Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Pro198Leu polymorphism affects the selenium status and GPx activity in response to Brazil nut intake
Bárbara R.
Cardoso
*ab,
Alexandre L.
Busse
c,
Dominic J.
Hare
 bd,
Cristiane
Cominetti
e,
Maria A.
Horst
a,
Gawain
McColl
b,
Regina M.
Magaldi
c,
Wilson
Jacob-Filho
c and
Silvia M. F.
Cozzolino
a
bd,
Cristiane
Cominetti
e,
Maria A.
Horst
a,
Gawain
McColl
b,
Regina M.
Magaldi
c,
Wilson
Jacob-Filho
c and
Silvia M. F.
Cozzolino
a
aFaculty of Pharmaceutical Sciences, Department of Food and Experimental Nutrition, University of São Paulo, São Paulo, Brazil
bThe Florey Institute of Neuroscience and Mental Health, The University of Melbourne, 30 Royal Parade, Parkville, Victoria 3052, Australia. E-mail: barbara.rita@florey.edu.au; baritacardoso@gmail.com; Tel: +61 0450 633537
cGeriatrics Division, Department of Internal Medicine, University of São Paulo Medical School, São Paulo, Brazil
dElemental Bio-imaging Facility, University of Technology Sydney, Broadway, New South Wales, Australia
eFaculty of Nutrition, Federal University of Goiás, Goiânia, Goiás, Brazil
First published on 2nd December 2015
Abstract
Selenoproteins play important roles in antioxidant mechanisms, and are thus hypothesised to have some involvement in the pathology of certain types of dementia. Mild cognitive impairment (MCI) and Alzheimer's disease (AD) are both thought to involve impaired biological activity of certain selenoproteins. Previously, supplementation with a selenium-rich Brazil nut (Bertholletia excelsa) has shown potential in reducing cognitive decline in MCI patients, and could prove to be a safe and effective nutritional approach early in the disease process to slow decline. Here, we have conducted a pilot study that examined the effects of a range of single nucleotide polymorphisms (SNPs) in genes encoding the selenoproteins glutathione peroxidase (GPX1) and selenoprotein P (SEPP) in response to selenium supplementation via dietary Brazil nuts, including selenium status, oxidative stress parameters and GPX1 and SEPP gene expression. Our data suggest that GPX1 Pro198Leu rs1050450 genotypes may differentially affect the selenium status and GPx activity. Moreover, rs7579 and rs3877899 SNPs in SEPP gene, as well as GPX1 rs1050450 genotypes can influence the expression of GPX1 and SEPP mRNA in response to Brazil nuts intake. This small study gives cause for larger investigations into the role of these SNPs in both the selenium status and response to selenium dietary intake, especially in chronic degenerative conditions like MCI and AD.
Introduction
Despite significant research, strategies to slow the progression of cognitive decline and the onset of dementia are lacking. It is estimated that the number of people living with dementia, such as Alzheimer's disease which accounts as the main type, will reach 65.7 million in 2030 and nearly double in 2050.1 Addressing dementia prior to the onset of clinical symptoms is as important as the development of therapeutics for post-diagnostic use. Several risk factors linked to lifestyle have been identified, which include dietary-related conditions such as midlife obesity, diabetes and hypertension, and if they were to be reduced by 10–25%, 1.1–3.0 million new AD cases worldwide could be prevented.2 Accordingly, more attention is being paid to the role that nutrition plays in the progression of AD and dementia in general.3,4 However, as is the case in developing new therapies, diagnosing dementia in its early stages remains a significant obstacle in identifying beneficial nutritional habits that may reduce the aforementioned risks.Although some people do not exhibit the explicit clinical symptoms of dementia, they may develop cognitive impairment beyond what would be expected for normal ageing. This condition, mild cognitive impairment (MCI), is associated with an increased risk of dementia and particularly AD, as brains of MCI patients exhibit many pathological features mirroring those of AD subjects.5 There is a high degree of heritability for AD with estimates ranging from 58 to 79%,6 with a number of genetic risk factors shared with MCI.7 Single nucleotide polymorphisms (SNPs) in amyloid precursor protein (APP) and presenilin-1 (PSEN1) and 2 (PSEN2) genes have been implicated in the predisposition of early-onset familial AD;8–11 and SNPs in the genes encoding apolipoprotein E (APOE) and α2-macroglobulin (A2M) are associated with late-onset AD.12,13 Genome wide association studies (GWAS) have identified several additional genes that appear to predict AD susceptibility,14 and the biological pathways in which they participate support several hypotheses regarding upstream AD pathology, including chronic inflammation, lipid transport and β-amyloid processing. Considering that oxidative stress has a central role in neurodegeneration, cognitive decline and AD in particular, it is important to understand the role of SNPs involved with antioxidant enzymes.
Selenium (Se) is an essential element in the central nervous system and deficiency has been associated with cognitive decline.15–17 Selenium deficiency may be compounded by SNPs in genes encoding selenoproteins and negatively influence the normally protective roles against oxidative stress played by many of the 25 identified Se-containing proteins.18–20 Jablonska et al.21 and Cominetti et al.22 suggested that the SNP Pro198Leu (reference SNP identification number rs1050450) in the glutathione peroxidase 1 (GPX1) gene might affect the antioxidant activity. Additionally, a differential correlation between erythrocyte GPx activity and erythrocyte Se levels according to Pro198Leu genotype has been observed,23 and Paz-Y-Miño et al.24 reported an association between the variant allele and increased risk for AD. The gene encoding selenoprotein P (SEPP) has several known SNPs, of which rs7579 (G/A substitution in the 3′-untranslated region) and rs3877899 were reported to have functional consequences on both the protein level and function.25–28
Brazil nuts (Bertholletia excelsa) are the richest food source of Se, and inclusion of this nut in the diet may be a viable strategy to increase dietary intake of Se. The high Se content in Brazil nuts is mainly as selenomethionine, the most bioavailable form of dietary Se.29 Sufficient Se intake and associated Se levels may be related to a decreased risk for AD.30–32 However, the aforementioned SNPs in genes encoding selenoproteins may influence the response to a dietary intervention,33 and thus, it is critical to understand the interaction between Se intake and genetic variation in selenoprotein homeostasis. The aims of this study were to evaluate the implications of GPX1 rs1050450 and SEPP rs7579 and rs3877899 SNPs on Se status and oxidative stress biomarkers in MCI patients, and to determine if these SNPs affect the response to supplemental Se intake from Brazil nuts.
Experimental
Study design
All procedures followed in this study have been performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments. They were reviewed and approved by the Ethics Committee of Faculty of Pharmaceutical Sciences at the University of São Paulo – Brazil (protocol number: 568). Informed consent was obtained from all individual participants and included in the study. The trial was registered at ClinicalTrials.gov under identification number NCT02121457.The study was conducted on patients diagnosed with MCI according to the criteria proposed by the International Working Group on Mild Cognitive Impairment.34 They attended the Memory and Aging Unit of the Geriatrics Division at the University of São Paulo Medical School (Brazil) during the period May 2011 to August 2012. Among the patients, 31 fulfilled the following inclusion criteria: age 60+ years; fluent in Portuguese language; absence of any other significant neurologic or psychiatric diseases; no regular intake of Brazil nuts or Se-containing supplements; and no intolerance to oleaginous food.
To investigate if the SNPs influenced the response to daily Brazil nut intake, we examined a randomised subgroup with 11 participants who consumed one Brazil nut daily for 6 months, as described elsewhere.32 The nuts were offered every 2 months in a bag containing 60 nuts. Compliance was monitored by counting the number of nuts returned at the conclusion of the study. A subject was considered compliant if ≥85% of the Brazil nuts were consumed. All subjects were instructed to maintain their normal diet and to avoid additional Brazil nuts during the study. The nutritional information of Brazil nuts used is given in Table 1, considering that the mean weight of one nut was 5 g, and each nut provided about 288.8 μg of Se.32
| Carbohydrate (mg g−1) | 108.9 |
| Protein (mg g−1) | 162.7 |
| Lipids (mg g−1) | 673.5 |
| Se (μg g−1) | 57.75 |
| Mean Se per nut (μg) | 288.8 |
Biochemical assays
Erythrocyte GPx activity was measured according to the method described by Paglia and Valentine35 with the Ransel 505 kit (RANDOX Laboratories, Crumlin, UK) using a biochemical analyser (Liasys® MAS, Rome, Italy). Malondialdehyde (MDA) was measured in plasma by high performance liquid chromatography (HPLC) on a Shimadzu (Kyoto, Japan) instrument with a Phenomenex (Torrance, CA, USA) reverse-phase C18 column according to the method described by Hong et al.36 The instrument was calibrated with a MDA standard stock solution in the following concentrations: 0.0, 0.25, 0.5, 1.0, 2.0, 4.0, 6.0 and 12.0 μM. Oxygen radical absorbance capacity (ORAC) was measured following the method described by Prior et al.37 using a Synergy H1 multi-mode microplate reader (Biotek Instruments, Winooski, VT, USA). The instrument was calibrated with Trolox prepared with phosphate buffer in the following concentrations: 100 μM, 50 μM, 25 μM, 12.5 μM, 6.25 μM. Samples were analysed in triplicate.Selenium levels in plasma and erythrocytes were analysed using hydride generation atomic absorption spectroscopy (AAS).38 Samples were prepared in duplicate and measurements were made in triplicate, equaling six readings per person. Method validity was maintained by a measured Se recovery of at least 85% of the certified content of a lyophilised human reference control (Seronorm Trace Elements Serum and Whole Blood; Sero AS, Billingstad, Norway).
Genotyping of the rs1050450, rs7579 and rs3877899 SNPs
Isolation of DNA from whole blood was carried out using a PureLink Genomic DNA kit (Invitrogen, Life Technologies Inc., Carlsbad, CA, USA), and the concentration was measured using a NanoDrop ND 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).Polymorphisms of GPX1 (rs1050450) and SEPP (rs7579 and rs3877899) genes were determined by real-time polymerase chain reaction (RT-PCR) with TaqMan SNP Genotyping assays (Life Technologies, Foster City, CA, USA). The assays were obtained as pre-designed from Applied Biosystems for rs7579 and rs3877899 (ID Assays C___8806056_10 and C___2841533_10, respectively) and custom-made through Custom TaqMan® Genomic Assays service for rs1050450. The RT-PCR reaction contained 12.5 μL of 1× TaqMan Genotyping Master Mix (Life Technologies), 1.25 μL of the 20× SNP
Genotyping Assay (Life Technologies) and 20 ng of genomic DNA in a 20 μL total reaction volume.
Samples were assayed along with no-template and HapMap controls and run on a StepOne Real-Time PCR System under the following conditions: an initial enzyme activation step at 94 °C for 10 min followed by 40 cycles at 92 °C for 15 s and 60 °C for 1 min for annealing and extension.
GPX1 and SEPP gene expression
GPX1 and SEPP gene expression were assessed in the subgroup composed of 11 participants who consumed Brazil nuts in order to evaluate their effect. mRNA was extracted from whole blood by using the RiboPure™ – Blood Kit (Life Technologies) and treated with DNase I to avoid DNA contamination. RNA was stored under −80 °C for further analysis. The final concentration was measured using a NanoDrop ND 1000 spectrophotometer (Thermo Scientific), and RNA integrity was considered acceptable when the absorbance ratios in 260 and 280 nm wavelengths were between 1.8 and 2. SuperScript® III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA, Cat #18080093) was used for reverse transcription, and the cDNA was adjusted to a concentration of 20 ng μL−1 in a final volume of 20 μL.Analysis of GPX1 and SEPP mRNA expression was carried out for each sample by real-time quantitative PCR (qPCR) assay on a StepOne Real-Time PCR System using specific hydrolysis probes (TaqMan® Gene Expression Assays – ID Assays Hs00829989_gH and Hs01032845_m1, respectively for GPX1 and SEPP) (Life Technologies). The qPCR were performed in duplicate and the samples contained 4 μL of cDNA, 10 μL of TaqMan® Universal Master Mix II (Life Technologies), 1 μL of the 20× TaqMan® Gene Expression Assay and 5 μL of RNase-free water in a 20 μL final volume. β-Actin was used as a reference gene as its expression was very stable among participants. A non-template (without cDNA) reaction was included with each qPCR run to serve as a negative control. The cycling qPCR for each sample started with a denaturation step at 50 °C for 2 min, followed by 45 cycles at 95 °C for 15 s, and 1 min at 60 °C for hybridisation and extension.
To evaluate the differential expression of GPX1 and SEPP among different genotypes for rs1050450, rs7579 and rs3877899, the ΔCq was normalised to the internal control gene β-actin, and so the results are expressed by [Cq target gene − Cq reference gene].
Assessment of selenium intake
Selenium intake was evaluated using a 3-day (2 weekdays and 1 weekend day) non-consecutive dietary food record. The selenium content in the diet was based on the levels determined by Ferreira et al.39 from the analysis of Se concentrations in Brazilian foods. These records were analysed using the NutWin Software (Escola Paulista de Medicina/UNIFESP/Brazil).Statistical analysis
Individuals who scored homozygous and heterozygous for allele variants were pooled and compared against wild types for all statistical analyses. Descriptive statistical analysis was performed, and all continuous variables are expressed as mean ± standard deviation (SD). Categorical variables were expressed using both a number and percentage. Variable distribution was evaluated using the Shapiro–Wilk W test. Allele frequencies were estimated using the gene-counting method.Differences between genotypes were assessed using Student's t-test for independent samples. Hierarchical multiple linear regression analysis was used to examine the association between Se status and genotypes for rs1050450, rs7579 and rs3877899 while controlling the other potentially confounding variables. Consequently, we designed models in which the genotype was used as the primary independent variable and plasma and erythrocyte Se levels were alternately used as the dependent variable. Covariates were entered in three sequential steps to examine their incremental validity: in the first step, the genotype was entered without covariates; in the second step, age and sex were entered; in the third step, Se intake was assessed. To evaluate the association between the genotype and oxidative stress parameters for the SNPs, ORAC, MDA and GPx activity were used alternately as dependent variables in discrete regression models. In these models, genotypes were also used as the primary independent variables, and covariates were entered as follows: (i) genotype without covariates; (ii) erythrocyte Se; and (iii) socio-demographic variables (sex and age) entered as a block. Pearson's correlation coefficient was used to estimate the correlation between GPx activity and the Se level in erythrocytes.
To describe the relationship between aspects of food consumption and biochemical characteristics independent of energy intake, Se intake was adjusted by energy intake according to Willet40 using linear regression (linear regression of nutrient intake on total caloric intake) and the addition of a constant (mean energy intake of the group).
The post-supplementation data were compared with the baseline data using a paired Student's t-test or Wilcoxon test as appropriate. The differences in the outcome measures between genotype groups over time were analysed using an independent sample Student's t-test when the data presented normal distribution or were continuous. Alternatively, a Mann–Whitney U-test for non-parametric variables was conducted.
All statistical analyses were performed using the Statistical Package for the Social Sciences software Version 20.0 for Windows (SPSS, Chicago, IL, USA), and a p value <0.05 was considered to be statistically significant. Figures displaying statistical analysis were produced using Prism 6 (GraphPad, La Jolla, CA, USA).
Results
Twenty participants fulfilled the aforementioned criteria and were included in this study. The mean age was 77.7 ± 5.3 (range = 70.0–92.0) years, and 30% were male. These characteristics of study population were not different among genotype groups. Genotype distribution and variant allele frequencies for the GPX1 rs1050450, SEPP rs7579 and SEPP rs3877899 polymorphisms are shown in Table 2. The selenium status and intake, GPx activity, ORAC and MDA levels on the baseline for all genotypes are shown in Table 3. No significant differences were observed regarding biochemical parameters among different genotypes.| Polymorphism | Wild type genotype | Heterozygous genotype | Variant genotype | Heterozygous + variant genotypes | Minor allele |
|---|---|---|---|---|---|
| GPX1 | |||||
| rs1050450 (C > T) | 8 (40%) | 7 (35%) | 5 (25%) | 12 (60%) | 0.425 |
| SEPP | |||||
| rs7579 (G > A) | 11 (55%) | 6 (30%) | 3 (15%) | 9 (45%) | 0.300 |
| rs3877899 (G > A) | 12 (60%) | 8 (40%) | 0 (0%) | 8 (40%) | 0.200 |
| Genotype | Age (years) | Sexa (male) | Plasma Se (μg L−1) | Erythrocyte Se (μg L−1) | Se intake (μg d−1) | GPx activity (U gHb−1) | ORAC (μmol TE mL−1) | MDA (μmol L−1) |
|---|---|---|---|---|---|---|---|---|
| a n (%). b p < 0.05. | ||||||||
| GPX1 | ||||||||
| rs1050450 | ||||||||
| CC | 77.3 ± 7.1 | 1 (12.5) | 47.36 ± 9.60 | 51.48 ± 23.22 | 26.77 ± 6.83 | 47.45 ± 20.16 | 1.00 ± 0.55 | 0.44 ± 0.88 |
| CT + TT | 77.83 ± 4.1 | 5 (41.7) | 57.51 ± 19.86 | 58.40 ± 19.46 | 36.01 ± 7.33 | 38.71 ± 11.79 | 0.89 ± 0.31 | 0.46 ± 0.07 |
| p = 0.856 | p = 0.371 | p = 0.147 | p = 0.499 | p = 0.030 | p = 0.294 | p = 0.602 | p = 0.599 | |
| SEPP | ||||||||
| rs7579 | ||||||||
| GG | 77.4 ± 4.4 | 5 (45.5) | 55.65 ± 19.76 | 53.76 ± 19.91 | 32.51 ± 10.06 | 41.33 ± 17.69 | 0.94 ± 0.49 | 0.44 ± 0.80 |
| GA + AA | 78.0 ± 6.5 | 1 (11.1) | 50.76 ± 13.47 | 57.91 ± 22.69 | 33.92 ± 5.77 | 43.24 ± 14.12 | 0.93 ± 0.32 | 0.46 ± 0.081 |
| p = 0.798 | p = 0.655 | p = 0.520 | p = 0.673 | p = 0.731 | p = 0.789 | p = 0.924 | p = 0.617 | |
| rs3877899 | ||||||||
| GG | 76.7 ± 3.6 | 4 (33.3) | 53.88 ± 11.41 | 57.53 ± 17.33 | 33.83 ± 8.69 | 43.79 ± 17.17 | 0.89 ± 0.35 | 0.46 ± 0.09 |
| GA | 79.1 ± 7.2 | 2 (25.0) | 52.80 ± 24.02 | 52.77 ± 26.07 | 31.02 ± 7.40 | 39.83 ± 14.27 | 1.00 ± 0.52 | 0.43 ± 0.06 |
| p = 0.393 | p = 0.371 | p = 0.909 | p = 0.659 | p = 0.574 | p = 0.583 | p = 0.610 | p = 0.343 | |
Linear regression models suggest that the presence of one or two variant alleles (CT or TT genotypes) for GPX1 rs1050450 was associated with higher Se plasma levels (+0.613 μg L−1 SD per allele; p < 0.05). The rs7579 and rs3877899 polymorphisms were not associated with the Se status, and none of the evaluated genotypes showed associations with changes in either MDA or ORAC oxidative stress markers. In contrast, age was associated with lower GPx activity when adjusted by genotype, sex and erythrocyte Se levels (β coefficient = −0.403, −0.449, −0.452, respectively for rs1050450, rs7579 and rs3877899). Although a t-test revealed higher Se intake in T-allele carriers (rs1050450), regression models showed no influence of Se intake on the measured biochemical parameters.
The correlation between GPx activity and erythrocyte Se concentration varied among the tested genotypes. For the GPX1 genotypes, this correlation was observed only in CC genotypes (r = 0.803, p < 0.05), but not in T-allele carriers (r = 0.531, p = 0.08). The same pattern of correlation was verified in subjects carrying the GG genotype regarding SEPP rs3877899 (r = 0.645, p < 0.05) in contrast to the GA individuals (r = 0.568, p = 0.14). When the correlation between GPx activity and erythrocyte Se content was analysed according to rs7579 genotype groups, we observed significance only for the GA + AA genotypes (r = 0.939, p < 0.001).
Among the 20 participants of the study, 11 were randomly assigned to receive one Brazil nut daily for six months. We reported previously that the intake of this nut increased GPx activity with a corresponding elevation in plasma and erythrocyte Se levels, while not influencing the levels of ORAC and MDA.32 In this experimental group, we found that: (i) according to genotypes before and after baseline measurement, we observed highly significant post-supplementation differences compared to pre-treatment for Se-related variables (blood Se and GPx activity); and (ii) an intragroup (between genotypes) comparison demonstrates that the genotype does not influence these variables (Table 4).
| Pre ± SD | Post ± SD | Change (95% CI) | p-Value for between genotype comparisona | ||
|---|---|---|---|---|---|
| a Unpaired Student's t-test. *Significantly different from baseline; p < 0.05 (Student's t-test). | |||||
| Selenium plasma (μg L −1 ) | |||||
| GPX1 rs1050450 | CC | 49.90 ± 10.31 | 246.19 ± 53.99 | 196.29* (110.40–282.18) | 0.140 |
| CT + TT | 59.86 ± 21.56 | 315.92 ± 75.80 | 256.06* (197.37–314.75) | ||
| SEPP rs7579 | GG | 59.62 ± 21.32 | 277.79 ± 62.09 | 218.16* (174.44–261.87) | 0.405 |
| GA + AA | 50.32 ± 11.73 | 312.93 ± 98.88 | 262.60* (121.58–406.62) | ||
| SEPP rs3877899 | GG | 52.77 ± 11.97 | 299.91 ± 68.68 | 247.13* (186.35–307.92) | 0.423 |
| GA | 62.31 ± 27.51 | 274.21 ± 92.40 | 211.91*(107.48–316.33) | ||
| Selenium erythrocyte (μg L −1 ) | |||||
| GPX1 rs1050450 | CC | 65.11 ± 13.62 | 519.23 ± 239.80 | 454.12* (84.03–824.22) | 0.290 |
| CT + TT | 56.37 ± 24.20 | 606.18 ± 151.23 | 549.81* (429.04–670.57) | ||
| SEPP rs7579 | GG | 58.17 ± 22.53 | 536.64 ± 116.86 | 478.47* (379.64–577.30) | 0.372 |
| GA + AA | 61.96 ± 19.83 | 640.93 ± 270.41 | 578.97* (174.67–983.27) | ||
| SEPP rs3877899 | GG | 57.14 ± 16.96 | 607.44 ± 177.57 | 550.29* (397.63–702.96) | 0.416 |
| GA | 63.75 ± 28.43 | 517.02 ± 199.14 | 453.27* (162.11–744.43) | ||
| GPx activity (U g −1 Hb) | |||||
| GPX1 rs1050450 | CC | 54.00 ± 10.29 | 67.67 ± 8.93 | 13.66* (4.67–22.65) | 0.555 |
| CT + TT | 33.15 ± 12.15 | 54.91 ± 24.73 | 21.76* (−9.38–52.91) | ||
| SEPP rs7579 | GG | 36.39 ± 15.08 | 64.92 ± 20.30 | 28.53* (6.03–51.03) | 0.122 |
| GA + AA | 48.33 ± 13.94 | 50.15 ± 20.79 | 1.81 (−36.08–39.72) | ||
| SEPP rs3877899 | GG | 39.64 ± 16.26 | 62.33 ± 24.76 | 22.68 (−8.15–53.52) | 0.440 |
| GA | 42.64 ± 15.28 | 54.68 ± 12.88 | 12.04* (2.45–21.64) | ||
| MDA (μmol L −1 ) | |||||
| GPX1 rs1050450 | CC | 0.38 ± 0.04 | 0.47 ± 0.14 | 0.09 (−0.09–0.28) | 0.926 |
| CT + TT | 0.44 ± 0.05 | 0.54 ± 0.12 | 0.10 (−0.06–0.26) | ||
| SEPP rs7579 | GG | 0.42 ± 0.03 | 0.51 ± 0.13 | 0.09 (−0.39–0.21) | 0.746 |
| GA + AA | 0.41 ± 0.08 | 0.53 ± 0.13 | 0.12 (−0.15–0.39) | ||
| SEPP rs3877899 | GG | 0.41 ± 0.06 | 0.50 ± 0.14 | 0.09 (−0.05–0.22) | 0.597 |
| GA | 0.42 ± 0.02 | 0.55 ± 0.07 | 0.13 (−0.09–0.35) | ||
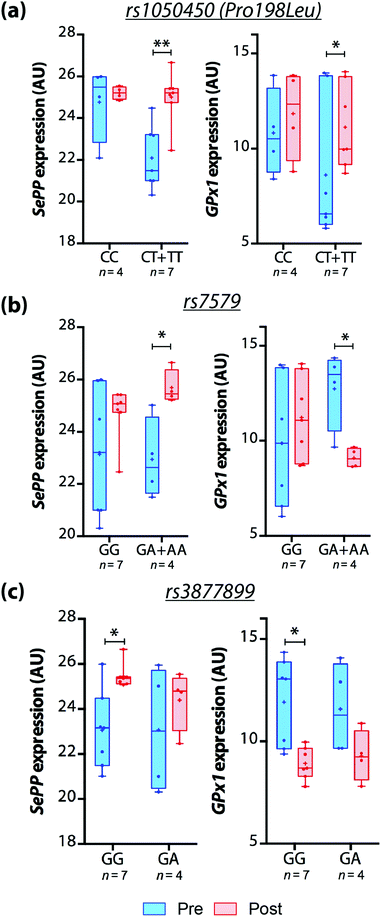
However, we observed that GPX1 and SEPP mRNA expression in response to Brazil nut intake was different among genotypes. In carriers of a variant allele (CT + TT) for rs1050450, GPX1 and SEPP mRNA expression increased over time and no significant differences were observed for CC carriers. SEPP mRNA expression also increased after treatment in A-carriers for rs7579 and GG genotype for rs3877899. On the other hand, GPX1 mRNA expression reduced significantly in A-carriers for rs7579 and GG carriers of rs3877899 (Fig. 1).
Discussion
This pilot study is the first to analyse the association between the Se status and oxidative stress markers in MCI patients with respect to selenoprotein SNPs. We hypothesised that the GPX1 rs1050450 and SEPP rs7579 and rs3877899 polymorphisms would influence Se parameters and oxidative stress biomarkers in these patients, as well as the response to Se intake via Brazil nuts.The rs1050450 polymorphism has been associated with an increased risk for AD,24 although the precise mechanism by which this SNP influences AD and MCI pathogenesis is unclear. Here, we observed that none of the GPX1 genotype groups had different plasma and erythrocyte Se levels; however, in fully adjusted logistic regression models the presence of T allele of GPX1 was associated with higher plasma Se. These results suggest that this SNP might affect the way in which the body metabolises and utilises Se, although they contrast with previous studies that did not observe differences in the Se status as a result of the Pro198Leu polymorphism.22,23 As observed in other studies,41–44 oxidative stress parameters were not affected by Pro198Leu genotypes. However, studies have associated the presence of the T allele with decreased GPx activity,27,45–47 and we observed that T carriers had 20% lower GPx activity in comparison with the CC genotype, although this difference was not statistically significant. Moreover, the correlation between GPx activity and erythrocyte Se concentration was significant only in TT genotypes, which allow us to hypothesise that post-transcription regulation might be occurring due to Pro198Leu genotypes. Studies have shown that GPx activity may plateau over certain Se levels.47 However, we observed a positive correlation between erythrocyte GPx activity and erythrocyte Se level only in CC genotype, suggesting that the GPx activity plateau varies according to the genotype. Considering the known association between decreased GPx activity in AD,18 these data suggest that, at least in MCI patients, the ability of the body to uptake Se and incorporate it into selenoproteins is influenced by the Pro198Leu genotype, which may explain the findings of Paz-y-Miño et al.,24 who reported an association between T allele and the risk of AD. This further supports a potential role for GPx in cognitive decline and the benefits of Se supplementation. Further investigation is required to confirm the effects of Pro198Leu on Se metabolism and requirements.
The SEPP polymorphism rs7579 is located in a region corresponding to the 3′-UTR,26 where a UGA codon is read as a selenocysteine (Sec) codon during selenoprotein synthesis.48 This SNP may alter the efficiency of Sec incorporation into SePP.49 The rs3877899 polymorphism is located in the coding region of the SEPP gene and may regulate the stability of SePP protein and cellular Se uptake.26 In our study, the rs7579 and rs3877899 SNPs neither influenced the Se status, nor oxidative stress parameters, although both have been associated with a change in the proportion of 50- and 60-kDa SePP isoforms in plasma, which may affect Se supply for the synthesis of different selenoproteins, including antioxidant enzymes, in different tissues.26
As oxidative stress has a central role in neurodegeneration and cognitive decline, inclusion of foods with antioxidant characteristics may be a worthwhile strategy to improve antioxidant capacity and reduce the risk of dementia with no otherwise ill-effects. Brazil nuts are unique due to their high Se content50,51 and potentially beneficial phenolic compounds.52 Following our previous evidence that regular Brazil nuts intake increased Se levels in MCI patients,32 we observed here that GPX1 or SEPP genotypes did not influence biomarkers of Se status. However, we did observe that the change in GPX1 and SEPP expression over time was not equivalent across genotype groups. Carriers of T-allele of Pro198Leu were more responsive to Brazil nuts intake regarding the increment of GPX1 and SEPP mRNA expression. Moreover, although not significant, we observed that CT + TT genotypes for Pro198Leu had higher GPx activity over time in comparison with the CC genotype. Taking together, these observations build up the hypothesis that Pro198Leu genotypes should be considered in studies that assess both the selenium nutritional status and the effect of selenium intake. The rs7579 and rs3877899 SNPs in SEPP gene also resulted in different changes on mRNA expression over time: a-allele of rs7579 was associated with decreased GPX1 and increased SEPP mRNA expression over time, and the same variation was observed for the GG genotype of rs3877899. Previous studies have shown that Se supplementation increased GPX153–57 and SEPP49,53,56,57 expression. However, we highlight that these studies assessed gene expression in enterocytes and hepatocytes in cell cultures and in vivo animal studies, and did not consider genotypes. Thus, we emphasise the need for more studies to understand the as-yet unknown mechanisms that possibly interact during selenoprotein synthesis, as changes in SePP synthesis can affect not only the function of the protein itself but also other selenoproteins that are involved in selenium hierarchy.33
Different environmental characteristics can be related to Se status and metabolism.58 In our study, age was negatively correlated with GPx activity, but not with the Se status, corroborating the results of Maurya et al.59 and contrasting those of Ferguson et al.47 The real association between age and selenoprotein metabolism should be further investigated, because it is hypothesised that GPx variation with age could be due to variability of Se availability.60 Body mass index can also affect Se metabolism, as obese people with the GG genotype are more responsive to Se supplementation.26 In our study, no differences in body mass index (BMI) were observed between groups (p = 0.107), and only one participant presented a BMI > 30, which precluded such analysis. It was not possible to see the effect of sex on biochemical parameters due to the small size of this pilot study. It is also important to note that multiple variants in selenoprotein genes and in the genes of other antioxidant enzymes may act synergistically to generate a different downstream response; as a result, the effects of a given SNP may be altered by SNPs in other genes.33 Therefore, larger studies building on our pilot data that examine SNPs in selenoprotein genes and other genes in relationship to different environmental and dietary factors will be invaluable.
Conclusions
Our findings suggest that the GPX1 rs1050450 polymorphism may influence Se status and selenoprotein synthesis, although it did not affect the direct response to Brazil nut intake with respect to Se levels, as well as observed in rs7579 and rs3877899 genotypes. However, we found that both GPX1 and SEPP expression in response to Brazil nut intake was affected by rs1050450, rs7579 and rs3877899 genotypes. This pilot data justifies a need for further studies to better understand the effects of these SNPs in response to dietary Se and determine the mechanism by which Se slows cognitive decline, and thus we aim to enlarge the sample size and generate more data moving forward.Notes and references
- Alzheimer's Disease International, Alzheimer's disease international report, The International Federation of Alzheimer's Disease and Related Disorders Societies, Inc., London, 2009 Search PubMed.
- D. E. Barnes and K. Yaffe, Lancet Neurol., 2011, 10, 819–828 CrossRef PubMed.
- N. D. Barnard, A. I. Bush, A. Ceccarelli, J. Cooper, C. A. de Jager, K. I. Erickson, G. Fraser, S. Kesler, S. M. Levin, B. Lucey, M. C. Morris and R. Squitti, Neurobiol. Aging, 2014, 35, S74–S78 CrossRef PubMed.
- D. R. Gustafson, M. Clare Morris, N. Scarmeas, R. C. Shah, J. Sijben, K. Yaffe and X. Zhu, J. Alzheimers Dis., 2015, 46, 1111–1127 CrossRef CAS PubMed.
- E. J. Mufson, L. Binder, S. E. Counts, S. T. DeKosky, L. deTolledo-Morrell, S. D. Ginsberg, M. D. Ikonomovic, S. E. Perez and S. W. Scheff, Acta Neuropathol., 2012, 123, 13–30 CrossRef CAS PubMed.
- M. Gatz, C. A. Reynolds, L. Fratiglioni, B. Johansson, J. A. Mortimer, S. Berg, A. Fiske and N. L. Pedersen, Arch. Gen. Psychiatry, 2006, 63, 168–174 CrossRef PubMed.
- A. J. Saykin, L. Shen, X. Yao, S. Kim, K. Nho, S. L. Risacher, V. K. Ramanan, T. M. Foroud, K. M. Faber, N. Sarwar, L. M. Munsie, X. Hu, H. D. Soares, S. G. Potkin, P. M. Thompson, J. S. K. Kauwe, R. Kaddurah-Daouk, R. C. Green, A. W. Toga and M. W. Weiner, Alzheimer's Dementia, 2015, 11, 792–814 CrossRef PubMed.
- A. Tandon and P. Fraser, Genome Biol., 2002, 3, 3014 CrossRef.
- B. De Strooper, P. Saftig, K. Craessaerts, H. Vanderstichele, G. Guhde, W. Annaert, K. Von Figura and F. Van Leuven, Nature, 1998, 391, 387–390 CrossRef CAS PubMed.
- M. Citron, D. Westaway, W. Xia, G. Carlson, T. Diehl, G. Levesque, K. Johnson-Wood, M. Lee, P. Seubert, A. Davis, D. Kholodenko, R. Motter, R. Sherrington, B. Perry, H. Yao, R. Strome, I. Lieberburg, J. Rommens, S. Kim, D. Schenk, P. Fraser, P. St George Hyslop and D. J. Selkoe, Nat. Med., 1997, 3, 67–72 CrossRef CAS PubMed.
- J. B. Kwok, Q. X. Li, M. Hallupp, S. Whyte, D. Ames, K. Beyreuther, C. L. Masters and P. R. Schofield, Ann. Neurol., 2000, 47, 249–253 CrossRef CAS PubMed.
- Y. Christen, Am. J. Clin. Nutr., 2000, 71, 621s–629s CAS.
- L. Bertram and R. E. Tanzi, Pharmacol. Res., 2004, 50, 385–396 CrossRef CAS PubMed.
- K. Sleegers, J.-C. Lambert, L. Bertram, M. Cruts, P. Amouyel and C. Van Broeckhoven, Trends Genet., 2010, 26, 84–93 CrossRef CAS PubMed.
- C. Berr, B. Balansard, J. Arnaud, A. M. Roussel and A. Alperovitch, J. Am. Geriatr. Soc., 2000, 48, 1285–1291 CrossRef CAS PubMed.
- B. R. Cardoso, T. P. Ong, W. Jacob-Filho, O. Jaluul, M. I. Freitas and S. M. Cozzolino, Br. J. Nutr., 2010, 103, 803–806 CrossRef CAS PubMed.
- S. Gao, Y. Jin, F. W. Unverzagt, C. Liang, K. S. Hall, J. Cao, F. Ma, J. R. Murrell, Y. Cheng, P. Li, J. Bian and H. C. Hendrie, BMC Psychiatry, 2012, 12, 72 CrossRef PubMed.
- B. R. Cardoso, B. R. Roberts, A. I. Bush and D. J. Hare, Metallomics, 2015, 7, 1213–1228 RSC.
- C. Méplan, V. Pagmantidis and J. Hesketh, in Nutritional Genomics: Impact in Health and Disease, ed. R. Brigélius-Flohé and H. Joost, Wiley-VCH Verlag GmbH & Co, Weinheim, 2006, pp. 132–158 Search PubMed.
- T. Shinkai, D. J. Muller, V. De Luca, S. Shaikh, C. Matsumoto, R. Hwang, N. King, J. Trakalo, N. Potapova, G. Zai, H. Hori, O. Ohmori, H. Y. Meltzer, J. Nakamura and J. L. Kennedy, Psychiatry Res., 2006, 141, 123–128 CrossRef CAS PubMed.
- E. Jablonska, J. Gromadzinska, E. Reszka, W. Wasowicz, W. Sobala, N. Szeszenia-Dabrowska and P. Boffetta, Eur. J. Nutr., 2009, 48, 383–386 CrossRef CAS PubMed.
- C. Cominetti, M. C. de Bortoli, E. Purgatto, T. P. Ong, F. S. Moreno, A. B. Garrido Jr. and S. M. Cozzolino, Nutrition, 2011, 27, 891–896 CrossRef CAS PubMed.
- B. R. Cardoso, T. P. Ong, W. Jacob-Filho, O. Jaluul, M. I. Freitas, C. Cominetti and S. M. Cozzolino, J. Nutrigenet. Nutrigenomics, 2012, 5, 72–80 CrossRef CAS PubMed.
- C. Paz-y-Mino, C. Carrera, A. Lopez-Cortes, M. J. Munoz, N. Cumbal, B. Castro, A. Cabrera and M. E. Sanchez, Am. J. Med. Sci., 2010, 340, 373–377 CrossRef PubMed.
- O. H. Al-Taie, N. Uceyler, U. Eubner, F. Jakob, H. Mork, M. Scheurlen, R. Brigelius-Flohe, K. Schottker, J. Abel, A. Thalheimer, T. Katzenberger, B. Illert, R. Melcher and J. Kohrle, Nutr. Cancer, 2004, 48, 6–14 CrossRef CAS PubMed.
- C. Meplan, L. K. Crosley, F. Nicol, G. J. Beckett, A. F. Howie, K. E. Hill, G. Horgan, J. C. Mathers, J. R. Arthur and J. E. Hesketh, FASEB J., 2007, 21, 3063–3074 CrossRef CAS PubMed.
- N. Karunasinghe, D. Y. Han, S. Zhu, J. Yu, K. Lange, H. Duan, R. Medhora, N. Singh, J. Kan, W. Alzaher, B. Chen, S. Ko, C. M. Triggs and L. R. Ferguson, Genes Nutr., 2012, 7, 179–190 CrossRef CAS PubMed.
- J. N. Hellwege, N. D. Palmer, J. T. Ziegler, C. D. Langefeld, C. Lorenzo, J. M. Norris, T. Takamura and D. W. Bowden, Gene, 2014, 534, 33–39 CrossRef CAS PubMed.
- I. B. G. Martens, B. R. Cardoso, D. J. Hare, M. M. Niedzwiecki, F. M. Lajolo, A. Martens and S. M. F. Cozzolino, Nutrition, 2015, 31, 1339–1343 CrossRef CAS PubMed.
- M. B. Stockler-Pinto, D. Mafra, N. E. Farage, G. T. Boaventura and S. M. Cozzolino, Nutrition, 2010, 26, 1065–1069 CrossRef CAS PubMed.
- C. Cominetti, M. C. de Bortoli, A. B. Garrido Jr. and S. M. Cozzolino, Nutr. Res., 2012, 32, 403–407 CrossRef CAS PubMed.
- B. R. Cardoso, D. Apolinario, V. da Silva Bandeira, A. L. Busse, R. M. Magaldi, W. Jacob-Filho and S. M. Cozzolino, Eur. J. Nutr., 2015 DOI:10.1007/s00394-014-0829-2.
- J. Hesketh and C. Meplan, Proc. Nutr. Soc., 2011, 70, 365–373 CrossRef.
- B. Winblad, K. Palmer, M. Kivipelto, V. Jelic, L. Fratiglioni, L. O. Wahlund, A. Nordberg, L. Backman, M. Albert, O. Almkvist, H. Arai, H. Basun, K. Blennow, M. de Leon, C. DeCarli, T. Erkinjuntti, E. Giacobini, C. Graff, J. Hardy, C. Jack, A. Jorm, K. Ritchie, C. van Duijn, P. Visser and R. C. Petersen, J. Intern. Med., 2004, 256, 240–246 CrossRef CAS PubMed.
- D. E. Paglia and W. N. Valentine, J. Lab. Clin. Med., 1967, 70, 158–169 CAS.
- Y. L. Hong, S. L. Yeh, C. Y. Chang and M. L. Hu, Clin. Biochem., 2000, 33, 619–625 CrossRef CAS PubMed.
- R. L. Prior, H. Hoang, L. Gu, X. Wu, M. Bacchiocca, L. Howard, M. Hampsch-Woodill, D. Huang, B. Ou and R. Jacob, J. Agric. Food Chem., 2003, 51, 3273–3279 CrossRef CAS PubMed.
- D. Q. Hao, G. H. Xie, Y. M. Zhang and G. J. Tian, Talanta, 1996, 43, 595–600 CrossRef CAS PubMed.
- K. Ferreira, J. Gomes, C. Bellato and C. Jordão, Rev. Panam. Salud Publica, 2002, 11(3), 172–177 Search PubMed.
- W. Willett, Nutritional Epidemiology, Oxford University Press, New York, 1998 Search PubMed.
- J. C. Charniot, A. Sutton, D. Bonnefont-Rousselot, C. Cosson, R. Khani-Bittar, P. Giral, N. Charnaux and J. P. Albertini, Free Radical Res., 2011, 45, 379–388 CrossRef CAS PubMed.
- T. S. Tang, S. L. Prior, K. W. Li, H. A. Ireland, S. C. Bain, S. J. Hurel, J. A. Cooper, S. E. Humphries and J. W. Stephens, Nutr., Metab. Cardiovasc. Dis., 2012, 22, 417–425 CrossRef CAS PubMed.
- U. Al-Alem, P. H. Gann, J. Dahl, R. B. van Breemen, V. Mistry, P. M. Lam, M. D. Evans, L. Van Horn and M. E. Wright, Genes Nutr., 2012, 7, 191–195 CrossRef CAS PubMed.
- Y. Takata, I. B. King, J. W. Lampe, R. F. Burk, K. E. Hill, R. M. Santella, A. R. Kristal, D. J. Duggan, T. L. Vaughan and U. Peters, J. Nutr., 2012, 142, 419–426 CrossRef CAS PubMed.
- G. Ravn-Haren, A. Olsen, A. Tjonneland, L. O. Dragsted, B. A. Nexo, H. Wallin, K. Overvad, O. Raaschou-Nielsen and U. Vogel, Carcinogenesis, 2006, 27, 820–825 CrossRef CAS PubMed.
- Y. M. Xiong, X. Y. Mo, X. Z. Zou, R. X. Song, W. Y. Sun, W. Lu, Q. Chen, Y. X. Yu and W. J. Zang, Osteoarthritis Cartilage, 2010, 18, 817–824 CrossRef CAS PubMed.
- L. R. Ferguson, N. Karunasinghe, S. Zhu, D. Y. Han, C. M. Triggs, A. X. A. H. Wang and J. G. Masters, Curr. Pharmacogenomics Pers. Med., 2012, 10, 204–216 CrossRef CAS.
- D. L. Hatfield, P. A. Tsuji, B. A. Carlson and V. N. Gladyshev, Trends Biochem. Sci., 2014, 39, 112–120 CrossRef CAS PubMed.
- C. Meplan, F. Nicol, B. T. Burtle, L. K. Crosley, J. R. Arthur, J. C. Mathers and J. E. Hesketh, Antioxid. Redox Signaling, 2009, 11, 2631–2640 CrossRef CAS PubMed.
- E. T. Bodo, Z. Stefanka, I. Ipolyi, C. Soros, M. Dernovics and P. Fodor, Anal. Bioanal. Chem., 2003, 377, 32–38 CrossRef CAS PubMed.
- E. G. da Silva, L. R. Mataveli and M. A. Arruda, Talanta, 2013, 110, 53–57 CrossRef CAS PubMed.
- L. Pires, A. Silva, L. Alencar, J. Pimentel, J. Mancini Filho and S. Cozzolino, Nutrire, 2011, 36s, 7s Search PubMed.
- Y. Hu, G. H. McIntosh, R. K. Le Leu, J. M. Upton, R. J. Woodman and G. P. Young, Br. J. Nutr., 2011, 106, 572–582 CrossRef CAS PubMed.
- A. Jerome-Morais, S. Bera, W. Rachidi, P. H. Gann and A. M. Diamond, Biochim. Biophys. Acta, 2013, 1830, 3399–3406 CrossRef CAS PubMed.
- Z. Xu, Z. Wang, J. J. Li, C. Chen, P. C. Zhang, L. Dong, J. H. Chen, Q. Chen, X. T. Zhang and Z. L. Wang, Food Chem. Toxicol., 2013, 58, 1–7 CrossRef CAS PubMed.
- K. Jotty, M. L. Ojeda, F. Nogales, M. L. Murillo and O. Carreras, Alcohol, 2013, 47, 545–552 CrossRef CAS PubMed.
- T. Maseko, K. Howell, F. R. Dunshea and K. Ng, Food Chem., 2014, 146, 327–333 CrossRef CAS PubMed.
- L. R. Ferguson, N. Karunasinghe, S. Zhu and A. H. Wang, Mutat. Res., 2012, 733, 100–110 CrossRef CAS PubMed.
- P. K. Maurya, P. Kumar, N. Siddiqui, P. Tripathi and S. I. Rizvi, Indian J. Biochem. Biophys., 2010, 47, 319–321 CAS.
- J. M. Beckett and M. J. Ball, Br. J. Nutr., 2011, 106, 718–724 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |