Open Access Article
Open Access ArticleCollection of amino acids and DNA from fingerprints using hydrogels
Ward
van Helmond†
ab,
Vincent
O'Brien†
c,
Robin
de Jong
b,
Jan
van Esch
 c,
Sander
Oldenhof
*bc and
Marcel
de Puit
c,
Sander
Oldenhof
*bc and
Marcel
de Puit
 *bc
*bc
aForensic Science, Amsterdam University of Applied Sciences, Weesperzijde 190, 1097 DZ Amsterdam, The Netherlands
bDigital Technology and Biometrics, Netherlands Forensic Institute, Laan van Ypenburg 6, 2497 GB, The Hague, The Netherlands. E-mail: s.oldenhof@nfi.minvenj.nl; m.de.puit@nfi.minvenj.nl
cAdvanced Soft Matter group, Chemical Engineering Department, Delft University of Technology, Van der Maasweg 9, 2629 HZ Delft, The Netherlands
First published on 4th January 2018
Abstract
The amino acid profile obtained from a fingerprint may provide valuable information on its donor. For forensic scientists, recovering evidence relating to the amino acid profile of a suspect can potentially be valuable for identification and exclusion purposes. Herein we detail the use of cross-linkable solutions of dextran-methacrylate to form hydrogels capable of collecting amino acids from surfaces followed by extraction and quantification with UPLC-MS. This method allows for the amino acid profile analysis of fingerprints while allowing for their increased visualisation at a later stage using the standard method of cyanoacrylation. We will demonstrate this method to also be capable of collecting DNA from fingerprints with a 20–60% yield in comparison to using a conventional cotton swab.
Introduction
The amino acid profile of an individual may provide information such as the sex, health and age of a donor.1,2 Common methods of amino acid profile analysis require access to either the blood, urine, saliva, faeces or cerebrospinal fluid to generate a donor profile.3 Interest in methods using sweat deposits in fingerprints for the analysis of amino acid levels has increased in recent years as analytical methods have been developed that allow for the rapid and accurate quantification of amino acid concentrations using GC, CE and (U)HPLC-MS.4–9 Sweat as a diagnostic biofluid has several advantages as it can be easily collected from patients without the need for invasive procedures and is safer for diagnosticians to handle as saliva or blood may contain viruses and must be treated with extra care.10,11Previous studies, where sweat has been analysed for amino acid profiling, have relied on the use of sweat collection devices that need to be attached to an individual's skin in order to collect adequate quantities of sweat for analysis, or via fingerprints by completely dissolving the fingerprint.12,13 For the sweat collection studies large volumes of sweat are required alongside specialist equipment to produce an amino acid profile. In the case of fingerprint analysis there are issues relating to the complex matrix of the fingerprints that limit the amount of amino acids that can be successfully extracted from fingerprints.4
In the field of forensic science, an investigator will primarily use swabs or lifting tape to collect evidence from a crime scene. Swabs are a destructive method of analyte collection and while they excel at absorbing analytes they show poor release of the trapped analytes that are intended for analysis.14 Lifting tape or gel lifters are a common way to recover fingerprints from surfaces, but these techniques do not facilitate the extraction of chemical components without destruction of the ridge detail. Ideally a collection material is non-marking (i.e. it does not physically mark the evidence although chemical components are extracted) to the surface it is applied on and shows rapid adsorption and release of collected analytes.
A method which collects amino acids from a surface or complex matrix like a fingerprint could be of interest for forensic investigators, but also for diagnostic clinical purposes. While methods exist for the direct chemical analysis of fingerprint residues, they rely on the introduction of the entire fingerprint into either a solvent or an analytical instrument in order to detect the presence of certain analytes on a fingerprint.15 This is not ideal for forensic investigators as it requires destruction of the fingerprint ridge detail, or specialised equipment to perform surface analysis at a crime scene. Hydrogels would provide a flexible solution to sample collection as they can be applied to non-porous surfaces. Hydrogels are 3D networks of hydrophilic polymers in which water is the dispersion medium.16 They have been found to be useful in a variety of areas from drug delivery and tissue engineering to cell culturing.17–19 Their capacity to absorb and hold water potentially makes them suitable as extraction media for water-soluble analytes from a surface.
In this paper, we demonstrate the possibility of using hydrogels as a means of collecting amino acids from fingerprints and their comparative performance for the use of direct extraction with a solvent for amino acid analysis. Both the collection of amino acid deposits on a glass surface and from fingerprints using hydrogels will be described. Additionally, since the collection of DNA from fingerprints is increasingly adapted in forensic casework, we investigated the use of hydrogels to collect DNA from fingerprints.
Experimental
Pre-hydrogel and amino acid solutions
Dextran-methacrylate was chosen as a hydrophilic polymer as it is conveniently cross-linked by photo-initiated radical polymerization forming a hydrogel. The method described by De Smedt et al. was followed to prepare dextran-methacrylate with a substitution degree of 2.5.20 The structure of the material was confirmed using 1H-NMR. The photo-initiator used, lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), was prepared according to the method described by Fairbanks et al.21 The structure of the material was confirmed using 1H-, 13C-, and 31P-NMR. Pre-hydrogel solutions were prepared by dissolving 1% w/v LAP and 10% w/v dextran-methacrylate in distilled water (1 mL, pH 6.5) under sonication for 20 minutes at room temperature. Amino acid and isotopically labelled internal standard (IS) solutions of glycine, L-alanine, L-serine, L-proline, L-valine, L-threonine, L-cysteine, hydroxy-L-proline, L-isoleucine, L-leucine, L-asparagine, L-ornithine, L-aspartic acid, L-glutamine, L-lysine, L-glutamic acid, L-methionine, L-histidine, L-phenylalanine, L-arginine, L-tyrosine, L-tryptophan and L-cystine of 2.0 mg L−1 were prepared as described previously.9Glass cover slip functionalization
Borosilicate microscope cover slips of dimensions 25 × 25 mm were purchased from Fisher Scientific. The slides were activated prior to functionalization using a Harrick's plasma cleaner for 240 seconds. They were transferred to a vacuum chamber containing 100 μL of 3-(trichlorosilyl)-propyl-methacrylate and left under dynamic vacuum for 4 hours to functionalize the surface.Fingerprint deposition
Fingerprints were deposited using a custom made fingerprinting device (Fig. 1) based on a similar device reported by Fieldhouse.22 This device maximizes a uniform application of force on the fingerprint and only allows a certain area of the fingerprint to be exposed to the substrate surface. Fingerprints were produced on glass slides. The hands of three males and two females, all ranging between 20 and 26 years old, were washed before returning to normal work activities. After one hour the fingerprints were deposited on the glass slides. After drying in air for one hour, the fingerprints were extracted either by hydrogel extraction or direct solvent extraction. Fingerprints for DNA extraction and quantification were produced as follows: 3 donors (2 male and 1 female, aged 21 to 28) were asked to rub their fingers against their forehead and hair for 3 seconds to generate more shed skin cells and subsequently deposit their fingerprints on glass slides. | ||
| Fig. 1 Custom made fingerprinting device applied to maximize reproducibility in fingerprint deposition. | ||
Amino acid standard samples
To determine the hydrogel extraction efficiency, amino acid working solution (25 μL) was deposited on a glass slide and then dried in air for 30 minutes. The amino acids were then extracted using hydrogels, or were extracted directly with a solvent.Analyte absorption experiments
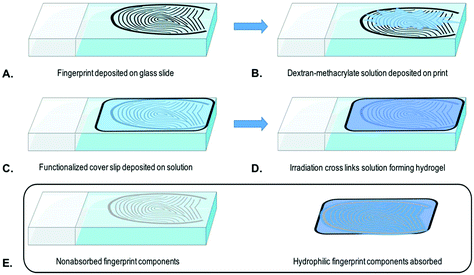
In a typical extraction experiment, a freshly prepared solution (20 μL) of dextran-methacrylate (10% w/v) and LAP (1% w/v) is applied to a surface containing analytes of interest (Fig. 2A and B). Thereafter, a methacrylate-functionalized glass cover slip is placed over the solution to be able to collect the hydrogel (Fig. 2C). After 3 minutes, the sample is irradiated for 30 seconds, using a simple 405 nm laser pen (1 mW), generating the hydrogel (Fig. 2D). During cross-linking the methacrylate groups of the dextran-methacrylate and the cover slide will react, hereby covalently binding the hydrogel to the glass cover. By careful removal of the cover slide, the hydrogel containing the available hydrophilic analytes is taken away, leaving nonabsorbed components behind (Fig. 2E).Amino acid extraction
Hydrogels were applied onto a surface of interest as described above and subsequently transferred to a beaker and the analytes were extracted with MeOH/H2O (3 mL, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) under sonication for 30 min. IS solution (10 μL) was added and the extract was then transferred to a 15 mL polypropylene conical tube. The sample solution was then evaporated under nitrogen flow, and the material was dissolved in MeOH containing 5% v/v formic acid (50 μL) and transferred into an injection vial. Solvent extracted samples, which were prepared for comparison, were processed similarly, except that the glass slides containing the amino acids were transferred to a beaker directly.
1) under sonication for 30 min. IS solution (10 μL) was added and the extract was then transferred to a 15 mL polypropylene conical tube. The sample solution was then evaporated under nitrogen flow, and the material was dissolved in MeOH containing 5% v/v formic acid (50 μL) and transferred into an injection vial. Solvent extracted samples, which were prepared for comparison, were processed similarly, except that the glass slides containing the amino acids were transferred to a beaker directly.
Amino acid analysis by UPLC-MS
Amino acids were quantified using UPLC-MS, as described previously.9 In brief, for each amino acid a calibration curve ranging from 0.10 to 1.60 mg L−1 was produced, corresponding to 5–80 ng of a single amino acid in the sample volume (50 μL) for a fingerprint. These solutions were prepared by adding respectively 2.5, 5.0, 10, 15, 30, and 40 μL of 2.0 mg L−1 amino acid working solution into a vial. IS solution (10 μL), composed of isotopically labelled amino acids, was added to each vial, and MeOH containing 5% v/v formic acid was added to make a total volume of 50 μL. The samples were then analysed using UPLC-MS using a 150 mm UPLC amide BEH column (Waters, Milford, MA, USA). A Waters Acquity I-class UPLC autosampler and a binary solvent pump were used to inject and separate the sample solution. The effluent of the column was analysed by means of electrospray ionisation time-of-flight MS (ESI-TOF-MS) using an accurate mass TOF with a dual electrospray source (6220 Agilent, Santa Clara, CA, USA). Data were processed and quantified using Agilent Mass Hunter Qualitative Analysis software (version B.05.00) and Quantitative Analysis software (version B.05.00).DNA extraction and quantification
DNA was collected from fingerprints using either a cotton swab or via our hydrogel method. When using the cotton swab, the complete fingerprint was swabbed with a dry cotton swab and subsequently the cotton swab was transferred to a 1.5 mL aliquot containing 300 μL ATL buffer (Qiagen, Inc.). Hydrogel lifting of the fingerprints was performed as described above. After lifting, the hydrogels were transferred to a 1.5 mL aliquot containing 300 μL ATL buffer. The concentration of human DNA in the samples was determined using real-time PCR as described by Nicklas and Buel.23Results
Amino acid standard solution
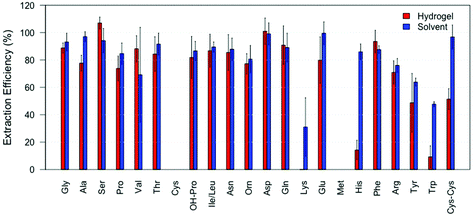
Initial experiments were performed using the standard amino acid solution containing 22 amino acids. After absorption and subsequent extraction from the hydrogel, 19 amino acids out of 22 (all but L-cysteine, L-lysine and L-methionine) were successfully detected and quantified while 20 were found in the samples directly extracted with solvent (all but L-cysteine and L-methionine). Fig. 3 shows the extraction efficiency (IS corrected) of each amino acid for both methods. The extraction efficiency varied between amino acids and extraction methods and, as expected, was in general below 100%. For all the detected and quantified amino acids, the extraction efficiency was similar using either extraction method, except for L-lysine, L-histidine, L-tryptophan and L-cystine. From Fig. 3 we can see that L-methionine and L-cysteine are not found in any samples collected using either extraction method. The lack of L-cysteine may be caused by the formation of cysteic acid, which is indicated by the detection of a mass at 170.012 m/z, the theoretical mass ([M + H]+) of cysteic acid. L-Methionine may undergo oxidation to yield methionine sulfoxide, as a mass of 166.052 m/z is detected, which corresponds to the theoretical mass ([M + H]+). L-Tryptophan is extracted with 50% efficiency when solvent extraction is used while it is extracted with 10% efficiency when the hydrogel is applied. As we have reported earlier, the analysis of L-tryptophan, when extracted from fingerprints, is problematic.12 It is possible that the radical photoinitiator, used in the gel, attacks the conjugated ring system of L-tryptophan reducing its concentration, making the analysis after gel extraction more complicated. However, it is clear from Fig. 3 that the hydrogel is not only capable of collecting the amino acids from a surface, but that subsequent extraction and detection from the hydrogel is also possible. While values are relatively similar across both methods it must be noted that the hydrogel includes the extra step of collecting the analytes from the surface.Amino acids recovered from fingerprints
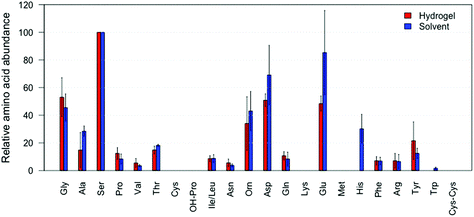
After successfully applying our hydrogel system in amino acid absorption from a glass surface, we investigated its ability to absorb amino acids from fingerprints in a non-marking way, see Fig. 4 (amino acids reported in relative abundances to serine). We found that the application of the hydrogel to an actual fingerprint led to the positive identification of 15 different amino acids. This indicates that at least 15 amino acids were absorbed into the hydrogel, and could subsequently be identified and quantified, see Fig. 4 (red bars). Importantly, visualization of the fingerprints after hydrogel treatment was still successful, as can be seen in Fig. 5. In comparison, 17 amino acids were detected from fingerprints that were extracted with a solvent and completely dissolved for analysis, see Fig. 4 (blue bars). As can be seen in Fig. 4, the hydrogel extracted fingerprints yielded largely similar amino acid profiles to the solvent extracted fingerprints, except for L-histidine and L-tryptophan abundances. L-Histidine and L-tryptophan were not detected using the hydrogel, which can be explained by the lower extraction efficiency of these amino acids using the hydrogels, as described above.The large variance in the recovered amino acid profiles likely originates from the natural variation of amino acid concentration in fingerprints and the relatively low amount of fingerprints included in this proof of principle. While a large effort was made to create reproducible fingerprints, variation between donors (inter-variability) cannot be avoided.24 In terms of absolute amino acid extraction, the hydrogel lifted fingerprints on average yielded 532 ng of amino acid compared to 807 ng in the solvent extracted fingerprints. Despite this, the relative amino acid concentrations obtained with the hydrogels coincided with the relative concentrations collected via direct solvent extraction. L-Tryptophan is not detected likely due to its low abundances in fingerprints,1 and/or the potential radical degradation. L-Cysteine and L-methionine are not detected, which concurs with the results obtained when extracted from amino acid standard samples as described above. To the best of our knowledge our method is the first that allows the extraction of amino acids from a fingerprint while retaining the ability for subsequent visualisation of the fingerprint.
Visualisation of lifted fingerprints
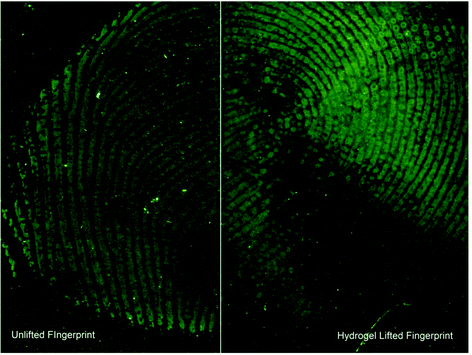
Central to this method is its non-marking nature. As the primary solvent is water the hydrophilic constituents of the fingerprint are collected selectively, leaving enough hydrophobic material to visualise the fingerprint using cyanoacrylation followed by dyeing with basic yellow. Fig. 5 is a split fingerprint example of the comparison in quality between a standard cyanoacrylate and basic yellow treatment and lifting using the hydrogel prior to cyanoacrylate and basic yellow treatment. It is clear that after lifting the hydrogel, including the absorbed hydrophilic solutes, there is sufficient material for further development using standard fingerprint visualization techniques.The majority of the ridge detail is still present with slight degradation of the fine details. The likely cause of the “smudges” is localised variances in the absorption of analytes. As fingerprints vary in their composition the likelihood for one area to be slightly more hydrophobic and therefore repel the penetration effects of the hydrogel is higher. However, the method presented here is unique in that it can successfully collect analytes of a hydrophilic nature of interest from a fingerprint surface in similar quantities as traditional methods and allow for the subsequent visualisation of the fingerprint.
DNA recovered from fingerprints
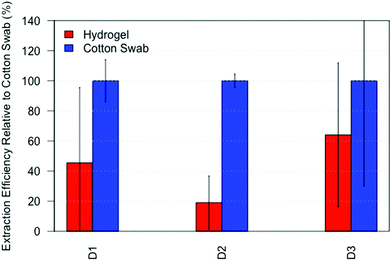
After applying the hydrogel to extract amino acids from fingerprints, we investigated the ability of the hydrogels to absorb DNA from fingerprints. Compared to conventional DNA sampling using a cotton swab, the hydrogels roughly yielded between 20–60% of the DNA quantity, see Fig. 6. Although this is significantly lower than the swab, we still believe hydrogels to be an addition to the current workflow, for instance when sampling unequal surfaces such as gun grips. Moreover, DNA could be recovered from fingerprints while leaving sufficient material behind for visualization. The larger variation in DNA yielded by the hydrogels is most likely due to the extra steps in the sample preparation, especially the transfer of the lifted hydrogel to an aliquot, indicating that further optimization of the sample preparation is necessary.Conclusion
We have shown that cross-linked hydrogels can be applied to absorb amino acids from both fingerprints and amino acid solution deposits on glass, and extracted the amino acids from the hydrogel for their qualitative and quantitative detection using UPLC-MS. These results show the selective uptake of the water solubles, leaving sufficient material for further investigations of the fingerprint. Post-collection visualisation of the fingerprints is also shown to be possible using cyanoacrylate and immersion in a basic yellow dye. Moreover, the hydrogels were able to collect DNA from fingerprints with a 20–60% yield compared to the conventional cotton swab. Together these results show that hydrogels are promising materials for evidence collection as they combine both the dissolving power of a solution with the physical lifting capacity of a gel. Combined with the rapid release of analytes when immersed in common solvents they present themselves as a possible replacement or complementary method for analyte collection.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge the following funding scheme: V. O. Erasmus+ grant funding, S. O. Ministry of Security and Justice, Innovation grant, W. v. H. NWO research grant no. 2014-01-124PRO (RAAK-PRO). The authors would also like to thank Shermayne Siem-Gorré for lending her hand for Fig. 1 and Frits de Haan for supplying the image. The authors thank Josja Graafland and Francisca Duijs for executing the DNA quantification.References
- C. Huynh, E. Brunelle, L. Halámková, J. Agudelo and J. Halámek, Anal. Chem., 2015, 87, 11531–11536 CrossRef CAS PubMed
.
- S. Ohtani, Y. Matsushima, Y. Kobayashi and K. Kishi, J. Forensic Sci., 1998, 43, 949–953 CAS
.
-
S. M. Rutherfurd and G. S. Gilani, inCurrent Protocols in Protein Science, John Wiley & Sons, Inc., 2001 Search PubMed
.
-
S. Dawling, S. Jickels and A. Negrusz, Clarke's Analysis of Drugs and Poisons, 2004, vol. 3, 425–499 Search PubMed
.
- M. M. Delgado-Povedano, M. Calderón-Santiago, F. Priego-Capote and M. D. Luque de Castro, Anal. Chim. Acta, 2016, 905, 115–125 CrossRef CAS PubMed
.
- T. Atherton, R. Croxton, M. Baron, J. Gonzalez-Rodriguez, L. Gámiz-Gracia and A. M. García-Campaña, J. Sep. Sci., 2012, 35, 2994–2999 CrossRef CAS PubMed
.
- N. E. Archer, Y. Charles, J. A. Elliott and S. Jickells, Forensic Sci. Int., 2005, 154, 224–239 CrossRef CAS PubMed
.
- K. A. Mountfort, H. Bronstein, N. Archer and S. M. Jickells, Anal. Chem., 2007, 79, 2650–2657 CrossRef CAS PubMed
.
- W. van Helmond, C. J. Kuijpers, E. van Diejen, J. Spiering, B. Maagdelijn and M. de Puit, Anal. Methods, 2017, 9, 5697–5702 RSC
.
- CDC Blodborne Exposure Guide, https://www.cdc.gov/oralhealth/infectioncontrol/faq/bloodborne_exposures.htm, (accessed 26/08/2016, 2016).
- S. Jadoon, S. Karim, M. R. Akram, A. Kalsoom Khan, M. A. Zia, A. R. Siddiqi and G. Murtaza, Int. J. Anal. Chem., 2015, 2015, 7 Search PubMed
.
- M. de Puit, M. Ismail and X. Xu, J. Forensic Sci., 2014, 59, 364–370 CrossRef CAS PubMed
.
- M. M. Delgado-Povedano, M. Calderón-Santiago, F. Priego-Capote and M. D. Luque de Castro, Talanta, 2016, 146, 310–317 CrossRef CAS PubMed
.
- M. S. Adamowicz, D. M. Stasulli, E. M. Sobestanovich and T. W. Bille, PLoS One, 2014, 9, e116351 Search PubMed
.
- M. J. Bailey, R. Bradshaw, S. Francese, T. L. Salter, C. Costa, M. Ismail, R. P. Webb, I. Bosman, K. Wolff and M. de Puit, Analyst, 2015, 140, 6254–6259 RSC
.
- E. M. Ahmed, J. Adv. Res., 2015, 6, 105–121 CrossRef CAS PubMed
.
- N. A. Peppas, P. Bures, W. Leobandung and H. Ichikawa, Eur. J. Pharm. Biopharm., 2000, 50, 27–46 CrossRef CAS PubMed
.
- J. A. Benton, C. A. DeForest, V. Vivekanandan and K. S. Anseth, Tissue Eng., Part A, 2009, 15, 3221–3230 CrossRef CAS PubMed
.
- J. A. Rowley, G. Madlambayan and D. J. Mooney, Biomaterials, 1999, 20, 45–53 CrossRef CAS PubMed
.
- S. C. De Smedt, A. Lauwers, J. Demeester, M. J. Van Steenbergen, W. E. Hennink and S. P. F. M. Roefs, Macromolecules, 1995, 28, 5082–5088 CrossRef CAS
.
- B. D. Fairbanks, M. P. Schwartz, C. N. Bowman and K. S. Anseth, Biomaterials, 2009, 30, 6702–6707 CrossRef CAS PubMed
.
- S. Fieldhouse, Forensic Sci. Int., 2011, 207, 96–100 CrossRef PubMed
.
- J. A. Nicklas and E. Buel, J. Forensic Sci., 2006, 51, 1005–1015 CrossRef CAS PubMed
.
- A. Girod, R. Ramotowski and C. Weyermann, Forensic Sci. Int., 2012, 223, 10–24 CrossRef CAS PubMed
.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |