 2017, 18, 529 CrossRef PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- M. Linkert, C. T. Rueden, C. Allan, J.-M. Burel, W. Moore, A. Patterson, B. Loranger, J. Moore, C. Neves, D. MacDonald, A. Tarkowska, C. Sticco, E. Hill, M. Rossner, K. W. Eliceiri and J. R. Swedlow, J. Cell Biol., 2010, 189, 777–782 CrossRef CAS PubMed.
- D. Zhao and J. S. Moore, Org. Biomol. Chem., 2003, 1, 3471–3491 RSC.
- P. A. Korevaar, S. J. George, A. J. Markvoort, M. M. J. Smulders, P. A. J. Hilbers, A. P. H. J. Schenning, T. F. A. De Greef and E. W. Meijer, Nature, 2012, 481, 492–496 CrossRef CAS PubMed.
- J. Cairns, Cold Spring Harbor Symp. Quant. Biol., 1962, 27, 311–318 CrossRef CAS PubMed.
- M. J. Waring, J. Mol. Biol., 1965, 13, 269–282 CrossRef CAS PubMed.
- J. B. Lepecq and C. Paoletti, J. Mol. Biol., 1967, 27, 87–106 CrossRef CAS PubMed.
- J. Olmsted and D. R. Kearns, Biochemistry, 1977, 16, 3647–3654 CrossRef CAS PubMed.
- L. S. Lerman, J. Mol. Biol., 1961, 3, 18–30 CrossRef CAS PubMed.
- G. M. Blackburn, M. J. Gait, D. Loakes and D. M. Williams, Nucleic Acids in Chemistry and Biology, Royal Society of Chemistry, Cambridge, 2006 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, NMR spectra, additional UV-vis and fluorescence spectra, additional cryo-EM images, AFM images, fluorescence quantum yield determination. See DOI: 10.1039/d0nr04103c |
| This journal is © The Royal Society of Chemistry 2020 |