Inhibition of SARS coronavirus helicase by bismuth complexes†
Nan
Yang
a,
Julian A.
Tanner
b,
Zai
Wang
b,
Jian-Dong
Huang
b,
Bo-Jian
Zheng
c,
Nianyong
Zhu
a and
Hongzhe
Sun
*a
aDepartment of Chemistry, The University of Hong Kong, Pokfulam Road, Hong Kong, P.R. China. E-mail: hsun@hku.hk; Fax: (+852)2857 1586
bDepartment of Biochemistry, The University of Hong Kong, Pokfulam Road, Hong Kong, P.R. China
cDepartment of Microbiology, The University of Hong Kong, Pokfulam Road, Hong Kong, P.R. China
First published on 16th August 2007
Abstract
A series of bismuth complexes were synthesized and characterized, and most of them exhibited inhibition against the SARS coronavirus helicase ATPase and duplex-unwinding activities at micromolar concentrations.
Bismuth compounds have been used clinically as medicines for centuries in the treatment of a variety of gastrointestinal disorders.1–7 Radioactive bismuth isotopes (212Bi and 213Bi) have also been demonstrated as targeted radio-therapeutic agents for cancer therapy.5,8 Recently, the application of bismuth drugs was extended to the antiviral field. We have previously found that the bismuth compounds efficiently inhibit the growth of severe acute respiratory syndrome coronavirus (SARS-CoV) in vivo.9
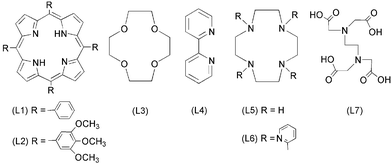
SARS caused by SARS-CoV, killed hundreds of people world-wide when it broke out in 2003.9 Previous studies demonstrated that the NTPase/helicase protein from SARS-CoV is likely to play crucial roles in the viral life cycle, making it an attractive target for anti-SARS therapy.10–12 As bismuth is known to bind strongly to metallothionein through the formation of Bi–S bonds,13 we hypothesized that bismuth ions might be able to inhibit the enzymatic activities of the SARS-CoV helicase by binding to its N-terminal metal binding domain (MBD), a cysteine-rich region. Bismuth complexes such as ranitidine bismuth citrate were subsequently found to inhibit SARS-CoV helicase ATPase and duplex-unwinding activities.9 Herein, we synthesized a series of bismuth compounds with various N, O-containing chelate ligands (Scheme 1) and their activities against the SARS-CoV helicase were examined and compared.
 | ||
| Scheme 1 The ligands of bismuth and zinc complexes. | ||
The bismuth porphyrin complexes 1 ([Bi(L1)(NO3)]·H2O, L1 = 5,10,15,20-tetraphenyl-21H,23H-porphine) and 2 ([Bi(L2)(NO3)]·H2O, L2 = 5,10,15,20-tetra(1,2,3-trimethoxyphenyl)-21H,23H-porphine) were synthesized in pyridine according to a modified method.‡ On standing 2 in a CH3OH–CHCl3 solution at room temperature for several days, dark green crystals of 2, suitable for X-ray analysis, formed.§ X-ray crystallography revealed that Bi3+ coordinates to four nitrogen atoms from the porphyrin ring and an oxygen atom from a nitrate in a distorted pyramidal geometry (Fig. 1). The metal lies 1.339 Å above the porphyrin plane, similarly to other Bi-porphyrin complexes.14,15 The crystal structure of 2 shows that the complex lies about an inversion centre and that the Bi atom has 50% occupancy and is disordered over two sites above and below the porphyrin plane with Bi–Bi* distance of 2.679 Å (where Bi* is the Bi atom at equivalent position). The nitrate group is also disordered over two inversion related sites and has 50% occupancy (nitrate and nitrate* in Fig. 1).
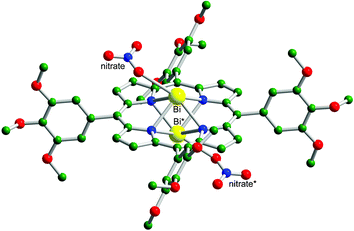 | ||
| Fig. 1 The crystal structure of 2. The hydrogen atoms have been omitted for clarity. Bi*, 50% occupancy of bismuth atom in the complex; nitrate*, 50% occupancy of nitrate group in 2. Color code: Bi, yellow; O, red; N, blue; C, green. | ||
Complexes 3 ([Bi(NO3)3(L3)], L3 = 12-crown-4),164 ([BiCl3(L4)1.5], L4 = 2,2′-bipyridine),175a ([Bi(L5)(H2O)(ClO4)3], L5 = 1,4,7,10-tetraazacyclododecane),185b ([Zn(L5)(H2O)](ClO4)2),196 ([Bi(NO3)4(L6)], L6 = 1,4,7,10-tetrakis(2-pyridylmethyl)-1,4,7,10-tetraazacyclododecane),20 and 7 ([Bi(L7)], L7 = ethylenediaminetetraacetic acid)21 were synthesized according to the literature and had satisfactory elemental analysis. Complexes 1, 2, 3, 4 and 5a had poor solubility in water but can be readily dissolved in DMSO, whereas 5b, 6 and 7 were highly water soluble.
The UV-vis spectrum of 2 showed one major absorbance at 468 nm in DMSO with little changes observed for over two days, indicating that the complex was stable in DMSO. In contrast, the UV-vis absorbance of 2 at 468 nm in Tris-HCl buffer at pH 6.8 decreased whereas the absorbance at 425 nm increased in intensity (Fig. 2(a)). The two absorption peaks at 468 and 425 nm were assigned to Bi-bound porphyrin and free porphyrin ligand (L2), respectively.15 The transposition of predominance of Bi-bound porphyrin complex and free porphyrin ligand (L2) suggests the release of Bi3+ from the complex. The absorption at 468 nm decreased exponentially with a half-life of ca. 56 min (Fig. 2(b)), corresponding to the half-life of demetalation of 2. Similar behaviour was observed for 1 except that the complex 1 decomposed more slowly than 2 (half-life of demetalation >5 h). Ligand L6 and Bi3+ forms a relative “strong” complex (6) which shows almost no decomposition overnight under identical conditions (ESI,† Fig. S1).
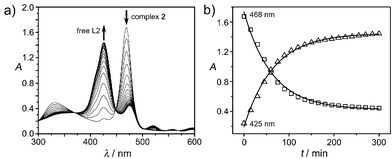 | ||
| Fig. 2 The demetalation reaction of 2 in 50 mM Tris-HCl, pH 6.8. (a) The UV-vis spectra of 2 at different times; (b) the timecourse of the UV absorbance at 425 and 468 nm. | ||
The SARS-CoV helicase (nsp13-pp1ab, accession number NP_828870, originally denoted as nsp10) was cloned and purified as previously described.9 The inhibition of bismuth complexes against SARS helicase ATPase activity was measured by a phosphate release assay.¶ As shown in Fig. 3(a) and Table 1, most bismuth complexes exhibit inhibition activity against the enzyme. Among them, two bismuth porphyrin complexes exhibited the highest inhibition activities against helicase ATPase with IC50 values of 0.6 ± 0.1 (1) and 0.5 ± 0.1 µM (2). Complexes 3, 4 and 5a exhibited slightly lower inhibition activities with IC50 values of 4.7–7.4 µM. A zinc complex of L5 was synthesized (5b) and its inhibition activity towards ATPase was examined for comparison to 5a. Interestingly, 5b exhibited no activity against the ATPase even at a concentration of 5 mM, indicating that bismuth ions play the key role in the inhibitory effects. Complexes 6 and 7 were surprisingly poor inhibitors compared with other bismuth complexes, probably attributed to tight binding of the complex ligands.
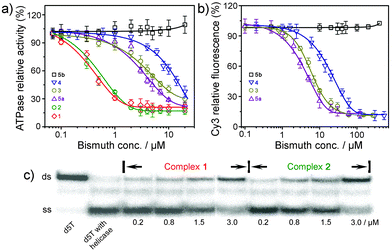 | ||
| Fig. 3 (a) Inhibition of ATPase activity of the SARS-CoV helicase by bismuth and zinc complexes; (b) titration of the ds-DNA unwinding activity of the SCV helicase with increasing concentrations of bismuth and zinc compounds as observed by FRET assay. Both 6 and 7 have similar results to 5b and have been omitted for clarity; (c) titration of the ds-DNA unwinding activity of the SCV helicase with complexes 1 and 2 observed by radiography of labeled duplex. Lane 1, ds-DNA (d5T); lane 2, d5T with helicase in the absence of bismuth complex; lanes 3–10, with the addition of increasing concentrations of complexes 1 and 2. | ||
| Complex | IC50/µM | |
|---|---|---|
| ATPase activity | Helicase activitya | |
| a Helicase activity of 3, 4, 5a, 5b, 6 and 7 were measured by FRET-based assay; helicase activity of 1 and 2 were measured by radiography of labeled duplex after acrylamide gel electrophoresis (Fig. 3(c)). | ||
| 5b | >5000 | >5000 |
| 7 | >5000 | >5000 |
| 6 | >5000 | >5000 |
| 4 | 7.4 ± 1.1 | 11.0 ± 1.8 |
| 3 | 6.4 ± 1.0 | 7.2 ± 0.8 |
| 5a | 4.7 ± 0.6 | 5.0 ± 0.8 |
| 1 | 0.6 ± 0.1 | <3.0 |
| 2 | 0.5 ± 0.1 | <3.0 |
The inhibition of bismuth and zinc complexes against the SARS helicase duplex-unwinding activity was measured by FRET-based assays as previously described,9 modified to use oligomers suitable for a 5′ to 3′ helicase. The principle behind the assay is thus: two oligomers are designed so that they anneal with a 5′ overhang. At the 3′ end of the longer oligomer (that has the 5′ overhang) there is a Cy3 fluorophore. At the 5′ end of the shorter oligomer there is a BHQ-2 quencher. When the two oligomers are annealed, the Cy3 fluorescence is quenched by the FRET effect due to the close proximity of BHQ-2. If the oligomers are dissociated through action of the helicase, then the strong fluorescent signal of Cy3 is restored. Two oligomers were first annealed and then incubated with purified helicase and increasing concentrations of bismuth and zinc complexes in a 96-well microplate.|| Similar to the helicase ATPase assay, both 6 and 7 exhibited almost no inhibition to helicase duplex-unwinding activity even at a concentration of 5 mM. The IC50 values of 3, 4 and 5a were 7.2 ± 0.8, 11.0 ± 1.8 and 5.0 ± 0.8 µM respectively, comparable to the IC50 values for the inhibition of ATPase activity of the SARS-CoV helicase (Fig. 3(b)). As expected, 5b exhibited no activity against helicase. The helicase duplex-unwinding inhibition activity of two bismuth porphyrin complexes (1 and 2) were measured by a previously developed radioassay because of the emission overlay of Cy3 fluorophore with the complex itself when using the FRET-based assay.**12 The duplex (d5T) and released strands were resolved by polyacrylamide gel electrophoresis and visualized by phosphor-imaging (Fig. 3(c)). We observed that the duplex was almost fully unwound by the enzyme in the absence of bismuth complexes (lane 2), and the helicase unwinding activity to the duplex was inhibited gradually upon the addition of increasing concentrations of 1 or 2 (Fig. 3(c) and Table 1). Less than 50% of duplex was unwound by helicase when the concentration of complex 1 or 2 reached to 3.0 µM (the intensities of ds-DNA band of Fig. 3(c) were analyzed by program Image J (Version 1.36b)), indicating that the IC50 values of both complexes 1 and 2 were less than 3.0 µM (ESI,† Fig. S2).
The combined inhibition results of bismuth and zinc complexes to helicase ATPase and duplex-unwinding activities demonstrated that the bismuth complexes exhibited the same order of inhibition with 1 ≈ 2 > 5a > 3 > 4 > 6 ≈ 7 ≈ 5b in both assays (Table 1). The different inhibition activities of these complexes may be related to their different coordination environments. This verified that Bi3+ ions play a critical role in the inhibition of SARS helicase biological activities. The treatment of SARS-CoV infected cells by bismuth-containing complexes or biomolecules such as ranitidine bismuth citrate and Bi2-hTF (hTF = human transferrin) confirmed that the bismuth plays an inhibitory role during later stages of the replicative cycle, consistent with the helicase being the target in vivo.9
In summary, we have synthesized several bismuth complexes with a bismuth porphyrin complex being structurally characterized. These complexes exhibit activities against both SARS-CoV helicase ATPase and duplex-unwinding activities. Further trials will be required in animal models to demonstrate whether these complexes (e.g., 1 and 2) are effective in inhibiting the SARS-CoV in vivo and whether these complexes are able to inhibit other viruses (e.g. HCV) in which the helicase is also important in the viral life cycle.
This work was supported by the Research Grants Committee of Hong Kong (HKU7040/05P and HKU7546/06M), Welfare and Food Bureau under the Research Fund for the Control of Infectious Diseases (01030182 and 02040192) and Area of Excellence on Chemical Biology.
Notes and references
- (a) M. J. Abrams and B. A. Murrer, Science, 1993, 261, 725–730 CrossRef CAS; (b) K. H. Thompson and C. Orvig, Science, 2003, 300, 936–939 CrossRef CAS.
- J. R. Lambert and P. Midolo, Aliment. Pharmacol. Ther., 1997, 11, S27–S33.
- H. Sun, L. Zhang and K. Y. Szeto, Met. Ions Biol. Syst., 2004, 41, 333–378 CAS.
- G. G. Briand and N. Burford, Chem. Rev., 1999, 99, 2601–2657 CrossRef CAS.
- S. Hassfjell and M. W. Brechbiel, Chem. Rev., 2001, 101, 2019–2036 CrossRef CAS.
- Z. Guo and P. J. Sadler, Angew. Chem., Int. Ed., 1999, 38, 1512–1531 CrossRef CAS.
- R. Ge and H. Sun, Acc. Chem. Res., 2007, 40, 267–274 CrossRef CAS.
- N. Yang and H. Sun, Coord. Chem. Rev., 2007 DOI:10.1016/j.ccr.2007.03.003.
- N. Yang, J. A. Tanner, B. J. Zheng, R. M. Watt, M. L. He, L. Y. Lu, J. Q. Jiang, K. T. Shum, Y. P. Lin, K. L. Wong, M. C. M. Lin, H. F. Kung, H. Sun and J. D. Huang, Angew. Chem., Int. Ed., 2007 DOI:10.1002/anie.200701021.
- J. A. Tanner, B. J. Zheng, J. Zhou, R. M. Watt, J. Q. Jiang, K. L. Wong, Y. P. Lin, L. Y. Lu, M. L. He, H. F. Kung, A. J. Kesel and J. D. Huang, Chem. Biol., 2005, 12, 303–311 CrossRef CAS.
- A. Bernini, O. Spiga, V. Venditti, F. Prischi, L. Bracci, J. D. Huang, J. A. Tanner and N. Niccolai, Biochem. Biophys. Res. Commun., 2006, 343, 1101–1104 CrossRef CAS.
- J. A. Tanner, R. M. Watt, Y. B. Chai, L. Y. Lu, M. C. Lin, J. S. Peiris, L. L. Poon, H. F. Kung and J. D. Huang, J. Biol. Chem., 2003, 278, 39578–39582 CrossRef CAS.
- H. Sun, H. Li, I. Harvey and P. J. Sadler, J. Biol. Chem., 1999, 274, 29094–29101 CrossRef CAS.
- Z. Halime, L. Michaudet, M. Razavet, C. Ruzié and B. Boitrel, Dalton Trans., 2003, 4250–4254 RSC.
- Z. Halime, M. Lachkar, E. Furet, J. F. Halet and B. Boitrel, Inorg. Chem., 2006, 45, 10661–10669 CrossRef CAS.
- R. D. Rogers, A. H. Bond and S. Aguinaga, J. Am. Chem. Soc., 1992, 114, 2960–2967 CrossRef CAS.
- G. A. Bowmaker, F. M. M. Hannaway, P. C. Junk, A. M. Lee, B. W. Skelton and A. H. White, Aust. J. Chem., 1998, 51, 325–330 CAS.
- (a) R. Luckay, I. Cukrowski, J. Mashishi, J. H. Reibenspies, A. H. Bond, R. D. Rogers and R. D. Hancock, J. Chem. Soc., Dalton Trans., 1997, 901–908 RSC; (b) I. Cukrowski and R. C. Luckay, Anal. Chim. Acta, 1998, 372, 323–331 CrossRef CAS.
- (a) M. Shionoya, E. Kimura and M. Shiro, J. Am. Chem. Soc., 1993, 115, 6730–6737 CrossRef CAS; (b) M. Shionoya, T. Ikeda, E. Kimura and M. Shiro, J. Am. Chem. Soc., 1994, 116, 3848–3859 CrossRef CAS.
- (a) X. H. Bu, X. C. Cao, W. Chen, R. H. Zhang and C. Thomas, Polyhedron, 1998, 17, 289–293 CrossRef CAS; (b) X. Wang, X. Zhang, J. Lin, J. Chen, Q. Xu and Z. Guo, Dalton Trans., 2003, 2379–2380 RSC.
- S. P. Summers, K. A. Abboud, S. R. Farrah and G. J. Palenik, Inorg. Chem., 1994, 33, 88–92 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig S1: The demetalation reaction of 6 in aqueous solution. Fig S2: The measurement of intensities of ds-DNA (d5T) bands of complexes 1 and 2 after being resolved by polyacrylamide gel electrophoresis and visualized by phosphor-imaging. See DOI: 10.1039/b709515e |
| ‡ Synthesis: To 50 ml of refluxing pyridine containing 0.1 g of L2 was added 1.0 g of Bi(NO3)3, and a further 1.0 g was introduced 2 h later. After 5 h, the mixture was freeze dried in hood overnight and subsequently dissolved in about 1.0 ml CHCl3 and loaded to the silica gel column. The product was obtained in 35% yield by washing the column with CHCl3–CH3OH. |
§ Crystal data: C56H56BiN5O17, M = 1280.04, triclinic, P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) (no. 2), a = 7.715(15), b = 13.208(3), c = 14.116(3) Å, α = 92.43(3), β = 100.53(3), γ = 92.84(3)°, V = 1408.5(5) Å3, Z = 1, Dc = 1.509 g cm−3, μ(Mo-Kα) = 3.207 mm−1, F(000) = 646, λ = 0.71073 Å, T = 253 K. CCDC 654367. For crystallographic data in CIF or other electronic format see DOI: 10.1039/b709515e (no. 2), a = 7.715(15), b = 13.208(3), c = 14.116(3) Å, α = 92.43(3), β = 100.53(3), γ = 92.84(3)°, V = 1408.5(5) Å3, Z = 1, Dc = 1.509 g cm−3, μ(Mo-Kα) = 3.207 mm−1, F(000) = 646, λ = 0.71073 Å, T = 253 K. CCDC 654367. For crystallographic data in CIF or other electronic format see DOI: 10.1039/b709515e |
| ¶ ATPase assays were performed by measuring the release of phosphate using a colorimetric method based on complexation with malachite green and molybdate (AM/MG reagent). In a 50 µl volume, typically 0.2–0.7 pmol of protein, 50 mM Tris-HCl, pH 6.8, 5 mM MgCl2, 1 mM ATP and 25 µg ml−1 poly(U) (Amersham Biosciences, s20,ω = 4.8), were incubated for 10 min at 25 °C. The reaction was stopped by the addition of 10 µl of 0.5 M EDTA, and then the color was developed for 5 min following the addition of 200 µl of AM/MG reagent (0.034% malachite green, 1.05% ammonium molybdate, 0.04% Tween20, in 1 M HCl). 40 µl of 34% (w/v) trisodium citrate was then added and the absorbance at 630 nm, typically using a 96-well plate reader, was reported. The same amount of DMSO was added to the assay as a control. |
| || Two oligomers were synthesized and purified by HPLC: DT20Cy3 (5′-TTTTTTTTTTTTTTTTTTTTCGAGCACCGCTGCGGCTGCACC(Cy3)-3′), and ReleaseBHQ (5′-(BHQ2)GGTGCAGCCGCAGCGGTGCTCG-3′). The two oligomers were annealed (DT20Cy3/ReleaseBHQ = 1/1.2) at a concentration of 8.2 µM (of DT20Cy3) in 10 mM Tris-HCl pH 8.5, heating to 90 °C, then cooling slowly to 40 °C over 1 h. The reaction was carried out in a 150 µl volume of 5 nM DT20Cy3-ReleaseBHQ, 10 nM Release oligomer (5′-GGTGCAGCCGCAGCGGTGCTCG-3′), 0.5 mM ATP, 0.1 mg ml−1 BSA, 2 nM SCV helicase, 5 mM MgCl2, 50 mM Tris-HCl pH 6.8 at 25 °C. The change in fluorescence (λex = 550 nm and λem = 570 nm) after 5 min was used to monitor the extent of unwinding of the duplex. The same volume of DMSO solution was added to the reaction as a control. |
| ** The synthetic released oligonucleotide (5′-GGTGCAGCCGCAGCGGTGCTCG-3′) was labeled with [γ-32P]ATP (3000 Ci mmol−1) using T4 polynucleotide kinase. The labelled released oligo (100 pmol) was annealed to 150 pmol of oligos “5T20” (5′-TTTTTTTTTTTTTTTTTTTTCGAGCACCGCTGCGGCTGCACC-3′) in 100 µl of Annealing Buffer (25 mM Hepes, pH 7.4, 1 mM EDTA, 25 mM NaCl) by heating to 95 °C for 5 min, followed by gradual cooling to 30 °C at 0.5–1 °C min−1, to form substrate DNA duplex d5T. Purified helicase protein (ca. 0.3 pmol) was incubated with the d5T (ca. 600 fmol) in 30 µl of Reaction Buffer (20 mM Hepes, pH 7.4, 5 mM magnesium acetate, 2.5 mM ATP, 2 mM dithiothreitol, 0.1 mg ml−1 bovine serum albumin, 10% glycerol) for 5 min at 25 °C. Reactions were halted by the addition of 15 µl of gel loading buffer (2% SDS, 100 mM EDTA, 15% w/v Ficoll, 0.1% xylene cyanol, 0.05% bromophenol blue), and the products (40 µl) were immediately resolved on 15% polyacrylamide gels (29 : 1 acrylamide–bisacrylamide in (×1) TBE (89 mM Tris base, 89 mM boric acid, 10 mM EDTA)). Gels were analyzed on Storm 860 Phosphor-Imager using ImageQuaNT software. |
| This journal is © The Royal Society of Chemistry 2007 |
