Exosome-mediated microRNA-497 delivery for anti-cancer therapy in a microfluidic 3D lung cancer model†
Kyeongsoo
Jeong‡
a,
Yeong Jun
Yu‡
b,
Jae Young
You
a,
Won Jong
Rhee
 *ac and
Jeong Ah
Kim
*ac and
Jeong Ah
Kim
 *bd
*bd
aDepartment of Bioengineering and Nano-Bioengineering, Incheon National University, Incheon 22012, Republic of Korea. E-mail: wjrhee@inu.ac.kr
bResearch Center for Bioconvergence Analysis, Korea Basic Science Institute, Chungbuk 28119, Republic of Korea. E-mail: jakim98@kbsi.ac.kr
cDivision of Bioengineering, Incheon National University, Incheon 22012, Republic of Korea
dDepartment of Bio-Analytical Science, University of Science and Technology, Daejeon 34113, Republic of Korea
First published on 7th January 2020
Abstract
Non-small cell lung cancer (NSCLC) is one of the leading causes of death from cancer worldwide. The delivery and controlled regulation of miRNAs via exosomes is known as a potential therapeutic approach in the treatment of cancer. In this study, human cell-derived exosomes were used as delivery vehicles for miRNAs, and we investigated their anti-tumor and anti-angiogenic effects on NSCLCs that were cultured in 2D and 3D microfluidic devices. We demonstrated that exosomes that contained miRNA-497 (miR-497) effectively suppressed tumor growth and the expression of their associated genes, i.e., yes-associated protein 1 (YAP1), hepatoma-derived growth factor (HDGF), cyclin E1 (CCNE1), and vascular endothelial growth factor-A (VEGF-A), in A549 cells. Also, the level of VEGF-A-mediated angiogenic sprouting was decreased drastically in human umbilical vein endothelial cells (HUVECs) cultured in a microfluidic device. To mimic the in vivo-like tumor microenvironment of NSCLC, A549 cells were co-cultured with HUVECs in a single device, and miR-497-loaded exosomes were delivered to both types of cells. As a result, both the tube formation of endothelial cells and the migration of tumor decreased dramatically compared to the control. This indicated that miR-497 has synergistic inhibitory effects that target tumor growth and angiogenesis, so exosome-mediated miRNA therapeutics combined with the microfluidic technology could be a predictive, cost-efficient translational tool for the development of targeted cancer therapy.
Introduction
MiRNA delivery to modulate cancer-associated gene expression is currently under investigation as a promising tool for cancer therapy.1–3 MiRNAs are known to be key players in cancer-related processes, including proliferation, apoptosis, invasion, and metastasis.4,5 One strategy for anti-cancer therapy is to deliver miRNA-like molecules that bind complementarily to tumorigenic and angiogenic factors to suppress their expression. Despite the high potential of miRNAs for gene therapy, they have many challenges to overcome to become therapeutically successful. For example, it is difficult for them to pass through cell membranes because they are negatively charged and hydrophilic.6For these reasons, exosomes have attracted significant attention because of their potential use for delivering exogenous therapeutic bioactive molecules to a target in vivo.7–10 Unlike other chemical and biological vehicles,11–13 exosomes are natural nanocarriers derived from cells, which facilitates intracellular molecular transport. In addition, their robust exosomal membrane can protect the miRNAs from degradation, and they are taken up rapidly by the target cells, which enables efficient gene delivery with lower immunogenicity/toxicity. The biocompatible and specific lipid/protein composition of the exosomal membrane that is capable of fusion with cells is one of their unique characteristics, allowing them to deliver target molecules directly into the cytosol.
To better predict the therapeutic effects of the miRNAs delivered to the target region through exosomes, an in vitro system is required as a translational tool to bridge the gap between preclinical models and clinical outcomes. Such an in vitro system should mimic the high complexity and spatial heterogeneity of an in vivo microenvironment. This will allow the validation of the potential therapeutic effect of the miRNAs delivered to the target site or the recipient cell, through visualization or by in-depth studies at the gene and protein levels.
With advances in microfabrication technology, 3D microfluidic systems with in vivo-friendly microenvironments are considered to be promising alternative tools for preclinical evaluations, as well as mechanism studies.14–20 Because 3D microfluidic models can recapitulate the structure, function, and physiological features of tissues, as well as dynamic three-dimensional environments in vivo, these models are better suited for the therapeutic testing of exosomal drugs. These models are often designed to accommodate cancer cells and vascular networks, enabling the fundamental mechanism studies associated with angiogenesis, invasion, and metastasis.20 In particular, gel-supported 3D cell culture devices that employ an extracellular matrix, e.g., collagen, have allowed the study of sprouting and migration in response to the chemical-factor gradient controlled by microfluidic channels.21
Many reports have shown that miR-497 binds to tumorigenic and angiogenic factors in lung cancer cells and vascular endothelial cells; thus, it blocks the protein expression associated with tumor growth and angiogenesis by inhibiting the translation.22–27 Based on these findings, we developed miR-497 exosomes as novel therapeutics for the treatment of NSCLC, which is the most important and prevalent subtype of lung cancer, and tested their effectiveness using a microfluidic device.
