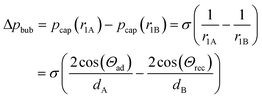Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Digital droplet PCR on disk†
Friedrich
Schuler
*ab,
Martin
Trotter
a,
Marcel
Geltman
a,
Frank
Schwemmer
b,
Simon
Wadle
ab,
Elena
Domínguez-Garrido
d,
María
López
d,
Cristina
Cervera-Acedo
d,
Paula
Santibáñez
d,
Felix
von Stetten
ab,
Roland
Zengerle
abc and
Nils
Paust
ab
aHahn-Schickard, Georges-Koehler-Allee 103, 79110 Freiburg, Germany
bLaboratory for MEMS Applications, IMTEK – Department of Microsystems Engineering, University of Freiburg, Georges-Koehler-Allee 103, 79110 Freiburg, Germany. E-mail: Friedrich.Schuler@imtek.uni-freiburg.de
cBIOSS – Centre for Biological Signalling Studies, University of Freiburg, Freiburg, Germany
dMolecular Diagnostics Laboratory, Rioja Salud-Foundation, La Rioja, Spain
First published on 9th November 2015
Abstract
Existing systems for digital droplet PCR (ddPCR) either suffer from low integration or are difficult to introduce to mass fabrication. Here we present an integrated system that is compatible to mass fabrication and combines emulsification, PCR, and fluorescence readout in a single chamber within a disposable cartridge (disk). Droplets are generated by injecting the sample into fluorinated oil via centrifugal step emulsification. The resulting emulsion is aligned in the PCR and readout zone by capillary action. During thermocycling, gas bubbles generated by degassing are removed by capillary driven transport through tapered regions in the PCR chamber. Thereby, the positioning of the emulsion within the readout zone of the PCR chamber is ensured at any time and no bubbles are present during readout. Manual handling of the disk solely requires pipetting of oil and PCR mix into the inlet structures, placing the disk into the thermocycler and subsequently into a microarray scanner. The functionality of the ddPCR process chain is demonstrated by quantitative detection of the cystic fibrosis causing mutation p.Phe508del, which is of interest for non-invasive prenatal testing (NIPT). The mutation was detected in a concentration range spanning four orders of magnitude. We envision that this work will lay the base for the development of highly integrated sample-to-digital-answer PCR systems that can be employed in routine clinical diagnosis.
Introduction
Digital droplet polymerase chain reaction (ddPCR) is a versatile tool that enables absolute quantification of nucleic acids without the need for any calibrants. It features single molecule sensitivity and is relatively tolerant towards inhibitors.1 To perform a ddPCR, the sample containing PCR mix and target DNA must first be emulsified. Other dPCR systems use physical compartments on stationary2 or centrifugal systems3 for digital PCR. In particular for centrifugal systems, it is difficult to integrate a sufficient number (>1000) of physical compartments, to ensure evenly sized, sufficiently small aliquots.In droplet digital PCR, typically more than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets in the nano- to picoliter range are produced, submerged in fluorinated oil.4 This emulsion is then thermocycled to perform a PCR in the droplets. Afterwards, the droplets are read out by fluorescence measurements. Commercialization of ddPCR5 drastically increased the use of the technique. However, the current methods require many handling steps and multiple pieces of specialized machinery and disposables (e.g. 4 machines and 2 disposables are needed when using one of the currently commercially available systems6). This in turn comes with relatively high costs and the need for specially trained personnel. To simplify this process, a higher integration of emulsification, thermocycling and readout is needed. Systems have been proposed to introduce the produced droplets into a shallow chamber instead of a tube, so that a monolayer of droplets forms.7,8 This monolayer is then thermocycled with subsequent readout in the same chamber. A major problem for microfluidic PCR in general9 and specifically when performed in shallow chambers is the development of bubbles, mainly caused by degassing of dissolved gas within the oil phase. This also leads to unfavourable evaporation of oil. Emerging bubbles may displace the emulsion from the reaction chamber, or exert shear forces on the droplets that can lead to merging of the same, posing problems during readout.8 It is therefore important to employ methods which either avoid bubble formation entirely or guide the bubbles away from the PCR and readout zone. Fully closed systems7 generate an internal pressure when bubbles are produced. This pressure prevents the generation of further bubbles. Such a fully closed system is more difficult to handle while still not eliminating bubble formation entirely. To counter this problem, pressurized systems with an active external pressure generation have been demonstrated to completely avoid bubble generation.9,10 However, these solutions need a bulky pressure source and interfaces that connect the chamber to a pressure source. Hatch et al.8 used mineral oil in PDMS/glass composite chips to avoid bubbles in most cases. Flushing the chip with oil was required as a pre-treatment to reduce bubbles. Still in about 5% of the reported experiments, bubble formation inhibited the experiment. In addition, the use of PDMS for microfluidic structures in general and specifically for droplet microfluidics has a number of disadvantages12 with the major one being the difficulty to scale up production.13 Compared to fluorinated oil, the use of mineral oils is inferior with regard to droplet stability. Thus, ddPCR systems should be compatible with fluorinated oils. However, the high gas solubility of fluorinated oils14 may lead to severe bubble problems. As a possible solution, degassing of the oil prior to use needs specialized equipment and requires the oil to be used directly afterwards since the uptake of gas from the surrounding air is relatively fast.
