Open Access Article
Open Access ArticleIn vitro and environmental toxicity of reduced graphene oxide as an additive in automotive lubricants†
Margarita
Esquivel-Gaon‡
 a,
Nhung H. A.
Nguyen‡
b,
Mauro F.
Sgroi
c,
Daniele
Pullini
c,
Flavia
Gili
c,
Davide
Mangherini
c,
Alina Iuliana
Pruna
de,
Petra
Rosicka
b,
Alena
Sevcu
a,
Nhung H. A.
Nguyen‡
b,
Mauro F.
Sgroi
c,
Daniele
Pullini
c,
Flavia
Gili
c,
Davide
Mangherini
c,
Alina Iuliana
Pruna
de,
Petra
Rosicka
b,
Alena
Sevcu
 b and
Valentina
Castagnola
b and
Valentina
Castagnola
 *a
*a
aCentre for BioNano Interactions, School of Chemistry and Chemical Biology, University College Dublin, Belfield, Dublin 4, Dublin, Ireland. E-mail: valentina.castagnola@cbni.ucd.ie
bTechnical University of Liberec, Institute for Nanomaterials, Advanced Technologies and Innovation, Bendlova 1409/7 CZ-461 17, Liberec, Czech Republic
cCentro Ricerche FIAT, Strada Torino 50, 10043 Orbassano (TO), Italy
dGemmate Technologies, via Reano 31, 10090 Buttigliera Alta (TO), Italy
eCenter for Surface Science and Nanotechnology, Polytechnic University of Bucharest, 313 Splaiul Independentei, 060042 Bucharest, Romania
First published on 26th March 2018
Abstract
Despite the ground-breaking potential of nanomaterials, their safe and sustainable incorporation into an array of industrial markets prompts a deep and clear understanding of their potential toxicity for both humans and the environment. Among the many materials with great potential, graphene has shown promise in a variety of applications; however, the impact of graphene based products on living systems remains poorly understood. In this paper, we illustrate that via exploiting the tribological properties of graphene nanosheets, we can successfully improve both the frictional behaviour and the anti-wear capacity of lubricant oil for mechanical transmission. By virtue of reducing friction and enhancing lubricant lifetimes, we can forecast a reduction in friction based energy loss, in addition to a decrease in the carbon footprint of vehicles. The aforementioned positive environmental impact is further strengthened considering the lack of acute toxicity found in our extensive in vitro investigation, in which both eukaryotic and prokaryotic cells were tested. Collectively, our body of work suggests that by the use of safe nanoadditives we could contribute to reducing the environmental impact of transportation and therein take a positive step towards a more sustainable automotive sector. The workflow proposed here for the evaluation of human and environmental toxicity will allow for the study of nanosized bare graphene material and can be broadly applied to the translation of graphene-based nanomaterials into the market.
Introduction
Due to a worldwide initiative to stabilise the worsening condition of global warming, industries and households across the planet, have been tasked with reducing their output of CO2 and other greenhouse emissions as a vital step to preserve our planet.1,2 In addition to reducing our carbon footprint through moving to clean and sustainable energy sources, further effort has been made to reduce energy loss or wastage in our existing technologies. Friction is, for example, a common energy loss problem in standard combustion engines and many other industrial processes, and hinders these systems by generation of excess heat, which is then required to be removed via radiators, leading to a large net loss of energy.3 One of the most effective approaches in addressing energy loss is to prevent initial heat generation by reducing the friction forces between moving parts via lubricants in the liquid or solid form. Currently, in the automotive field, lubricants act in reducing frictional forces by preventing sliding contact interfaces of severe metal-to-metal contacts, by forming a durable low-shear boundary film on rubbing surfaces and/or by acting as a hydrodynamic layer, mainly working as a heat transfer agent. However, it is clear that current lubrication formulas help to reduce, but do not eliminate friction, as a variety of wear patterns are common in the moving parts of combustion engines. Collectively, these wearing events not only lead to local heat generation and energy loss, but also physical damage within the system. Therefore, the development of new advanced lubricants integrating additive packages with higher anti-wear and anti-friction performances is of paramount importance in order to improve the energetic efficiency of modern internal combustion engines (and other industrial processes), while retaining the integrity of moving parts for longer periods.Over the last 20 years, a growing interest has arisen in the use of nanoparticles as friction modifiers or anti-wear additives in lubricants, by virtue of potential interesting tribological properties shown by some nanomaterials.4–7 The most promising tribological properties were found in tungsten and molybdenum dichalcogenides with onion-like microstructures, known as inorganic fullerenes, and graphene derived materials.8–13 Among them, graphene is a particularly promising candidate and it also possesses exceptional mechanical strength,14 which is highly desirable for wear protection, as well as a potentially low cost of production. Because of their improved heat conductivity and promising lubricating properties, a number of studies on graphene-based nanolubricants have been recently published.15–18 It is therefore reasonable to assume that graphene and other nano-additives will be utilized as a part of numerous modern lubrication systems very soon.
However, the introduction of graphene-based nanomaterials in the value chain of lubricants leads to concerns on potential exposure for humans and the environment.19–21
Nanomaterials could indeed accidentally contaminate the environment and/or the operators during manufacturing, transportation, usage and disposal of lubricants. These aspects are not usually taken into consideration in studies aimed at a wider application of graphene based nanolubricants, however they are of paramount importance for a responsible translation of these new technologies on the market.
In the context of the EU-FP7 Project FutureNanoNeeds, much effort has been devoted to translating those general exposure scenarios that are considered to be the most important along the value chain and life cycle of future nanomaterials into an understanding of in situ exposure conditions.
Therefore, it is essential to consider not only realistic exposure scenarios of the nanolubricants, but also a possible release of the nanostructures from the matrix/matrices as well as the transformation pathways into the environment.
In this work, enhanced tribological properties of a mineral base oil containing an antifriction nanoadditive based on reduced graphene oxide (rGO) nanosheets are reported. The so-composed nanolubricant exploits a green process for the reduction of graphene oxide making use of ascorbic acid. Concurrently, the toxicity and the ecotoxicity of the pristine material in a variety of dispersion scenarios were evaluated. In evaluating the toxicity of rGO the need for chemical functionalization is often reported to limit adhesion to the cell surface induced by high hydrophobicity and the consequent acute toxicity.22–25 In this work, protein-stabilized rGO nanosheets were studied in addition to the original nanolubricant material in order to prevent aggregation and to assess the effects of the nanosize on the uptake, without affecting the graphene surface properties.