First, we optimized the method for isolating miR-497-loaded exosomes, which enabled a high yield and purity, and we achieved efficient cellular delivery and stable protection of miR-497 using exosomes. Next, A549 cells and HUVECs were used to mimic the microenvironment of the tumor as a model of NSCLC. The complex in vivo-like microenvironment that provides cell–cell communication was constructed using a microfluidic device to verify the anti-cancer and anti-angiogenic effects of the exosomes loaded with miR-497. We demonstrated the versatile capability of these exosomes to reduce tumor angiogenesis in both cancer cells and endothelial cells, and we proved that the miRNA-loaded exosomes can synergistically and simultaneously regulate tumor growth and angiogenesis.
Results and discussion
Overall research strategy
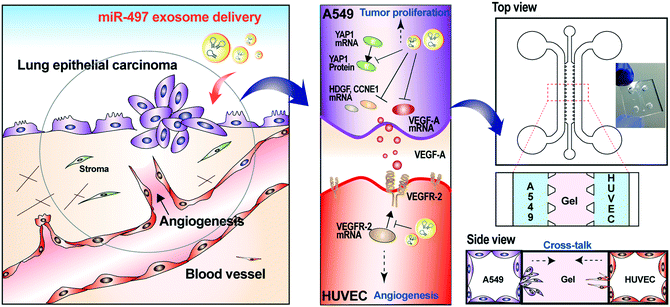
Non-small cell lung cancer (NSCLC) has one of the highest mortality rates among cancer-related diseases, and the 5-year survival rate of patients with NSCLC is very low. Recently, it has been shown that targeting a set of oncogenic genes or pathways simultaneously by miRNAs can trigger synergistic therapeutic effects.3,23 MiRNA can also target tumor-promoting stromal cells in addition to tumor cells. The angiogenesis by endothelial cells strongly affects the tumor formation, progression, and metastasis. Cancer cells actively release various angiogenic factors to form blood vessels and receive oxygen and nutrients. Among them, VEGF-A is the most potent trigger of angiogenesis, and it activates the angiogenic signaling pathway by binding to VEGFR-2, which is present in the membranes of endothelial cells. Several miRNAs are involved in modulating the gene expression associated with tumorigenesis and angiogenesis. Among them, miR-497 has been known to bind directly to the mRNA of VEGF-A, YAP1, HDGF, CCNE1, and VEGFR-2, and regulates their translations.28 There have been reports that NSCLC has a significantly higher ratio of VEGF-A protein over its mRNA, and a significantly lower level of miR-497 compared to other cancers.22 This suggests that NSCLC might have post-transcriptional control of VEGF-A. Thus, our strategy is that miR-497 is delivered through the exosome in the NSCLC cells and that it binds directly to tumor growth-related genes and VEGF-A mRNAs, which regulate the downstream signaling pathway, thereby inhibiting the expression of VEGF-A and the growth of tumors. Also, we investigated whether the delivered miR-497 also affected VEGFR-2 expression in HUVECs. To precisely investigate the phenomena that occur in the NSCLC microenvironment, we mimicked the local NSCLC microenvironment by co-culturing NSCLC cells (A549 cells) and endothelial cells (HUVECs) in a 3D microfluidic device, and we monitored the effects of miR-497 delivered by exosomes on tumorigenesis and angiogenesis (Fig. 1).Isolation, purification and characterization of exosomes
HEK293T cells were chosen to generate exosomal vehicles for loading miRNAs. Some researchers have reported that the exosomes derived from HEK293T cells contain minimal intrinsic cargos and there are no disease-associated pathway inducing molecules in the cells, so that any unwanted molecular delivery is minimized.29–31 Accordingly, HEK293T cell-derived exosomes are suitable for the delivery of exogenous miRNAs with a low possibility of adverse effects after administration. We collected a conditioned medium of HEK293T to obtain exosomes, and then they were subjected to the ExoQuick-TC™ precipitation (EXQ) or differential ultracentrifugation (UC) method. Generally, differential ultracentrifugation has been viewed as the gold standard method to obtain high-purity exosomes, but this method results in significantly greater losses of exosomes than the EXQ method. However, even though the EXQ method is a well-established, commercialized method, it has the critical problem of high protein impurities. Thus, after isolation using EXQ, we performed an additional step, i.e., ultrafiltration, to remove any unnecessary proteins in the exosome samples, and we compared the level of protein contamination relative to UC isolation. By doing so, we found that EXQ–UF reduced the protein contamination by about 39.1% relative to the method that only used EXQ (Fig. S1A†), and this purity was comparable to that of UC. However, UC–UF did not improve the purity any further, which presumably was due to the protein concentration of UC being low enough that the additional step of UF had no significant effect on the purity of the exosomes. In addition, we confirmed that this additional step using UF did not cause unwanted loss of the exosomes. The total number of exosomes after EXQ–UF was still similar to that of EXQ, i.e., it was 93.3% of the result of EXQ (Fig. S1B†). Thus, we concluded that the use of EXQ–UF was an efficient way to obtain a high yield of exosomes and low protein contamination. Accordingly, we optimized the isolation and purification of the exosomes for additional experiments as follows. First, the exosomes were isolated from the conditioned medium using EXQ and transfected with miRNAs, followed by purification by UF (Fig. S1C†). Fig. S1D† shows that no miR-497 was detected in the samples after UF without exosome loading, which indicated that UF also contributed to the complete removal of any free-miRNA that was not transfected in the exosomes. By doing this, only 6.7 mL of the conditioned medium were required to obtain 1011 exosomes using EXQ–UF, whereas about 74 mL of the conditioned medium were required using the UC-based method (Fig. S2A†).After transfection with miRNAs in the isolated exosomes, we analyzed the size distribution (Fig. S1E†) and zeta potential of these particles (Fig. S1F†), and it was found that the sizes and the surface charges of the transfected exosomes (miRNA exo) were similar to those of their original exosomes (CTRL exo). This indicated that the miRNA loading did not alter the physical properties of the exosomes significantly. Also, the TEM analysis of the exosomes showed a round morphology with sizes ranging from 30–100 nm (Fig. S1G†), which indicated that the isolated and transfected exosomes were still intact. The TEM analysis indicated that the sizes of the exosomes were relatively smaller than the sizes based on the NTA analysis because the exosomes might have shrunk during the sample preparation for TEM.32 In addition, western blot analysis was performed with the cell lysate, exosomes isolated by EXQ, and miRNA-loaded exosomes isolated by EXQ–UF against exosomal markers, such as Alix, TSG101, and CD81, and against calnexin, a non-exosomal marker. Western blot analysis showed that Alix, TSG101, and CD81 were detected in the HEK293T cell-derived exosomes and the miRNA-loaded exosomes, and calnexin was detected only in the HEK293T cell lysate (Fig. S1H†). No apparent differences in the amounts of exosomal surface proteins were observed, and this indicated that the integrity of the exosomes was maintained even after loading miRNA.
Loading efficiency and stability of exosomes
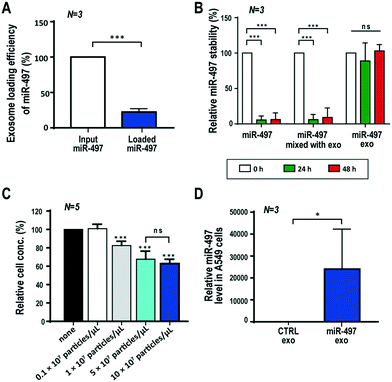
The efficiency of loading drugs is one of the key parameters in determining the efficacy of drug delivery systems. qRT-PCR was performed to assess how efficiently the miRNAs were loaded into the exosomes after using UF to remove unloaded miR-497. The result showed that the loading efficiency of miR-497 that was transfected in the exosomes was 22.4% of the input amount of miR-497 (Fig. 2A). In addition, we investigated the extent to which the exosomes protected the miRNAs within the membranes. The same amounts of free miR-497, a mixture of free miR-497 and exosomes, and miR-497 exosomes were incubated at 37 °C for 48 h, and then the miR-497 level was quantified after miRNA isolation from each sample. In the case of the free miRNAs and their mixture with exosomes, the miR-497 dramatically disappeared even after 24 h, meaning that most of the miR-497s were degraded. In contrast, the miR-497 loaded in the exosomes remained stable inside the exosomes for the most part, and the level was more than 90% of the original level. This indicated that exosomes are suitable vehicles for efficient and stable delivery of miRNAs (Fig. 2B).Determination of dose and cytotoxicity test
In order to determine the effective dose of miR-497 exosomes to treat cells, the dose-dependent effect of miR-497 exosomes on the proliferation of A549 cells (KCLB, Republic of Korea) was assessed using a WST-1 assay. The cells were seeded on a 96-well plate, and the exosomes were delivered with a wide range of concentrations, i.e., 0 to 10 × 107 particles per μL. Fig. 2C shows that the proliferation of A549 cells began to decrease from the treatment with 1 × 107 particles per μL, and it decreased further as the concentration of the miR-497 exosomes increased. There was no statistically significant difference between 5 × 107 and 10 × 107 particles per μL of miR-497 exosomes. To demonstrate that there was no cytotoxic effect caused by the exosomes, the cytotoxicity was assessed in A549 cells and HUVECs (C2517A, Lonza, Switzerland) after treatment of the control or miR-NC exosomes in a dose-dependent manner. However, no significant decrease of the viability of either type of cells was observed within this concentration range of exosomes, meaning that the exosomes did not exert any cytotoxic effect on the cells (Fig. S3†). Overall, the concentration of exosomes that was chosen as the optimal concentration was 5 × 107 particles per μL, and this concentration was used for additional experiments concerning how best to take advantage of miR-497 exosomes' anti-cancer effects.To verify the intracellular delivery of miR-497 by exosomes, qRT-PCR was conducted with lysates from A549 cells after treatment of miR-497 exosomes. The results showed that the intracellular miR-497 level was approximately 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times greater in the cells treated with miR-497 exosomes than it was in the cells treated with control exosomes (Fig. 2D). This result indicated that a large quantity of miR-497 was delivered successfully into the cells.
000 times greater in the cells treated with miR-497 exosomes than it was in the cells treated with control exosomes (Fig. 2D). This result indicated that a large quantity of miR-497 was delivered successfully into the cells.
Effect of miR-497 exosomes on A549 cell proliferation and migration
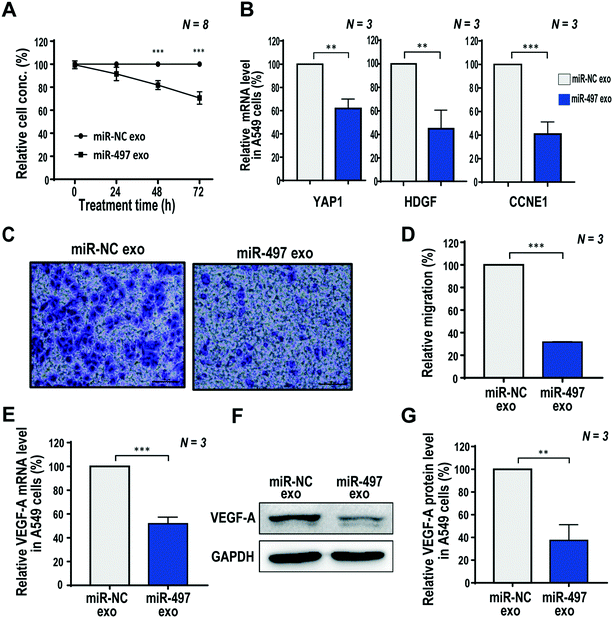
Based on the optimized concentration of exosomes shown above, we investigated the therapeutic effects of miR-497 exosomes, including their proliferation, migration, and angiogenic factor expression in NSCLC cells. First, the proliferation of cells was analyzed by the WST-1 assay at 0, 24, 48, and 72 h after the exosome treatment. There was no significant difference in the cell concentrations of miR-NC and miR-497 exosomes 24 h after treatment. Fig. 3A shows that, at 72 h after treatment, the concentration of cells treated with miR-497 exosomes was 70.7% of the cells treated with miR-NC exosomes. The proliferation inhibitory effect of the exosomes was almost the same as that of the exosomes purified by the UC-based method, as shown in Fig. S2B,† which indicated that the proliferation of A549 cells was suppressed significantly by the miR-497 delivered by the exosomes irrespective of the methods used to isolate the exosomes. MiR-497 has been reported to be associated with tumor growth, colony formation, invasion, angiogenesis, and epithelial mesenchymal transition (EMT) by directly targeting multiple genes. In NSCLC, miR-497 has been reported to suppress tumor growth by downregulating HDGF,33 CCNE1,23 and YAP1. In our results, the mRNA level of YAP1 decreased by 39.1% in the A549 cells (Fig. 3B) after the miR-497 exosome treatment, compared to that of miR-NC exosome-treated cells. The HDGF and CCNE1 levels also decreased by 55.8% and 59.2%, respectively (Fig. 3B). This demonstrated that miR-497 delivered by exosomes inhibited the tumor growth by successfully hybridizing to tumor growth-related mRNA in A549 cells, and blocking its translation. In addition, the downregulated CCNE1 is known to be involved in cell-cycle arrest, contributing to the inhibition of cell proliferation.34,35Next, we investigated the effect of the miR-497 exosome treatment on A549 cell migration. NSCLCs are known to grow very fast, and spread easily to other tissues. The active migration of tumor cells is essential for cancer metastasis.36 In order to determine whether these exosomes have the potential to inhibit tumor metastasis, the inhibitory effect of the exosomes on A549 cell migration was measured using a Transwell system. The miR-497 exosomes showed a significant inhibitory effect in that the relative migration decreased by 69.2% compared to the miR-NC exosomes in A549 cells (Fig. 3C and D). In a previous study,37 YAP1 has been found to be one of the key drivers of NSCLC metastasis because it promotes the EMT program. In our RT-PCR results, we identified that the miR-497 exosomes increased the level of epithelial markers, ZO-1 and E-cadherin, whereas they decreased the level of the mesenchymal marker, Snail. Another mesenchymal marker, vimentin, did not decrease (Fig. S4†). We suggest that the reduced cell migration was caused by the differential expression of EMT-related markers, which may be associated with the downregulated YAP1 by the miR-497 exosome treatment.
Since VEGF-A mRNA is the downstream target of miR-497, qRT-PCR and western blot analysis were performed to investigate the effect of miR-497 exosomes on the inhibition of the VEGF-A expression in A549 cells. VEGF-A is well-known as a molecule that induces angiogenesis, and it is the most potent angiogenic factor that stimulates the migration of tip cells from a blood vessel. This is responsible for most of angiogenesis due to the direct interaction with the VEGFR-2 of endothelial cells.38–40 As a result, A549 cells treated with miR-497 exosomes showed a dramatic decrease in VEGF-A mRNA and protein levels to 51.8% (Fig. 3E) and 37.4% (Fig. 3F and G), respectively, compared to cells treated with miR-NC exosomes. Thus, it is predicted that the miR-497 transferred into the cells through exosomes were bound directly to the VEGF-A mRNA and cleaved or degraded mRNA, thereby regulating the downstream signaling pathway, and affecting VEGF-A expression. Based on these results, miR-497 was delivered successfully to the cells via exosomes, resulting in the significant inhibition of the expression of VEGF-A, which is a potent angiogenic factor in NSCLCs.
Effect of miR-497 exosomes on angiogenesis of HUVECs
Angiogenesis, the recruitment of new blood vessels, is a very complex process that is regulated by a variety of signaling factors. Generally, this process accompanies the tip cell migration and the proliferation of stalk cells. Therefore, the proliferation rate of endothelial cells might have a significant effect on the spread and extent of vascular growth compared to the migration rate.39 In this context, the growth of endothelial cells was observed using a WST-1 assay to determine whether the delivery of miR-497 to the cells via exosomes affected the proliferation of HUVECs. First, we confirmed that a large quantity of miR-497 was delivered successfully into the HUVECs after treatment of exosomes (Fig. 4A). Also, Fig. 4B shows that the growth of HUVECs was suppressed significantly to 86.0% and 56.2% at 48 h and 72 h, respectively, after miR-497 exosome treatment, compared to the cells treated with miR-NC exosomes.Since VEGFR-2 is the most important VEGF-A receptor in the tumor microenvironment, the effect of miR-497 exosomes on VEGFR-2 expression was assessed. In the western blot result for VEGFR-2, its protein level decreased significantly to 67.2% in the cells treated with miR-497 exosomes as compared to those with miR-NC exosomes (Fig. 4C and D). Thus, it is obvious that miR-497 delivered by exosomes into HUVECs were bound to VEGFR-2 mRNA, effectively blocking its translation into protein. Subsequently, this will reduce the response of endothelial cells to the VEGF-A produced by cancer cells. It is noteworthy that miR-497 exosomes can inhibit VEGF-A expression in cancer cells (Fig. 3F). Thus, miR-497 exosomes may exert synergistic anti-angiogenesis effects on both NSCLC cells and endothelial cells by suppressing VEGF-A and VEGFR-2 expressions, respectively, which provides highly promising therapeutic opportunities for treating NSCLC.
Effect of miR-497 exosomes on sprouting angiogenesis of HUVECs and migration of A549 cells in a 3D microfluidic device
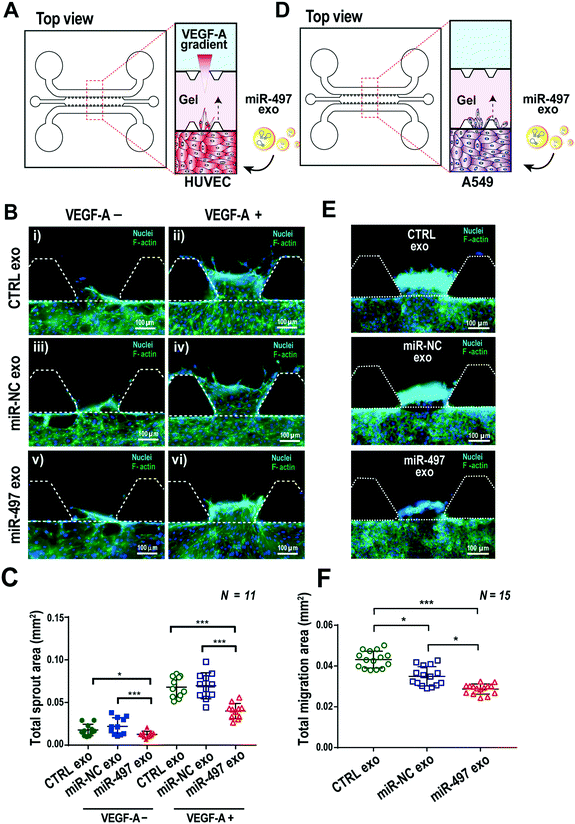
Angiogenesis is associated with several angiogenic factors and ligands/receptors with an elegant mechanism. Among them, VEGF-induced angiogenesis is the most well-known process, and the proper balance and regulation of the VEGF receptor level is important in orchestrating angiogenic sprouting. We used a PDMS-based 3D microfluidic device to validate whether miR-497 exosomes can regulate angiogenic sprouting of HUVECs induced by the VEGF gradient. Type-1 collagen was used as a scaffold material in the central channel of the device, and HUVECs were cultured in three dimensions in one side of the channel, and then VEGF-A was added at a concentration of 100 ng mL−1 in the opposite side of the channel to generate a VEGF-A concentration gradient across the gel (Fig. 5A). MiR-497 exosomes and miR-NC exosomes were added and replaced daily in the microchannel while culturing HUVECs for 3 days. The HUVECs started to sprout as soon as they were confluent on the surfaces of the channels. Even in the absence of VEGF-A, there was a basal level of sprouting in cells supplemented with CTRL or miR-NC exosomes. A slight decrease of sprouting was observed in cells treated with miR-497 exosomes (Fig. 5B and C). In the presence of a VEGF-A gradient, a severe level of sprouting was induced, resulting in a significant increase in the total sprouting in both the control exosome and the miR-NC exosome-treated HUVECs. Notably, the sprouting induced by VEGF-A clearly was suppressed when the HUVECs were treated with miR-497 exosomes (Fig. 5B and C). The time-dependent analysis of sprouting after VEGF-A treatment also demonstrated that there were significant decreases of sprout areas beginning on day 3, when the cells were treated with miR-497 exosomes (Fig. S5A and B†). Also, the number of tip cells and their activity seemed higher in the control exosome and the miR-NC exosome-treated cells (Fig. S5A†). The results were supported by the fact that the miR-497 exosomes suppressed the proliferation and expression of VEGFR-2 in the HUVECs (Fig. 4). Thus, it was concluded that the miR-497 exosomes suppressed the cell proliferation and migration of endothelial cells and that VEGFR-2 was suppressed, resulting in the reduction of angiogenic sprouting.The anti-migration activity of miR-497 exosomes on NSCLCs was also investigated in a microfluidic device (Fig. 5D). A549 cells were also cultured in a microfluidic channel, and we monitored their migration toward the 3D gels, which was induced by the relatively high concentration of nutrition on the opposite side of the channel (Fig. 5D). As expected, the migration of miR-497 cancer cells was inhibited significantly by miR-497 exosomes as compared to the control and the miR-NC exosomes (Fig. 5E and F). It is apparent that the miR-497 exosomes have anti-angiogenic and anti-migratory effects on both endothelial cells and tumor cells, which provides an enhanced therapeutic opportunity for the treatment of NSCLC.
Effect of miR-497 exosomes on anti-cancer therapy in a microfluidic 3D lung cancer model
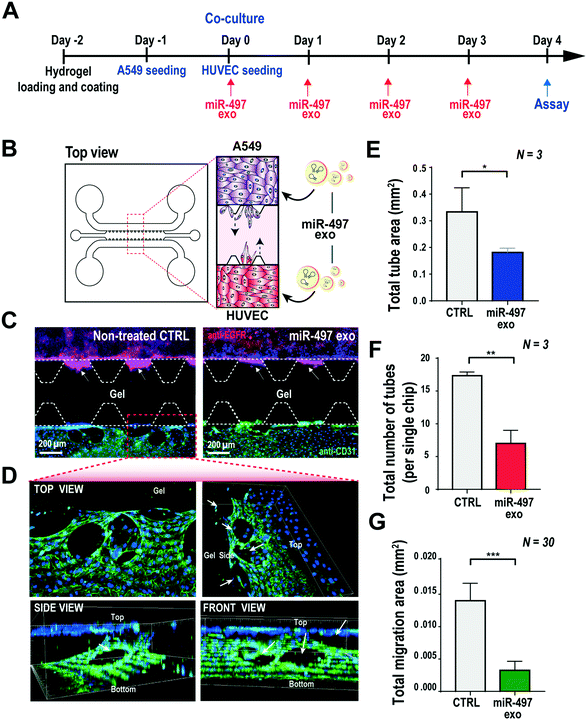
Based on the results shown above, the key advantage of our miR-497 exosomes is that their therapeutic effects are beneficial in targeting both cancer and endothelial cells. In this context, we developed a microfluidic 3D model that mimics the lung cancer microenvironment in order to investigate these simultaneous and synergistic anti-cancer effects. The cancer microenvironment is associated with various cells, including cancer and neighboring stromal cells, and they actively communicate with each other to regulate or promote tumor growth and metastasis.41 Since 2D mono-culture systems have limitations that prevent the precise understanding of these complex biological phenomena, it is very important to regulate the spatial and temporal conditions in a defined microenvironment. Thus, the microfluidic 3D lung cancer model was constructed to explore the anti-cancer effect of miR-497-loaded exosomes on lung cancer and endothelial cells by co-culturing both cells in the same microenvironment. A549 cells and HUVECs were cultured on each side of the channel across the gel channel, respectively. Therefore, the two types of cells can cross-talk through the gel, using the secreted cytokines from the cells cultured on each side of the channels, because the gelled collagen is a porous matrix. One day prior to HUVEC seeding, the A549 cells were seeded and cultured so that they could secrete VEGF-A as soon as the seeding of the HUVECs was done. The MiR-497 exosomes were added to both types of cells by replacing the medium containing miR-497 exosomes every day for 4 days (Fig. 6A and B). Four days after co-culturing, the cells were fixed and stained with an endothelial marker and a cancer marker, i.e., anti-CD31 and anti-EGFR, respectively, to assess the effects of miR-497 exosomes on angiogenesis and migration. When the endothelial cells were co-cultured with A549 lung cancer cells, the HUVECs began to align themselves and form lumen-containing tubules in the absence of the miR-497 exosomes (Fig. 6C), which were not observed in the mono-cultured HUVECs. The sprouting angiogenesis was not observed in this experiment, and this probably was due to the different signaling factors secreted from the cancer cells required for the HUVECs to induce angiogenesis.42 In addition, confocal microscopy analysis showed that the tubules were three-dimensionally formed on the top and the bottom surface of the channel, and they were even formed on the side surface facing the gel (Fig. 6D). However, when the co-cultured HUVECs were treated with miR-497 exosomes, we observed only a small number of tubes that were small in size (Fig. 6C), and the number of tubes was significantly less than that of the control (Fig. 6E and F). Concurrently, a huge migration of A549 cells cultured on the opposite side of the channel was observed in the absence of the treatment with miR-497 exosomes (Fig. 6C and G). However, cell migration was suppressed drastically by the treatment with the miR-497 exosomes, such that the total migration area was reduced by 76.7% compared to the control experiment. Overall, miR-497 exosomes have high therapeutic effects on both anti-angiogenesis and anti-migration even in the in vivo-like tumor microenvironment.Conclusions
NSCLC is one of the leading causes of cancer death worldwide, so there is a pressing need to develop a drug with enhanced safety and beneficial therapeutic effects. MiRNA-based drugs are valuable in clinical settings where precise and synergistic control of the progress of cancer is desired. In this research, exosomes were introduced to mediate miRNA delivery to provide a safe and effective treatment of NSCLC. The miR-497-encapsulated, exosome-based therapeutics was developed and assessed in NSCLC models using a 3D microfluidic device to verify its versatile function, including the stable protection and efficient delivery of miR-497 and the reduction of tumor angiogenesis both in NSCLC and endothelial cells. The results clearly demonstrated that miR-497 exosomes can successfully regulate the growth, migration, and angiogenesis of tumors, suggesting that miR-497 exosomes might also have a synergistic effect on an in vivo NSCLC system. Thus, we concluded that the exosome-based miRNA delivery combined with the microfluidic technology developed in this study will contribute to the development of highly-effective NSCLC therapy that targets tumorigenesis and angiogenesis.Author contributions
J. A. K. and W. J. R. conceived the study and designed the experiments. K. J., Y. J. Y., and J. Y. Y. performed the experiments and analyzed the results. K. J. and J. A. K. wrote the manuscript. J. A. K. and W. J. R. supervised the work and revised the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) (NRF-2017R1C1B2002377, NRF-2016R1A5A1010148, and NRF-2019R1A2C1003111) funded by the Ministry of Science and ICT (MSIT). This research was also supported by grants from the Korea Basic Science Institute (D010600). HEK293T cells were gifted by Prof. Kwon at Yonsei University in Republic of Korea.References
- G. A. Calin and C. M. Croce, Nat. Rev. Cancer, 2006, 6, 857–866 CrossRef CAS PubMed.
- S. E. Lee, H. R. Park, H. D. Yun, J.-J. Cho, H.-J. Ahn, C.-S. Park and Y. S. Park, BioChip J., 2017, 11, 188–195 CrossRef CAS.
- A. L. Kasinski, K. Kelnar, C. Stahlhut, E. Orellana, J. Zhao, E. Shimer, S. Dysart, X. Chen, A. G. Bader and F. J. Slack, Oncogene, 2015, 34, 3547–3555 CrossRef CAS PubMed.
- W. Lou, J. Liu, Y. Gao, G. Zhong, D. Chen, J. Shen, C. Bao, L. Xu, J. Pan and J. Cheng, Oncotarget, 2017, 8, 115787–115802 Search PubMed.
- C. J. Cheng, R. Bahal, I. A. Babar, Z. Pincus, F. Barrera, C. Liu, A. Svoronos, D. T. Braddock, P. M. Glazer and D. M. Engelman, Nature, 2015, 518, 107–110 CrossRef CAS PubMed.
- Y. Chen, D.-Y. Gao and L. Huang, Adv. Drug Delivery Rev., 2015, 81, 128–141 CrossRef CAS PubMed.
- N. Kosaka, F. Takeshita, Y. Yoshioka, K. Hagiwara, T. Katsuda, M. Ono and T. Ochiya, Adv. Drug Delivery Rev., 2013, 65, 376–382 CrossRef CAS PubMed.
- X. Zhang, X. Yuan, H. Shi, L. Wu, H. Qian and W. Xu, J. Hematol. Oncol., 2015, 8, 83 CrossRef PubMed.
- K. B. Johnsen, J. M. Gudbergsson, M. N. Skov, L. Pilgaard, T. Moos and M. Duroux, Biochim. Biophys. Acta, Rev. Cancer, 2014, 1846, 75–87 CrossRef CAS PubMed.
- A. A. Patil and W. J. Rhee, Biotechnol. Bioprocess Eng., 2019, 24, 689–701 CrossRef CAS.
- S. Höbel and A. Aigner, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5, 484–501 Search PubMed.
- C. Liu, J. Wen, Y. Meng, K. Zhang, J. Zhu, Y. Ren, X. Qian, X. Yuan, Y. Lu and C. Kang, Adv. Mater., 2015, 27, 292–297 CrossRef CAS PubMed.
- Y. Wu, M. Crawford, Y. Mao, R. J. Lee, I. C. Davis, T. S. Elton, L. J. Lee and S. P. Nana-Sinkam, Mol. Ther.--Nucleic Acids, 2013, 2, e84 CrossRef PubMed.
- V. van Duinen, S. J. Trietsch, J. Joore, P. Vulto and T. Hankemeier, Curr. Opin. Biotechnol., 2015, 35, 118–126 CrossRef CAS PubMed.
- D. Liu, F. Fontana, J. T. Hirvonen and H. A. Santos, Adv. Drug Delivery Rev., 2018, 128, 54–83 CrossRef CAS PubMed.
- J. H. Sung, J. Koo and M. Shuler, BioChip J., 2019, 13, 115–126 CrossRef CAS.
- M. Cirit and C. L. Stokes, Lab Chip, 2018, 18, 1831–1837 RSC.
- M. Rothbauer, H. Zirath and P. Ertl, Lab Chip, 2018, 18, 249–270 RSC.
- S. Shang, R. R. Soon, C. T. Lim, B. L. Khoo and J. Han, Lab Chip, 2019, 19, 369–386 RSC.
- S. J. Hachey and C. C. W. Hughes, Lab Chip, 2018, 18, 2893–2912 RSC.
- G. S. Jeong, S. Han, Y. Shin, G. H. Kwon, R. D. Kamm, S. H. Lee and S. Chung, Anal. Chem., 2011, 83, 8454–8459 CrossRef CAS PubMed.
- A. Gu, J. Lu, W. Wang, C. Shi, B. Han and M. Yao, Int. J. Biochem. Cell Biol., 2016, 70, 118–125 CrossRef CAS PubMed.
- Z. Han, Y. Zhang, Q. Yang, B. Liu, J. Wu, Y. Zhang, C. Yang and Y. Jiang, Oncotarget, 2015, 6, 13149–13163 Search PubMed.
- C. Huang, R. Ma, J. Yue, N. Li, Z. Li and D. Qi, Cell. Physiol. Biochem., 2015, 37, 342–352 CrossRef CAS PubMed.
- Y. Tu, L. Liu, D. Zhao, Y. Liu, X. Ma, Y. Fan, L. Wan, T. Huang, Z. Cheng and B. Shen, Sci. Rep., 2015, 5, 13827 CrossRef PubMed.
- Y. Tu, L. Liu, D. Zhao, Y. Liu, X. Ma, Y. Fan, L. Wan, T. Huang, Z. Cheng and B. Shen, Sci. Rep., 2016, 6, 21221 CrossRef CAS PubMed.
- G. Yang, G. Xiong, Z. Cao, S. Zheng, L. You, T. Zhang and Y. Zhao, Oncotarget, 2016, 7, 55900–55911 Search PubMed.
- J. Zhuang, Y. Ye, G. Wang, J. Ni, S. He, C. Hu, W. Xia and Z. Lv, Mol. Med. Rep., 2017, 16, 5815–5822 CrossRef CAS PubMed.
- J. Li, X. Chen, J. Yi, Y. Liu, D. Li, J. Wang, D. Hou, X. Jiang, J. Zhang and J. Wang, PLoS One, 2016, 11, e0163043 CrossRef PubMed.
- T. N. Lamichhane, R. S. Raiker and S. M. Jay, Mol. Pharmaceutics, 2015, 12, 3650–3657 CrossRef CAS PubMed.
- X. Zhu, M. Badawi, S. Pomeroy, D. S. Sutaria, Z. Xie, A. Baek, J. Jiang, O. A. Elgamal, X. Mo and K. L. Perle, J. Extracell. Vesicles, 2017, 6, 1324730 CrossRef PubMed.
- W. Zhang, P. Peng, Y. Kuang, J. Yang, D. Cao, Y. You and K. Shen, Tumor Biol., 2016, 37, 4213–4221 CrossRef CAS PubMed.
- W. Y. Zhao, Y. Wang, Z. J. An, C. G. Shi, G. A. Zhu, B. Wang, M. Y. Lu, C. K. Pan and P. Chen, Biochem. Biophys. Res. Commun., 2013, 435, 466–471 CrossRef CAS PubMed.
- Q. Luo, X. Li, Y. Gao, Y. Long, L. Chen, Y. Huang and L. Fang, Cancer Cell Int., 2013, 13, 95 CrossRef CAS PubMed.
- M. Furuta, K. Kozaki, K. Tanimoto, S. Tanaka, S. Arii, T. Shimamura, A. Niida, S. Miyano and J. Inazawa, PLoS One, 2013, 8, e60155 CrossRef CAS PubMed.
- M. Reck, S. Popat, N. Reinmuth, D. De Ruysscher, K. Kerr and S. Peters, Ann. Oncol., 2014, 25, iii27–iii39 CrossRef PubMed.
- M. Yu, Y. Chen, X. Li, R. Yang, L. Zhang, L. Huangfu, N. Zheng, X. Zhang, L. Lv, Y. Hong, H. Liang and H. Shan, Cell Death Dis., 2018, 9, 464 CrossRef PubMed.
- I. Lobov, R. Renard, N. Papadopoulos, N. Gale, G. Thurston, G. Yancopoulos and S. Wiegand, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 3219–3224 CrossRef CAS PubMed.
- K.-A. Norton and A. S. Popel, Sci. Rep., 2016, 6, 36992 CrossRef CAS PubMed.
- H. Gerhardt, M. Golding, M. Fruttiger, C. Ruhrberg, A. Lundkvist, A. Abramsson, M. Jeltsch, C. Mitchell, K. Alitalo and D. Shima, J. Cell Biol., 2003, 161, 1163–1177 CrossRef CAS PubMed.
- X. Ding, J. Ding, K. Wu, W. Wen, C. Liu, H. Yan, C. Chen, S. Wang, H. Tang and C. Gao, Oncogene, 2012, 31, 2899 CrossRef CAS PubMed.
- H.-W. Cheng, Y.-F. Chen, J.-M. Wong, C.-W. Weng, H.-Y. Chen, S.-L. Yu, H.-W. Chen, A. Yuan and J. J. Chen, J. Exp. Clin. Cancer Res., 2017, 36, 27 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9lc00958b |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2020 |