000 droplets in the nano- to picoliter range are produced, submerged in fluorinated oil.4 This emulsion is then thermocycled to perform a PCR in the droplets. Afterwards, the droplets are read out by fluorescence measurements. Commercialization of ddPCR5 drastically increased the use of the technique. However, the current methods require many handling steps and multiple pieces of specialized machinery and disposables (e.g. 4 machines and 2 disposables are needed when using one of the currently commercially available systems6). This in turn comes with relatively high costs and the need for specially trained personnel. To simplify this process, a higher integration of emulsification, thermocycling and readout is needed. Systems have been proposed to introduce the produced droplets into a shallow chamber instead of a tube, so that a monolayer of droplets forms.7,8 This monolayer is then thermocycled with subsequent readout in the same chamber. A major problem for microfluidic PCR in general9 and specifically when performed in shallow chambers is the development of bubbles, mainly caused by degassing of dissolved gas within the oil phase. This also leads to unfavourable evaporation of oil. Emerging bubbles may displace the emulsion from the reaction chamber, or exert shear forces on the droplets that can lead to merging of the same, posing problems during readout.8 It is therefore important to employ methods which either avoid bubble formation entirely or guide the bubbles away from the PCR and readout zone. Fully closed systems7 generate an internal pressure when bubbles are produced. This pressure prevents the generation of further bubbles. Such a fully closed system is more difficult to handle while still not eliminating bubble formation entirely. To counter this problem, pressurized systems with an active external pressure generation have been demonstrated to completely avoid bubble generation.9,10 However, these solutions need a bulky pressure source and interfaces that connect the chamber to a pressure source. Hatch et al.8 used mineral oil in PDMS/glass composite chips to avoid bubbles in most cases. Flushing the chip with oil was required as a pre-treatment to reduce bubbles. Still in about 5% of the reported experiments, bubble formation inhibited the experiment. In addition, the use of PDMS for microfluidic structures in general and specifically for droplet microfluidics has a number of disadvantages12 with the major one being the difficulty to scale up production.13 Compared to fluorinated oil, the use of mineral oils is inferior with regard to droplet stability. Thus, ddPCR systems should be compatible with fluorinated oils. However, the high gas solubility of fluorinated oils14 may lead to severe bubble problems. As a possible solution, degassing of the oil prior to use needs specialized equipment and requires the oil to be used directly afterwards since the uptake of gas from the surrounding air is relatively fast.
The above mentioned shortcomings highlight the need for an integrated and low cost ddPCR system that reliably removes bubbles from shallow chambers, is compatible with mass production and fluorinated oils and easy to handle for the end user. Here, we present such a cartridge (disk) that integrates a centrifugal microfluidic process chain for ddPCR analysis comprising the unit operations emulsification, capillary driven self-alignment and capillary driven bubble removal during PCR amplification and subsequent readout in one single chamber of a disposable disk. In this context, as introduced recently,15 the term process chain is used to provide standardized implementations of a complete laboratory workflow (here ddPCR) on a higher level of integration as compared to using unit operations. The integrated ddPCR system is applied for a proof-of-principle ddPCR for non-invasive prenatal testing (NIPT).
System description
The chip (a disk shaped disposable) is made from a thermoplast (COC) and is compatible to scalable manufacturing techniques such as injection moulding. The ddPCR process chain comprises centrifugal step emulsification11 to produce aqueous microdroplets in fluorinated oil. The emulsion is then positioned in the PCR chamber by capillary driven self-alignment. The tapered shape of the PCR chamber is also used to completely remove any bubbles formed during thermocycling, again by capillary driven bubble transport.16 Since droplets remain in a stable monolayer throughout the PCR, the subsequent fluorescence readout can be performed in the same chamber. User interaction is limited to 3 pipetting steps and one transfer of the disk. Automated emulsification and PCR are performed by the same centrifugal processing device. A post-PCR fluorescence scan is done in a commercial microarray scanner.Working principle
The centrifugal microfluidic disk comprises one minimalistic structure for emulsification, capillary driven self-alignment of the emulsions in the PCR and readout zone, and capillary driven bubble removal. The latter two are both new unit operations with respect to the implementation of ddPCR. A sketch with explanations is depicted in Fig. 1. | ||
| Fig. 1 Schematic outline of the all-in-one digital droplet PCR disk. A) Section of a circular disk with embedded microfluidic structures (top view), the centre of rotation is on the top. An inlet (red) is connected to the PCR chamber (green) by a channel (red). The channel leads to a centrifugal step emulsification nozzle (see inset C)). The emulsification chamber is connected to the inlet by an air vent. The PCR chamber features two zones, the PCR and readout zone (light green) and the surrounding bubble drains (dark green). Both, the inlet and the PCR chamber have inlets (black) to allow the loading of liquids to the disk. B) A cross section of the tapered PCR chamber. The largest part of the chamber is occupied by a pyramid with inclination angel α. The chamber is sealed with a sealing foil (blue) so that the chamber has a varying depth d. d0 should be larger than the droplet size, while the maximum depth should be less than twice the droplet size, so that a continuous monolayer forms. C) Centrifugal step emulsification nozzle (top view) as reported in our previous work.11 The feed channel (red) opens to a terrace (red) that has the same depth as the feed channel. The dark green area north of the red terrace is significantly deeper than the terrace. | ||
Centrifugal step emulsification
Droplets are generated by centrifugal step emulsification as shown recently.11 The process of droplet generation is illustrated in Fig. 2. In brief, an inlet is connected to a PCR chamber by a microfluidic channel. A backward facing step is located at the junction of the channel and the PCR chamber (i.e. the line between the red and green region in Fig. 1C) is the backward facing step). First, the disk is loaded with oil; afterwards PCR mix is introduced to the inlet and spun down the channel by centrifugal forces. Monodisperse droplets form at the backward facing step. The droplet production is exclusively determined by surface tension and the size of the channel. The droplets rise in the fluorinated oil towards the centre of rotation, since the droplets (composed of PCR mix, ρ = 1.0 g ml−1) are less dense than the surrounding fluorinated oil (ρ = 1.6 g ml−1). | ||
| Fig. 2 Principle of centrifugal step emulsification amended from.11 The top view shows the step emulsification nozzle. The inlet channel coming from below opens to both sides forming a terrace. After that a sudden change in depth (step, dashed line) occurs that leads to the PCR and readout chamber. The PCR mix gets pushed through the inlet channel by centrifugal pressure. When it reaches the step a droplet is formed by capillary forces. The droplet rises in the surrounding oil due to buoyancy. | ||
Capillary driven self-alignment
During centrifugation, the emulsion is located in the radially outer part of the chamber touching the radially outer side of the pyramid. Two counteracting forces act on the emulsion (see Fig. 3). First, the centrifugal force that pushes the emulsion radially outward. Second, the capillary force acting on the oil/gas interface located in the shallow area between the sealing foil and the side of the pyramid. The capillary force pulls the emulsion radially inward onto the shallow region.When spinning the disk at relatively high rotational frequencies, the centrifugal force dominates and a mensicus almost concentric to the center of rotation is formed. A small deviation from concentric shape is due to a slightly higher capillary force in the shallow region of the pyramide. Decreasing the speed of rotation leads to a decreased centrifugal force. The capillary force starts to take over and pulls the emulsion onto the shallow areas until it completely detaches from the radially outer part of the chamber and is located in the shallow area.
Capillary driven bubble removal
In order to remove any disturbing bubbles, capillary driven bubble transport is employed which has been used previously for removal of bubbles in methanol fuel cells.17,18 While this principle can be used to support the formation of droplets,19 here it is applied to the capillary driven bubble movement. The transport of bubbles away from the PCR zone is achieved by tilting the bottom of the chamber relative to the ceiling. The bottom of the PCR chamber thus forms a pyramid with its tip located in the centre (compare Fig. 1). The angle between the pyramid sides (i.e. the bottom of the chamber) and the ceiling (α) enables the transport of bubbles away from the centre of the pyramid to its sides as described in the following. Bubbles trapped between the bottom and ceiling of a shallow chamber are deformed as soon as the bubble diameter exceeds the chamber height (Fig. 4). Since bottom and ceiling are tilted with respect to each other at an angle α (compare Fig. 1B), different curvatures of the interfaces of the bubble form with the higher curvature located in the more shallow region, as schematically depicted in Fig. 4B). | ||
| Fig. 4 Schematic of capillary driven bubble removal. A) Top view of an air bubble (grey) amidst PCR droplets (blue) in oil (yellow). From top, the bubble has a circular shape. B) Cross section through the bubble. The bubble is trapped between ceiling and bottom of the chamber. The ceiling and bottom are tilted with respect to each other at an angle α. The pressure in the oil/air is depicted below. The pressure is constant in the oil, increases sharply at the interface and is static in the air volume again (otherwise there would be air flow to equate the pressure differences). The second pressure jump across the interface is smaller since the radius r1B is larger than r1A. C) Cross section through the chamber (compare Fig. 1B)). Sequence from 1 to 6: an air bubble is forming and transported away to the bubble drain on the right. The emulsion flows around the bubble. | ||
According to the Young–Laplace equation, the pressure at the interface in non-spherical bubbles is defined as:
where pcap is the capillary pressure at the interface of the bubble in the shallow region (A) and the deeper region (B), respectively. r1A and r1B are the mean curvature radii in the cross-section plane of the bubble and dA and dB are the chamber depth at the two interface positions A and B. Θad and Θrec are the advancing and receding contact angles of the oil at the interface to gas, respectively.
The pressure difference across the bubble causes a pressure increase in the continuous liquid phase (oil) from shallow to the deep (Fig. 4B, bottom). Consequently, an oil flow sets on from deep to shallow regions around the bubble. This pushes the bubble in the opposite direction of the oil flow towards the opening of the tilted bottom and ceiling, until equilibrium is reached. This equilibrium is achieved when the bubble is of perfectly spherical shape. In a perfect spherical shape, all three radii r1A, r1B, and r2 are the same and no pressure differences remain.
Experimental
Production of disks
The microfluidic disks were fabricated by Hahn-Schickard Lab-on-a-Chip Design & Foundry Service.20 First, a master mold was produced by micro milling in polymethyl methacrylate (PMMA) (Maertin, Freiburg, Germany) using a Kern Evo (Kern Micro- und Feinwerktechnik GmbH & Co. KG, Eschenlohe, Germany). Quality control was performed by using a Zeiss AX10 microscope equipped with an AxioCam ICc1 and an Imager M2m (Zeiss, Jena, Germany). Afterwards, a PDMS master was produced by pouring a mixture of Elastosil 607 and Elastosil 675 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, with monomer/crosslinker ratios of 9
1, with monomer/crosslinker ratios of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 respectively) into the milled substrate and applying centrifugal force (Hettich Zentrifuge Rotanta 460 R). A hot stamp (Wickert WMP-30) was used to produce replicates of the master mold in COC (Topas 5013). Inlet holes were opened with a drill and the thickness reduced to 1 mm with the Kern Evo. Pressure sensitive adhesive (PSA) foil (#900 320, HJ Bioanalytik, Erkelenz, Germany) was used to seal the structures with aid of a PCS 30 lamination tool (Jakob Weiß & Söhne, Sinsheim, Germany). The channel was 90 μm wide and 60 μm deep, the terrace was 100 μm wide. The slope angle α was varied from 1–6°, the width and length of the pyramidal structures was varied between 12.6 mm and 4.2 mm. The tip of the pyramid was located 180 μm beneath the sealing foil.
1 respectively) into the milled substrate and applying centrifugal force (Hettich Zentrifuge Rotanta 460 R). A hot stamp (Wickert WMP-30) was used to produce replicates of the master mold in COC (Topas 5013). Inlet holes were opened with a drill and the thickness reduced to 1 mm with the Kern Evo. Pressure sensitive adhesive (PSA) foil (#900 320, HJ Bioanalytik, Erkelenz, Germany) was used to seal the structures with aid of a PCS 30 lamination tool (Jakob Weiß & Söhne, Sinsheim, Germany). The channel was 90 μm wide and 60 μm deep, the terrace was 100 μm wide. The slope angle α was varied from 1–6°, the width and length of the pyramidal structures was varied between 12.6 mm and 4.2 mm. The tip of the pyramid was located 180 μm beneath the sealing foil.
PCR
The disk was processed in a LabDisk player prototype (Qiagen Lake Constance, Stockach, Germany) that allows for rotation of the disks while heating and cooling with ambient air. For PCR, the inlet was filled with 15 μl of Novec7500 with 5 wt% of Picosurf-1 (Dolomite, Royston, UK). The fluorinated oil was transferred to the PCR chamber by centrifugal force (15 Hz for 1 min). Reaction mix was prepared using Qiagen HotStar TaqPlus polymerase with 3 mM Mg2+ final concentration in the reaction mix and dNTPs used according to manufacturer's protocol (Qiagen, Hilde, Germany). Primer (300 nM final concentration), probes (200 nM final concentration) and target DNA (for sequences see below) were added to the mix and 20 μl were then introduced in the inlet chamber. Emulsification took place by centrifugation (10 Hz for 8 min). Then, 20 μl of FC-40 with 5 wt% of Picosurf-1 (Dolomite, Royston, UK) was introduced in the PCR chamber directly. FC-40 was introduced since the higher boiling point reduces the evaporation of oil during PCR. However, the disk needs to be filled with Novec7500 initially since droplet production is more reliable in this oil. The upper inlet holes were sealed with an air permeable filter (PTFE Pet Backed 0.2 μm, Millipore, Tullagreen, Ireland), while the lower was sealed with PSA foil. The thermal protocol for the PCR was 5 min at 95 °C, followed by 45 cycles of 15 s at 95 °C and 60 s at 58 °C. The disk was rotated at 0.5 Hz. Sequences of primers, probes and DNA all in 5′→3′ direction (Life Technologies, Darmstadt, Germany):Forward primer: GATTATGCCTGGCACCATTAAAG
Backward primer: GCATAATCAAAAAGTTTTCAC
Probe: FAM- CACCAATGATATTTT- MGB
Amplicon sequence: TGCCTGGCACCATTAAAGAAAATATCATTGGTGTTTCCTATGATGAATATAGATACAGAAGCGTCATCAAAGCATGCCAACTAGAAGAGGTAAGAAACTATGTGAAAACTTTTTGATTATGC (CFTR gene, OMIM 602421)
A dilution series of target DNA was prepared using LoBind tubes (Eppendorf, Hamburg, Germany) and dilution buffer containing 0.2× TE buffer and 10 ng μl−1 herring sperm DNA. DNAse/RNAse free water (Thermo Fisher Scientific, Waltham, US) was used for the preparation of the buffers.
Imaging
For imaging of the bubble generation during PCR conditions, a modified LabDisk player with a stroboscopic setup21 was used. Fluorescence imaging was performed using a LaVision Bioanalyzer (4/4S, LaVision BioTec GmbH, Bielefeld, Germany) with a custom made holder at 475 nm emission and 530 nm emission wavelength and 800 ms integration time. ImageJ22 was used for evaluation of the images. Circular regions of interests were placed over the droplets manually and the grey value was measured. A threshold was defined manually. More than 500 droplets were evaluated per sample. To increase the sensitivity, more droplets would need to be counted which could be accomplished in future using an automated image recognition routine.Results and discussion
Centrifugal step emulsification
The droplets have a diameter of 147 μm with a CV of 1.4% (for details see Fig. S1†). A sample of 20 μl DNA-solution yields approximately 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets of 1.8 nl, each. A dead volume of <1% of the sample remains trapped in the channel. If needed, it can be removed by flushing the channel with oil to reduce the dead volume to zero.
000 droplets of 1.8 nl, each. A dead volume of <1% of the sample remains trapped in the channel. If needed, it can be removed by flushing the channel with oil to reduce the dead volume to zero.
Capillary driven self-alignment
The process of capillary driven self-alignment of the emulsion in the PCR and readout zone is shown in Fig. 5 and in Movie S1.† The self-alignment process of the emulsion transports >99% of the emulsion to the PCR and readout zone with only very few droplets remaining in the edges of the chamber. Moreover, no merging or coalescence of droplets due to shear forces could be observed. The total time needed for the alignment of the emulsion can be as low as 8 s. Since the disk surface is not perfectly smooth but features some grooves from micro milling, some pinning of the emulsion can be observed. It can be seen in Movie S1† that the pinning leads to a somewhat uneven propagation of the interface onto the shallow region. An inhibition of positioning by pinning however, has never been observed in more than 50 experiments. The emulsion is now perfectly aligned in the PCR zone for cycling and readout.Capillary driven bubble removal
The gas solubility of liquids decreases with increasing temperature. Since the gas solubilities of FC-40 and HFE7500 are very high (0.27 and 0.48 ml air/ml oil, respectively),14 the absolute amount of gas set free during heating from 25 °C to 95 °C is high as well. Some of this gas volume can be released by diffusion at the oil/air interface, while a large part will be released in form of bubbles which need to be transported away. The amount of gas set free in form of bubbles was estimated to be no lower than 18% of the emulsion volume.With respect to the geometry of the pyramid for capillary driven bubble transport, different configurations were studied with inclination angles α of 1°, 2°, 4°, and 6°. All of these configurations removed all bubbles without any problems and allowed for precise positioning of the emulsion. This is likely due to the fact that the air bubbles in the emulsion have no direct contact to either bottom or ceiling. The fact that bubble transport is independent of inclination angle α supports the assumption that the advancing and receding contact angles Θad and Θrec at the left and right border of the moving bubble, respectively (compare Fig. 4B)) are zero or very close to it. This is due to the fact that a thin film of (highly wetting) fluorinated oil remains between the air and the polymer. The air bubble therefore never touches the polymer wall. This is also in line with the theory.19,23
The bubble transport works for all bubbles that are larger than the height of the chamber (see Movie S2†). This is important since the size of the bubbles varies. Very small bubbles that do not touch bottom and ceiling dissolve completely in the oil when the temperature is reduced (e.g. during the annealing step of a PCR) and the gas uptake capacity of the oil increases. It can be concluded that no bubble remains in the emulsion after PCR since all bubbles are either transported away by the capillary action or small enough to dissolve in the oil. In all cases, no matter if small or large bubbles (largest observed bubbles were up to 10% of the total surface of PCR zone) are transported, the emulsion was not affected. This means that no merging of droplets with each other or unwanted transport of droplets to the air vents was observed. The bubbles were transported with an average speed of at least 8 mm s−1 (for α = 2°). Bubble speed was measured by comparing the location of bubbles in pictures taken at defined time intervals with a stroboscopic setup in the LabDisk Player under rotation. Since the spinning frequency was low (0.5 Hz), time intervals were large (2 s). Therefore, the number can only give a lower limit.
The size and number of pyramids situated in one chamber was varied as well, with up to 3 pyramids in one chamber and pyramid sizes of 4.2 mm to 12.6 mm in length (for detailed measurements compare Table S1†). All of them worked without any problems. Smaller pyramids led to a decrease in overall bubble production. This can be attributed to the higher surface to volume ratio with a much larger oil/gas interface and a smaller distance from the centre of the pyramid to the next interface. As a consequence, for smaller pyramids, diffusion of dissolved gas to the oil/air interface is enhanced. Therefore, the contribution of diffusion as a mean of dissolved gas removal increases compared to convective transport in the form of bubbles.
It should be noted that some evaporation of oil and droplets takes place during PCR. This leads to an increasing concentration of salt in the shrinking droplets. The increase in salt concentration might lead to changes or even breaks in the structure of the probe. A break of the probe leads to an increase of fluorescence which lead to edge effects at the emulsion/air interface. In these regions, droplets cannot be evaluated (<10% of all droplets). The edge effects are illustrated in Fig. 5C). In order to keep these regions small, the emulsion/air interface should be reduced as far as possible. Consequently, the larger pyramids are beneficial. However, care must be taken that distance between sealing foil and bottom of the chamber does not allow the droplets to form a double layer of droplets. The larger the pyramid is, the deeper the chamber gets (at a given angle α). As a compromise, a structure with 2 parallel pyramids was chosen (Fig. 5C) and structure 3 in Table S1†) to accommodate a high number of droplets, while keeping edge effects at a comparatively low level.
Digital droplet PCR
To show the feasibility of the system, a digital droplet PCR targeting the mutation p.Phe508del was performed in the disk. p.Phe508del is a mutation present in more than 70% of patients suffering from cystic fibrosis.24 A serial dilution of four 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10-dilutions of target DNA was tested in four different chambers on one disk. The number of positive reactions (i.e. of positive droplets) was counted and the number of target molecules initially present in the sample calculated by using Poisson statistics.25 The results are shown in Fig. 6. The number of expected copies was estimated from qPCR measurements of highest concentration before dilution. As can be seen from the graph, the decrease in measured copy numbers is linear but has an offset from the expected values. The offset might be due to the fact that estimation of absolute copy numbers by qPCR is not very precise. Another reason for the offset could be that the sample was stored and shipped in between the qPCR and ddPCR measurements. It is likely that some DNA degraded yielding lower measurements in the ddPCR. However, in order study the possibility for absolute quantification of copy numbers by the ddPCR system, another ddPCR has been performed using pTYB1 vector DNA as target. The results are provided in the ESI† (see Fig. S2). Here, a good correlation of measured and expected copy numbers proves the capability of the system to absolutely quantify DNA. The limit of detection of the ddPCR system is in the order of 5 cp. μl−1. At this concentration, DNA molecules can still be detected, albeit with a relatively large error (compare Fig. 6A) and Fig. S2†).
10-dilutions of target DNA was tested in four different chambers on one disk. The number of positive reactions (i.e. of positive droplets) was counted and the number of target molecules initially present in the sample calculated by using Poisson statistics.25 The results are shown in Fig. 6. The number of expected copies was estimated from qPCR measurements of highest concentration before dilution. As can be seen from the graph, the decrease in measured copy numbers is linear but has an offset from the expected values. The offset might be due to the fact that estimation of absolute copy numbers by qPCR is not very precise. Another reason for the offset could be that the sample was stored and shipped in between the qPCR and ddPCR measurements. It is likely that some DNA degraded yielding lower measurements in the ddPCR. However, in order study the possibility for absolute quantification of copy numbers by the ddPCR system, another ddPCR has been performed using pTYB1 vector DNA as target. The results are provided in the ESI† (see Fig. S2). Here, a good correlation of measured and expected copy numbers proves the capability of the system to absolutely quantify DNA. The limit of detection of the ddPCR system is in the order of 5 cp. μl−1. At this concentration, DNA molecules can still be detected, albeit with a relatively large error (compare Fig. 6A) and Fig. S2†).
 | ||
| Fig. 6 A) Measured copy numbers corrected by Poisson statistics are depicted on the y-axis. The expected copy numbers were estimated from qPCR measurements (see ESI†) and plotted on the x-axis. The solid red line is a linear fit (R2 > 99.7%) while the dashed blue line represents the ideal curve. The deviation between them is most likely due to an uncertainty when estimating copy numbers with qPCR and due to some DNA degradation during transport of the studied sample. The errors are 1σ-errors including sample and Poisson errors. B) False colour fluorescent images of droplets after ddPCR. The subcaptions of the images give the expected copy numbers as derived from the qPCR measurement. For an expected copy number of 100.7, an area was chosen to show at least 3 positive droplets. The real droplet count for a larger area was relatively lower. | ||
Conclusion
We presented an all-in-one digital droplet PCR disk with an integrated centrifugal microfluidic process chain comprising emulsification, capillary driven self-alignment of the emulsion within the readout zone, and capillary driven bubble transport for bubble removal in shallow chambers during ddPCR. Capillary driven bubble transport proved to be effective and efficient by quickly removing 100% of the bubbles and throughout PCR, no merging of the droplets due to shear forces induced by the bubbles was observed.The ddPCR system requires minimal user interaction with only 3 pipetting steps. Fabrication of the disks is compatible to mass fabrication by injection moulding.
As compared to previous digital isothermal reactions using centrifugal systems,11 centrifugal step emulsification combined with capillary driven bubble removal is now able to perform ddPCR experiments. A disposable disk was developed and used to perform a ddPCR targeting p.Phe508del mutation. Quantifying the absolute number of DNA molecules containing p.Phe508del mutation is important for non-invasive prenatal testing (NIPT).
Compared to existing ddPCR solutions e.g. the Bio-Rad machine, our system offers easier handling and higher integration. However, the sensitivity is lower at the current stage, since fewer droplets are produced and evaluated. To increase sensitivity, an automated image recognition routine would help in evaluating droplets quickly. More droplets can be generated if the droplet size is decreased, this can be done by changing the geometry of the channel as explained earlier.11
In future, we envision that the development of the single chamber droplet digital PCR process chain will facilitate the development of fully integrated and highly automated sample-to-digital-answer systems. Reagents could be pre-stored lyophilized or in liquid form in stick packs26 on the disposable disk. For an application in a clinical setting, the DNA would need to be extracted from the sample matrix which requires additional steps, before the ddPCR. For that purpose, upstream sample preparation could be integrated using established centrifugal microfluidic unit operations.15,27
Acknowledgements
We gratefully acknowledge financial support from EU Framework 7 project “ANGELab” #317635. We want to thank Michael Lehnert for helpful discussions. We furthermore want to thank the Lab-on-a-Chip Design & Foundry Service of Hahn-Schickard for manufacturing of disks and the custom made disk holder for the LaVision Bioanalyzer, as well as Oliver Barth and Benjamin Rutschinski for helpful discussions.Notes and references
- (a) G. Nixon, J. A. Garson, P. Grant, E. Nastouli, C. A. Foy and J. F. Huggett, Anal. Chem., 2014, 86, 4387–4394 CrossRef CAS PubMed; (b) M. T. Guo, A. Rotem, J. A. Heyman and D. A. Weitz, Lab Chip, 2012, 12, 2146–2155 RSC.
- F. Shen, W. Du, J. E. Kreutz, A. Fok and R. F. Ismagilov, Lab Chip, 2010, 10, 2666–2672 RSC.
- S. O. Sundberg, C. T. Wittwer, C. Gao and B. K. Gale, Anal. Chem., 2010, 82, 1546–1550 CrossRef CAS PubMed.
- United States of America Pat., US8535889B2, 2011 Search PubMed.
- J. M. Perkel, Life Science Technologies, 2014, pp. 212–214, http://www.sciencemag.org/site/products/lst_20140411.pdf Search PubMed.
- Bio-Rad, QX200™ Droplet Digital™ PCR System, Bio-Rad, Bulletin 6311 Rev C.
- K. Kistrup, C. E. Poulsen, M. F. Hansen and A. Wolff, Lab Chip, 2015, 15, 1998–2001 RSC.
- A. C. Hatch, J. S. Fisher, A. R. Tovar, A. T. Hsieh, R. Lin, S. L. Pentoney, D. L. Yang and A. P. Lee, Lab Chip, 2011, 11, 3838–3845 RSC.
- J. M. Karlsson, T. Haraldsson, S. Laakso, A. Virtanen, M. Maki, G. Ronan and W. van der Wijngaart, in TRANSDUCERS 2011 - 2011 16th International Solid-State Sensors, Actuators and Microsystems Conference, pp. 2215–2218 Search PubMed.
- L. Clime, D. Brassard, M. Geissler and T. Veres, Lab Chip, 2015, 15, 2400–2411 RSC.
- F. Schuler, F. Schwemmer, M. Trotter, S. Wadle, R. Zengerle, F. von Stetten and N. Paust, Lab Chip, 2015, 15, 2759–2766 RSC.
- (a) R. Mukhopadhyay, Anal. Chem., 2007, 79, 3248–3253 CrossRef CAS PubMed; (b) E. K. Sackmann, A. L. Fulton and D. J. Beebe, Nature, 2014, 507, 181–189 CrossRef CAS PubMed; (c) R. Seemann, M. Brinkmann, T. Pfohl and S. Herminghaus, Reports on progress in physics, Physical Society, Great Britain, 2012, vol. 75, p. 016601 Search PubMed; (d) N. Wu, Y. Zhu, S. Brown, J. Oakeshott, T. S. Peat, R. Surjadi, C. Easton, P. W. Leech and B. A. Sexton, Lab Chip, 2009, 9, 3391–3398 RSC.
- H. Becker, Lab Chip, 2009, 9, 2759–2762 RSC.
- Dolomite, Fluoridrop 40, available at: http://www.dolomite-microfluidics.com/images/stories/PDFs/datasheets/fluoridrop_40_datasheet.pdf, accessed 19 June 2015.
- O. Strohmeier, M. Keller, F. Schwemmer, S. Zehnle, D. Mark, F. von Stetten, R. Zengerle and N. Paust, Chem. Soc. Rev., 2015, 44, 6187–6229 RSC.
- T. Metz, N. Paust, R. Zengerle and P. Koltay, Microfluid. Nanofluid., 2010, 9, 341–355 CrossRef CAS.
- N. Paust, C. Litterst, T. Metz, M. Eck, C. Ziegler, R. Zengerle and P. Koltay, Microfluid. Nanofluid., 2009, 7, 531–543 CrossRef CAS.
- C. Litterst, S. Eccarius, C. Hebling, R. Zengerle and P. Koltay, J. Micromech. Microeng., 2006, 16, S248 CrossRef CAS.
- R. Dangla, S. C. Kayi and C. N. Baroud, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 853–858 CrossRef CAS PubMed.
- D. Mark, Lab-on-a-Chip Design + Foundry Service, available at: http://www.hahn-schickard.de/fertigung/lab-on-a-chip-design-foundry-service/, accessed 1 July 2015 Search PubMed.
- M. Grumann, T. Brenner, C. Beer, R. Zengerle and J. Ducrée, Rev. Sci. Instrum., 2005, 76, 025101 CrossRef.
- M. D. Abramoff, P. J. Magalhães and S. J. Ram, Biophotonics International, 2004, 11, 36–42 Search PubMed.
- T. Horozov, Curr. Opin. Colloid Interface Sci., 2008, 13, 134–140 CrossRef CAS.
- F. van Goor, S. Hadida, P. D. J. Grootenhuis, B. Burton, J. H. Stack, K. S. Straley, C. J. Decker, M. Miller, J. McCartney, E. R. Olson, J. J. Wine, R. A. Frizzell, M. Ashlock and P. A. Negulescu, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 18843–18848 CrossRef CAS PubMed.
- Quantitative Real-Time PCR: Methods and Protocols. Chapter 4 - Introduction to Digital PCR, ed. R. Biassoni and A. Raso, Springer, New York, 2014 Search PubMed.
- T. van Oordt, Y. Barb, J. Smetana, R. Zengerle and F. von Stetten, Lab Chip, 2013, 13, 2888–2892 RSC.
- (a) S. Hugo, K. Land, M. Madou and H. Kido, S. Afr. J. Sci., 2014, 110, 1–7 CrossRef; (b) B. S. Lee, Y. U. Lee, H.-S. Kim, T.-H. Kim, J. Park, J.-G. Lee, J. Kim, H. Kim, W. G. Lee and Y.-K. Cho, Lab Chip, 2011, 11, 70–78 RSC; (c) T.-H. Kim, J. Park, C.-J. Kim and Y.-K. Cho, Anal. Chem., 2014, 86, 3841–3848 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5lc01068c |
| This journal is © The Royal Society of Chemistry 2016 |