In particular, in vitro tests on eukaryotic cells were carried out using a high throughput screening approach26 in which a myriad of toxicity endpoints were assessed simultaneously; the toxicity parameters included: cell count, mitochondrial membrane potential, lysosomal acidification, and cell membrane integrity in a cell line associated with lung exposure models. We proposed and validated this method as a convenient label-free, high throughput tool to indirectly assess internalization and trafficking pathways of rGO.
In addition, the effect of rGO nanosheets dispersed in different media was tested on the common soil bacteria, P. putida, by determining its growth, morphology, and DNA fragmentation. Certain amounts of rGO nanosheets may enter municipal or industrial wastewater treatment plants, and potentially have an effect on bacterial consortia in activated sludge, an important component of biomineralization processes.27
Nitrifying bacteria play a key role in nitrogen cycling through ammonia/ammonium removal from water that has higher concentrations of these compounds, such as municipal wastewaters or polluted rivers and lakes. Oil, if accidentally spilled, decomposes very slowly and reduces the oxygen supply to the nitrifying bacteria that rely on aerobic metabolism. The influence of rGO nanosheets on the metabolic activity of nitrifying bacteria involved in ammonia oxidation in activated sludge is still rather unknown and has been evaluated in this study for the first time. The standard test (ISO 9408:1999) used for evaluation of aerobic biodegradability of organic compounds in aqueous medium was used in this study, slightly modified for activated sludge.
The reported full characterization, not only on the physical–chemical properties and additive efficacy of graphene, but also on its biological and environmental impact, is a fundamental first step to promote the transition of nanomaterials for early development into the industrial value chain.
Results and discussion
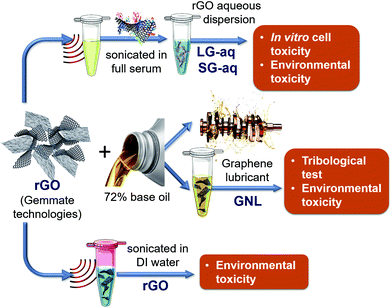
The pristine graphene (rGO) nanomaterial has been fully characterised in its solid state, as reported in the ESI (Fig. S-1–S-5†). The graphene-based nanolubricant (GNL) comprised of rGO dispersed in a base oil (about 72%, as described in the Materials and methods section) has been used for tribology. A biological aqueous dispersion (G-aq) of the pristine rGO, obtained as described in the Materials and methods section, was also prepared in order to study the toxicity of the nanomaterial in vitro. This dispersion was obtained by using full serum protein as a stabilization layer around the graphene nanosheets.28 In this way we were also able to take into account the effect of the presence of proteins, and therefore a more realistic scenario for a nanomaterial interacting with human cells.29–36 The aqueous dispersion can be separated into subpopulations of different sizes as shown in Fig. S-6a† by Differential Centrifugal sedimentation (DCS). The subpopulations were named SG-aq (subpopulation with smaller size distribution of nanosheets) and LG-aq (subpopulation with larger size distribution of nanosheets). For the environmental toxicity studies three different graphene dispersions were used including GNL, pristine rGO powder sonicated for 30 min in deionized water (DI), SG-aq and LG-aq. The dispersions are schematised in Fig. 1.The stability of the dispersion in bacterial growth media was characterised by Differential Centrifugal Sedimentation (DCS), as reported in Fig. S-6b.†
Tribology study
Many nanoparticles were used as lubricant additives. In particular, different types of oxides were tested, such as TiO2, Al2O3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37,38 or SiO2
37,38 or SiO2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 or metal dichalcogenides with inorganic fullerene structures such as WS2 and MoS2.9,11,40,41 However, in the last 5 years, graphene has emerged as a more convenient candidate for this application.12,13,42 The main advantages in the use of this nanomaterial are related to the potential low cost of production (by using graphite as a raw material) and the possibility of “tuning” the “degree of reduction”, obtaining materials with very different surface properties (e.g. the presence of –OH and other functional groups that allow dispersion of the material in different lubricant base oils). Moreover the surface properties also influence heavily the interaction of rubbing surfaces.
39 or metal dichalcogenides with inorganic fullerene structures such as WS2 and MoS2.9,11,40,41 However, in the last 5 years, graphene has emerged as a more convenient candidate for this application.12,13,42 The main advantages in the use of this nanomaterial are related to the potential low cost of production (by using graphite as a raw material) and the possibility of “tuning” the “degree of reduction”, obtaining materials with very different surface properties (e.g. the presence of –OH and other functional groups that allow dispersion of the material in different lubricant base oils). Moreover the surface properties also influence heavily the interaction of rubbing surfaces.
The Stribeck curves of the base oil and the GNL are shown in Fig. 2.
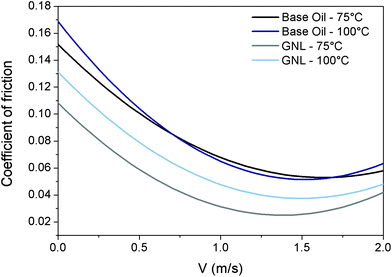 | ||
| Fig. 2 Tribological properties of rGO. Stribeck curves for base oil at 75 °C (black) and 100 °C (blue) and GNL at 75 °C (grey) and 100 °C (light blue). | ||
From this graph it can be seen how the addition of rGO is able to reduce the Coefficient of Friction (CoF) by approximately 40% over all the lubrication regimes, and therefore could help to reduce fuel consumption and CO2 emission by vehicles. The reduction in friction coefficient has been attributed in the literature to the adhesion of graphene sheets to the sliding surfaces during shear contact. This creates a protective tribo-film able to prevent the sliding surfaces from coming in contact with each other directly.43
The tests were performed at two different temperatures (75 °C and 100 °C) to cover a realistic temperature range typical for automotive applications.
Significant reduction in the coefficient of friction at both temperatures was achieved at different velocities corresponding to different tribological regimes (boundary, mixed and elasto-hydrodynamic). This was probably due to the ability of rGO to form bonds with the rubbing surfaces and protect them.
The effect of the testing temperature was found more pronounced in the case of GNL. This phenomenon has no obvious explanation but it could be related to the fact that the viscosity and density of the base oil are reduced by the temperature therefore rGO can precipitate/segregate more easily.44
In vitro toxicology: uptake, mitochondrial activity and high content analysis
Graphene and mostly graphene oxide nanosheet uptake has been studied in a range of different cellular models including: the lungs,45,46 liver and colorectal epithelial cells,47,48 macrophages49–51 and neuronal cells.22 Collectively, the data show a broad range of interactions between graphene and cells, ranging from no observable cellular uptake or membrane association to full internalisation, with evidence of graphene nanoparticles found in cytoplasmic compartments such as vacuoles and lysosomes. The cytotoxicity of graphene derived materials remains controversial and is dependent on a series of physical chemical parameters.52In the literature, macrophages have been shown to have a higher graphene uptake compared to lung epithelial cells. Additionally, differential nanomaterial uptake may be due to the intrinsic characteristics of the material tested within each study, including size and agglomeration state which can define the mechanism of uptake used by the cells. For example, agglomerated nanoparticles may be more susceptible to internalisation by macrophages via phagocytosis while, smaller size graphene flakes may enter through clathrin mediated mechanisms.53
The lungs are one of the major portals of entry for nanomaterials to gain access into the human body. In this context, it is very important for graphene nanoparticles – most of which are fabricated as dry powders – to be tested in a lung inhalation exposure model.
Here, an A549 human lung epithelial cell line and RAW 264.7 murine macrophages were used for in vitro testing.
In recent studies, the toxicity of graphene oxide, with a lateral size between 160 nm and 780 nm, has been evaluated using the same lung epithelial cell line (A549). These data suggested that cell viability was affected in a negative way but only with a 20% growth reduction overall by 200 μg mL−1 of graphene oxide when compared to the untreated control.45
Further studies have appeared in the literature which used pristine graphene in an alternative lung model which used murine macrophages (RAW 264.7 cells).50 In this case, the results suggested that cellular viability was impacted by 25%, even at low concentrations (25 μg mL−1) via measuring mitochondrial membrane potential depletion.50
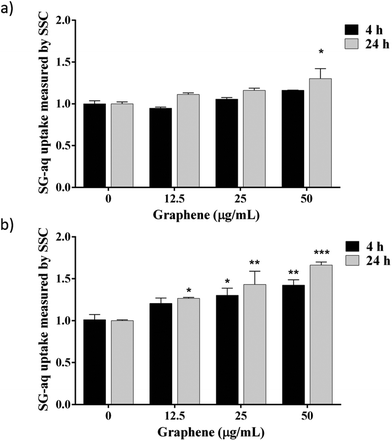
In this study, a first attempt was made using GNL, and no graphene nanosheets were found in contact with cells: the nanomaterial never came into contact with the aqueous phase, being well dispersed in the base oil (data not shown). However, two possible exposure scenarios along the value chain must be taken into account: (i) inhalation during GNL production and (ii) skin contact in case of accidental spilling. In both cases, it is important to know whether rGO nanosheets, once in contact with the cells via inhalation or skin penetration, could be taken up and internalised by the cells. To this aim, biological dispersions LG-aq and SG-aq were tested. The two dispersions did not lead to significantly different results therefore we report here only the results related to SG-aq. The cells were exposed to increasing concentrations of SG-aq for 4 h and 24 h and were analysed by flow cytometry (Fig. 3). Cell side scattering was used as a readout signal since an increase in graphene uptake leads to increasing granularity of cells, and therefore increasing side scattering.48,54,55 Transmission electron microscopy (TEM) on cell sections after exposure to G-aq confirmed the actual internalization of the nanosheets detected by flow cytometry (see Fig. S-7†). The results shown in Fig. 3 indicate that the complexity/granularity increased after exposure to SG-aq, this effect being time, concentration and cell type dependent, can be associated with nanoparticle uptake. In agreement with the literature, an increase in uptake by the RAW 264.7 cells compared with the A549 cells is observed, likely due to their origin, as these cells are mature macrophages and epithelial cells, respectively.
Cells exposed to increasing concentrations of SG-aq also showed no decrease in mitochondrial activity after 24 h of exposure (see Fig. S-8†). However, after 72 h of exposure, the mitochondrial activity showed a decrease of 10% in the A549 cells and 20% in the RAW 264.7 cells in a concentration dependent manner.
In our study such effects were not observed after 24 h of exposure to graphene as reported in other studies.50 These differences in the cellular response may be due to the presence of proteins used to disperse rGO as the protein corona could act as an attenuator of the cytotoxic effects of nanoparticles.33,46 However, it is realistic to think that the in vivo concentration of proteins is large compared to the one used for classic cell culture.
Subsequently a high content analysis-fluorescence microscopy based in vitro toxicity screening26 was carried out using SG-aq. Live cells were analysed after 24 h of exposure to SG-aq. Epifluorescence images are shown in the ESI (Fig. S-9†).
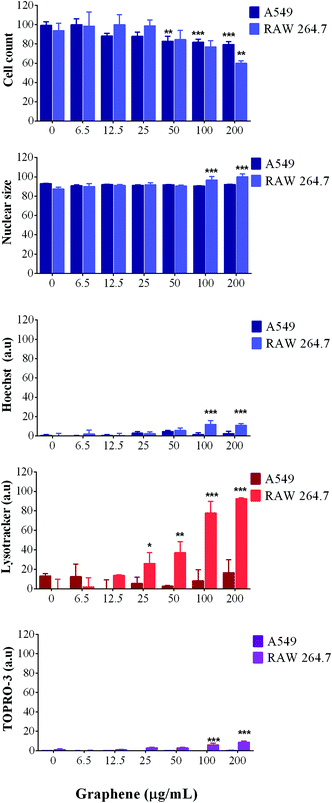
For A549 cells, image analysis showed no significant decrease in the number of cells, suggesting that the cells were able to progress through their cell cycle as normal; meanwhile, analysis of their nuclear morphology showed no swelling, condensation or increase in fluorescence intensity, which are all hallmarks of cell death.
Furthermore, LysoTracker red was used to measure the lysosomal activity and the results showed an increase in lysosomal activity with increasing concentrations of SG-aq (Fig. 4) suggesting that lysosomal trafficking pathways are activated in a concentration dependent manner.
Moreover, cell membrane integrity (stained with TOPRO-3) was not affected at any concentration or time point, thus indicating that SG-aq does not strongly impact cellular toxicity at the levels tested within this study (Fig. 4).
In contrast to A549 cells, RAW 264.7 cells showed a reduction in the number of cells present, in a concentration dependent manner when incubated with SG-aq. Additionally, LysoTracker red fluorescence intensity increased after exposure to nanoparticles, which would suggest phagocytosis by the macrophages due to their natural role in the removal of unwanted material in biological systems. Finally, cell membrane integrity was also affected only at the highest concentrations of SG-aq (200 μg mL−1), consistent with the results observed by nuclear analysis.
Ecotoxicity evaluation: bacterial growth, morphology, DNA fragmentation and respiration
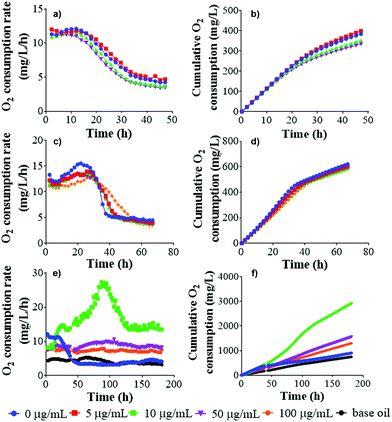
Graphene as well as other nanomaterials have often exhibited antibacterial properties.56–60Here, GNL, pristine rGO, SG-aq and LG-aq were exposed to P. putida culture in order to determine their effect on bacterial growth (6 h of exposure), bacterial morphology and DNA fragmentation (24 h of exposure). Moreover, the effect of rGO on natural nitrifying microorganisms was determined in activated sludge from a wastewater treatment plant (48 h of exposure) based on O2 consumption. Very few examples of these studies can be found in the literature for graphene oxide (GO),61 where some concentration-dependent toxicity was reported, and graphene nanoplatelets.62 Carbon nanotubes were also reported to be toxic to microorganisms from activated sludge.63,64 However, this is, to the best of our knowledge, the first study of the effect of protein-stabilized rGO nanosheets of different sizes on nitrifying bacteria, therefore we are unable to directly compare our results with other studies.
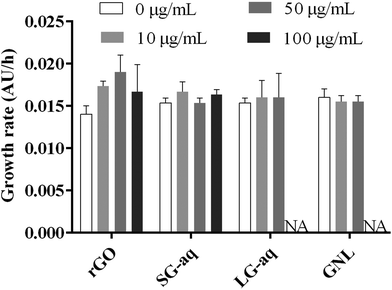
From our results, pristine rGO and SG-aq did not affect bacterial growth at concentrations up to 100 μg mL−1 (Fig. 5). Similarly, LG-aq and GNL had no effect on concentrations up to 50 μg mL−1. The test with the highest concentration of 100 μg mL−1 could not be included because aggregation of rGO in LG-aq and formation of clouds of base oil in GNL interfered with the measurement of absorbance values at 600 nm. Furthermore, no direct effect on bacterial cell morphology, examined using Scanning Electron Microscopy (SEM), was observed after P. putida exposure to all tested rGO dispersions over 24 h (Fig. S-10†).
DNA fragmentation was determined from the same P. putida samples prepared for the inspection of cell morphology, in order to evaluate the potential genotoxicity of rGO nanosheets. There was no visual difference in cell morphology between P. putida exposed to rGO dispersions and the control. Similarly, DNA fragmentation of Gram-negative P. aeruginosa did not show significant effects when exposed to rGO for 24 h (Fig. S-11†).65 However, it was reported that rGO inhibited E. coli growth at 85 μg mL−1 and even showed antibacterial properties via several mechanisms such as (1) direct interaction and degradation of outer and inner cell membranes of bacteria and (2) bacteria entrapment by rGO nanosheets.66
To determine the effect of rGO nanosheets on nitrifying bacteria, the O2 consumption rate was measured alongside cumulative O2 uptake, showing the amount of O2 consumed or cumulated by the bacteria during oxidation of the substrate. Any effect of pristine rGO and LG-aq on nitrifying bacteria was observed over 48 h-monitoring of O2 (Fig. 6). In the case of GNL, the measurement of respiration was rather challenging. As we already described for the in vitro test, the oil phase remained on the surface of activated sludge and therefore effective mixing was not possible. This interfered with the exchange of gases across the oily surface and consequently the activated sludge decomposed.
Conclusions
Tribological results demonstrated that rGO is a promising additive for the production of nano-lubricants with improved anti-friction properties. The low cost and relatively straight forward, environmentally friendly synthesis renders rGO an excellent candidate component for next generation formulation of lubricants.In this paper we propose a workflow for the evaluation of rGO-based nanolubricant human and environmental toxicity taking into account different steps of the material value chain and different rGO dispersions. In particular, the protein stabilised rGO dispersion allowed us to maintain nanomaterial stability in biological media, and therefore analyse the effect of the nanometric size without affecting the surface with chemical functionalization.
The use of dispersions that contain a dramatic range in graphene sheet sizes and sedimentation characteristics can lead to misleading results, in particular when comparing different cell lines (macrophages and epithelial cells) used in inhalation exposure models. In this study, by controlling the graphene nanosheet size range, the dispersions remained stable for periods longer than that of the experimental exposure model conditions avoiding the sedimentation of larger sheets (which might result in an enhanced graphene concentration at the cellular interface). This allowed for appropriate nanoflake–cellular receptor interaction and accurate nanoflake uptake and toxicity observations.
Here, we suggest the use of high content analysis as a rapid, cost effective, and high throughput method for the evaluation of rGO cell internalization and we report the study of the effect of rGO dispersions on bacterial growth, morphology and DNA fragmentation on P. putida and on nitrifying bacteria by measuring the O2 consumption rate. The here proposed workflow can be widely applied in view of the translation of graphene-based nanomaterials to the market.
On exposing human epithelial cells to increasing concentrations of rGO, no significant decrease was observed in cell viability up to 100 μg mL−1 after 24 h exposure; lysosomal activity increased in a concentration dependent manner suggesting the activation of endocytic pathways. No significant effect of rGO was observed on P. putida or on nitrifying microorganisms at a concentration of 100 mg L−1. It must be noted that nanomaterial toxicity is required to be monitored over long periods in order to understand chronic effects at sublethal concentrations. However, no clear acute toxicity of industrial grade rGO nanomaterial has emerged for the conditions applied in this study.
Materials and methods
Synthesis of chemically reduced graphene oxide and preparation of materials
rGO was provided by Gemmate Technologies. The material was obtained by using a modified Hummers’ method as described previously.67 Basically, graphite powder was oxidized in two steps with H2SO4 and KMnO4. The chemical conversion of GO into rGO was performed using ascorbic acid as a reducing agent.For the toxicity study, rGO was dispersed by sonication in (1) DI water (stock concentration of 10 mg mL−1) and (2) in full serum: SG-aq (stock concentration of 0.2 mg mL−1), and LG-aq (stock concentration of 1.8 mg mL−1). LG-aq was prepared in a larger quantity to obtain enough material for respirometry.
GNL was prepared by mixing rGO with the base oil and the dispersant through sonication. A sonic ultrasound source with a maximum power of 750 W was used. The time and power of ultrasound waves and the amount of dispersant were optimized to obtain a complete and stable dispersion of rGO in the base oil.
Tribological testing
Tribological tests were performed using a Bruker-UMT for Stribeck curve determination. The Stribeck curve relates the coefficient of friction (CoF) to the Stribeck number (product between the viscosity of the fluid and relative velocity of sliding surfaces divided by the applied load) and is used to describe the frictional characteristics of a liquid lubricant over conditions usually spanning the boundary, mixed and hydrodynamic regimes.68 To obtain the Stribeck curves we kept two variables fixed (load and viscosity) and varied the third (velocity) over a suitable range so that the contact interface went through the region of asperity contact (boundary), as well as full fluid-film separation (hydrodynamic). The tribological contact was a ball on disc under pure sliding conditions. The raw CoF data were averaged over three repetitions and then interpolated by a polynomial function. The measurements were performed at 75 °C and 100 °C, to cover a realistic temperature range typical for automotive applications.Biological dispersion
The pristine rGO (at the starting concentration of 10 mg mL−1) was dispersed in a solution of complete foetal bovine serum (FBS) at a concentration of 50% v/v in phosphate buffered saline (PBS) under ultrasonication for 1 h. The undispersed sheets were discarded by centrifugation at 1500 rpm for 15 min and only the supernatant was retained for washing. Excess protein was washed by three sub-sequential centrifugations at maximum speed (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) for 20 min, and the supernatant was discarded each time and replaced with fresh PBS. A tip sonicator SONICS (vibra cell) with an ultrasonic processor GEX130 was used at 30% of power to disperse the rGO in complete serum. Foetal bovine serum (FBS) was purchased from GIBCO.
000 rpm) for 20 min, and the supernatant was discarded each time and replaced with fresh PBS. A tip sonicator SONICS (vibra cell) with an ultrasonic processor GEX130 was used at 30% of power to disperse the rGO in complete serum. Foetal bovine serum (FBS) was purchased from GIBCO.
Dispersion characterization
Differential centrifugal sedimentation (DSC) experiments were performed with a CPS Disc Centrifuge DC24000 (CPS Instruments). 100 μL of sample were injected into an 8–24% PBS based sucrose gradient. Density values of 1.75 g mL−1, refractive index of 2.377 and non-sphericity factor of 3 were used. The rotational speed of the disk was set to 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–20
000–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm.
000 rpm.
Cell culture
The A549 lung epithelial cell line was obtained from the Leibniz Institute DSMZ German Collection of Microorganisms and Cell Cultures. A549 cells were cultured in MEM medium. RAW 264.7 murine macrophage cell lines were obtained from the American Type Culture Collection (ATCC). RAW 264.7 cells were cultured in DMEM cell culture. Both cell culture media were supplemented with 10% (v/v) Foetal Bovine Serum (FBS) and 1% penicillin–streptomycin and the cells were maintained at 37 °C with 5% CO2.High content analysis
A549 and RAW 264.7 cells were seeded in 96-well plates and incubated for 24 h and 72 h. rGO nanoparticles were added to achieve a final concentration of 0, 6.25, 12.5, 25, 50 and 100 μg mL−1 in cell culture media and incubated for 24 h and 72 h. Thereafter a cocktail of fluorescent probes was added to the cell culture and incubated for an additional 30 minutes at 37 °C. The cocktail included: 400 nM Hoechst 33342, 200 nM of LysoTracker® red and 800 nM of TOPRO®-3. The dyes were purchased from Life Technologies. The cells were analysed by high content analysis using an Arrayscan VTI 740 (Thermo Scientific). The fluorescence intensity was collected using a combination of excitation/emission filters. Hoechst 33342 was visualized in the blue channel, LysoTracker® red in the red channel while TOPRO®-3 in the far red channel. Cell count, nuclear size and nuclear intensity were generated from Hoechst 33342 object count, mean object area and average intensity, respectively. The LysoTracker® red intensity indicates the acidic organelles while TOPRO®-3 was used to assess cellular membrane integrity, as it is a membrane impermeable dye.26 The results have been normalized against control cells under the same conditions. The graphs show the mean ± standard error of a representative experiment in triplicate.Cell viability assays
A549 and RAW 264.7 cell viability were determined 24 and 72 h post treatment using (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) and MTS (Promega Corporation, USA). The MTS assay indirectly indicated the number of living cells by measuring the mitochondrial activity of viable cells, after treatment with rGO particles. Briefly, the cells were seeded onto 96-well plates and exposed to increasing concentrations of rGO (6.25–100 μg mL−1). The absorbance was recorded on a microplate reader Varioskan Flash (Thermo Scientific, USA).Nanoparticle uptake
An easy and reliable approach to evaluate nanoparticle uptake was employed using light side scattering by flow cytometry, which essentially measures the increase in cellular granularity as a result of material uptake.69 Briefly, A549 cells were seeded in a 24-well plate and incubated for 24 h before adding rGO nanoparticles. Three concentrations of nanoparticles were incubated for 4 or 24 h and thereafter, the cells were washed three times with PBS and then cells were collected. The side-scatter light was collected using an Accuri C6 flow cytometer. Samples were normalized against the control (0 μg mL−1). The results were analysed by two-way ANOVA and Dunnett's multiple comparison test. The significance levels were *P < 0.05, **P < 0.01, ***P < 0.001.Bacterial culture
Bacterial strain of Gram-negative soil bacterium Pseudomonas putida was obtained from the Czech Collection of Microorganisms, Masaryk University, Czech Republic. The bacterial inoculum was always prepared fresh before ecotoxicity testing by growing a single colony overnight in a soy nutrient broth (Sigma-Aldrich) at 27 °C. The culture was adjusted to 0.01 (±0.002) optical density at 600 nm (OD600) using a UV-Vis spectrophotometer (Hach Lange DR6000, Germany) right before performing the test.Transmission electron microscopy imaging
For the sample preparation firstly 2.5% glutaraldehyde solution was added to the cells overnight to allow fixing. 1% osmium tetroxide (OsO4) was added to stain cell monolayers for 1 hour. After removal of OsO4, Sorensen's phosphate buffer was applied for washing for 10 min. Next, dehydration of cells was carried out with ethanol at different concentrations as follows: 30% ethanol for 10 min; 50% ethanol for 10 min; 70% ethanol for 10 min; 90% ethanol for 10 min; and finally, 100% ethanol for 20 min three times.Cells were pre-embedded with a mixture of epoxy resin and ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) for 1 hour, followed by 2 hour embedding in full epoxy resin (100%) at 37 °C. Later, epoxy resin was removed completely and replaced with fresh resin. Resin is then polymerised in an oven at 65 °C for 24 hours.
1 v/v) for 1 hour, followed by 2 hour embedding in full epoxy resin (100%) at 37 °C. Later, epoxy resin was removed completely and replaced with fresh resin. Resin is then polymerised in an oven at 65 °C for 24 hours.
A diamond knife was used for sectioning; 80 nm sections were obtained. For mounting of sections, copper grids of 3.05 mm diameter were used with 200 square meshes and with a coating of choice. Copper grids were stained with uranyl acetate (2%) for 20 min, and washed with lead citrate (0.4%) for 10 min.
For imaging, a transmission electron microscope (TEM, TECNAI 12) was operated at 120 kV.
Characterization of GO in bacterial growth medium
All materials used for ecotoxicology tests were vortexed with bacterial growth medium (soy broth) and the particle size was determined after 24 h using a Disc Centrifuge Model DC24000 (CPS Instruments, UK) at the maximum operational speed of 8142 rpm. The parameters for CPS were operated with a maximum diameter of 1 μm and minimum diameter of 0.001 μm. Particle density was 1.75 g mL−1, refractive index was 2.377, particle absorption was 1.656 K and non-sphericity factor was 3.Bacterial growth rate
rGO suspensions (rGO, SG-aq and LG-aq) were added to the bacterial culture to obtain the following final concentrations: 10, 50 and 100 μg mL−1. From each sample, 1 mL aliquot was transferred to each well of a 24-well plate. Control samples were run in parallel: (i) bacterial culture without rGO suspension as a negative control and (ii) with added GNL as a reference control. Each sample was prepared in triplicate. Samples were incubated at 27 °C and measured at an optical density of 600 nm (OD600) for every 2 hours for 6 h using a multi-mode reader (Synergy HTX, BioTek, USA).The bacterial growth rate (μ) was defined by R linear regression of cell density (absorbance units, AU) versus incubation time (hours) (AU h−1) as described in earlier publications.70,71 The results were analysed by ordinary ANOVA and Dunnett's multiple comparison test. The significance levels were *P < 0.05, **P < 0.01, ***P < 0.001.
Bacterial morphology
Samples from the bacterial growth test were collected after 24 h by centrifugation for 10 minutes at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 rpm (miniSpin, Eppendorf, Germany). The pellet was washed one time with DI water to remove media. A drop of sample was fixed on a silica surface and allowed to dry naturally for 20 minutes before SEM analysis by using a field-emission scanning electron microscope (SEM; Zeiss Ultra Plus, Zeiss, Germany). SEM images were acquired at an accelerating voltage of 5 kV at a low probe current (about 15 pA) using an InLens secondary electron detector with SmartSEM software.
500 rpm (miniSpin, Eppendorf, Germany). The pellet was washed one time with DI water to remove media. A drop of sample was fixed on a silica surface and allowed to dry naturally for 20 minutes before SEM analysis by using a field-emission scanning electron microscope (SEM; Zeiss Ultra Plus, Zeiss, Germany). SEM images were acquired at an accelerating voltage of 5 kV at a low probe current (about 15 pA) using an InLens secondary electron detector with SmartSEM software.
DNA fragmentation
The cells of 24 h from the growth rate test were lysed in 250 μL of cell lysis buffer containing 50 mM of Tris-HCl, 10 mM ethylenediaminetetraacetic acid, 0.1 M NaCl, and 0.5% SDS at pH 8.0. The lysate was incubated with 0.5 mg mL−1 RNase A at 37 °C for 1 h, and then with 0.2 mg mL−1 proteinase K at 50 °C overnight. Phenol extraction of this mixture was performed, and DNA in the aqueous phase was precipitated by using 25 mL (1/10 vol) of 7.5 M ammonium acetate and 250 mL (1/1 vol) of isopropanol. DNA electrophoresis was performed in 1% agarose gel containing 1 mg L−1 of ethidium bromide at 70 V, and the DNA fragments were visualized by exposing the gel to UV light, followed by photography.65Respiration test
The respiration activity of nitrifying microorganisms in activated sludge was monitored during 48 h or up to 180 h at 21 °C when exposed to rGO and LG-aq and GNL. The medium was prepared according to the standard methodology of the EN ISO 9408 with minor modifications: the solution with nutrients was diluted with a filtrate from the activated sludge instead of deionized water (DI) to achieve better conditions for microorganisms. For each respirometer run, fresh activated sludge was used at a concentration of 0.5 g L−1 based on dry matter. NH4Cl as a nutrient for nitrifying microorganisms was added into each sample with a final concentration of 50 mg L−1 for nitrogen. The total volume of samples was 50 mL in sterilized glass bottles. The standard methodology of EN ISO 9408 was followed with minor modifications: organic compound was replaced with rGO suspensions. rGO and LG-aq were added in the following final concentrations: rGO – 5, 10 and 50 μg mL−1, LG-aq – 5, 10, 50 and 100 μg mL−1 and GNL – 10, 50 and 100 μg mL−1, respectively. Activated sludge at a concentration of 0.5 g L−1 with medium served as a negative control and a referenced control was activated sludge at a concentration of 0.5 g L−1 with a 10 mL base oil. The oxygen consumption rate (mg O2 per L per h) and cumulative oxygen consumption (mg O2 per L) were continually monitored using a Micro-Oxymax respirometer (Columbus Instruments International, USA). Each sample was prepared in duplicate because the amount of material needed for this test is significantly high. The results were analysed by ordinary ANOVA and Dunnett's multiple comparison test. The significance levels were *P < 0.05, **P < 0.01, ***P < 0.001.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project has received funding from the the European Union FP7 FutureNanoNeeds project (NMP/2013/1.3-3) under the grant agreement no. 604602. The authors acknowledge Jingji Li for the help with TEM imaging.M. E.-G. acknowledges the support of funding from the Irish Research Council under the Enterprise Partnership PostDoctoral Fellowship Scheme 2015 (EPSPD/2015/37).
V. C. acknowledges the support of funding from the European Union's Horizon 2020 research and innovation programme Graphene Core 1 under grant agreement no. 696656, the Irish Research Council under the Government of Ireland Postdoctoral Fellowship scheme (GOIPD/2016/128) and Ing. Olindo Boselli for the useful discussions. A. S. and N. H. A. N. acknowledge the support provided by the Research Infrastructure NanoEnviCz, supported by the Ministry of Education, Youth and Sports of the Czech Republic (LM2015073). This work was partly supported by the Ministry of Education, Youth and Sports under the project LO1201 in the framework of the targeted support of the “National Programme for Sustainability I”.
References
- A. Yamasaki, J. Chem. Eng. Jpn., 2003, 36, 361–375 CrossRef CAS.
- P. M. Cox, R. A. Betts, C. D. Jones, S. A. Spall and I. J. Totterdell, Nature, 2000, 408, 184 CrossRef CAS PubMed.
- K. Holmberg, P. Andersson and A. Erdemir, Tribol. Int., 2012, 47, 221–234 CrossRef.
- K. Lee, Y. Hwang, S. Cheong, L. Kwon, S. Kim and J. Lee, Curr. Appl. Phys., 2009, 9, e128–e131 CrossRef.
- L. Rapoport, Y. Bilik, Y. Feldman and M. Homyonfer, Nature, 1997, 387, 791 CrossRef CAS.
- M. Chhowalla and G. A. Amaratunga, Nature, 2000, 407, 164–167 CrossRef CAS PubMed.
- M. Gulzar, H. Masjuki, M. Kalam, M. Varman, N. Zulkifli, R. Mufti and R. Zahid, J. Nanopart. Res., 2016, 18, 223 CrossRef.
- I. Lahouij, B. Vacher, J.-M. Martin and F. Dassenoy, Wear, 2012, 296, 558–567 CrossRef CAS.
- L. Yadgarov, V. Petrone, R. Rosentsveig, Y. Feldman, R. Tenne and A. Senatore, Wear, 2013, 297, 1103–1110 CrossRef CAS.
- M. Sgroi, M. Asti, F. Gili, F. A. Deorsola, S. Bensaid, D. Fino, G. Kraft, I. Garcia and F. Dassenoy, Tribol. Int., 2017, 105, 317–325 CrossRef CAS.
- M. Sgroi, F. Gili, D. Mangherini, I. Lahouij, F. Dassenoy, I. Garcia, I. Odriozola and G. Kraft, Tribol. Trans., 2015, 58, 207–214 CrossRef CAS.
- D. Berman, A. Erdemir and A. V. Sumant, Mater. Today, 2014, 17, 31–42 CrossRef CAS.
- J. Zhao, Y. Li, Y. Wang, J. Mao, Y. He and J. Luo, RSC Adv., 2017, 7, 1766–1770 RSC.
- C. Lee, Q. Li, W. Kalb, X.-Z. Liu, H. Berger, R. W. Carpick and J. Hone, science, 2010, 328, 76–80 CrossRef CAS PubMed.
- W. Azmi, M. Sharif, T. Yusof, R. Mamat and A. Redhwan, Renewable Sustainable Energy Rev., 2017, 69, 415–428 CrossRef CAS.
- A. Rasheed, M. Khalid, A. Javeed, W. Rashmi, T. Gupta and A. Chan, Tribol. Int., 2016, 103, 504–515 CrossRef CAS.
- A. K. Rasheed, M. Khalid, R. Walvekar, T. C. S. M. Gupta and A. Chan, J. Mater. Res., 2016, 31, 1939–1946 CrossRef CAS.
- H. Huang, J. Tu, L. Gan and C. Li, Wear, 2006, 261, 140–144 CrossRef CAS.
- A. Bianco, Angew. Chem., Int. Ed., 2013, 52, 4986–4997 CrossRef CAS PubMed.
- D. Bradley, Mater. Today, 2012, 15, 230 CrossRef.
- C. Bussy, H. Ali-Boucetta and K. Kostarelos, Acc. Chem. Res., 2012, 46, 692–701 CrossRef PubMed.
- Y. Zhang, S. F. Ali, E. Dervishi, Y. Xu, Z. Li, D. Casciano and A. S. Biris, ACS Nano, 2010, 4, 3181–3186 CrossRef CAS PubMed.
- A. Sasidharan, L. Panchakarla, P. Chandran, D. Menon, S. Nair, C. Rao and M. Koyakutty, Nanoscale, 2011, 3, 2461–2464 RSC.
- N. Chatterjee, H.-J. Eom and J. Choi, Biomaterials, 2014, 35, 1109–1127 CrossRef CAS PubMed.
- C. Cheng, S. Nie, S. Li, H. Peng, H. Yang, L. Ma, S. Sun and C. Zhao, J. Mater. Chem. B, 2013, 1, 265–275 RSC.
- S. Anguissola, D. Garry, A. Salvati, P. J. O'Brien and K. A. Dawson, PLoS One, 2014, 9, e108025 Search PubMed.
- D. Ahmed, X. Mao, B. K. Juluri and T. J. Huang, Microfluid. Nanofluid., 2009, 7, 727–731 CrossRef CAS.
- P. Laaksonen, M. Kainlauri, T. Laaksonen, A. Shchepetov, H. Jiang, J. Ahopelto and M. B. Linder, Angew. Chem., Int. Ed., 2010, 49, 4946–4949 CrossRef CAS PubMed.
- C. Corbo, R. Molinaro, M. Tabatabaei, O. C. Farokhzad and M. Mahmoudi, Biomater. Sci., 2017, 5(3), 378–387 RSC.
- C. Gunawan, M. Lim, C. P. Marquis and R. Amal, J. Mater. Chem. B, 2014, 2, 2060–2083 RSC.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Åberg, K. A. Dawson and A. Salvati, ACS Nano, 2012, 6, 5845–5857 CrossRef CAS PubMed.
- A. Verma and F. Stellacci, Small, 2010, 6, 12–21 CrossRef CAS PubMed.
- A. Verma, O. Uzun, Y. Hu, Y. Hu, H.-S. Han, N. Watson, S. Chen, D. J. Irvine and F. Stellacci, Nat. Mater., 2008, 7, 588–595 CrossRef CAS PubMed.
- B. Pelaz, P. del Pino, P. Maffre, R. Hartmann, M. Gallego, S. Rivera-Fernandez, J. M. de la Fuente, G. U. Nienhaus and W. J. Parak, ACS Nano, 2015, 9, 6996–7008 CrossRef CAS PubMed.
- L. Peña-Parás, J. Taha-Tijerina, L. Garza, D. Maldonado-Cortés, R. Michalczewski and C. Lapray, Wear, 2015, 332, 1256–1261 CrossRef.
- T. Luo, X. Wei, X. Huang, L. Huang and F. Yang, Ceram. Int., 2014, 40, 7143–7149 CrossRef CAS.
- D.-X. Peng, C.-H. Chen, Y. Kang, Y.-P. Chang and S.-Y. Chang, Ind. Lubr. Tribol., 2010, 62, 111–120 CrossRef.
- V. An, Y. Irtegov and C. d. Izarra, J. Nanomater., 2014, 2014, 188 Search PubMed.
- I. Lahouij, E. Bucholz, B. Vacher, S. Sinnott, J. Martin and F. Dassenoy, Nanotechnology, 2012, 23, 375701 CrossRef CAS PubMed.
- S. S. K. Kiu, S. Yusup, C. V. Soon, T. Arpin, S. Samion and R. N. M. Kamil, J. Phys. Sci., 2017, 28, 257 CrossRef.
- Y. Li, J. Zhao, C. Tang, Y. He, Y. Wang, J. Chen, J. Mao, Q. Zhou, B. Wang and F. Wei, Adv. Mater. Interfaces, 2016, 3(22), 1600700 CrossRef.
- A. Senatore, V. D'Agostino, V. Petrone, P. Ciambelli and M. Sarno, ISRN Tribol., 2013, 2013 Search PubMed , 425809.
- Y. Chang, S.-T. Yang, J.-H. Liu, E. Dong, Y. Wang, A. Cao, Y. Liu and H. Wang, Toxicol. Lett., 2011, 200, 201–210 CrossRef CAS PubMed.
- W. Hu, C. Peng, M. Lv, X. Li, Y. Zhang, N. Chen, C. Fan and Q. Huang, ACS Nano, 2011, 5, 3693–3700 CrossRef CAS PubMed.
- T. Lammel, P. Boisseaux, M.-L. Fernández-Cruz and J. M. Navas, Part. Fibre Toxicol., 2013, 10, 27 CrossRef CAS PubMed.
- M. Kucki, L. Diener, N. Bohmer, C. Hirsch, H. F. Krug, V. Palermo and P. Wick, J. Nanobiotechnol., 2017, 15, 46 CrossRef PubMed.
- J. Ma, R. Liu, X. Wang, Q. Liu, Y. Chen, R. P. Valle, Y. Y. Zuo, T. Xia and S. Liu, ACS Nano, 2015, 9, 10498–10515 CrossRef CAS PubMed.
- Y. Li, Y. Liu, Y. Fu, T. Wei, L. Le Guyader, G. Gao, R.-S. Liu, Y.-Z. Chang and C. Chen, Biomaterials, 2012, 33, 402–411 CrossRef CAS PubMed.
- R. G. Mendes, B. Koch, A. Bachmatiuk, X. Ma, S. Sanchez, C. Damm, O. G. Schmidt, T. Gemming, J. Eckert and M. H. Rümmeli, J. Mater. Chem. B, 2015, 3, 2522–2529 RSC.
- V. C. Sanchez, A. Jachak, R. H. Hurt and A. B. Kane, Chem. Res. Toxicol., 2011, 25, 15–34 CrossRef PubMed.
- Q. Mu, G. Su, L. Li, B. O. Gilbertson, L. H. Yu, Q. Zhang, Y.-P. Sun and B. Yan, ACS Appl. Mater. Interfaces, 2012, 4, 2259–2266 CAS.
- H. Suzuki, T. Toyooka and Y. Ibuki, Environ. Sci. Technol., 2007, 41, 3018–3024 CrossRef CAS PubMed.
- C. Greulich, J. Diendorf, T. Simon, G. Eggeler, M. Epple and M. Köller, Acta Biomater., 2011, 7, 347–354 CrossRef CAS PubMed.
- M. J. Hajipour, K. M. Fromm, A. A. Ashkarran, D. J. de Aberasturi, I. R. de Larramendi, T. Rojo, V. Serpooshan, W. J. Parak and M. Mahmoudi, Trends Biotechnol., 2012, 30, 499–511 CrossRef CAS PubMed.
- A. Panáček, L. Kvítek, R. Prucek, M. Kolář, R. Večeřová, N. Pizúrová, V. K. Sharma, T. j. Nevěčná and R. Zbořil, J. Phys. Chem. B, 2006, 110, 16248–16253 CrossRef PubMed.
- S. Liu, T. H. Zeng, M. Hofmann, E. Burcombe, J. Wei, R. Jiang, J. Kong and Y. Chen, ACS Nano, 2011, 5, 6971–6980 CrossRef CAS PubMed.
- S. S. Nanda, D. K. Yi and K. Kim, Sci. Rep., 2016, 6, 28443 CrossRef CAS PubMed.
- S. C. Smith and D. F. Rodrigues, Carbon, 2015, 91, 122–143 CrossRef CAS.
- F. Ahmed and D. F. Rodrigues, J. Hazard. Mater., 2013, 256, 33–39 CrossRef PubMed.
- H. N. Nguyen, S. L. Castro-Wallace and D. F. Rodrigues, Environ. Sci.: Nano, 2017, 4, 160–169 RSC.
- L. A. Luongo and X. J. Zhang, J. Hazard. Mater., 2010, 178, 356–362 CrossRef CAS PubMed.
- Y. Yin and X. Zhang, Water Sci. Technol., 2008, 58, 623–628 CrossRef CAS PubMed.
- S. Gurunathan, J. W. Han, A. A. Dayem, V. Eppakayala and J.-H. Kim, Int. J. Nanomed., 2012, 7, 5901 CrossRef CAS PubMed.
- W. Hu, C. Peng, W. Luo, M. Lv, X. Li, D. Li, Q. Huang and C. Fan, ACS Nano, 2010, 4, 4317–4323 CrossRef CAS PubMed.
- A. Pruna, D. Tamvakos, M. Sgroi, D. Pullini, E. A. Nieto and D. Busquets-Mataix, Int. J. Mater. Res., 2015, 106, 398–405 CrossRef CAS.
- H. Czichos and K.-H. Habig, Tribologie-Handbuch: Tribometrie, Tribomaterialien, Tribotechnik, Springer-Verlag, 2010 Search PubMed.
- R. Zucker, E. Massaro, K. Sanders, L. Degn and W. Boyes, Cytometry, Part A, 2010, 77, 677–685 CrossRef CAS PubMed.
- M. S. Darwish, N. H. Nguyen, A. Ševců and I. Stibor, J. Nanomater., 2015, 16, 89 Search PubMed.
- M. S. Darwish, N. H. Nguyen, A. Ševců, I. Stibor and S. K. Smoukov, Mater. Sci. Eng., C, 2016, 63, 88–95 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7nr08597d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |